Abstract
The PML-RARα oncogene is the central effector of acute promyelocytic leukemia (APL). PML-RARα physically interacts with epigenetic-modifying enzymes including DNA methyltransferases (Dnmt) to suppress critical downstream targets. Here, we show that increased expression of Dnmt3a1 cooperates with PML-RARα in vivo to promote early lethality secondary to myeloid expansion and dysfunction in primary mice. Bone marrow cells from these mice cause leukemogenesis with a shortened latency and a higher penetrance on transplantation into irradiated recipients. Furthermore, leukemic cells overexpressing PML-RARα and Dnmt3a1 display increased methylation at a target promoter compared with PML-RARα or Dnmt3a1 controls. Our findings show a cooperation between the PML-RARα oncogene and the Dnmt3a1 enzyme in vivo and that Dnmt levels can be rate limiting in APL progression.
Introduction
Acute promyelocytic leukemia (APL) is characterized by the balanced reciprocal chromosomal rearrangement t(15;17), which results in the juxtaposition of the promyelocytic leukemia gene (PML) and the retinoic acid receptor α gene (RARA) to form the oncogenic fusion protein PML-RARα (1). This is accompanied by a block in differentiation resulting in an overproduction of immature myeloid cells, called promyelocytes, in the bone marrow (2, 3). Transgenic mice expressing PML-RARα under the control of the myeloid-specific promoter, cathepsin G, develop myeloid hyperplasia, with around 15% to 30% of mice progressing to leukemia after a latency period of 9–12 months (4). Additional genetic changes cooperate with PML-RARα to reduce the latency and increase the penetrance of APL development, suggestive of a requirement for secondary cooperative events (5, 6).
It has been shown that PML-RARα recruits epigenetic enzymes such as DNA methyltransferases (Dnmt) to specific target gene promoters, resulting in CpG island methylation and transcriptional repression in cultured human APL cells (7). Additionally, in vitro experiments reveal that PML-RARα also interacts with other epigenetic proteins such as histone deacetylases (8, 9), the methyl-CpG–binding protein MBD1 (10), and the Polycomb repressive complex 2 (PRC2) to silence target genes, including those normally regulated by RARα (11).
APL cells can be forced to differentiate in the presence of superphysiologic doses of all-trans retinoic acid (ATRA). Interestingly, treatment of APL cells with a combination of Dnmt inhibitors and ATRA seems to enhance the differentiation of APL cells, suggesting a cooperative role for Dnmts and PML-RARα in APL maintenance (7). However, a role for Dnmts or other epigenetic enzymes in the development of leukemia has not been shown. Therefore, we tested the ability of the epigenetic enzyme Dnmt3a1 to cooperate with PML-RARα in inducing APL in mice. We predicted that over-expression of Dnmt3a1 along with PML-RARα would result in enhanced silencing of PML-RARα targets and enhance leukemogenesis. Indeed, transplantation of cells from the bone marrow of PML-RARα+Dnmt3a1 mice into irradiated recipients resulted in the development of leukemia with a greater penetrance and shorter latency compared with cells obtained from PML-RARα mice. Additionally, leukemic cells from PML-RARα+Dnmt3a1 mice displayed enhanced methylation at a target gene promoter. Together, our results show that PML-RARα and Dnmt3a1 cooperate in vivo to promote oncogene-specific target methylation and development of APL.
Materials and Methods
Mouse strains
hCG-PML-RARα (4) mice were crossed to Rosa26rtTA mice (12). Progeny of these crosses were bred to the TRE-Dnmt3a1 mice (13) to obtain triple transgenic mice and the relevant controls. Mice were maintained on 2 mg/mL doxycycline supplemented with 10 mg/mL sucrose in their drinking water from 3 weeks of age onwards. The following primers were used for genotyping:
For Rosa26rtTA:
Rosa 26a: AAAGTCGCTTCTGAGTTGTTAT
Rosa 26b: GCGAAGAGTTTGCCTCAACC
Rosa 26c: GAGGGGAGAAATGGATAT
For hCG-PML-RARα:
hCGFwd: GGCCTGACCTCATCCCATAG
hCGRev: GCCCTTTTCCCCATCCTAGG
For TRE-Dnmt3a1:
ColA: GCACAGCATTGCGGACATGC
ColB: CCCTCCATGTGTGACCAAGG
ColC: GCAGAAGCGCGGCCGTCTGG
Mice were bred and maintained at the University of California at San Francisco (UCSF), and their care was in accordance with UCSF guidelines.
Survival analysis
Mice of the noted genotypes were followed over a period of 288 days, with percentage of survivors calculated at the end of this period.
Methylcellulose colony formation
Total bone marrow cells (5,000) were cultured in Iscove’s modified Dulbecco’s medium–based methylcellulose medium (Methocult M3100; Stemcell Technologies) and supplemented as previously described (14). Cells were replated every 7 days on fresh methylcellulose medium.
