Abstract
A complete description of the serological response following exposure of humans to complex pathogens is lacking and approaches suitable for accomplishing this are limited. Here we report, using malaria as a model, a method which elucidates the profile of antibodies that develop after natural or experimental infection or after vaccination with attenuated organisms, and which identifies immunoreactive antigens of interest for vaccine development or other applications. Expression vectors encoding 250 Plasmodium falciparum (Pf) proteins were generated by PCR/recombination cloning; the proteins were individually expressed with >90% efficiency in E. coli cell-free in vitro transcription and translation reactions, and printed directly without purification onto microarray slides. The protein microarrays were probed with human sera from one of four groups which differed in immune status: sterile immunity or no immunity against experimental challenge following vaccination with radiation-attenuated Pf sporozoites, partial immunity acquired by natural exposure, and no previous exposure to Pf. Overall, 72 highly reactive Pf antigens were identified. Proteomic features associated with immunoreactivity were identified. Importantly, antibody profiles were distinct for each donor group. Information obtained from such analyses will facilitate identifying antigens for vaccine development, dissecting the molecular basis of immunity, monitoring the outcome of whole-organism vaccine trials, and identifying immune correlates of protection.
Keywords: Plasmodium falciparum, malaria, antigen identification, high throughput, immune screening, proteomics, protein microarray, proteome microarray, protein chip, vaccine, diagnostics
INTRODUCTION
The Plasmodium falciparum (Pf) genome encodes an estimated 5,268 putative proteins [1]. The parasite has a complex multi-stage life cycle. After an individual is bitten by a Plasmodium infected female Anopheles spp. mosquito, sporozoites in the peripheral circulation invade the liver and develop into schizonts containing as many as 30,000 merozoites each. The liver schizonts then rupture, releasing the merozoites into the bloodstream where each can subsequently invade an erythrocyte. This initiates a cycle of intra-erythrocytic stage, development, rupture, and re-invasion, resulting in a 15–30 fold increase in the numbers of parasites in the bloodstream every 48 hours. These asexual erythrocytic-stage parasites are responsible for the clinical manifestations and pathology of malaria.
Decades of research in the pre-genomic era has identified no more than a score of promising Pf vaccine or diagnostic targets, representing less than 0.5% of the entire genome. With the recent completion of the genomic sequence of Pf and elucidation of the Pf proteome [1–7] we have an opportunity to implement high throughput approaches to identify novel Pf antigens for vaccine, diagnostic or other applications and to better understand the complex host-parasite relationship. However, there is currently no in silico algorithm that can be used effectively to identify serodiagnostic immune profiles or antigens that confer protective immunity from genomic sequence data alone. Various approaches have been proposed for antigen and epitope identification, including expression cloning [8], elution and mass spectrometry sequencing of naturally processed MHC-bound peptides [9–11], in vitro testing of pools of overlapping peptides [12–14], and reverse immunogenetics [15, 16]. Unfortunately, these methods underestimate the complexity of responses, and none can be applied for high throughput analysis of large amounts of genomic sequence data or very large numbers of patient or animal samples.
Herein, we use protein microarrays [17–19] for identifying immunodominant antigens and defining immunoreactivity profiles amongst distinct donor groups of differing malaria immune status, including individuals who are demonstrably protected from malaria. We show that these protein microarrays identify characteristic immunoreactive antigen profiles recognized by serum antibodies from distinct donor groups of individuals exposed to P. falciparum, and identify immunodominant antigens which may represent promising targets for vaccine development.
MATERIALS AND METHODS
Gene / Open Reading Frame selection
A set of open reading frames (ORFs) derived from the Pf genomic sequence database (<http://www.plasmodb.org/plasmo/home.jsp>)[20] and representing 250 putative Pf proteins (4.75% of the entire genome) was targeted for cloning, expression, and protein microarray chip printing. The genes were selected according to specific sets of criteria, including pattern of stage-specific gene or protein expression deduced from genomic or proteomic datasets, subcellular localization, secondary structure, and known immunogenicity or antigenicity in human and animal models. Since the study was designed to include evaluation of samples from volunteers experimentally immunized with radiation attenuated Pf sporozoites, the gene panel included putative Pf proteins expressed in the sporozoite and/or liver stage of the parasite life cycle. Each gene was classified within one of nine categories (Supplementary Table S1).
To manage the Pf sequence information, we developed a database and a web-interface (http://contact14.ics.uci.edu/virus/mal_index.php) for accessing the sequence of each ORF from the Pf genome. The following information is provided in an index view: chromosome number, gene ID, strand direction, exon number, section number, 5-prime primer, 3-prime primer, size of segment (nucleotides, amino acids, molecular weight), and a flag for whether or not the section contains internal stop codons.
