Abstract
Lateral gene transfer (LGT) is an important mechanism of natural variation among prokaryotes. Over the full course of evolution, most or all of the genes resident in a given prokaryotic genome have been affected by LGT, yet the frequency of LGT can vary greatly across genes and across prokaryotic groups. The proteobacteria are among the most diverse of prokaryotic taxa. The prevalence of LGT in their genome evolution calls for the application of network-based methods instead of tree-based methods to investigate the relationships among these species. Here, we report networks that capture both vertical and horizontal components of evolutionary history among 1,207,272 proteins distributed across 329 sequenced proteobacterial genomes. The network of shared proteins reveals modularity structure that does not correspond to current classification schemes. On the basis of shared protein-coding genes, the five classes of proteobacteria fall into two main modules, one including the alpha-, delta-, and epsilonproteobacteria and the other including beta- and gammaproteobacteria. The first module is stable over different protein identity thresholds. The second shows more plasticity with regard to the sequence conservation of proteins sampled, with the gammaproteobacteria showing the most chameleon-like evolutionary characteristics within the present sample. Using a minimal lateral network approach, we compared LGT rates at different phylogenetic depths. In general, gene evolution by LGT within proteobacteria is very common. At least one LGT event was inferred to have occurred in at least 75% of the protein families. The average LGT rate at the species and class depth is about one LGT event per protein family, the rate doubling at the phylum level to an average of two LGT events per protein family. Hence, our results indicate that the rate of gene acquisition per protein family is similar at the level of species (by recombination) and at the level of classes (by LGT). The frequency of LGT per genome strongly depends on the species lifestyle, with endosymbionts showing far lower LGT frequencies than free-living species. Moreover, the nature of the transferred genes suggests that gene transfer in proteobacteria is frequently mediated by conjugation.
Keywords: horizontal gene transfer, microbial evolution, symbionts
Introduction
Lateral gene transfer (LGT or horizontal gene transfer) is the process by which prokaryotes acquire DNA and incorporate it into their genome. Mechanisms for LGT entail transformation, transduction, conjugation, and gene transfer agents (Thomas and Nielsen 2005; Lang and Beatty 2007). LGT has a major role in shaping the distribution of genes across genomes during prokaryote evolution (Doolittle and Bapteste 2007) with only few genes that are resistant to it in the laboratory (McInerney and Pisani 2007; Sorek et al. 2007) and probably none that are resistant to it over the full course of evolutionary time (Bapteste et al. 2009). The fate of the DNA acquired by the different transfer mechanisms can vary in the laboratory. For example, DNA transferred by conjugation in Escherichia coli is recombined into the genome and can survive there for a few generations or longer (Babic et al. 2008), whereas DNA transferred by phage during transduction may be stably integrated into the genome or degraded by bacterial antiviral defense mechanisms, CRISPRs (Marraffini and Sontheimer 2008; Horvath and Barrangou 2010).
Phylogenetic inference of LGT frequency during prokaryote evolution—that is, estimating LGT by looking for discordant branching patterns in trees—provides a wide range of estimates that anywhere from about 20% of all genes are affected by LGT (Snel et al. 2002; Beiko et al. 2005), to perhaps 40% (Kunin et al. 2005) or up to 90% or more of all genes have been affected at some point in their past (Mirkin et al. 2003). This large range of estimates stems to no small extent from inherent difficulties of sequence alignment and phylogenetic inference using highly divergent and/or poorly conserved sequences (Roettger et al. 2009), which comprise the vast majority of data from sequenced genomes. Estimates of the proportion of recently acquired genes per genome using nucleotide patterns or codon bias deliver similar results, showing that on average about 14% of the genes in each genome are recently acquired by lateral transfer (Ochman et al. 2000; Nakamura et al. 2004). Once adapted within the genome, acquired functional genes can then be inherited vertically from generation to generation (Babic et al. 2008) or donated once again at a later time. The modest quantity of 14% recent acquisitions at a given point in time thus accumulates over geological timescales, such that minimum estimates based on network approaches indicate that on average 81 ± 15% of the genes in each prokaryotic genome have been affected by LGT at some stage during evolution (Dagan et al. 2008).
Prokaryotic genome content and size reflect prokaryotic lifestyle (Moran and Wernegreen 2000; Podar et al. 2008), and the frequency of acquired genes is positively correlated with genome size (Nakamura et al. 2004; Cordero and Hogeweg 2009). Yet differences between different bacterial taxonomic groups hint that this is not the only factor affecting the amount of acquired genes within a genome. Recent LGT within the genome of E. coli, having about 4,500 open reading frames (ORFs), was estimated by aberrant codon usage to affect 18% of the gene families (Lawrence and Ochman 1998). In cyanobacteria, having an average of 2,500 ORFs, about 50% of the protein families were inferred to evolve by LGT (Zhaxybayeva et al. 2006; Shi and Falkowski 2008), the high frequency of LGT in cyanobacteria possibly relates to their specific ecological niche and the need to adapt quickly to a dynamic environment (Dufresne et al. 2008; Shi and Falkowski 2008).
Proteobacteria comprise the largest phylum-level group of prokaryotes, including 56,948 currently identified species (44% of all eubacterial species according to NCBI Taxonomy in August 2009). The phylum was named after the Greek god Proteus, who can assume many different shapes, to reflect the enormous biochemical and phenotypic diversity within this group (Stackebrandt et al. 1988). The majority of known proteobacteria are mesophilic, with some exception of thermophilic species (e.g., Thimonas thermosulfata) and psychrophilic (e.g., Polaromonas hydrogenivorans). Most of the known proteobacteria are free living, and some can dominate in certain marine environments, such as members of the Roseobacter clade (Brinkhoff et al. 2008). Some are found in symbiotic association, either mutualistic like the Bradyrhizobium japonicum (a symbiont of rice) or aggressive parasites, such as the Rickettsiae. Others are predatory proteobacteria that feed upon other prokaryotes (Davidov and Jurkevitch 2009). Energy metabolism in proteobacteria is extremely diverse, including chemoorganotrophs (e.g., E. coli), chemolithotrophs (e.g., the sulfur-oxidizing bacteria Thiobacillus aquaesulis and the ammonia-oxidizing bacteria Nitrosomonas europaea), or phototrophs (e.g., Rhodospirillum rubrum) (Kersters et al. 2006). Based on phylogenetic reconstruction of 23S ribosomal RNA (rRNA) and other genetic markers, the phylum was originally divided into four classes: alpha-, beta-, gamma-, and deltaproteobacteria (Stackebrandt et al. 1988), the epsilonproteobacteria (Campylobacterales in some schemes) being a later addition (Gupta 2006). Reflecting their diversity, proteobacteria currently comprise almost half (826 of 1,749 as of January 2010) of all completely sequenced genomes (Markowitz et al. 2010).
Acquisition of new and adaptatively suitable genes from distantly related species by LGT is an evolutionarily quick alternative to modifying preexisting genes via point mutations. For example, the genome of the eubacterium Salinibacter ruber that resides in the extremely halophilic habitat of saltern crystallizer ponds, harbors many genes shared with haloarchaeal species, probably as a result of niche-specific acquisitions (Mongodin et al. 2005). Shared gene content following LGT is found also between species having similar symbiotic relation with similar host, as in the case of the genus Bradyrizobium (alphaproteobacteria) and Ralstonia solanacearum (betaproteobacteria), both of which are soil bacteria undergoing symbiosis, either mutualistic or parasitic, with plants (Kunin et al. 2005).
Networks of shared genes are a useful tool to recover common gene content across many bacterial genomes (Beiko et al. 2005; Kunin et al. 2005; Fukami-Kobayashi et al. 2007; Dagan et al. 2008; Halary et al. 2010). Among the proteobacteria, phylogeny for specific groups has been examined using tree-based methods, for example, in the gamma- (Lerat et al. 2005), the epsilon- (Gupta 2006), and the alphaproteobacteria (Wu et al. 2004; Ettema and Andersson 2009). However, phylogenies only depict the evolutionary history of one or few genes within a genome, not for the whole genome. Network approaches to study genome evolution within the proteobacteria, where genome sizes can range from under 160 kb (Nakabachi et al. 2006) to over 9 Mb (Kaneko et al. 2002), have not been reported to date. Here, we investigate genome evolution within proteobacteria using a network approach.
Materials and Methods
Data
Sequenced genomes of 329 proteobacteria and their taxonomical classification were downloaded from NCBI web site (http://www.ncbi.nlm.nih.gov/; version of April 2008). All proteins were clustered by identity into gene families using the reciprocal best Blast hit (BBH) approach (Tatusov et al. 2000). Each protein was Blasted against each of the genomes. Pairs of proteins that resulted as reciprocal BBHs of E-value < 1×10−10 were aligned using ClustalW (Thompson et al. 1994). Pairwise protein sequence identity was calculated by the number of identical amino acids divided by alignment length. Protein pairs with above the amino acid identity threshold (T30–T70) were clustered into protein families of ≥2 members using the MCL algorithm setting the inflation parameter, I, to 2.0 (Enright et al. 2002). Previous work has shown that I values between 1.8 and 2.2 has little influence on the distribution of gene family size in this kind of analysis (Dagan et al. 2008). Protein families for genomes in specific class or species were extracted from the protein families of the total data set.
Reconstruction of Gene Trees
For the reconstruction of RPL31 and asparaginyl-tRNA synthetase phylogenies, protein sequences included in the protein family at the chosen threshold were aligned using ClustalW (Thompson et al. 1994). A maximum likelihood tree was reconstructed from the alignment using PHYML (Guindon and Gascuel 2003) with the default JTT substitution matrix (Jones et al. 1992).
Network of Shared Protein Families
Networks of shared proteins (NSPs) of all proteobacteria were reconstructed from protein families clustered at a given protein sequence identity threshold. The number of shared proteins between each genome pair was calculated as the number of protein families in which both genomes are present. A division of the network into modules was found by defining a modularity function of each bipartition of the network as the number of edges within a community minus the expected number of edges in the community. An optimal division is then found by maximizing this function over all possible divisions, using eigenspectrum analysis (Newman 2006; Dagan et al. 2008).