Transplantation studies
Congenic recipient mice were irradiated using a cesium source irradiator with lethal (1,200 rad) dose delivered in a split dose 3 hours apart and were given antibiotic-containing water for at least 4 weeks after irradiation. Mice were injected immediately after irradiation. For i.v. injections, 2 × 106 donor bone marrow cells were mixed with 300,000 Sca1-depleted congenic spleen cells, resuspended in a volume of 100 μL, and injected into the retro-orbital plexus. Donor and recipient cells were distinguished by expression of different allelic forms of CD45 (CD45.1 versus CD45.2). Throughout the experiment, transplanted recipients were maintained on doxycycline-containing water. Round 1 transplants were performed using fresh bone marrow cells, whereas round 2 transplants were performed with cryopreserved cells.
Bisulfite sequencing
We isolated genomic DNA from spleen cells. Bisulfite conversion was performed as previously described (15). Briefly, 3 μg of genomic DNA from spleen were digested with EcoRV. DNA was purified using phenol-chloroform extraction after restriction digestion followed by denaturation with NaOH. Bisulfite conversion was carried out overnight at 55°C followed by cleanup using the Promega Wizard Cleanup kit (using the manufacturer’s protocol). PCR was performed using primers (forward: GGTTTGGTTAGGAATAGGAGAGTAGA; reverse: AACAACCCTACAAAAACCTTCAAC) to amplify the RARβ promoter. PCR products were cloned into the PCR2.1 vector using the TOPO-TA kit (Invitrogen) and transformed into chemically competent bacteria. Individual colonies were picked, expanded, DNA extracted, and sequenced using the same primers.
Cell staining and flow cytometry
For flow cytometry, single-cell suspension of bone marrow or spleen cells was prepared in HBSS + 2% fetal bovine serum (FBS). The following mouse antibodies, c-kit, Sca1, CD34, FcγR, Flk2, Gr1, Mac1, Ter119, B220, CD19, CD8, CD4, and CD3, conjugated with fluorophores FITC, phycoerythrin (PE), Pacific Blue, Cy7PE, Cy5PE, and allophycocyanin (APC), were used for staining. These were added to cells at appropriate concentrations and incubated on ice for 20 minutes in the dark. Biotinylated antibodies were washed and incubated with streptavidin-Cy7PE. Cells were washed and resuspended in a final volume of 300 μL of HBSS + 2% FBS with propidium iodide for dead cell exclusion. Events (30,000) were collected for analyzing mature cell populations, whereas 1,000,000 events were collected for analyzing stem cells using a BD LSRII. Data were analyzed using FlowJo. Forward and side scatter were used to select gated cells for analysis.
Histopathology
Sternum, spleen, kidney, liver, lungs, and heart were fixed in 10% buffered formalin solution. Sternums were decalcified before embedding in paraffin. Sections were stained with H&E.
Oxidative burst
Inflammation was induced by injecting 1 mL of a 3% thioglycollate solution intraperitoneally. Peritoneal exudates were harvested 72 hours later by lavage with PBS. Cells were spun down and resuspended in 1.5 mL HBSS + 2% FBS. Cell suspension (300 μL) was aliquoted into each tube. Dihydrorhodamine-123 (3 μL of 29 nmol/L) was added and incubated at 37°C for 5 minutes. Phorbol myristate acetate (60 μL of 20 μg/mL) was added per tube and incubated at 37°C for 15 minutes. Samples were immediately analyzed on a FACSCalibur, and the shift in FL1 fluorescence was determined using FlowJo.
Results
Building of an inducible Dnmt3a1–PML-RARα APL model
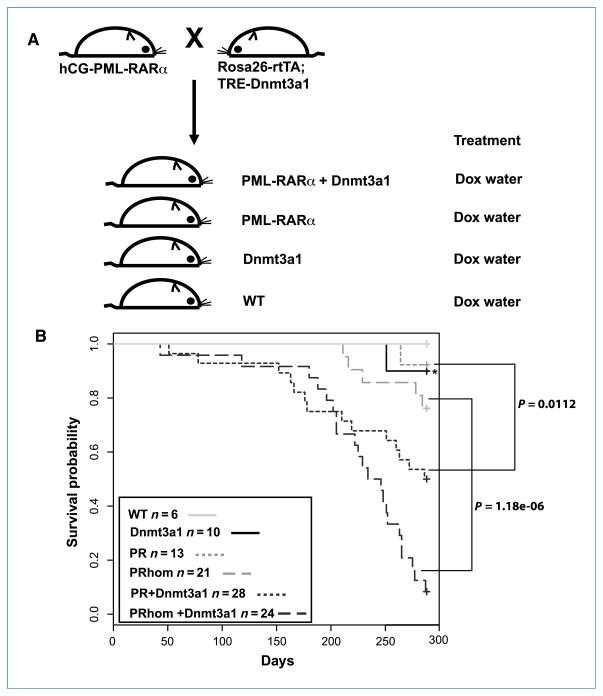
Dnmt3a forms a complex with PML-RARα at target promoters, resulting in their transcriptional silencing in human APL cells (7). To determine whether PML-RARα and Dnmt3a cooperate to induce leukemia in mice, we used an inducible system, whereby Dnmt3a1 is under the control of the tetracycline-inducible promoter element (TRE-Dnmt3a1; ref. 13). TRE-Dnmt3a1 mice were crossed to mice expressing the reverse tetracycline-controlled transactivator, rtTA, under the control of the ubiquitous promoter, Rosa26 (R26-rtTA; ref. 12), and PML-RARα (PR) under the control of the human myeloid-specific promoter, cathepsin G (hCG; ref. 4), to generate triple transgenic mice PR+rtTA+Dnmt3a1 (PR+Dnmt3a1; Fig. 1A). As we were unable to determine the copy number of the PML-RARα allele from these crosses, we later established a breeding strategy by which we obtained only homozygous PML-RARα mice (PRhom mice). Mice that carried the R26-rtTA and TRE-Dnmt3a1 alleles in addition to the homozygous PML-RARα were designated as PRhom+Dnmt3a1. Controls included in these studies include mice carrying both R26-rtTA and TRE-Dnmt3a1 alleles (Dnmt3a1 mice) and mice carrying either one or none of the alleles [wild-type (WT) mice; Fig. 1A]. All mice were maintained on doxycycline-containing water beginning at 3 wk of age. Dnmt3a1 mRNA and protein levels were induced, on the administration of doxycycline, only in mice that carried both Rosa26rtTA and TRE-Dnmt3a1 alleles (Supplementary Fig. S1A and C). Induction of Dnmt3a1 did not result in an alteration in PML-RARα expression levels (Supplementary Fig. S1B).