PCR amplification of linear acceptor vector
Plasmid pXT7 (3.2 kb, KanR) was previously described [21]; genes cloned into this vector by the methods described herein encode an N-terminal 10x histidine tag and C-terminal hemagglutinin tag. Plasmid pXT7 (10 µg) was linearized with BamHI (0.1 µg/µlDNA/0.1 mg/ml BSA/0.2 units/µl BamHI; 37°C for 4 hr; additional BamHI was added to 0.4 units/µl at 37°C overnight). The digest was purified using a PCR purification kit (Qiagen, Valencia, CA), quantified by fluorometry using Picogreen (Molecular Probes, Carlsbad, CA) according to the manufacturer's instructions, and verified by agarose gel electrophoresis (1 µg). One ng of this material was used to generate the linear acceptor vector in a 50-µl PCR using 0.5 µM each of primers 5'-CTACCCATACGATGTTCCGGATTAC and 5'-CTCGAGCATATGCTTGTCGTCGTCG, and 0.02 units/µl Taq DNA polymerase (Fisher Scientific, buffer A)/0.1 mg/ml gelatin (Porcine, Bloom 300; Sigma, G-1890)/0.2mM each dNTP with the following conditions: initial denaturation of 95°C for 5 min; 30 cycles of 95°C for 0.5 min, 50°C for 0.5 min, and 72°C for 3.5 min; and a final extension of 72°C for 10 min.
PCR amplification of ORF insert
A total of 1–10 ng of Pf genomic DNA (3D7 strain) was used as template in a 50-µl PCR. The following primers were used (0.5 µM each): 5'-CATATCGACGACGACGACAAGCATATGCTCGAG (20-mer ORF specific at the 5' end) and 5'-ATCTTAAGCGTAATCCGGAACATCGTATGGGTA (20-mer ORF specific at the 3' end). The Pf genome is the most A+T rich genome sequenced to date with an overall (A+T) composition of 80.6%, rising to ~90% in introns and intergenic regions [1]. Consequently, PCR amplification of Pf genes using genomic DNA template was problematic. Initially, PCR was carried out using regular Taq DNA polymerase: 0.02 units/µl TaqDNA polymerase (buffer A, Fisher Scientific)/0.1 mg/ml gelatin (Bloom 300, Porcine; G-1890, Sigma)/0.2 mM each dNTP. Conditions were as follows: initial denaturation of 95°C for 5 min; 30 cycles of 20 sec at 95°C, 30 sec at 50°C, and 60 sec/kb at 72°C (1–3 min on average, based on ORF size); and a final extension of 72°C for 10 min. PCR products that were more difficult to produce were amplified by using a 30 sec annealing time at 45°C or 40°C, instead of 30 sec at 50°C. Also, the extension temperature was decreased from 65–72°C to 50°C. Subsequently PCR products were obtained using a Taq polymerase with improved proof-reading characteristics (Triplemaster from Eppendorf), increasing the efficiency of the PCR step to 87%: 0.04 units/µl Triple Master PCR system (high-fidelity buffer, Eppendorf)/0.4 mM each dNTP (Eppendorf). Conditions were as follows: initial denaturation of 95°C for 3 min; 35 cycles of 15 sec at 95°C, 30 sec at 40°C, and 60 sec/kb at 50°C (1–3 min on average, based on ORF size); and a final extension of 50°C for 10 min. PCR products that were difficult were reamplified using 50 ng genomic DNA. The PCR product was visualized by agarose gel electrophoresis (3 µl). For quantification, the product was purified (PCR purification kit, Qiagen) and quantified by fluorometry (Picogreen, Molecular Probes). Since the reliability of producing the desired PCR product decreases as the length of the genomic DNA fragment increases, exons longer than 3,000 bp were divided into multiple overlapping sections, with 50 nucleotide overlaps.
In vivo recombination cloning
Competent cells were prepared in our laboratory by growing DH5α cells at 18°C in 500 ml of SOB (super optimal broth) medium (2% tryptone/0.5% yeast extract/10 mM NaCl/2.5 mM KCl/20 mM MgSO4) to an OD of 0.5–0.7. The cells were washed and suspended in 10 ml of pre-chilled PCKMS buffer (10 mM Pipes/15 mM CaCl2/250 mM KCl/55 mM MnCl2/5% sucrose, pH 6.7) on ice, and 735 µl of DMSO was added dropwisewith constant swirling. The competent cells were frozen on dry ice-ethanol in 100-µl aliquots and stored at −80°C. Each transformation consisted of the following: 10 µl of competent DH5α and 10 µl of DNA mixture (40 ng of PCR-generated linear vector/10 ng of PCR-generated ORF fragment; molar ratio, 1:1; vector, 1-kb ORF fragment). For transformation, the purification of PCR product was unnecessary. The mixture was incubated on ice for 45 min, heat shocked at 42°C for 1 min, and chilled on ice for 1 min; mixed with 250 µl of SOC (super optimal catabolizer) medium (2% tryptone/0.55% yeast extract/10 mM NaCl/10mM KCl/10 mM MgCl2/10 mM MgSO4/20 mM glucose); incubated at 37°C for 1 hr; diluted into 3 ml of LB medium supplemented with 50 µg of kanamycin per ml (LB Kan 50); and incubated with shaking overnight. The plasmid was isolated and purified from this culture, without colony selection.
In vitro protein expression
Plasmid templates used for in vitro transcription/translation were prepared by using QIAprep SpinMiniprep kits (Qiagen), including the "optional" step, which contains protein denaturants to deplete RNase activity. In vitro transcription/translation reactions (RTS 100 Escherichia coli HY kits; Roche) were set up in 25 µl PCR 12-well strip tubes and incubated for 5 h at 30°C, according to the manufacturer's instructions.