Reconstruction of a Reference Tree
A reference tree was constructed using rRNA operon (16S, 23S, and 5S) sequences within a taxonomically constrained framework invoking forced monophyly of classes (Dagan et al. 2008). Only one rRNA operon sequence per species was used. Bacterial genomes may contain several copies of the ribosomal operon. However, the intragenomic variation is commonly smaller than the intergenomic variation (Pei et al. 2010), hence the use of a single sequence per genome generates a reference gene tree for these taxa. The sequences of the three genes were first aligned using ClustalW (Thompson et al. 1994) for each of the main classes. The alignments of the three genes were concatenated, gapped sites were removed, and a maximum likelihood tree of each group was inferred using PHYML (Guindon and Gascuel 2003) with the default HKY substitution matrix (Hasegawa et al. 1985). From each group alignment, a consensus sequence was reconstructed by concatenating the most abundant nucleotide in each alignment column into a single sequence. The consensus sequences were used to infer the tree of groups with PHYML (Guindon and Gascuel 2003) and to root each neighboring group subtree. Leaves in the tree of groups were replaced with each rooted group subtree.
For the gene content reference tree, protein families at T30 cutoff were converted into a binary presence/absence pattern (PAP). The total PAP matrix for m protein families and n genomes is defined as aij, where i = 1, …, m, and j = 1, …, n. If protein family i is present in genome j, then aij = 1, otherwise aij = 0. The PAP matrix was converted into PHYLIP format using an in-house PERL script. The reconstruction of a gene content tree was performed using Wagner parsimony (Felsenstein 1983) as implemented in the MIX program of the PHYLIP package (Felsenstein 2004).
Reconstruction of a Minimal Lateral Network
Presence and absence of protein families were superimposed on the reference tree and LGTs inferred to yield gene origin for all protein families at internal nodes according to the different LGT allowance models as described by Dagan and Martin (2007). Briefly, this approach seeks the lower bound of LGT frequency during evolution of the genomes in question using the distribution of ancestral genome sizes as a constraint and optimization criterion. Different evolutionary models allowing incrementally increased LGT frequency across the reference tree each specify a different number of LGTs per protein family as required to account for the presence/absence pattern for each gene (Dagan and Martin 2007). Gene loss events are unpenalized. Using a recursive binary procedure, the evolutionary reconstruction allows for variable number of gene origins per protein family up to the maximum allowed by the given model (e.g., no LGT, one LGT, three LGTs etc.). Under a model allowing no LGT, all gene presence/absence patterns are attributed to loss only, with the result that all genes in the sample are scored as present in the genome ancestral to the species studied. The distribution of ancestral genome sizes at each node is scored for each model. Incrementally adding LGT reduces ancestral genome sizes, whereas models allowing excessive amounts of LGT during evolution make ancestral genomes too small. The preferred model (allowed max number of LGT events per gene) is determined as the one that brings the genome size distribution of ancestral genomes into best statistical agreement with that of contemporary species (Dagan and Martin 2007). All gene origins within each protein family are connected to form a clique; hence, the number of edges reconstructed for a protein family of k gene origins is k (k − 1)/2. Edges connecting the same two nodes for different protein families are joined to form a single edge that is weighted according to the number of protein families in which it appeared. The same procedure was repeated for each phylogenetic depth using a subtree of the class/species extracted from the rooted reference tree.
The minimal number of LGT events that is required to explain the gene origin distribution of a certain protein family of k gene origin is k − 1. However, in the minimal lateral network (MLN) approach, we have no concrete information regarding donors and recipients in the LGT event so that lateral edges are reconstructed to connect among all gene origins reconstructed per protein family. Thus, MLN data sets reconstructed from LGT allowance models that exceed a single LGT event per protein family (LGT1 model) contain more edges (or heavier edges) than the minimal frequency of LGT events required to explain gene distribution patterns in the data set. For example, a protein family for which two origins were inferred will include one lateral edge which corresponds to a single LGT event required in order to explain the distribution of the protein family. But a protein family for which three origins were inferred will include three lateral edges connecting all origins, whereas the minimum number of LGT required in this case is only two. To study the properties of LGT network, for each such data set, 1,000 MLN replicates (rMLN) were reconstructed where the sum of edge weight corresponds the number of gene transfers. For this purpose, randomly selected edges were deleted for each protein family of more than three origins, and all edges of all protein families were summarized into a single rMLN.
Identification of Recently Acquired Genes by Aberrant Nucleotide Pattern
Recently acquired genes are expected to have unusual codon usage and GC content when compared with the whole proteome. Therefore, GC content may be used to detect the foreign origin of a gene (Garcia-Vallve et al. 2000; Nakamura et al. 2004). The statistical analysis of GC content is favored over codon usage because it has better statistical power. Genes with atypical GC content are detected by comparing their GC content with the genomic GC using the χ2 test with a false discovery rate of 5% (Benjamini and Hochberg 1995).
Results and Discussion
Clustering of the 1,207,272 proteins within the 329 proteobacterial genomes using amino acid identity threshold of 30% (T30) resulted in 74,667 protein families of size ≥2 proteins. Only 14 of these families are universally present in all proteobacteria. These include mostly ribosomal proteins together with proteins involved in information processes, such as chaperonin GroEL (supplementary table S1, Supplementary Material online). A recent investigation into the quality of genome annotation in NCBI data set revealed frequent misannotation of core genes in gammaproteobacteria (Poptsova and Gogarten 2010), hence the number of universal genes reported here using the standard annotation might be underestimated. Using the T30 threshold results also in 140,333 (12% of the total) unclustered proteins. Singleton proteins—frequently named also ORFans (Fischer and Eisenberg 1999)—are genes for which no reciprocal BBH above T30 was found within the current genomes sample. These may be either novel genes that are specific to the genome or genes that are shared with genomes not present in our sample. To test the latter possibility, we first searched for homologs to these singletons within 97 proteobacterial genomes that were added to the NCBI database between April 2008 (our version) and January 2009, increasing by 30% the proteobacterial genome sample size. Of the 140,333 singletons, 10,880 (8%) proteins had reciprocal BBH within the larger sample at T30. This averages to a removal of 112 singletons with each additional proteobacterial genome that is sampled. The remaining 129,453 singletons were then searched for homologs within 335 nonproteobacterial prokaryote genomes in NCBI genomic database (April 2008 version). For 18,692 proteins, we found nonproteobacterial reciprocal BBH at T30. Hence, on average, each nonproteobacterial genome includes 55 homologs to proteobacterial singletons at that protein sequence identity threshold. The remainder of 110,491 (9%) singletons remains as such.
The search for homologs to the singletons in our sample supplies two observations. First, increasing the sample of searched genomes by 230% (761 genomes in total) reduced the percent of singletons by only a very modest proportion (from 12% to 9% of the proteobacterial gene repertoire). Second, the ratio of singletons found in newly sequenced proteobacterial genomes and nonproteobacterial genomes is roughly 2:1.
The Distribution of Shared Proteins among Proteobacteria
Shared gene content among prokaryotes may be the result of either common ancestry or LGT. Notwithstanding various factors affecting protein evolutionary rates (Graur and Li 2000), protein sequence identity among orthologs within protein families that evolve by vertical inheritance alone is expected to be roughly proportional to the divergence time of the compared species (Novichkov et al. 2004; Dagan et al. 2010). Protein-coding genes acquired by LGT are expected to have higher sequence identity among donor and acceptor groups than the expected for an average gene reflecting the reference sequence tree, assuming that the transfer event occurred after the divergence of the reference operon sequences. If all proteins were evolving by vertical inheritance alone (i.e., if they were all strictly coevolving, physically linked to the same rRNA operon in their current chromosome), then using ascending amino acid identity thresholds for the reconstruction of protein families would result in a strictly hierarchical genome (taxon) clustering of increasingly narrow taxon sample. Thus, low identity thresholds are expected to yield kingdom- or phylum-specific families, for example, whereas increasing identity thresholds will yield protein families that are specific to lower taxonomic ranks, such as class, order, genus, and finally species-specific protein families, etc. Exceptions to this rule (i.e., anomalously high sequence similarity) can indicate the workings LGT in the data.
To study gene distribution patterns over ascending protein similarities in proteobacteria, we repeated the clustering into protein families using ascending thresholds for the sequence similarity between reciprocal BBHs. Increased protein sequence identity thresholds resulted in larger numbers of protein families, each spanning fewer genomes. The number of protein families at T30 is 74,667 with 41,255 (55%) small protein families spanning ≤4 genomes. No universal families are recovered using T70, which results in 139,564 protein families and a larger number of smaller families 96,717 (69%) spanning ≤4 genomes. The frequency of universal protein families decreases with protein sequence identity threshold, leaving a single family at T55 (ATP-dependent Clp protease) and no universal families found above that threshold (table 1 and supplementary table S1, Supplementary Material online).
Table 1.