Figure 1.
Overexpression of Dnmt3a1 with hCG-PML-RARα cooperates to shorten survival of transgenic mice. A, generation of hCG-PML-RARα, Rosa26rtTA, and TRE-Dnmt3a1 mice and controls. Progeny of crosses between hCG-PML-RARα and Rosa26rtTA; TRE-Dnmt3a1 were maintained on doxycycline (Dox)–containing water from 3 wk of age. B, graph showing survival percentage of WT, Dnmt3a1, PR, PR+Dnmt3a1, PRhom, and PRhom+Dnmt3a1 mice maintained on doxycycline-containing water starting at ~3 wk of age. Survival was monitored over a period of 288 d. Asterisk indicates the single Dnmt3a1 mouse that died. Postmortem analysis of this mouse revealed normal myeloid compartment in both bone marrow and spleen.
PML-RARα and Dnmt3a1 cooperate to enhance lethality in mice
Mice expressing only PML-RARα under the control of the hCG promoter develop fatal leukemia with a long latency period and a low penetrance (4). To determine whether Dnmt3a1 cooperates with PML-RARα to enhance fatal leukemia, we monitored control and experimental mice for changes in their morbidity and mortality over a period of 288 days. WT (n = 6), Dnmt3a1 (n = 10), R (n = 13), PR+Dnmt3a1 (n = 28), PRhom (n = 21), and PRhom+Dnmt3a1 (n = 24) mice were maintained on doxycycline-containing water. Lethality was observed as early as 51 days for PR+Dnmt3a1 mice and 43 days for PRhom+Dnmt3a1 mice (Fig. 1B). This was significantly earlier than the lethality seen for PR mice (264 days) and for PRhom mice (211 days). No early lethality was observed in WT mice, whereas a single mouse died in the Dnmt3a1 group at 251 days. These data show that Dnmt3a1 cooperates with PML-RARα to shorten survival of mice.
Mice expressing PML-RARα alone and PML-RARα with Dnmt3a1 display similar levels of myeloid expansion in the bone marrow and spleen
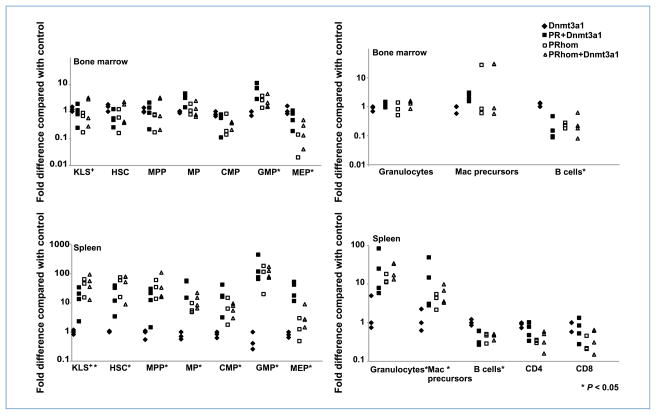
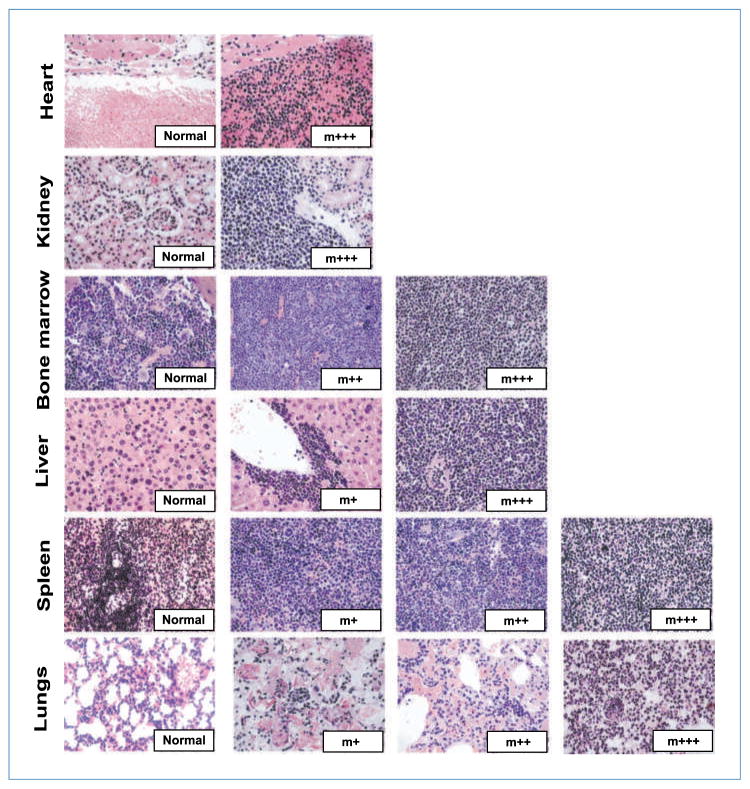
To determine the cause of morbidity and mortality in mice expressing PML-RARα and Dnmt3a1, immunophenotyping and population analysis was performed on bone marrow and spleen cells isolated from control Dnmt3a1, PRhom, and PRhom+Dnmt3a1 mice. Cells were separated based on well-characterized cell surface markers defining different hematopoietic populations including hematopoietic stem cells (HSC), multipotent progenitors (MPP), common myeloid progenitors (CMP), granulocyte monocyte progenitors (GMP), megakaryocyte erythrocyte progenitors (MEP), mature granulocytes, immature myeloid cells (c-kit+/Mac1+), CD4 T cells, CD8 T cells, and B cells. Dramatic changes were seen in several of these populations across the mutant mice (Fig. 2; Supplementary Fig. S2). Specifically, the bone marrow showed a significant increase in the fraction of GMP and decrease in the fraction of MEP and B cells in PRhom, PRhom+Dnmt3a1, and PR+Dnmt3a1 mice (Supplementary Fig. S2, top; Fig. 2). No significant change was observed in the other bone marrow cell populations. The spleens showed a broader spectrum of changes with increases