Immuno-dot blots
To assess relative efficiency of protein expression, 0.3 µl of whole rapid-translation system (RTS) reactions were spotted manually onto nitrocellulose and allowed to air dry before blocking in 5% nonfat milk powder in TBS containing 0.05% Tween 20. Blots were probed with hyperimmune sera diluted to 1:1,000 in blocking buffer with or without 10% E. coli lysate. Routinely, dot blots were stained with both mouse anti-poly-HIS mAb (clone, HIS-1; H-1029, Sigma) and rat anti-hemagglutinin (HA) mAb (clone, 3F10; 1 867 423, Roche), followed by alkaline phosphatase-conjugated goat anti-mouse IgG (H+L) (BioRad) or goat anti-rat IgG (H+L) (Jackson ImmunoResearch) secondary Abs, respectively. Bound human Abs were visualized with nitroblue tetrazolium (nitro-BT) developer to confirm the presence of recombinant protein.
Microarray chip printing
For microarrays, 10 µl of 0.125% Tween 20 was mixed with 15 µl of RTS reaction (to a final concentration of 0.05% Tween 20), and 15-µl volumes were transferred to 384-well plates. The plates were centrifuged at 1,600 × g to pellet any precipitate, and supernatant was printed without further purification onto nitrocellulose-coated FAST glass slides (Schleicher & Schuell) by using an OmniGrid 100 microarray printer (Genomic Solutions, Ann Arbor, MI). All ORFs were spotted in duplicate to enable statistical analysis of the data. Data values reported herein represent the average of pairs. In addition, each chip contained an area printed with controls consisting of RTS reaction using no DNA or empty T7 vector control.
Protein microarray screening
Microarray chips were probed with human serum that was first pre-absorbed against E. coli-lysate to block anti-E. coli antibodies as described previously [21]. This is necessary because high titers of anti-E. coli antibodies mask any protein-specific responses when using whole RTS reactions on dot blots and arrays. For all staining, slides were first blocked for 30 min in protein array-blocking buffer (Schleicher & Schuell) and then incubated in serum for 2 hr, at room temperature. Antibodies were visualized with Cy3-conjugated secondary Abs (biotinylated secondary followed by Streptavidin PBXL-3, for HIS-probing) (Jackson ImmunoResearch) and scanned in a ScanArray 4000 laser confocal scanner (GSI Lumonics, Billerica, MA). Fluorescence intensities were quantified by using QuantArray software (GSI Lumonics). Other studies in the UCI laboratory have established that the signal intensities of a given protein probed with different sera are proportional to relative antibody titers in the different sera [22]. Additionally, a good correlation between independent print runs was obtained when individual serum samples from different patients were compared [23].
ELISA
To validate the immunoreactivity detected by the protein microarrays, sera were also analyzed by ELISA against a known and well-characterized Pf pre-erythrocytic stage antigen (PfCSP) as previously described [24]. The mean OD readings of quadruplicate assays were recorded, and results reported as the OD value at each serum dilution and as endpoint dilution (defined as greater than the mean +/− 3 standard deviations of negative control sera).
Indirect Fluorescent Antibody test (IFAT)
Antibody recognition of Pf (NF54/3D7) sporozoite or blood stage parasites was evaluated by IFAT as described previously [25]. Reactivity was scored as positive when the immunofluorescence pattern of the parasite was recognized and when the fluorescence was above the background of the negative controls. IFAT results were expressed as the endpoint serum dilution at which positive fluorescence was detected.
Malaria-exposed donor groups
Individuals were selected for study on the basis of malaria history. Studies were conducted in compliance with all applicable regulations governing the protection of human subjects. The irradiated sporozoite study protocol was approved by the Naval Medical Research Center Committee for the Protection of Human Subjects, the Office of the Special Assistant for Human Subject Protections at the Naval Bureau of Medicine and Surgery, and the Human Subjects Research Review Board of the Army Surgeon General. The Kenyan samples were collected under a study protocol approved by the Naval Medical Research Institute’s Committee for the Protection of Human Subjects, the Walter Reed Army Institute of Research Human Use Committee, and the Kenya Medical Research Institute/National Ethical Review Committee. Written informed consent was obtained from all subjects. Subject details are provided as Supplementary Data (Supplementary Tables S2 and S3 online).
Sporozoite immunized volunteers (Supplementary Table S2)
Caucasian volunteers (n=10) were experimentally immunized with radiation-attenuated Pf sporozoites as previously described [26]. Subjects were challenged by the bites of 5 Pf infected Anopheline mosquitoes, and evaluated for the development of clinical malaria by daily monitoring of thick blood smears beginning 7 days after challenge and continuing until day 28, then weekly for 4 weeks if they became parasitemic or 8 weeks if they did not become parasitemic. Protection was defined as complete absence of blood-stage parasitemia (sterile protection). Six of the 10 immunized volunteers were protected against sporozoite challenge and were classified as sporozoite-immune; four were not protected and were classified as sporozoite-exposed but non-immune. Serum samples were collected from each volunteer prior to immunization (pre-bleed), at the completion of the immunization series and immediately prior to challenge (pre-challenge), and following challenge (post-challenge). Pre- and post-challenge IFAT titers against Pf sporozoites were: protected 613 (mean; range 160–1280) and 560 (mean; range 160–1280); and unprotected 170 (mean; range 40–320) and 240 (mean; range 80–640).