Number of Protein Families in the Various Thresholds and Characteristics of the Result Shared Protein Network.
| Protein Similarity Threshhold | No. of Families | Singletons | No. of Proteins | No. of Families ≤4 Species | No. of Universal Families | No. of Edges | Mean Edge Weight | Median Edge Weight | No. of Modules |
| T30 | 74,667 | 140,333 (12%) | 1,066,939 | 41,255 (55%) | 14 | 53,956 | 854 ± 527 | 762 | 4 |
| T35 | 83,740 | 165,256 (14%) | 1,042,016 | 47,670 (57%) | 10 | 53,956 | 743 ± 521 | 631 | 3 |
| T40 | 93,806 | 194,360 (16%) | 1,012,912 | 54,835 (58%) | 6 | 53,956 | 624 ± 515 | 497 | 3 |
| T45 | 104,420 | 228,996 (19%) | 978,276 | 62,957 (60%) | 4 | 53,956 | 505 ± 503 | 362 | 3 |
| T50 | 114,155 | 266,022 (22%) | 941,250 | 70,817 (62%) | 2 | 53,956 | 400 ± 489 | 251 | 3 |
| T55 | 123,386 | 307,825 (25%) | 899,447 | 79,076 (64%) | 1 | 53,956 | 304 ± 473 | 158 | 5 |
| T60 | 130,651 | 351,589 (29%) | 855,683 | 86,077 (66%) | 0 | 53,956 | 225 ± 453 | 92 | 6 |
| T65 | 136,199 | 398,264 (33%) | 809,008 | 92,094 (68%) | 0 | 53,956 | 164 ± 431 | 42 | 9 |
| T70 | 139,564 | 446,640 (37%) | 760,632 | 96,717 (69%) | 0 | 53,869 | 118 ± 407 | 17 | 11 |
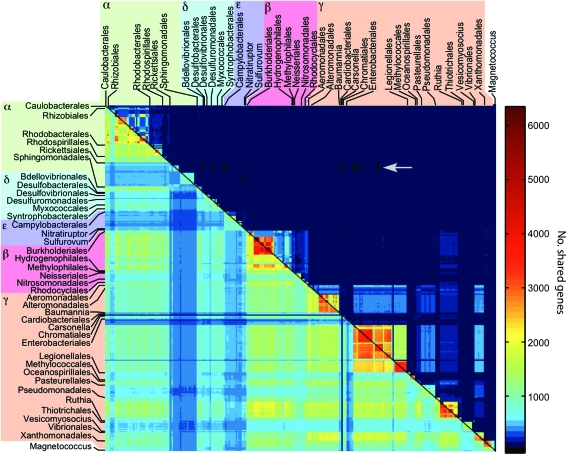
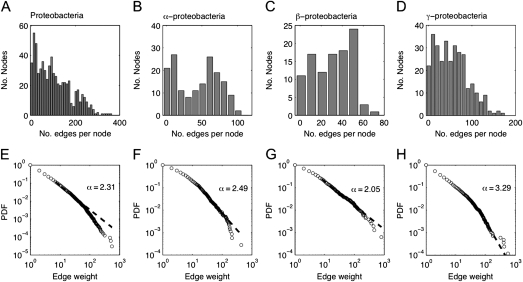
To summarize shared gene distribution patterns among proteobacteria in various protein sequence identity thresholds (Ti), we reconstructed an NSP for 30–70% protein sequence identity thresholds. The network includes 329 vertices (genomes) and a maximum of 53,956 edges (number of shared protein families). Edge weights in this network are calculated as the number of shared protein families between two connected genomes. The use of increasing protein sequence identity thresholds results in gradual decrease in common families among distantly related species and leads to a different network for each threshold. Using T30–T65, the NSP among proteobacteria is a clique where all genomes are connected with each other. Increasing protein sequence identity thresholds of T70 eliminates 87 edges from the NSP (table 1). A comparison of NSP at T65 and T70 shows that edges connected at one end at least to species having small genome size (below 1,500 genes) are the first to be disconnected from the network. Such species include the Rickettsiales (alphaproteobacteria), Zymomonas mobilis ZM4 (alphaproteobacteria), and Helicobacter pylori HPAG1 (epsilonproteobacteria; supplementary table S2, Supplementary Material online).
Although the connectivity distribution in the NSP is almost identical over different protein sequence identity thresholds, edge weights among the connected species changes considerably (fig. 1). The NSP at T30 reveals a clear taxonomic structure within gene distribution patterns across proteobacterial species. Closely related species within the same taxonomic class are connected by edges of higher weights (they share more protein families) in comparison with species from different classes. Clusters of highly connected species may be observed among different strains of the same genus, such as the Burkholderiales (betaproteobacteria), Enterobacteriales (gammaproteobacteria), and Pseudomonadales (gammaproteobacteria). Genera of small genome size are connected to other species with edges having lower weights. Such are the Rickettsiales (alphaproteobacteria) and Campylobacterales (epsilonproteobacteria). The background frequency of shared protein families at T30 has a median of 427 shared protein families between any pair of species.
FIG. 1.
The NSP families. A matrix representation of the NSPs in T30 (below the diagonal) and T70 (above the diagonal). The species are sorted by an alphabetical order of the order and genus. The color scale of cell aij in the matrix indicates the number of shared protein families between genomes i and j. An arrow at the upper diagonal points to genome pairs that are disconnected at T70.
At T70, several highly connected genera clusters within the NSP are clearly observed (fig. 1), and the median of shared protein families between any pair of species is 17. Edges of weight >2,000 are found almost exclusively among species from the same genus or class. However, even at the high identity threshold of 70%, the NSP is almost a clique, with 298 (90.6%) of the species still completely connected within the network. In total, 3,637 protein families are present in species from two classes or more; hence, they are distributed across wide taxonomic scale. These protein families are relatively small, 2,331 (64%) of them are present in ≤10 species. Such patchy protein families comprising orthologs from different classes, at the protein identity threshold where only strains are still highly connected, can be the result of vertical inheritance and widespread differential loss or LGT. If the former, then these are highly conserved proteins that originated in the proteobacterial LUCA and were lost during evolution in most of the species, except for the ones where they are still present. This argument is very problematic. First, because there are no proteobacterial universal proteins at T70 (supplementary table S3, Supplementary Material online) so that proteins of proteobacterial LUCA origin are more diverged than T70. Second, protein conservation and the propensity to be lost are negatively correlated (Krylov et al. 2003) so that such an abundant loss during evolution of those protein families would be highly improbable. Hence, orthologs in the highly patchy protein families are candidates for LGT among proteobacterial species.
To test the characteristics of these LGT-candidate protein families, we investigated the functional annotation of extra patchy protein families that are present in ≤10 genomes from two proteobacterial classes or more at T70 (2,430 families). Many of these proteins (729; 31%) are annotated as hypothetical proteins, mostly common to betaproteobacteria and gammaproteobacteria (214). Only one hypothetical protein is common to genomes from four different classes, found in Acidovorax JS42 (betaproteobacteria), Aeromonas hydrophila ATCC 7966 (gammaproteobacteria), Aeromonas salmonicida A449 (gammaproteobacteria), Bdellovibrio bacteriovorus (deltaproteobacteria), Herminiimonas arsenicoxydans (betaproteobacteria), Mesorhizobium loti (alphaproteobacteria), and Sorangium cellulosum str. So ce 56 (deltaproteobacteria). A Blast search in NCBI showed that this protein is annotated in other bacterial genomes as glyoxalase protein family. Proteins of this family are important for the detoxification of methylglyoxal (Sukdeo and Honek 2008).
Most of the annotated proteins are involved in metabolic and cellular processes, whereas the minority are informational genes. We find that 101 (4%) of these proteins are plasmid-related proteins, such as IS-elements transposase, integrases, and stabilization proteins. In contrast, we find that 44 (2%) protein families are phage-related proteins, such as phage tail proteins, prophage CP4-57 regulatory protein, and phage integrase. These frequencies may be used for inference about relative contribution of LGT by plasmids (conjugation) versus transduction in the present genome sample. These two modes of LGT are very different from each other in the distance that is required between donor and recipient. Conjugation may be viewed as a personal delivery, whereas transduction is more like long distance mail. The 2-fold higher frequency of plasmid-related genes in comparison with phage related in the very patchy gene distribution patterns suggests that much of the LGT in proteobacteria in this sample is mediated by conjugation, where donor and recipient cells are in close proximity (Halary et al. 2010).
The set of highly similar patchy protein families includes three ribosomal proteins and one tRNA synthetase. The T70 protein family of 50S ribosomal protein L31 groups six betaproteobacteria with Methylococcus capsulatus str. Bath, a gammaproteobacterium. A phylogenetic tree of this protein including all species clustered at T60 reveals that the same seven species are grouped together in a clade, indicating that M. capsulatus str. Bath, has acquired its ribosomal protein L31 from a betaproteobacterium (supplementary fig. S1, Supplementary Material online). The T70 cluster of asparaginyl-tRNA synthetase groups nine gammaproteobacteria with Myxococcus xanthus str. DK1622, a deltaproteobacterium. A phylogenetic tree of this protein as T55 results in a clade of the same nine species, indicating that M. xanthus str. DK1622 acquired its asparaginyl-tRNA synthetase from a gammaproteobacterium (supplementary fig. S2, Supplementary Material online). Both 50S ribosomal protein L31 and asparaginyl-tRNA synthetase are single-copy genes. Single-copy genes have been recently found to be more resistant to transfer into E. coli than multicopy genes (Sorek et al. 2007). But these two examples show that single-copy informational genes can be replaced via LGT, consistent with other reports in the literature (Chan et al. 2009).
Modules within the NSP
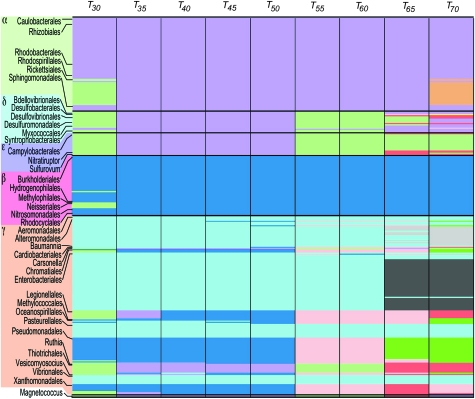
Using a modularity function that classifies the genomes into modules, we identified connectivity patterns across the NSP. These modules are groups of genomes more densely connected among themselves than with genomes outside the group (Newman 2006; Dagan et al. 2008). Across different identity thresholds (T30–T70), the modularity function applied to the NSP reveals a structure of genetic connectivity (shared genes) that does not strictly overlap with the proteobacteria classes as defined by traditional means, that is, their rRNA sequence (fig. 2). At T30, the NSP comprises four modules. The first module (purple) includes the majority of alphaproteobacteria and two deltaproteobacteria (M. xanthus str. DK1622, S. cellulosum str. So ce 56). The second module (green) includes alphaproteobacterial endosymbionts (Anaplasma, Ehrlichia, Rickettsia, and Wolbachia), the majority of deltaproteobacteria, all epsilonproteobacteria, two betaproteobacterial human pathogens (Neisseria meningitides and N. gonorrhoeae), and several gammaproteobacterial endosymbionts (Coxiella, Legionella, Francisella, and Xylella). The third module (blue) includes the majority of betaproteobacteria and few soil bacteria from the gammaproteobacteria, including Pseudomonas and Xanthomonas. The last module (cyan) is specific to gammaproteobacteria.