in the HSC, MPP, immature myeloid, and mature myeloid populations, accompanied by a decrease in the percentage of B cells (Supplementary Fig. S2, bottom; Fig. 2). Histologic examination of organs revealed a myeloid expansion in the bone marrow and spleen of PRhom, PRhom+Dnmt3a1, and PR+Dnmt3a1 mice, consistent with the flow cytometric analysis (Table 1; Fig. 3). Bone marrow cells from PRhom and PRhom +Dnmt3a1 mice cultured in vitro on methylcellulose showed similar proliferation and differentiation into myeloid cells (Supplementary Fig. S3). Together, these results show a similar myeloid hyperplasia phenotype in PML-RARα mice with or without Dnmt3a1 overexpression.
Figure 2.
Myeloid expansion in mice expressing PML-RARα and Dnmt3a1. Graphs represent fold differences in percentage of cells in mice analyzed compared with their experimental control (either WT or Dnmt3a1) from five independent experiments. Based on the expression of specific cell surface markers, cells of the bone marrow and spleen were classified into the following populations: (a) hematopoietic stem and progenitors (KLS+; c-kit+/Lin−/Sca1+); (b) HSCs (c-kit+/Lin−/Sca1+/Flk2−/CD34−); (c) MPPs (c-kit+/Lin−/Sca1+/Flk2+/CD34+); (d) myeloid progenitors (MP; c-kit+/Lin−/Sca1−); (e) granulocytes (Gr1+/Mac1+); (f) myeloid precursors (Mac1+/c-kit+); (g) B cells (B220+/CD19+); and (h) T cells (CD4+ or CD8+). MP cells were further subdivided into CMPs (CD34+/FcγR−), granulocyte/macrophage progenitors (GMP; CD34+/FcγR+), and megakaryocyte/erythrocyte progenitors (MEP; CD34−/FcγR−). Fold difference in the percentage of cells in each compartment is calculated based on the following formula: fold difference = (Exp %)/(Control %). Statistical analysis was performed using Kruskal-Wallis test. *, P < 0.05.
Table 1.
Summary of histopathology of organs retrieved from mice included in the immunophenotypic analysis
| Mouse no. | Genotype | Age | Spleen | BM | Liver | Kidney | Lungs | Heart |
|---|---|---|---|---|---|---|---|---|
| 218.16 | Dnmt3a1 | 112 | ONR | – | – | – | – | – |
| 279.10 | Dnmt3a1 | 133 | – | – | – | – | – | ONR |
| 58.9 | PR+Dnmt3a1 | 140 | ONR | m+++ | m+ | ONR | – | – |
| 156.5 | PR+Dnmt3a1 | 193 | ONR | m++ | – | ONR | c+ | – |
| 226.1 | PR+Dnmt3a1 | 101 | ONR | m++ | m++ | – | m+ | – |
| lymph+ | ||||||||
| c++ | ||||||||
| 237.4 | PR+Dnmt3a1 | 121 | ONR | m++ | m+ | – | m+ | – |
| c++ | ||||||||
| 218.4 | PR+Dnmt3a1 | 185 | m+ | m++ | – | – | – | – |
| 273.6 | PRhom | 132 | m+ | m++ | m+ | – | – | ONR |
| 254.2 | PRhom | 166 | m++ | m++ | m++ | – | m+ | – |
| 254.13 | PRhom | 162 | m+ | m++ | m+ | – | – | – |
| 254.1 | PRhom+Dnmt3a1 | 146 | m+ | m++ | m+ | – | m++ | – |
| lymph+ | ||||||||
| c+ | ||||||||
| 272.2 | PRhom+Dnmt3a1 | 133 | m+ | m++ | m+ | – | lymph+ | ONR |
| c+ | ||||||||
| 254.6 | PRhom+Dnmt3a1 | 166 | m+ | m++ | – | – | m+ | – |
| lymph+ | ||||||||
| c+ | ||||||||
| 254.11 | PRhom+Dnmt3a1 | 162 | m+ | m++ | m+ | – | m++ | – |
| c++ |
NOTE: –, normal.