Individuals naturally exposed to malaria (Supplementary Table S3)
Kenyan subjects (n=12) were residents of the Asembo Bay area of Kenya. In this area, the year round prevalence of Pf infection amongst children 6 months to 6 years of age has been documented as 94.4–97.8% [27–29]. Enrolled subjects reported an average of 2.1 episodes of clinical malaria within the previous year. The donor group derives from a subset of 192 volunteers previously enrolled in an immunoepidemiology study [30] and selected for the current study covering a range of sex, age, malaria history and recognition of native Pf sporozoites and parasitized erythrocytes by IFAT. Pf sporozoite and blood-stage IFAT titers for the pool of hyperimmune sera from these 192 individuals were 5,120 and 81,920, respectively.
Data and statistical analysis
A detailed description of the statistical treatment has been published elsewhere [31].
Analysis of individual array measurements
We first defined the background (true negative) signal as the average signal of the negative control spots on the array. This enabled us to compare each protein’s signal with the background signal to determine which antigens showed significant positive responses. Since comparisons needed to be conducted for groups of arrays as well, we transformed the raw signals using the vsn (asinh transformation, similar to log for higher intensities) method [32], shown to effectively calibrate array measurements through shifting and scaling and also to stabilize the variance in DNA microarray and 2D difference gel electrophoresis [33] data analyses. Each protein was spotted only twice per array. Because standard deviation estimates can be unreliable (artificially high or low) when there is low replication of measurements, we applied the Bayes-regularization technique described in Baldi and Long [34]. This technique derives more robust estimates of the variance of each protein as a weighted combination of the sample variance and the pooled variance of neighboring proteins with similar signal intensity. Given n replicates, the regularized estimate of the variance is given by the equation:
where υ0 is the confidence in the background variance of the neighboring genes, σ02 is the background variance equal to the average variance of the neighboring genes, and s2 is the empirical variance. The neighboring genes are defined in terms of a window size, after ranking the protein by mean levels of expression. υ0 can also be regarded as a form of “pseudo-counts”, as if υ0 virtual data points were added to the data, or a parameter that controls the relative strength of the prior distribution. Using these new regularized estimates for the standard deviation, we conducted a series of Bayes-regularized one-sided t-tests on proteins with higher mean signal than the defined control to reliably estimate the signal difference between each protein and control, and computed the corresponding p-values. It should be noted that this regularization approach has been used in several studies analyzing both DNA and protein microarray data [31, 35–38] and its effectiveness has been independently validated [39] by analyzing a DNA microarray dataset with known relative concentrations between spike-in and controls.
Analysis of groups of array measurements
In addition to determining the positive proteins recognized by each of the individual sera, we averaged replicated spot measurements per sera (arithmetic mean) and pooled the responses for each group, to identify the positive proteins while taking into account the biological variation within the sera in each group. Prior to the pooling of measurements, the signals were transformed using the vsn method [32] described earlier. Similar to the analysis of individual sera, we defined the true negative control using the mean control signal spotted on the arrays and performed Bayes-regularized t-tests were performed within each group to compare and rank the proteins with a higher mean signal than the mean group control. For the individual and group analysis, using the average standard deviation of 30 neighboring proteins along with a weight of υ0=5 “pseudocounts” for computing the Bayes-regularized variance was observed to achieve a moderate regularization effect. Given the large number of hypotheses being tested, we applied the method in Storey [40] and Allison et al. [41] to the set of p-values to estimate the experiment-wide false discovery rates (FDR). For the individual and group analyses of the 43 arrays, a p-value cutoff of 0.05 corresponded to an estimated FDR level of 0.06 – 0.065. With the additional criteria applied for determining a positive response as described below, we expect the actual FDR to be lower than this estimate.
Criteria of positivity
As well as analyzing the intensity of response (as above), we also assessed the frequency of response for each antigen = number of individuals within a given donor group for which that antigen was positive on the basis of normalized signal intensity relative to control. Final classification of antigen reactivity was made taking into account both magnitude of response (signal intensity) and frequency of recognition. The responses by a particular donor group were considered positive overall if all of the following (conservative) criteria were met (where ‘control’ refers to spots on array from coupled transcription/translation reactions lacking DNA template):
normalized signal intensity > 4.0 (ratio of signal intensity of test relative to control) > 4.0 (4-fold above control signal being a conservative cut-off that is 17 times the standard deviation of the controls),
response was statistically significant (p < 0.05) compared with control signal intensity, and
number of positive responses within donor group ≥ 2.0 (i.e. reproducible recognition)
RESULTS
Gene amplification and cloning
The set of ORFs was amplified and cloned using a high throughput PCR recombination cloning method developed in our laboratory [21]. The efficiencies of the overall process of PCR amplification and cloning are summarized in Table 1, and the results for F. tularensis (Ft) and M. tuberculosis (Mtb) are also tabulated for comparison. In all cases both the PCR and cloning steps were greater than 95% efficient. A subset of the cloned genes was sequenced to verify that the insertion matched the targeted ORF, the PCR fragment was in the correct orientation, and there were no mutations introduced in the overlapping region during homologous recombination. In >99% of the cases the correct insertions were verified (Table 1).