FIG. 2.
Modules in the NSP in the different protein sequence identity thresholds. Modules are shown as colored boxes within columns for thresholds from T30 to T70. Proteobacterial orders are indicated in rows for comparison. An expanded table of the panel containing all species names is given in supplementary table S4 (Supplementary Material online).
Reconstruction of modules from the NSP at T35–T50 results in only three modules. One module includes all alphaproteobacteria, epsilonproteobacteria, and deltaproteobacteria together with seven strains of Francisella tularensis and one F. philomiragia (gammaproteobacteria). Another module includes all betaproteobacteria together with many soil gammaproteobacteria, including Acinetobacter baumannii, seven Pseudomonas species, three species of Psychrobacter, and four species of Xanthomonas. The third module is unique to gammaproteobacteria. At T55–T60, the betaproteobacteria and gammaproteobacteria fall into three class-specific modules, epsilonproteobacteria and deltaproteobacteria are joined with Francisella (gammaproteobacteria), and all alphaproteobacteria are joined with S. cellulosum str. So ce 56 (deltaproteobacteria). At T65–T70, the alphaproteobacterial endosymbionts fall apart, with several modules that are common to alphaproteobacteria, deltaproteobacteria, and epsilonproteobacteria. The betaproteobacteria appear as a unique module, whereas gammaproteobacteria disarticulate into seven modules (supplementary table S4, Supplementary Material online).
A hefty debate is currently ablaze about the utility and meaning of the “tree of life” (see Doolittle and Bapteste 2007 vs. Galtier and Daubin 2008 cf. Bapteste et al. 2009), particularly in the context of the overall evolutionary history of prokaryotes. One could argue that the debate boils down to the difference between attempts to reconstruct the whole of the evolutionary process and attempts at organismal classification (Doolittle 1999). Proponents of the tree of life are arguing that one or a few genes serve as a useful and valid proxy for the evolution of the whole chromosome (Ciccarelli et al. 2006; Galtier and Daubin 2008). Dissidents are arguing that since only about 30 genes are demonstrably present across many genomes (but very often sharing less than 20% amino acid identity in most comparisons) the “tree of life” constructed by such means speaks for only about 1% of the data in genomes (Dagan and Martin 2006), which typically harbor about 3,000 genes. The modules of the present study point to issues concerning the concept of phylogeny within proteobacteria. Phylogeny usually refers to a hierarchical branching pattern, as in a phylogenetic tree. If we look at the modules that are identified here on the basis of shared genes (fig. 2), the classification of proteobacteria into alpha, beta, gamma, delta, and epsilon groups is not recovered for any threshold. Indeed, the only of the five classes that is recovered as a distinct module at any of the nine thresholds is the betaproteobacteria class at thresholds T55, T65, and T70 (fig. 2).
The modules of shared genes detected here do not reflect a hierarchical “phylogeny” of the proteobacterial classes as suggested by “tree of life” schemes based on a few concatenated genes. For example, Ciccarelli et al. (2006) reported a branching order of (((((γ,β),α),ϵ),δ),outgroup) for the proteobacterial classes. No such phylogenetic hierarchy is suggested by the modules of shared genes (fig. 2). This reinforces an earlier criticism that the phylogeny of a sample representing 1% of the genome is a poor proxy for what is to be found in the rest of the genome. We do observe, however, a module at T35–T50 grouping the (α,ϵ,δ) classes together with some γ-representatives, most notably the Thiotrichales, represented here by the deep-sea vent chemoautotroph Thiomicrospira (Scott et al. 2006) and strains of the intracellular pathogen F. tularensis (Oyston 2008) plus Magnetococcus. Modules within the gammaproteobacteria correspond to some extent to family-level classifications of this class, which are also poorly resolved with concatenated sequences (Gao et al. 2009).
Species included in the NSP modules at all protein sequence identity thresholds differ significantly in their genome size (P < 0.05 using the Kruskal–Wallis test; Zar 1999), hence genome size is not the prime determinant of module structure. Nonetheless, endosymbionts that are all characterized by very small genomes are grouped into common modules across taxonomic class boundaries, but this is because they tend to relinquish the same sets of genes (Pal et al. 2006; Moran 2007) not because the genomes are small per se. Moreover, betaproteobacteria and gammaproteobacteria whose habitat is mainly within the soil are clearly grouped together in varying protein sequence identity thresholds (figs. 1 and 2). This finding is in line with the observation that cooccurring microbes have similar genomes regardless (sometimes) of their phylogenetic relatedness (Chaffron et al. 2010) and the view that transfer might be more frequent between genomes of prokaryotes sharing similar habitats (Jain et al. 2003). Overall, community structure within the NSP appears to have a phylogenetic backbone but is also influenced by bacterial lifestyle and habitat.
Minimal Lateral Networks
Gene sharing patterns found here indicate that LGT is common among proteobacteria. But how frequent is frequent? To quantify the lower bound frequency of LGT at three phylogenetic depths within proteobacteria—phylum, class, and species—we constructed MLNs (Dagan et al. 2008). In that approach, LGT frequency is inferred against the criterion of ancestral genome size. An evolutionary model that entails no LGT during evolution results in untenably large ancestral genomes (Doolittle et al. 2003). Yet, if genome evolution in the past was not fundamentally different from todays, then ancestral genomes should have similar sizes to those of contemporary genomes. The approach is thus based on applying evolutionary models that allow increasing frequencies of LGT, until the distributions of ancestral and contemporary genome sizes are statistically reconciled (Dagan and Martin 2007). Phylogenetic inference by the MLN approach yields estimated ancestral genome sizes together with an inference of laterally shared gene families among species or groups of species, the gene distributions of which are better explained by LGT than the phylogenetic tree. These two outcomes can be graphically represented by a network in which the vertices are the nodes of the reference tree, and the edges are either vertical tree branches or inferred lateral gene sharing edges (Dagan et al. 2008).
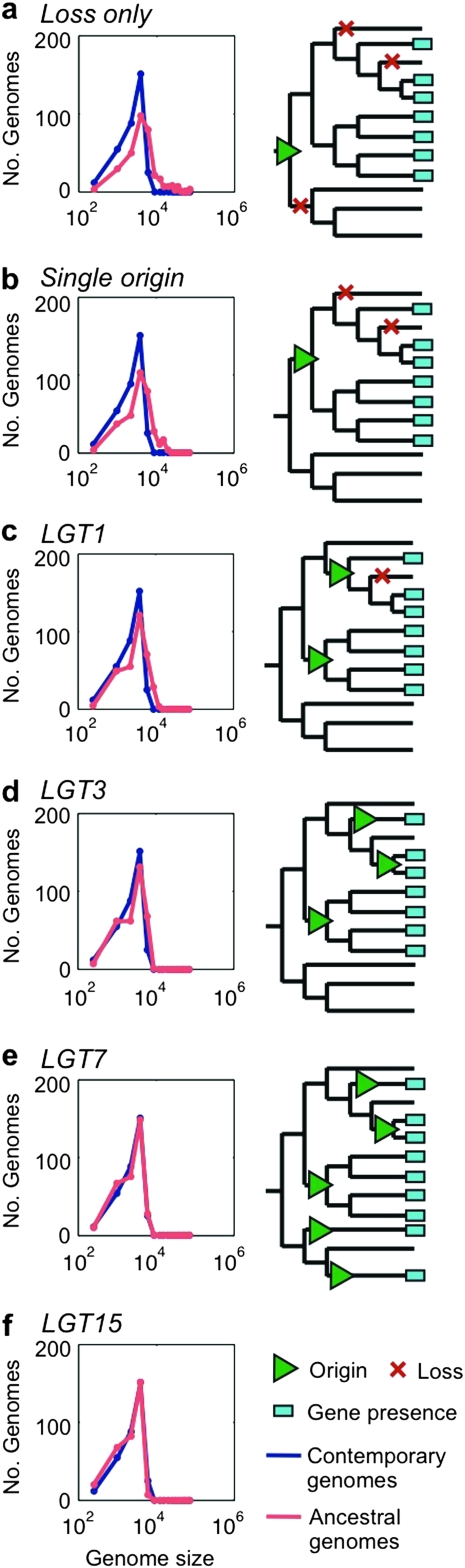
Our results suggest that LGT is more frequent at the phylum level than in the class or species level. For the data of all proteobacteria species, a model that allows up to seven LGTs per protein family (LGT7) was accepted (P = 0.44, using Wilcoxon test (Zar 1999; fig. 3). Although seven LGTs per family are allowed in this model, only a minority of the gene occurrence patterns require that amount. In most (28%) of the protein families whose evolution includes LGT, it occurred only once, whereas protein families whose evolution includes seven LGTs are very rare (0.78%; table 2A). The weighted mean LGT frequency within proteobacteria phylum is thus 1.9 LGTs per protein family.
FIG. 3.
Distribution of contemporary and ancestral genome sizes in phylum depth under the different LGT allowance models (left) and schematic representation of the evolutionary scenario implicated by the models (right). The models (A) loss only, (B), single origin, (C) LGT1, and (D) LGT3 result in significantly larger ancestral genome sizes in comparison to contemporary genome sizes (α = 0.05, using Kolmogorov–Smirnov test). The LGT7 model (E) results in similar distributions of ancestral and contemporary genome size (P = 0.44, using Wilcoxon test). The LGT15 model (F) results in significantly smaller ancestral genome sizes in comparison to contemporary genome sizes (α = 0.05, using Kolmogorov–Smirnov test).