Abbreviations: BM, bone marrow; m, myeloid expansion/infiltration [+, low; ++, intermediate; +++, extensive (leukemic)]; c, eosinophilic crystals (+, low; ++, intermediate; +++, extensive); lymph, lymphocyte infiltration (+, low; ++, intermediate; +++, extensive); ONR, organs could not be retrieved.
Figure 3.
Histologic characterization of mice. Representative images of organ sections of heart, kidney, bone marrow, liver, spleen, and lungs from mice included in the survival analysis or those included in the immunophenotypic analysis, stained with H&E. All images are taken at ×40 magnification. Representative images of normal tissue and tissue with myeloid expansion/infiltration (m) are shown. The degree of infiltration is represented as follows: +, low; ++, intermediate; +++, extensive. Summary of all mice analyzed is shown in Tables 1 and 2.
Mice expressing PML-RARα and Dnmt3a1 die at a stage before the development of leukemia
To determine whether PR mice overexpressing Dnmt3a1 were dying earlier due to premature development of leukemia, histopathologic analysis was performed on organs that were retrieved from mice shortly after death (Table 2; Fig. 3). Of the five PRhom mice that died, organs could be retrieved from three of these mice. Two of them had grossly enlarged spleens with complete loss of splenic architecture and infiltration of immature myeloid cells into the lungs, liver, and kidney. Together, these findings are consistent with previously described features of APL observed in the hCG-PML-RARα mice (4). However, the third mouse had a normal-sized spleen, although a myeloid expansion was observed in the bone marrow and spleen. In the PRhom+Dnmt3a1 group of mice, histologic examination was performed on 10 of the 22 mice that died. Nine of them had normal-sized spleens with a mild-to-moderate myeloid hyperplasia in the bone marrow and spleen. The livers of these mice showed a mild to moderate perivascular infiltration of well-differentiated myeloid cells. One mouse had an enlarged spleen with immature myeloid infiltration, as well as myeloid infiltration in the liver, heart, lungs, and kidney, consistent with leukemia development. We were able to retrieve organs from only a single PR+Dnmt3a1 mouse, which displayed mild-to-moderate myeloid hyperplasia in the bone marrow and spleen and a perivascular infiltration in the liver of mature myeloid cells. The single Dnmt3a1 mouse that died was found to have completely normal histology of all organs examined. Therefore, Dnmt3a1 overexpression in the PR background seemed to be inducing early lethality due to causes other than leukemia.
Table 2.
Summary of histopathology of organs retrieved from mice included in the survival analysis
| Mouse no. | Genotype | Age at death (d) | Spleen | BM | Liver | Kidney | Lungs | Heart |
|---|---|---|---|---|---|---|---|---|
| 485.5 | PRhom | 229 | m+++ | m+++ | m+ | – | m++ | – |
| 291.16 | PRhom | 278 | m+++ | m+++ | m+++ | m+ | m++ | – |
| 273.12 | PRhom | 284 | m+++ | m+++ | m+++ | – | c+ | m+++ |
| c++ | c+ | c+ | ||||||
| 291.10 | PRhom+ Dnmt3a1 | 202 | m++ | m++ | – | – | c+++ | – |
| 291.8 | PRhom+ Dnmt3a1 | 287 | m++ | m+++ | m+ | – | lymph+ | – |
| c++ | ||||||||
| 291.14 | PRhom+ Dnmt3a1 | 118 | m+++ | m+++ | m+++ | m+++ | m+++ | m+++ |
| 302.3 | PRhom+ Dnmt3a1 | 180 | m+ | m++ | m+ | – | m+ | – |
| lymph+ | ||||||||
| c+++ | ||||||||
| 291.18 | PRhom+ Dnmt3a1 | 265 | m++ | m+++ | m++ | – | lymph+ | – |
| c+++ | ||||||||
| 291.2 | PRhom+ Dnmt3a1 | 275 | ONR | ONR | ONR | ONR | c+++ | – |
| 272.6 | PRhom+ Dnmt3a1 | 263 | m+ | m++ | – | – | c++ | – |
| 291.12 | PRhom+ Dnmt3a1 | 265 | ONR | m+++ | m+ | – | m++ | – |
| c+++ | ||||||||
| 272.5 | PRhom+ Dnmt3a1 | 251 | m++ | m++ | m+ | – | c+ | – |
| 291.15 | PRhom+ Dnmt3a1 | 222 | ONR | m+++ | ONR | – | m+ | – |
| lymph+ | ||||||||
| c++ | ||||||||
| 291.9 | PRhom+ Dnmt3a1 | 277 | ONR | ONR | ONR | ONR | lymph++ | – |
| c++ | ||||||||
| 291.5 | PRhom+ Dnmt3a1 | 188 | m+ | m++ | m+ | – | lymph+ | – |
| c++ | ||||||||
| 234.9 | PR + Dnmt3a1 | 251 | m+ | m++ | m+ | – | lymph+ | – |
| 234.10 | Dnmt3a1 | 251 | – | – | – | – | – | – |
NOTE: –, normal.