Table 1.
Efficiency of P. falciparum PCR and cloning.
| PCR | Recombination Cloning | ||||||
|---|---|---|---|---|---|---|---|
| Target # |
Yield | Efficiency | Target # |
Yield | Efficiency | Sequence Verificationn |
|
| F. tularensis | 1933 | 1889 | 98% | 1889 | 1871 | 99% | >99% |
| M. tuberculosis | 3989 | 3890 | 98% | 3890 | 3836 | 98% | >99% |
| P. falciparum | 960 | 922 | 96% | 737 | 715 | 97% | >99% |
Individual genes were amplified by PCR using genomic DNA as template from each of the pathogens. The PCR efficiency was 96–98%. The individual PCR fragments were inserted into the T7 expression vector by recombination cloning with a successful yield of 97–99%. To verify cloning efficiency, about 25–30% of plasmids from the recombination cloning step were randomly selected and sequenced from both ends to check if 1) the insert matches targeted ORF, 2) PCR fragment is cloned in correct orientation and 3) there is no mutation(s) introduced in the overlapping region during homologous recombination. The numbers for Pf represent a panel of Pf genes which includes the ORFs evaluated in the current study.
Protein expression
As compared with other organisms, Pf proteins have been difficult to express by conventional methodologies in bacterial, yeast or insect expression systems [42–44]. We evaluated the efficiency of Pf protein expression in our E. coli based cell-free in vitro transcription/translation system using 295 different cloned, HIS- or HA-tagged Pf ORFs as templates (Figure 1). The 250 Pf proteins on the chip are represented as 295 ORFs because some of them were cloned as multiple exons and ORFs >3000 base pairs were cloned as overlapping segments. More than 90% of the proteins were positive for the HIS and HA tags. Some of the plasmids encoding Pf proteins had a stop codon before the HA tag; only 7% of those vectors expressed the C-terminal HA tag. This small amount of read-through verifies the use of the HA tag as a probe for full-length product (i.e. expression of full length proteins from the clones of interest) (Table 2). Efficiency of expression for vectors encoding M. tuberculosis and F. tularensis genes are shown for comparison. Uniformly high expression efficiencies >90% were obtained in all cases except when a stop codon was placed in front of the HA tag.
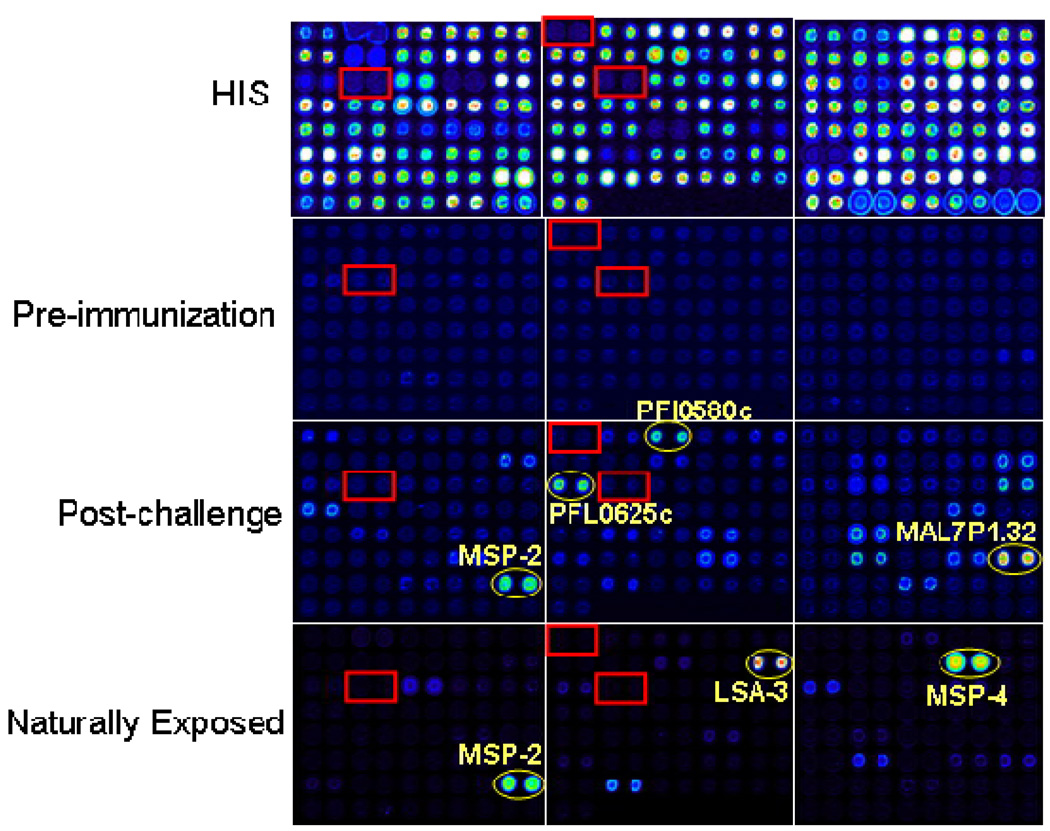
Figure 1. Probed Arrays.