Table 2.
Statistically Accepted LGT Allowance Models Using T30 Protein Families for the Different Data Sets with (A) rRNA and (B) Gene Content Reference Trees.
| Data Set | No. of Species | No. of Families | LGT Model | P Value | Mean LGT Frequency | 1 Origin | 2 Origin | 3 Origin | 4 Origin | 5 Origin | 6Origin | 7 Origin | 8 Origin | |
| A | ||||||||||||||
| Proteobacteria | 329 | 74,667 | LGT1 | 0.44 | 1.9 | 18,763 (25%) | 21,366 (29%) | 9,048 (13%) | 11,760 (16%) | 6,535 (9%) | 3,520 (5%) | 2,707 (4%) | 582 (1%) | |
| Alphaproteobacteria | 82 | 27,810 | LGT1 | 0.25 | 0.6 | 6,018 (25%) | 17,760 (75%) | |||||||
| LGT3 | 0.43* | 1.1 | 6,018 (25%) | 6,792 (29%) | 8,329 (35%) | 2,639 (11%) | ||||||||
| Betaproteobacteria | 52 | 25,199 | LGT1 | 0.26* | 0.7 | 3,830 (19%) | 16,492 (81%) | |||||||
| LGT3 | 0.14 | 1.1 | 3,830 (19%) | 5,816 (29%) | 9,014 (44%) | 1,662 (8%) | ||||||||
| Gammaproteobacteria | 157 | 40,327 | LGT3 | 0.46 | 1.2 | 9,179 (25%) | 10,253 (28%) | 13,089 (36%) | 3,669 (10%) | |||||
| Escherichia coli | 12 | 7,879 | LGT1 | 0.48 | 0.7 | 653 (10%) | 5,589 (90%) | |||||||
| Francisella tularensis | 7 | 1,840 | Origin | 0.11 | ||||||||||
| LGT1 | 0.47* | 0.3 | 1,255 (73%) | 462 (27%) | ||||||||||
| Yersinia pestis | 7 | 4,439 | LGT1 | 0.59 | 0.9 | 122 (3%) | 4,080 (97%) | |||||||
| B | ||||||||||||||
| Proteobacteria | 329 | 74,667 | LGT1 | 0.98 | 1.7 | 21,782 (29%) | 20,824 (28%) | 9,030 (12%) | 10,393 (14%) | 6,318 (8%) | 3,454 (5%) | 2,368 (3%) | 472 (1%) | |
| Alphaproteobacteria | 82 | 27,810 | LGT1 | 0.37* | 0.6 | 6,397 (27%) | 17,381 (73%) | |||||||
| LGT3 | 0.32 | 1.1 | 6,397 (27%) | 6,743 (28%) | 8,390 (35%) | 2.248 (9%) | ||||||||
| Betaproteobacteria | 52 | 25,199 | LGT1 | 0.1 | 0.6 | 4,707 (23%) | 15,615 (77%) | |||||||
| LGT3 | 0.14* | 11 | 4,707 (23%) | 6,116 (30%) | 7,117 (35%) | 2,382 (12%) | ||||||||
| Gammaproteobacteria | 157 | 40,327 | LGT3 | 0.47* | 1.5 | 10,751 (30%) | 9,627 (27%) | 4,241 (12%) | 6,532 (18%) | 3,093 (9%) | 1,436 (4%) | 482 (1%) | 28 (0%) | |
| E. coli | 12 | 7,879 | LGT1 | 0.19 | 0.3 | 3,714 (60%) | 2,453 (40%) | |||||||
| F. tularensis | 7 | 1,840 | Origin | 0.23 | ||||||||||
| LGT1 | 0.73* | 0.2 | 1,311 (76%) | 406 (24%) | ||||||||||
| LGT3 | 0.14 | 0.3 | 1,238 (72%) | 479 (28%) | ||||||||||
| Y. pestis | 7 | 4,439 | Origin | 0.1 | ||||||||||
| LGT1 | 0.95* | 0.2 | 3,400 (81%) | 802 (19%) | ||||||||||
| LGT3 | 0.07 | 0 3 | 3,400 (81%) | 280 (7%) | 522 (12%) | |||||||||
| LGT1 | 0.05 | 0.3 | 3,400 (81%) | 280 (7%) | 314 (7%) | 208 (5%) | ||||||||
* For data sets where more than one model was statistically accepted, the most probable model is marked by an asterisks.
Within the classes of proteobacteria, the LGT3 model was accepted for the alphaproteobacteria and gammaproteobacteria, with a weighted LGT frequency of 1.3 per protein family in both groups. The frequency of LGT events per protein family follows a similar distribution in alphaproteobacteria and gammaproteobacteria as well (table 2A). Within the betaproteobacteria, the LGT1 model was accepted, with a weighted mean LGT frequency of 0.8 per protein family (table 2A). None of the models was accepted for the deltaproteobacteria and epsilonproteobacteria. However, in both groups, resulting ancestral genome sizes from the origin-only model are significantly larger than contemporary genome sizes (P < 0.01, using Kolmogorov–Smirnov test [Zar 1999]; supplementary fig. S3, Supplementary Material online). Moreover, ancestral genome sizes resulting from the LGT1 model are significantly smaller than contemporary genome sizes (P < 0.01 using Kolmogorov–Smirnov test; supplementary fig. S3, Supplementary Material online). This suggests that deltaproteobacteria and epsilonproteobacterial gene distribution patterns, in combination with the rRNA reference tree topology, require an evolutionary model that is somewhere between origin-only and LGT1 models, allowing probably a single LGT event to only part of the protein families. However, our current MLN reconstruction approach applies uniform model choice to all protein families. A more complicated approach in which each protein family is fitted its own model would require an a priori assumption of gene origin to loss ratios (e.g., Kunin et al. 2005), these are regarded in the MLN approach (Dagan and Martin 2007) as a variable whose value is to be inferred rather than a user-defined parameter.
For the LGT frequency estimation at the species level, we selected three gammaproteobacterial species whose genome sample of sequenced strains is large enough to conduct our analysis. These include E. coli (12 genomes), F. tularensis (7 genomes), and Yersinia pestis (7 genomes). For these three data sets, the LGT1 model was accepted (P = 0.48, 0.47, and 0.49 respectively, using Wilcoxon test; supplementary fig. S3, Supplementary Material online) with a weighted mean LGT frequency of 0.7 LGTs per protein family in E. coli, 0.3 LGTs per protein family in F. tularensis, and 0.9 LGTs per protein family in Y. pestis. LGT at the species level is recombination. Hence, the LGT rates calculated here for the species data sets may be regarded a lower bound estimate for recombination rates. Because in our approach we analyze the presence/absence patterns of genes and not their sequences, our inference yields an estimate for the gene spread by recombination but largely underestimates overall recombination rates.
LGT Inference against a Gene Content Reference Tree
LGT frequencies inferred using the MLN approach are robust to different reference phylogenetic trees reconstructed from various protein families, yet they may be affected by the patchiness of the gene distribution patterns across the reference tree (Dagan and Martin 2007). This is because when the LGT allowance in the MLN approach is increased from none to two and then more gene origins (by gene birth or LGT), these are distributed in pairs to descendants of an ancestor for which a gene origin was reconstructed using the previous model. Moreover, by using a species tree reconstructed from the rRNA sequences, we assume that the phylogeny of a single operon truly represents the evolutionary history of proteobacteria. This assumption may be problematic for the evolution of prokaryotes that is reticulated by nature (Bapteste et al. 2009). Here, we test the robustness of the MLN approach to the patchiness of gene distribution patterns and the rRNA phylogenetic tree by using a gene content tree (Snel et al. 1999) as the reference tree. Such a reference tree is expected to minimize the patchy gene distribution patterns and thereby provide more conservative estimates of LGT among proteobacteria.
Gene content trees were reconstructed from the complete presence/absence data at T30 using Wagner parsimony approach (Felsenstein 1983). The gene content tree including all proteobacteria was rooted on the branch separating (α,δ,ϵ) from (β,γ). The resulting tree supports the monophyly of alphaproteobacteria, deltaproteobacteria, and epsilonproteobacteria (but not the position of the root, obviously). The betaproteobacteria branch with gammaproteobacteria in two groups, one includes N. meningitides (betaproteobacteria) and Polynucleobacter (gammaproteobacteria), whereas the other includes the rest of the species divided into two class-specific clades (supplementary fig. S4, Supplementary Material online). Reconstruction of the MLN for all proteobacteria using the gene content tree as the reference tree yielded the LGT7 model as the best fit between ancestral and contemporary genome sizes, whereas all other LGT allowance models were rejected (supplementary figs. S5 and S6, Supplementary Material online). Although it is the same LGT allowance model that was inferred using the rRNA reference tree, the mean LGT rate is lower—“but only slightly so”—using the gene content reference tree, with a weighted mean of 1.7 LGTs per protein family, in comparison to 1.9 with the rRNA tree. This somewhat lower rate is the result of reduced patchiness in gene distribution patterns using the gene content tree, leading to 29% monophyletic families (in comparison to 25% using the rRNA tree) whose distribution on the tree requires no LGT (table 2B). The small increment of average LGT rate from 1.9 to 1.7 using the gene content tree, where the patchiness criterion is used to cluster the genomes, simply reflects the patchiness of the data in total. In other words, the present data require a substantial amount of LGT to account for the observed gene distributions, any way one cuts the cake.