Abbreviations: BM, bone marrow; m, myeloid expansion/infiltration [+, low; ++, intermediate; +++, extensive (leukemic)]; c, eosinophilic crystals (+, low; ++, intermediate; +++, extensive); lymph, lymphocyte infiltration (+, low; ++, intermediate; +++, extensive); ONR, organs could not be retrieved.
Indeed, histologic examination of organs from preleukemic PRhom+Dnmt3a1 mice also revealed inflammatory infiltrates in the lungs, composed of mature granulocytes, lymphocytes, and macrophage-like cells filled with eosinophilic crystals in their cytoplasm (Supplementary Fig. S4; Tables 1 and 2), consistent with crystalline macrophage pneumonia. This condition has been observed in C57Bl/6 mice, 129Sv mice, and mice deficient for p47phox (16–18). It also occurs at low frequency and with less severity in PML-RARα alone mice (Table 1). The exact etiology of the observed cooperativity between PML-RARα and Dnmt3a1 is not clear. However, given the expression of PML-RARα in the myeloid lineage, an alteration in myeloid cell function is likely in part responsible. We did in fact observe a hyperresponsiveness in PML-RARα granulocytes, although Dnmt3a1 could not be shown to further affect this in an assay for release of reactive oxygen metabolites (Supplementary Fig. S5). Overall, it seems that Dnmt3a1 accentuates a propensity to crystalline macrophage pneumonia preexisting in PML-RARα mice. Importantly, the infiltrates consolidated large parts of the lungs in the PR+Dnmt3a1 mice, resulting in a dramatic reduction of airway space. Therefore, the pneumonia is the likely cause of early lethality in these animals, possibility not permitting enough time for the development of leukemia.
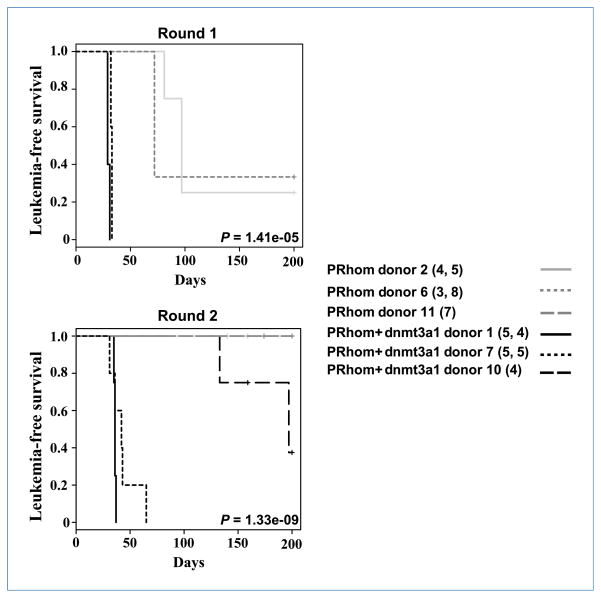
Transplantation of bone marrow cells expressing PML-RARα and Dnmt3a1 results in the development of leukemia
To overcome the difficulty of assessing the contribution of Dnmt3a1 to leukemogenesis due to the early nonleukemic lethality, we performed bone marrow transplants. We harvested bone marrows of littermate mice of PRhom and PRhom+Dnmt3a1 genotypes at 6 to 7 months of age and used the cells to reconstitute lethally irradiated recipient mice. Donors were at a similar stage of preleukemia as determined by splenic composition (Supplementary Fig. S6). Mice were monitored for leukemia development and survival over a period of 200 days (Fig. 4; Supplementary Fig. S7). Consistent with the hypothesis that Dnmt3a1 cooperates with PML-RARα in leukemogenesis, two PRhom+Dnmt3a1 donors rapidly gave rise to leukemias with features of APL in all recipients, whereas recipients of PRhom donor marrows developed leukemias with longer latencies (Fig. 4). Leukemia development was scored by the presence of infiltrating immature myeloid cells that resulted in the enlargement and loss of splenic architecture. The splenic phenotype was accompanied by infiltration into other organs such as the lungs, liver, kidneys, and heart. Repetition of this experiment using frozen cells replicated the transplantability and rapid lethality of cells from PRhom+Dnmt3a1 mice, whereas cryopreserved PRhom cells did not cause leukemia in recipients even at 200 days despite contribution to long-term hematopoiesis (data not shown). We tested a third cryopreserved mouse at 6.5 months of age of both PRhom and PRhom+Dnmt3a1 genotypes and observed leukemia only in the PRhom+Dnmt3a1 recipients. Collectively, mice receiving PRhom+Dnmt3a1 cells developed leukemia with a 91% penetrance (21 of 23 transplanted mice), with leukemias arising from all 3 donors. In contrast, only 18.5% (5 of 27) of mice transplanted with PRhom cells died of leukemia and at much later time points, with only 2 of 3 donors giving rise to leukemia (Fig. 4). Indeed, several PR-alone transplants survived the 200-day follow-up (Supplementary Fig. S7), and of those that died, many died of nonleukemic causes (compare Supplementary Fig. S7 with Fig. 4). In contrast, only one mouse transplanted with PRhom+Dnmt3a1 cells died from nonleukemic causes. These findings show that Dnmt3a1 overexpression does indeed cooperate with PML-RARαin promoting leukemogenesis.