Pf protein microarrays were generated as described in Methods. Three individual arrays displaying a subset of targets evaluated in this study were probed with sera as described below, followed by biotinylated secondary and streptavidin PBXL-3, read in a ScanArray 4000 confocal laser scanner, and the signal intensities quantified. Arrays were probed with (A) anti-polyhistidine mouse monoclonal antibody (B) sera from malaria-naïve individuals (pre-immunization specimens from volunteers subsequently immunized with radiation attenuated Pf sporozoites) (C) sera from irradiated sporozoite immunized and infected volunteers post-challenge, or (D) sera from adults naturally exposed to malaria in Africa. Control reactions that lacked vector template (boxes) and reactions of some well characterized Pf antigens (circles) were also spotted.
Table 2.
Efficiency of protein expression in E. coli cell-free transcription/translation system.
| Expression Efficiency | |||||
|---|---|---|---|---|---|
| Total | HIS | HA | |||
| # | # | % | # | % | |
| F. tularensis | 1614 | 1481 | 92% | 1506 | 93% |
| M. tuberculosis | 894 | 881 | 99% | 890 | 100% |
| P. falciparum (stop-HA) | 112 | 104 | 93% | 8 | 7% |
| P. falciparum (HA-stop) | 183 | 172 | 94% | 165 | 90% |
Protein microarray chips were printed and probed with anti-histidine antibody (against the N-terminal HIS tag) or anti-HA antibody (against the C-terminal HA tag). Spots with signal intensity > 4 fold above the negative control were scored as positive.
To validate the immuoreactivity of in vitro cell-free expressed Pf proteins compared with conventional protein production methods, the panel of all test sera (pre-immunization, pre-challenge and post-challenge sera from 10 irradiated sporozoite immunized volunteers, cross-sectional sera from 12 naturally exposed Kenyans; pool of hyperimmune Kenyan sera) was assayed by a conventional ELISA assay using high quality recombinant protein capture antigen for PfCSP; the same sera were used to probe the microarray chips. The correlation (r2) of the ELISA data with microarray chip data for Pf CSP was 0.76 (p< 4e-09). Other studies showing good correlations between ELISA and microarray data have been published by our group, in Lyme disease and vaccinia models [22, 23].
Immune screening
Pf protein microarrays were probed with sera from 12 subjects who were naturally exposed to malaria in a hyperendemic region of Kenya, and 10 subjects who were experimentally immunized with radiation attenuated Pf sporozoites and either protected (n=6) or not protected (n=4) against challenge with infectious sporozoites; pre-immunization pre-challenge and post-challenge sera were evaluated. Subject details are provided in Supplementary Tables S2 and S3. The corresponding region of several probed arrays displaying a subset of the targets under study, probed with sera from three of the four donor groups, is shown in Figure 1. The reactivity of an anti-histidine monoclonal antibody against the N-terminal HIS tagged proteins is shown in Figure 1a. Sera from malaria-naïve subjects showed little or no reactivity (Figure 1b), whereas sera from Pf exposed individuals (either experimentally or naturally infected) showed pronounced reactivity against numerous antigens (Figures 1c & 1d).
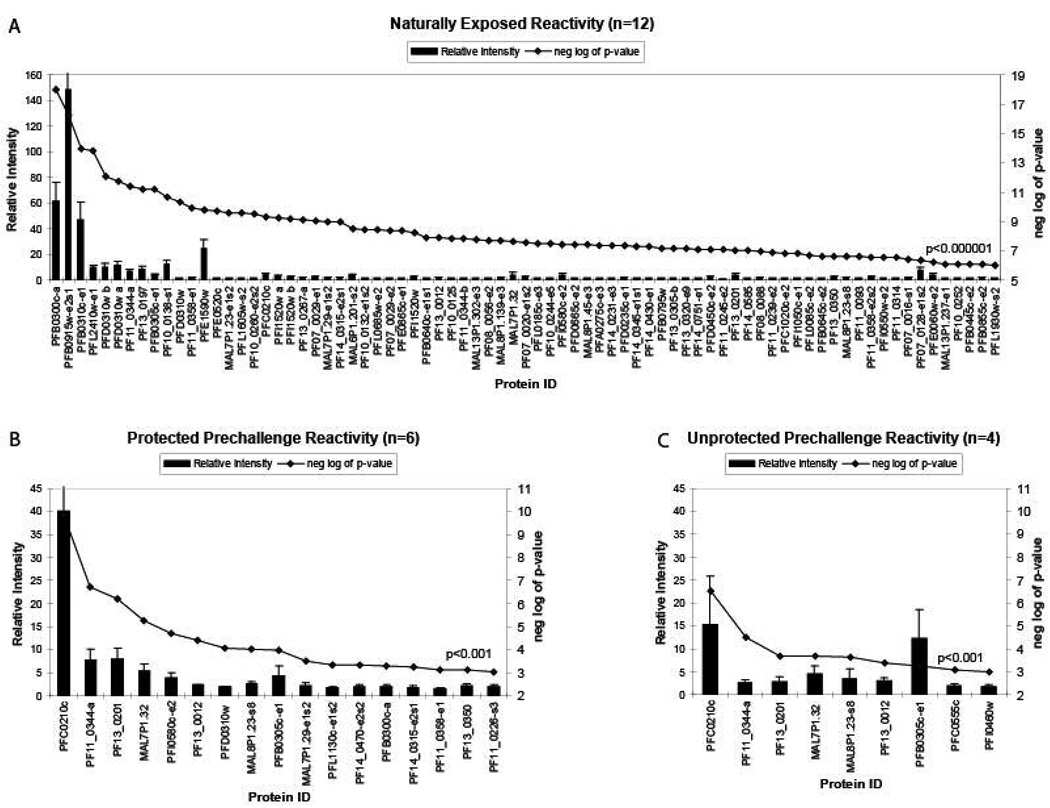