We repeated the same inference procedure for the class- and species-level data sets. At the class level, the best-fitting model using the gene content tree resulted in an inference of a lower LGT allowance for alphaproteobacteria (LGT1) and higher LGT allowance in betaproteobacteria (LGT3) and gammaproteobacteria (LGT7). As with the rRNA reference tree, no model was accepted for the deltaproteobacteria and epsilonproteobacteria, where the distribution of ancestral genome sizes shows that an allowance model between origin only and LGT1, had it existed in our approach, might be the most fitting for these classes (supplementary fig. S5, Supplementary Material online). In the three species-level data sets (E. coli, F. tularensis, and Y. pestis), the same LGT allowance model was accepted using the gene content reference tree, with slightly lower LGT rates (table 2B). Hence, our attempt to minimize LGT rate inference by reducing the patchiness of gene distribution patterns across the reference tree using the gene content tree resulted in more monophyletic protein families, yet the inferred LGT allowance models and average LGT rate were hardly changed and sometimes were even increased.
MLN Properties
The MLN reconstructed for all proteobacteria using T30 protein families, with the RNA reference tree, and the LGT7 model contains in total 657 nodes, with 329 external nodes (—operational taxonomic units [OTUs]) and 328 internal nodes (hypothetical taxonomic units [HTUs]), connected by 51,762 lateral edges (fig. 4). For protein families that have undergone more than one LGT, the number of lateral edges in the MLN exceeds the minimum number of LGTs required to account for the gene distribution. Hence, to address LGT network properties for the MLN, 1,000 rMLN were generated in which the number of lateral edges and the minimum number of LGTs for genes transferred more than once correspond exactly. Lateral edge frequency and edge weight distribution are similar among the rMLN networks. The number of lateral edges in the rMLNs is 3,345 ± 73 (coefficient of variation = 2%) on average. The connectivity (number of lateral edges per node) ranges between 0 and (344–384) with a mean between 100 and 102 and median between 85 and 91 (table 3). The connectivity distribution is semi-exponential with very few nodes that are highly connected (fig. 5A). Bigger genomes are generally more highly connected than smaller genomes, yet genome size explains only 16% of the variation in connectivity (P < 0.01, using Spearman correlation; Zar 1999).
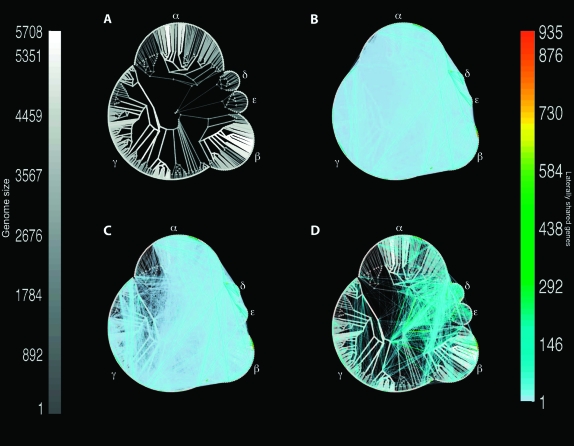
FIG. 4.
A minimal LGT network for 329 proteobacteria. (A) The reference tree used to ascribe vertical inheritance for inference of the MLN. (B) The MLN showing all 51,762 edges of weight ≥1 gene in the MLN. Vertical edges are indicated in gray, with both the width and the shading of the edge shown proportional to the number of inferred vertically inherited genes along the edge (see scale on the left). The lateral network is indicated by edges that do not map onto the vertical component, with number of genes per edge indicated in color (see scale on the right). (C) The MLN showing only the 13,632 edges of weight ≥5 genes. (D) The network showing only the 3,007 edges of weight ≥20 genes.
Table 3.
Statistical Properties of MLN/rMLNs in Phylum and Class Level.
| Prateobacteria | Alphaproteohacteria | Betaproteobacteria | Gammaproteobacteria | Escherichia coli | Francisella tularensis | Yersinia pestis | |
| No. edges | 33,457 ± 73 | 3,606 ± 17 | 1,595 | 8,447 ± 31 | 108 | 24 | 37 |
| Mean connectivity | 100–102 | 43–44 | 31 | 53–54 | 6 | 4 | 5 |
| Median connectivity | 85–91 | 46–52 | 33 | 47–52 | 5 | 5 | 6 |
| No. of edges > 20 | 982 ± 10 (3 ± 0.03%) | 313 ± 6 (9 ± 0.2%) | 145 (9%) | 409 ± 6 (5 ± 0.l%) | 29 (27%) | 4 (17%) | 7 (19%) |
| No. of edges ≥ 5 | 587 ± 30 (18 ± 0.l%) | 1,395 ± 12 (39 ± 0.4%) | 588 (37%) | 1,855 ± 15 (22 ± 0.2) | 60 (56%) | 15 (63%) | 18 (49%) |
| No. edges = 1 | 17,018 ± 91 (51 ± 0.2%) | l,017 ± 21 (28±0.5%) | 487 (30%) | 3,902 ± 42 (46 ± 0.4%) | 20 (19%) | 1 (4%) | 6 (16%) |
| No. of edges = 2 | 592 ± 61 (18 ± 0,2%) | 541 ± 18 (15 ± 0.5%) | 263 (16%) | 1,476 ± 29 (17 ± 0.3%) | 11 (10%) | 0 (0%) | 7 (19%) |
| No. of OTU–OTU edges | 1,188 ± 42(35 ± 0.l%) | l,206 ± 9+(33 ± 0.2%) | 555 (35%) | 2,832 ± 18 (34 ± 0.2%) | 41 (38%) | 9 (37%) | 18 (49%) |
| No. of HTU–HTU edges | 604 ± 39 (18 ± 0.l%) | 686 ± 9 (19 ± 0.2%) | 293 (18%) | 1,696 ± 16 (20 ± 0.2%) | 17 (16%) | 4 (17%) | 4 (11%) |
| No. of OTU–HTU edges | 1,553 ± 55 (46 ± 0.2%) | 1,713 ± 12 (47 ± 0.3%) | 747 (47%) | 3,919 ± 23 (46 ± 0.2%) | 50 (46%) | 11 (46%) | 15 (41%) |
OTU–OTU edges connect between two external nodes (contemporary species). HTU–HTU edges connect between two internal nodes (ancestral species). OTU–HTU edges connect between an external and an internal nodes.
FIG. 5.
Properties of the minimal LGT networks in phylum and class scales. Properties are shown for a randomly selected replicate. The coefficient of variation for the whole data was ∼2% (table 3). (A–D) Distribution of connectivity, the number of one-edge-distanced neighbors for each vertex, in the MLN. (E–H) Probability density function (PDF) of edge weight in the lateral component of the MLN.
The MLN reconstructed at the proteobacterial class level shows the distribution of laterally shared genes in higher resolution. Network properties for the alphaproteobacteria and gammaproteobacteria were calculated from 1,000 rMLN networks, the statistics of which show uniformity of lateral edge frequency and edge weight distribution (table 3). Data for the betaproteobacteria were extracted from the MLN directly because the best-fitting model was LGT1, which results in an MLN where the number of edges corresponds the minimum number of LGT events per protein family.
The connectivity distribution in the alphaproteobacterial MLN is bimodal, suggesting two groups of species that are either weakly or strongly connected within the lateral network (fig. 5B). The graphical representation of the MLN for that class reveals that the Rickettsiales comprise the weakly connected group (fig. 6A). In our data set, the order Rickettsiales includes 21 endosymbiotic parasites from the genera Anaplasma, Ehrlichia, and Rickettsia. The host-associated lifestyle of these species is a barrier to LGT in many cases and probably the reason for their low connectivity in the MLN. The connectivity distribution in the betaproteobacterial MLN is almost uniform (fig. 5C) with similar frequencies of nodes across the connectivity range (0–50 edges per node) and five more nodes whose connectivity is above this range. Clades of symbionts within the betaproteobacterial MLN, the Neisseriales and Nitrodomonadales, are weakly connected (fig. 6B). The Burkholderiales in our sample include 31 species of diverse lifestyles that account for the majority (60%) of betaproteobacterial species in the data. The overall gene distribution patterns are quite uniform across that order (fig. 1), yet the parasites among them (Ralstonia species) having lower connectivity than the free-living species (Burkholderia species; fig. 6B).
FIG. 6.
A minimal LGT network for proteobacterial classes alpha (A), beta (B), and gamma (C). Vertical edges are indicated in gray, with both the width and the shading of the edge shown proportional to the number of inferred vertically inherited genes along the edge (see scale bar). The lateral network is indicated by edges that do not map onto the vertical component, with number of genes per edge indicated in color (see scale bar). The MLN showing only edges of weight ≥5 genes.
The connectivity distribution in the gammaproteobacterial MLN is semi-exponential (fig. 5D). The graphical representation of the gammaproteobacterial MLN shows that symbionts, such as Pasteurellales, are weakly connected within the lateral network. The Enterobacteriales, comprising about third of the gammaproteobacteria in our sample (51 species) include four main genera, Escherichia, Shigella, Salmonella, and Yersinia. The MLN contains 1,326 (16%) lateral edges connecting among the nodes (internal and external) in this clade, suggesting abundant LGT among species in this group, with the exception of Yersinia, that like other pathogenic and symbiotic strains in our data set are relatively disconnected from the network (fig. 6C).
The distribution of lateral edge weight in the proteobacterial MLN is linear in log–log scale (fig. 5E), with a majority of single gene edges (51 ± 0.2%) and a minority of heavy edges weighing 20 genes or more (3 ± 0.03%). Similar edge weight distributions are observed within the alphaproteobacteria, betaproteobacteria, and gammaproteobacteria MLNs (table 3 and fig. 5F–H). This means that most of the LGT events among proteobacteria entail single genes rather than bulk transfers.