Figure 4.
PML-RARα cooperates with Dnmt3a1 to cause leukemia in irradiated recipients. Kaplan-Meier comparative survival analysis of irradiated congenic mice transplanted with cells from either PRhom or PRhom+Dnmt3a1 donors is shown. Leukemia-free survival is plotted against days after transplantation. Three donors were used for each genotype. Numbers in parenthesis indicate the number of recipients injected per donor, with the first number representing round 1 and the second number round 2. Single numbers indicate the number of recipients used in round 2.
PML-RARα and Dnmt3a1 cooperate to methylate a downstream target in leukemia development
The increased leukemia in PR+Dnmt3a1 recipient mice would suggest that these two proteins cooperate in vivo to methylate downstream targets and hence promote tumorigenesis. To test whether Dnmt3a1 promoted methylation of PML-RARα targets, we used bisulfite sequencing to analyze the DNA methylation status of the PML-RARα target gene RARβ in spleen cells (Fig. 5A). The RARβ promoter showed increased CpG methylation in leukemic recipient mice relative to the preleukemic donors. More importantly, PRhom+Dnmt3a1 leukemic mice showed consistently higher levels of DNA methylation than their PRhom counterparts (Fig. 5B). Dnmt3a1 alone mice showed no evidence of increased methylation. These findings show that overexpression of Dnmt3a1 cooperates with PML-RARα to promote hypermethylation of its target genes during the progression of leukemia. The increase in methylation with time is consistent with the methylated cells having a competitive advantage over their unmethylated counterparts.
Figure 5.
Methylation status of the RARβ promoter in spleen cells of donor and leukemic recipients. A, schematic of the region of the RARβ promoter. Each line with a circle at the end of it represents a CpG dinucleotide. Numbers indicate the positions relative to the transcription start site (+1). B, bisulfite genomic sequencing of the RARβ promoter for representative transplant donors and leukemic recipients. Each circle represents a CpG dinucleotide. Open circles are unmethylated and black filled circles are methylated CpGs. Each row represents an individual sequencing run. The percentage of methylated CpGs is denoted next to each sample.
Discussion
Our results show that inducible expression of Dnmt3a1 cooperates with PML-RARα to promote APL leukemogenesis. In primary mice, overexpression of PML-RARα and Dnmt3a1 resulted in early death secondary to an eosinophilic infiltrate that consolidated the lungs, leading to death before progression to leukemia. Such an infiltrate has been previously described in the PML-RARα model (19) and was seen infrequently in PR-alone mice in this study. However, it was dramatically enhanced in the PR+Dnmt3a1 mice. In secondary transplant experiments, recipient mice receiving PR+Dnmt3a1 cells progressed to leukemia with higher incidence and shorter latency than their PR-alone counterparts. Leukemic PR+Dnmt3a1 cells showed increased DNA methylation at a PML-RARα target relative to leukemic PR-alone cells. Therefore, we conclude that the levels of a de novo methyltransferase can be rate limiting in the methylation of PML-RARα targets and the progression to leukemia.
The requirement for secondary mutations in addition to PML-RARα for the development to APL is well documented. Activating mutations in Flt3 or in K-Ras increase the penetrance and shorten the latency of leukemia development in the PML-RARα mouse model (5, 6) and are commonly found in APL patients (20–23). Accumulation of epigenetic marks at appropriate targets could also function as cooperating events in conjunction with PML-RARα. Recent studies examining the role of epigenetic marks in human APL suggest that several target genes are heavily decorated by DNA methylation, histone methylation, and histone acetylation, suggesting that significant epigenetic changes occur during leukemia development (24). Previous work showed that Dnmt3b could enhance the development of intestinal tumor formation in an APCmin mutant background (13, 25). However, this cooperation likely occurs through parallel pathways, as there is no evidence that the mostly nuclear Dnmt3b enzyme interacts with the cytoplasmic APC protein. Therefore, our work is the first example to our knowledge to test cooperativity in vivo between an oncogene and epigenetic-modifying enzyme that physically interact. We speculate that a deeper exploration into the development of several cancers will highlight the ever-increasing importance of epigenetic enzymes in cooperating with oncogenes and tumor suppressor genes in cancer progression.