Data were analyzed by the ‘group average’ method as described in the Methods section. A detailed description of this statistical treatment has been previously published [31]. The reactivity against those antigens most strongly recognized by donor sera from each of three donor groups is plotted in Figure 2. For each antigen, the average signal intensity (SI) of test/negative control for all subjects within a donor group is presented as a histogram, and the statistical significance relative to control (negative log p-value, determined by Bayes regularized t-test) is indicated by dot plot. For the naturally exposed donor group, there were 156 antigens with p values < 1e-3; data for 77 antigens with p values < 1e-7 are shown in Figure 2a. For the sera from the irradiated sporozoite immunized groups collected immediately before challenge, there were 17 antigens with p values < 1e-3 in the protected group (Figure 2b) and 9 antigens with p values < 1e-3 in the unprotected group (Figure 2c). Background reactivity with sera from malaria-naïve donors was very low (only 2% [5/250] of the Pf antigens were recognized by more than one malaria-naïve serum sample and no antigens were recognized by more than two sera) (data not presented). The low p values indicate that the signals obtained from the chip are highly significant.
Figure 2. Antigen immunoreactivity profiles of donor groups of differing malaria immune status- statistical significance.
Pf protein microarrays were generated and screened as described in the legend to Figure 1. Primers were designed to amplify each exon separately; the exons of genes containing introns are designated with a small letter ‘e’, as e1, e2, etc. Large genes (and exons) greater than 3,000 base pairs were amplified in segments with each segment overlapping by 150 nucleotides; the segments are designated with a small letter ‘s’ as s1, s2, etc. Pf protein microarrays were probed with (A) sera from 12 individuals naturally exposed to hyperendemic malaria in Kenya, or with sera from 10 individuals experimentally immunized with radiation-attenuated Pf sporozoites and either (B) protected (n=6) or (C) not protected (n=4) against challenge with infectious Pf sporozoites. Average signal intensities for each donor group are shown. For each antigen, the average signal intensity (SI) of test/negative control (relative intensity) for all subjects within a donor group is presented as a histogram, and antigens are plotted in order of decreasing strength of recognition. The statistical significance relative to control (negative log p-value, determined by Bayes regularized t-test) is indicated by dot plot.
Overall, 72 highly reactive Pf antigens met the following criteria: i) normalized signal intensity (ratio of signal intensity of test relative to control > 4, where ‘control’ refers to spots on array from coupled transcription/translation reactions lacking DNA template), ii) p < 0.05, and iii) number of positive responses within a particular donor group ≥ 2 (Supplementary Table S4). Sixteen of the 23 most immunoreactive proteins are well-characterized Pf antigens, many of which are under clinical development and evaluation (www.who.int/vaccine_research/documents/en/malaria_current_11042003.pdf). Fifty-six of the immunoreactive Pf antigens are novel; three of these have been identified previously by MudPIT analysis of erythrocyte ghosts (PF11_0314, PFE0060w, PFE1590w) [45], three have been identified previously by ImmunoSense™ T cell screening (PF11_0226, high T cell reactivity; PF10_0179, intermediate T cell reactivity; PF14_0751, no T cell reactivity) [46], and 50 have not been previously described as immunologically reactive.