The MLN reconstruction for all species-level data sets, using both reference trees, prefers the LGT1 model with an average LGT frequency of about one LGT per protein family (table 2). The MLN reconstruction for the species level typically results in a heavy lateral edge that is found close to the root, between the first two nodes that diverge from it (supplementary fig. S7A–C, Supplementary Material online). Such a lateral edge means that many gene families of patchy distribution are shared between the two immediate descendants of the root node, the distribution of which cannot be explained by vertical inheritance alone. Reducing the patchiness of the gene distribution patterns by using a gene content reference tree resulted in similar MLNs (supplementary fig. S7D–F, Supplementary Material online). In species-level MLNs, the majority of nodes are connected by a lateral edge, exept two to four nodes (Supplementary fig. S8, Supplementary Material online). The distribution of edge weights is semi-linear in a log–log scale, hence most of the LGT events are of single genes, whereas individual transfer events involving many genes are rare.
The distribution of lateral edges within the proteobacteria rMLN shows that the probability for an intraclass lateral edge (9.4 ± 0.03) is similar to the probability for an interclass lateral edge (6.9 ± 0.02). However, the median edge weight of intraclass edges, which is two genes per edge in all rMLNs, is significantly larger (P < 0.05) than that of interclass edges, a single gene per edge in all rMLNs. This means that the probability for an LGT event within and outside the class is similar, yet more genes are transferred per LGT event between species from the same class. The probability for a lateral edge between the different classes reveals that LGT between alphaproteobacteria, deltaproteobacteria, and betaproteobacteria is similar, but LGT between epsilonproteobacteria or gammaproteobacteria and other classes is lower (table 4).
Table 4.
Frequency and Weight of Lateral Edges in Intraclass and Interclass Subsets.
| Alphaproteobacteria | Deltaproteobacteria | Epsilonproteobacteria | Betaproteobacteria | Gammaproteobacteria | |
| Alphaproteobacteria | 12.9 ± 0.07 (3–3) | ||||
| Deltaproteobacteria | 14 ± 1.5 (2–2) | 29 ± 0.3 (7–9) | |||
| Epsilonproteobacteria | 3.8 ± 0.1 (1–1) | 19 ± 0.3 (2–2) | 20.2 ± 0.03 (3–5) | ||
| Betaproteobacteria | 11 ± 0.08 (1–2) | 13.8 ± 0.1 (1–2) | 3.68 ± 0.1 (1–1) | 15.8 ± 0.1 (2–3) | |
| Gammaproteobacteria | 5.1 ± 0.04 (1–1) | 8.1 ± 0.09 (1–1) | 3.5 ± 0.06 (1–1) | 7.1 ± 0.05 (1–1) | 7.4 ± 0.04 (1–2) |
Numbers in parenthesis denote edge weight range.
Edge probability is calculated as the frequency of edges divided by the number of nodes in the group.
Proportion of Recent Gene Acquisition and Cumulative Impact of LGT
Most of the edges in the proteobacterial MLN (46 ± 0.2% of edges in the rMLN) connect between OTU nodes (contemporary genomes) and HTU nodes (ancestral genomes). Such edges are inferred for protein families that are shared among a group of species where all except one are grouped into one monophyletic clade. The reconstructed lateral edge connects the common ancestor of that clade and the OTU of the outsider species. Lateral edges connecting two OTU nodes are slightly less frequent (35 ± 0.1%), whereas edges connecting two HTU nodes are the minority (18 ± 0.1%; fig. 7). Similar ratios of lateral edge types were inferred for the classes and species data sets (table 3).
FIG. 7.
A three-dimensional projection of the MLN. Edges in the vertical component are shown in the same gray scale as in figure 3. Vertices inferred as gene origin in the same protein family are connected by a lateral edge signifying a laterally shared gene. Lateral edges are classified into three groups according to the types of vertices they connect within the vertical component (see table 3 for details): 11,941 OTU–OTU edges (magenta), 15,425 HTU–OTU edges (yellow), and 6,066 HTU–HTU edges (cyan).
Lateral edges connecting between two OTUs reflect recent LGT events. The proportion of protein families connected by an OTU–OTU edge per genome may serve as a lower bound estimate for the proportion of recently acquired genes within the genome. The average proportion of recent acquisitions per genome inferred from the MLN in phylum depth with the rRNA reference tree is 9.6% recently acquired genes per genome. Moreover, the frequency of recently acquired genes positively correlated with genome size (rs = 0.6, P < 0.01). Similar mean proportions of recently acquired genes are estimated for the three classes (7–9%; table 5). The estimated proportions in the species level are about 4% recent acquisitions (table 5). To test how our estimates are affected by the sample of species included in the MLN, we compared them for the same group of species, from the MLNs reconstructed in class and phylum phylogenetic depth. We find that larger sample size results in slightly higher proportions of recently acquired genes (0.1–4.2% difference; table 5). Hence, the phylogenetic depth (i.e., sample size) has little influence on the inferred proportions of recently acquired genes using the MLN approach.
Table 5.
Recently Acquired Genes and Cumulative Impact of LGT.
| Phvloqenetic depth | % Recent LGT by MLN |
% Recent LGT by Nucleotide Pattern | Ratio of MLN/Nucleotide Pattern | % Cumulative LGT by MLN |
||||
| Phylum | Class | Species | Phylum | Phylum | Class | Species | ||
| Proteobacteria | 9.7 ± 7.7 | 21.5 ± 8.9 | 0.5 ± 0.6 | 73.7 ± 10.9 | ||||
| Alpha | 9.6 ± 7.0 | 9.2 ± 8.5 | 16.6 ± 7.9 | 0.6 ± 0.5 | 69.1 ± 9.9 | 60.9 ± 12.3 | ||
| Beta | 11.1 ± 5.8 | 6.9 ± 6.0 | 26.8 ± 8.4 | 0.5 ± 0.3 | 75.2 ± 4.7 | 60.0 ± 8.3 | ||
| Gamma | 7.3 ± 5.1 | 7.2 ± 6.9 | 21.4 ± 7.8 | 0.4 ± 0.4 | 78.6 ± 9.4 | 76.6 ± 10.2 | ||
| Escherichia coli | 5.0 ± 3.8 | 4.1 ± 3.1 | 3.3 ± 2.7 | 28.5 ± 2.1 | 0.2 ± 0.1 | 85.2 ± 3.8 | 84.6 ± 3.8 | 26.8 ± 3.5 |
| Francisella tularensis | 5.0 ± 5.3 | 4.1 ± 5.2 | 4.4 ± 6.9 | 17.5 ± 0.7 | 0.3 ± 0.3 | 67.4 ± 2.1 | 65.1 ± 2.0 | 17.0 ± 2.0 |
| Yersinia pestis | 4.0 ± 3.3 | 3.1 ± 2.6 | 3.9 ± 7.3 | 26.7 ± 0.8 | 0.2 ± 0.1 | 86.0 ± 2.0 | 84.6 ± 2.0 | 17.5 ± 3.4 |
The MLN, comprising of both phylogenetic tree for the vertically inherited genes and lateral network for the laterally transferred genes, enables us to estimate the cumulative impact of LGT during microbial evolution. The proportion of protein families within each genome that is connected by a lateral edge reflects the proportion of genes within the genome that was affected by LGT during their history. Within the phylum depth using the rRNA reference tree, we find that, on average, 73% of the genes per genome are affected by LGT at some point during evolution. A similar proportion is observed with the class depth for gammaproteobacteria, whereas in alphaproteobacteria and betaproteobacteria, we find lower cumulative impact of LGT (60%; table 5). The same inference in species depth yields significantly lower proportions (17–26%; table 5). To test if the cumulative impact of LGT in species depth is indeed lower or rather an outcome of smaller sample size, we compared the inference for the same species using the phylum and class depth data sets. We find that the proportion of genes affected by LGT during evolution inferred in species depth is much lower than the inference using the class or phylum data sets.
How Severely Does the MLN Underestimate LGT?
The estimated proportion of recently acquired genes per genome using the MLN is 9.7% of each genome in the phylum depth on average, that is, lower than the proportion of recent LGT inferred using aberrant nucleotide patterns that in earlier studies was between 14% and 18% per genome (Lawrence and Ochman 1998; Nakamura et al. 2004). The MLN approach is expected to yield lower bound minimum estimates mainly because it relies on gene presence/absence patterns that are uninformative for evolutionary events, such as allele recombination and gene replacement by LGT (e.g., Andam et al. 2010), and because it conservatively does not count all LGT events that might be detected by tree comparisons (Dagan et al. 2008). How severely does the MLN underestimate LGT? In order to ascertain this, we compared the proportions of recent LGT per genome using the MLN approach with that determined on the basis of aberrant nucleotide patterns by detecting all genes having significantly different GC content in comparison to their genome. The GC content method preferentially reveals recently acquired genes that exhibit an atypical codon usage indicating their foreign origin (Lawrence and Ochman 1998; Nakamura et al. 2004). Across the phylogenetic samples studied, the frequency of genes detected as recently acquired using the two methods is positively correlated (rs = 0.55, P < 0.01) (table 5). However, the GC method detects an average of 21% recent acquisitions per genome in the proteobacterial phylum sample or roughly twice the value estimated by MLN, whereby the degree to which the MLN approach underestimates recent LGT increases to about a factor of six as the sample approaches the species level (table 5). Both effects—MLN underestimation and its increase toward the species level—are attributable to the circumstance that two kinds of genes are excluded from the MLN approach. First, the GC content approach can identify acquisitions from any donor genome, whereas the MLN only identifies LGTs involving genomes within the sequenced set. Second, the GC content approach identifies LGT among singletons, whereas the MLN does not. Both effects become more severe with smaller and more closely related genome samples. Thus, although the graphical representation of the MLN (fig. 7) might appear quite complex in terms of lateral edges, it still represents a minimum estimate, not an optimal estimate, of gene sharing among these genomes.
Conclusions
Network analyses of proteobacterial genomes reveal frequent LGT among members of the phylum. The main trends in proteobacterial gene sharing are observed among species from different taxa inhabiting the same habitat. Together with the high content of plasmid proteins in laterally shared protein families, this suggests that most of the LGT in proteobacteria occurs over short physical distances, where donor and recipient are proximate. Our analysis shows that higher LGT rates are inferred within the phylum level than the species level; yet, LGT is more probable among similar species from the same class, so that modules of shared protein families are similar to traditional proteobacterial classification schemes but lacking the traditional hierarchy.