Supplementary Material
Acknowledgments
We thank S. Espejel-Carbajal and S. Sevcikova for technical assistance with the oxidative burst assay, Rudolf Jaenisch for sharing the Dnmt3a1-inducible mice, Tim Ley for sharing the hCG-PML-RARα mice, and members of the Blelloch laboratory for critically reading the manuscript.
Grant Support
University of California Cancer Research Coordinating Committee and NIH grant K08 NS48118 (R. Blelloch).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR α fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–84. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 2.Bennett JM, Catovsky D, Daniel MT, et al. Proposals for the classification of the acute leukaemias. French-American-British (FAB) Co-operative Group. Br J Haematol. 1976;33:451–8. doi: 10.1111/j.1365-2141.1976.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 3.Bennett JM, Catovsky D, Daniel MT, et al. A variant form of hypergranular promyelocytic leukemia (M3) Ann Intern Med. 1980;92:261. doi: 10.7326/0003-4819-92-2-261. [DOI] [PubMed] [Google Scholar]
- 4.Grisolano JL, Wesselschmidt RL, Pelicci PG, Ley TJ. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RAR α under control of cathepsin G regulatory sequences. Blood. 1997;89:376–87. [PubMed] [Google Scholar]
- 5.Chan IT, Kutok JL, Williams IR, et al. Oncogenic K-ras cooperates with PML-RAR α to induce an acute promyelocytic leukemia-like disease. Blood. 2006;108:1708–15. doi: 10.1182/blood-2006-04-015040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sohal J, Phan VT, Chan PV, et al. A model of APL with FLT3 mutation is responsive to retinoic acid and a receptor tyrosine kinase inhibitor, SU11657. Blood. 2003;101:3188–97. doi: 10.1182/blood-2002-06-1800. [DOI] [PubMed] [Google Scholar]
- 7.Di Croce L, Raker VA, Corsaro M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–82. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 8.Grignani F, De Matteis S, Nervi C, et al. Fusion proteins of the retinoic acid receptor-α recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–8. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 9.Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–4. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 10.Villa R, Morey L, Raker VA, et al. The methyl-CpG binding protein MBD1 is required for PML-RARα function. Proc Natl Acad Sci U S A. 2006;103:1400–5. doi: 10.1073/pnas.0509343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villa R, Pasini D, Gutierrez A, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–25. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–8. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 13.Linhart HG, Lin H, Yamada Y, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–22. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–7. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 15.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoenerhoff MJ, Starost MF, Ward JM. Eosinophilic crystalline pneumonia as a major cause of death in 129S4/SvJae mice. Vet Pathol. 2006;43:682–8. doi: 10.1354/vp.43-5-682. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Cheng LI, Yi L, et al. p47phox deficiency induces macrophage dysfunction resulting in progressive crystalline macrophage pneumonia. Am J Pathol. 2009;174:153–63. doi: 10.2353/ajpath.2009.080555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray AB, Luz A. Acidophilic macrophage pneumonia in laboratory mice. Vet Pathol. 1990;27:274–81. doi: 10.1177/030098589002700409. [DOI] [PubMed] [Google Scholar]
- 19.Marchesi F, Monestiroli SV, Capillo M, et al. Eosinophilic crystals as a distinctive morphologic feature of a hyaline droplet nephropathy in a mouse model of acute myelogenous leukaemia. J Vet Med A Physiol Pathol Clin Med. 2003;50:103–7. doi: 10.1046/j.1439-0442.2003.00486.x. [DOI] [PubMed] [Google Scholar]
- 20.Callens C, Chevret S, Cayuela JM, et al. Prognostic implication of FLT3 and Ras gene mutations in patients with acute promyelocytic leukemia (APL): a retrospective study from the European APL Group. Leukemia. 2005;19:1153–60. doi: 10.1038/sj.leu.2403790. [DOI] [PubMed] [Google Scholar]
- 21.Kiyoi H, Naoe T, Yokota S, et al. Internal tandem duplication of FLT3 associated with leukocytosis in acute promyelocytic leukemia. Leukemia Study Group of the Ministry of Health and Welfare (Kohseisho) Leukemia. 1997;11:1447–52. doi: 10.1038/sj.leu.2400756. [DOI] [PubMed] [Google Scholar]
- 22.Longo L, Trecca D, Biondi A, et al. Frequency of RAS and p53 mutations in acute promyelocytic leukemias. Leuk Lymphoma. 1993;11:405–10. doi: 10.3109/10428199309067933. [DOI] [PubMed] [Google Scholar]
- 23.Yokota S, Kiyoi H, Nakao M, et al. Internal tandem duplication of the FLT3 gene is preferentially seen in acute myeloid leukemia and myelodysplastic syndrome among various hematological malignancies. A study on a large series of patients and cell lines. Leukemia. 1997;11:1605–9. doi: 10.1038/sj.leu.2400812. [DOI] [PubMed] [Google Scholar]
- 24.Martens JH, Brinkman AB, Simmer F, et al. PML-RARα/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell. 2010;17:173–85. doi: 10.1016/j.ccr.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 25.Lin H, Yamada Y, Nguyen S, et al. Suppression of intestinal neoplasia by deletion of Dnmt3b. Mol Cell Biol. 2006;26:2976–83. doi: 10.1128/MCB.26.8.2976-2983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.