Supplementary Material
Supplementary tables S1–S5 and figures S1–S8 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Shijulal Nelson-Sathi for critically reading the manuscript. This study was supported by the German Research Foundation (T.K., T.D., and W.M.), the German Federal Ministry of Education and Research (O.P., T.D., and W.M.), and an European Research Council grant NETWORKORIGINS (W.M.). Computational support and infrastructure were provided by the “Zentrum fuer Informations- und Medientechnologie” at the Heinrich Heine University of Duesseldorf. Raw data and phylogenetic trees reconstructed in this study are publicly available at www.molevol.de/resources.
References
- Andam CP, Williams D, Gogarten JP. Biased gene transfer mimics patterns created through shared ancestry. Proc Natl Acad Sci U S A. 2010;107:10679–10684. doi: 10.1073/pnas.1001418107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M. Direct visualization of horizontal gene transfer. Science. 2008;319:1533–1536. doi: 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- Bapteste E, O'Malley MA, Beiko RG, et al. (11 co-authors) Prokaryotic evolution and the tree of life are two different things. Biol Direct. 2009;4:34. doi: 10.1186/1745-6150-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci U S A. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Brinkhoff T, Giebel HA, Simon M. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol. 2008;189:531–539. doi: 10.1007/s00203-008-0353-y. [DOI] [PubMed] [Google Scholar]
- Chaffron S, Rehrauer H, Pernthaler J, von Mering C. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 2010;20:947–959. doi: 10.1101/gr.104521.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Beiko R, Darling A, Ragan M. Lateral transfer of genes and gene fragments in prokaryotes. Genome Biol Evol. 2009;1:439–448. doi: 10.1093/gbe/evp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Cordero OX, Hogeweg P. The impact of long-distance horizontal gene transfer on prokaryotic genome size. Proc Natl Acad Sci U S A. 2009;106:21748–21753. doi: 10.1073/pnas.0907584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci U S A. 2008;105:10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W. Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc Natl Acad Sci U S A. 2007;104:870–875. doi: 10.1073/pnas.0606318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Roettger M, Bryant D, Martin W. Genome networks root the tree of life between prokaryotic domains. Genome Biol Evol. 2010;2:379–392. doi: 10.1093/gbe/evq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidov Y, Jurkevitch E. Predation between prokaryotes and the origin of eukaryotes. Bioessays. 2009;31:748–757. doi: 10.1002/bies.200900018. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc Natl Acad Sci U S A. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Boucher Y, Nesbo CL, Douady CJ, Andersson JO, Roger AJ. How big is the iceberg of which organellar genes in nuclear genomes are but the tip? Philos Trans R Soc Lond B Biol Sci. 2003;358:39–57. doi: 10.1098/rstb.2002.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A, Ostrowski M, Scanlan DJ, et al. (15 co-authors) Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 2008;9:R90. doi: 10.1186/gb-2008-9-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema TJG, Andersson SGE. The alpha-proteobacteria: the Darwin finches of the bacterial world. Biol Lett. 2009;5:429–432. doi: 10.1098/rsbl.2008.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Parsimony in systematics—biological and statistical issues. Annu Rev Ecol Syst. 1983;14:313–333. [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) Seattle (WA): Department of Genome Sciences, University of Washington; 2004. [Google Scholar]
- Fischer D, Eisenberg D. Finding families for genomic ORFans. Bioinformatics. 1999;15:759–762. doi: 10.1093/bioinformatics/15.9.759. [DOI] [PubMed] [Google Scholar]
- Fukami-Kobayashi K, Minezaki Y, Tateno Y, Nishikawa K. A tree of life based on protein domain organizations. Mol Biol Evol. 2007;24:1181–1189. doi: 10.1093/molbev/msm034. [DOI] [PubMed] [Google Scholar]
- Galtier N, Daubin V. Dealing with incongruence in phylogenomic analyses. Philos Trans R Soc Lond B Biol Sci. 2008;363:4023–4029. doi: 10.1098/rstb.2008.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Mohan R, Gupta RS. Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria. Int J Syst Evol Microbiol. 2009;59:234–247. doi: 10.1099/ijs.0.002741-0. [DOI] [PubMed] [Google Scholar]
- Garcia-Vallve S, Romeu A, Palau J. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 2000;10:1719–1725. doi: 10.1101/gr.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D, Li WH. Fundamentals of molecular evolution. 2nd ed. Chapter 4. Sunderland (MA): Sinauer Associates; 2000. pp. 99–164. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Molecular signatures (unique proteins and conserved indels) that are specific for the epsilon proteobacteria (Campylobacterales) Bmc Genomics. 2006;7:167. doi: 10.1186/1471-2164-7-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. Network analyses structure genetic diversity in independent genetic worlds. Proc Natl Acad Sci U S A. 2010;107:127–132. doi: 10.1073/pnas.0908978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Jain R, Rivera MC, Moore JE, Lake JA. Horizontal gene transfer accelerates genome innovation and evolution. Mol Biol Evol. 2003;20:1598–1602. doi: 10.1093/molbev/msg154. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Sato S, Tabata S. Complete genomic sequences and comparative analyses of cyanobacteria. Plant Cell Physiol. 2002;43:S123. [Google Scholar]
- Kersters K, Vos PD, Gillis M, Swings J, Vandamme P, Stackebrandt E. Introduction to the Proteobacteria. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The prokaryotes volume 5: Proteobacteria: alpha and beta subclasses. New York: Springer; 2006. pp. 3–37. [Google Scholar]
- Krylov DM, Wolf YI, Rogozin IB, Koonin EV. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Goldovsky L, Darzentas N, Ouzounis CA. The net of life: reconstructing the microbial phylogenetic network. Genome Res. 2005;15:954–959. doi: 10.1101/gr.3666505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AS, Beatty JT. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15:54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci U S A. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:807–814. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IM, Palaniappan K, et al. (12 co-authors) The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2010;38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney JO, Pisani D. Genetics—paradigm for life. Science. 2007;318:1390–1391. doi: 10.1126/science.1151657. [DOI] [PubMed] [Google Scholar]
- Mirkin BG, Fenner TI, Galperin MY, Koonin EV. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongodin EF, Nelson KE, Daugherty S, et al. (18 co-authors) The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci U S A. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci U S A. 2007;104:8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Wernegreen JJ. Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol Evol. 2000;15:321–326. doi: 10.1016/s0169-5347(00)01902-9. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- Newman MEJ. Finding community structure in networks using the eigenvectors of matrices. Phys Rev. 2006:E74. doi: 10.1103/PhysRevE.74.036104. [DOI] [PubMed] [Google Scholar]
- Novichkov PS, Omelchenko MV, Gelfand MS, Mironov AA, Wolf YI, Koonin EV. Genome-wide molecular clock and horizontal gene transfer in bacterial evolution. J Bacteriol. 2004;186:6575–6585. doi: 10.1128/JB.186.19.6575-6585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Oyston PCF. Francisella tularensis: unravelling the secrets of an intracellular pathogen. J Med Microbiol. 2008;57:921–930. doi: 10.1099/jmm.0.2008/000653-0. [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, Lercher MJ, Csermely P, Oliver SG, Hurst LD. Chance and necessity in the evolution of minimal metabolic networks. Nature. 2006;440:667–670. doi: 10.1038/nature04568. [DOI] [PubMed] [Google Scholar]
- Pei AY, Oberdorf WE, Nossa CW, et al. (16 co-authors) Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl Environ Microbiol. 2010;76:3886–3897. doi: 10.1128/AEM.02953-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Anderson I, Makarova KS, et al. (27 co-authors) A genomic analysis of the archaeal system Ignicoccus hospitalis-Nanoarchaeum equitans. Genome Biol. 2008;9:R158. doi: 10.1186/gb-2008-9-11-r158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poptsova MS, Gogarten JP. Using comparative genome analysis to identify problems in annotated microbial genomes. Microbiology. 2010;156:1909–1917. doi: 10.1099/mic.0.033811-0. [DOI] [PubMed] [Google Scholar]
- Roettger M, Martin W, Dagan T. A machine-learning approach reveals that alignment properties alone can accurately predict inference of lateral gene transfer from discordant phylogenies. Mol Biol Evol. 2009;26:1931–1939. doi: 10.1093/molbev/msp105. [DOI] [PubMed] [Google Scholar]
- Scott KM, Sievert SM, Abril FN, et al. (43 co-authors) The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLoS Biol. 2006;4:2196–2212. doi: 10.1371/journal.pbio.0040383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Falkowski PG. Genome evolution in cyanobacteria: the stable core and the variable shell. Proc Natl Acad Sci U S A. 2008;105:2510–2515. doi: 10.1073/pnas.0711165105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12:17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- Sorek R, Zhu YW, Creevey CJ, Francino MP, Bork P, Rubin EM. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318:1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Murray RGE, Truper HG. Proteobacteria-classis nov, a name for the phylogenetic taxon that includes the purple bacteria and their relatives. Int J Syst Bacteriol. 1988;38:321–325. [Google Scholar]
- Sukdeo N, Honek JF. Microbial glyoxalase enzymes: metalloenzymes controlling cellular levels of methylglyoxal. Drug Metabol Drug Interact. 2008;23:29–50. doi: 10.1515/dmdi.2008.23.1-2.29. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Sun LV, Vamathevan J, et al. (30 co-authors) Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:327–341. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar JH. Biostatistical analysis. Upper Saddle River (NJ): Prentice Hall; 1999. [Google Scholar]
- Zhaxybayeva O, Gogarten JP, Charlebois RL, Doolittle WF, Papke RT. Phylogenetic analyses of cyanobacterial genomes: quantification of horizontal gene transfer events. Genome Res. 2006;16:1099–1108. doi: 10.1101/gr.5322306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.