Profile of antigen recognition between donor groups
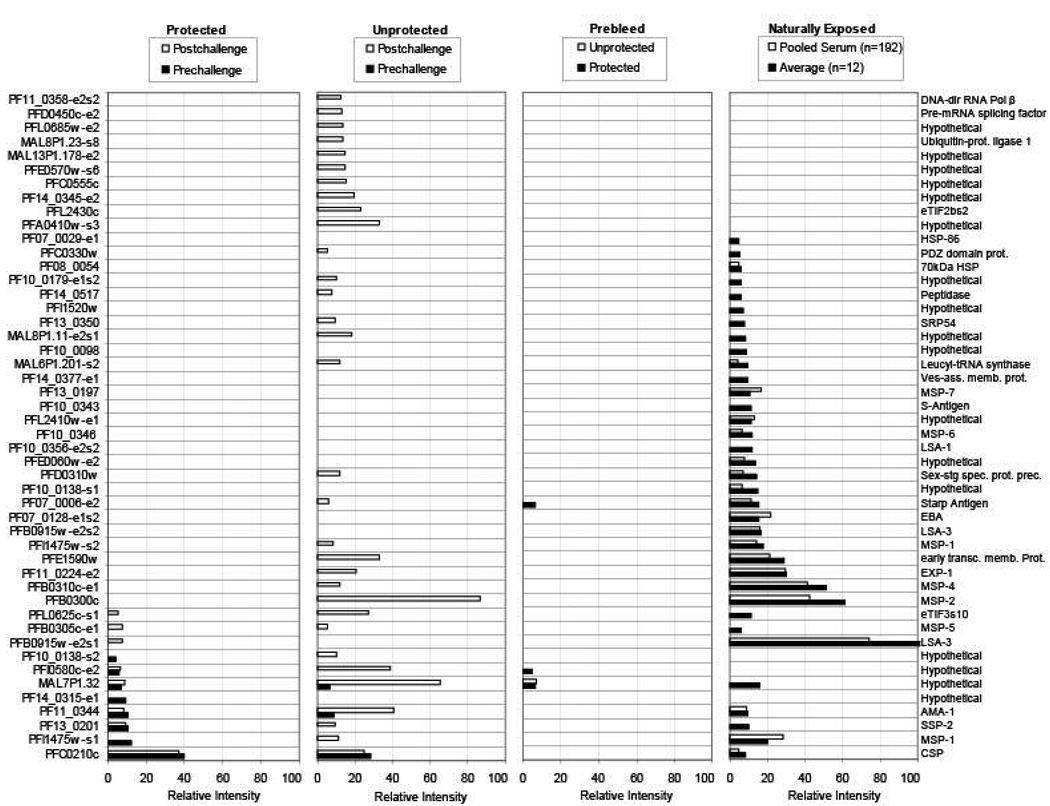
Characteristic profiles of antigen reactivity were noted for each of the distinct donor groups. The profiles of immunoreactivity against 48 ORFs representing the 45 top ranked (immunodominant) antigens are shown in Figures 3 and 4. Three reactive high molecular weight antigens (PFB0915w, LSA3; PFI1475w, MSP1; PF10_0138, hypothetical) were printed twice as overlapping segments and each segment was reactive. In Figure 3, the signal intensities for recognition of each antigen by individual serum samples are shown using a colorized matrix. The relative intensities of the average normalized signal intensity for each antigen by the averaged response of each donor group are plotted in Figure 4. Additional details for other antigens are provided as Supplementary Data (Supplementary Table S4).
Figure 3. Immunoreactivity profiles per subject.
Pf protein microarrays were probed with sera as described in Figure 2. The colored squares represent the signal intensity for recognition of 48 highly reactive ORFs. Antigens were selected according to the following criteria, i) normalized signal intensity (ratio of signal intensity of test relative to control) > 4, ii) p < 0.05, and iii) frequency ≥ 2. The p values were determined by the Bayes-regularized t-test for ‘individual arrays’ as described in the methods section. The scale represents the normalized SI (intensity/negative control without template DNA) for each subject sera against each antigen.
Figure 4. Immunoreactivity profiles per donor group.
Pf protein microarrays were probed with sera as described in Figure 2 and SI's normalized against the negative control as described in Figure 3. Histograms represent the normalized SI's for recognition for each of the top 48 most reactive antigens divided by the averaged response of each donor group. Antigens are the same as shown in Figure 3. In the Naturally Exposed group, ‘Pooled Serum’ is from 192 hyperimmune donors from Kenya, and ‘Average’ is the average of the signals from 12 individual sera from the same region.
The sera from naturally exposed subjects gave a substantially different immunoreactivity profile than the sporozoite immunized group. Naturally exposed subjects reacted more strongly to a larger number of antigens than the sporozoite immunized subjects, and the subset of sporozoite immunized volunteers who were protected against sporozoite challenge reacted more strongly to a larger number of antigens than those who were not protected.
Sera from naturally exposed individuals recognized 33 antigens (16 characterized, 17 novel). Twenty-eight proteins were specific for the naturally exposed group. Five proteins (CSP [PFC0210c], SSP2/TRAP [PF13_0201], AMA1 [PF11_0344], MSP1 [PFI1475w], and a previously uncharacterized Pf protein MAL7P1.32) reacted with sera from both naturally exposed and sporozoite immunized groups. One protein (PFI0580c) was specific for the irradiated sporozoite immunized groups. Of the known antigens, naturally exposed individuals preferentially recognized blood stage antigens (MSP1 [PFI1475w], MSP2 [PFB0300c], MSP4 [PFB0310c], MSP5 [PFB0305c], MSP7 [PF13_0197], Exp1 [PF11_0224], EBA175 [PF07_0128], and LSA3 [PFB0915w] (originally thought by some researchers to be a liver-stage antigen), whereas sporozoite immunized volunteers responded predominantly to the immunodominant sporozoite surface protein, CSP [PFC0210c] as well as SSP2/TRAP [PF13_0201] and AMA1 [PF11_0344]. Interestingly, CSP was the only protein recognized by all irradiated sporozoite immunized volunteers (Figure 3).
Eight antigens were reproducibly recognized prior to sporozoite challenge in irradiated sporozoite immunized subjects who were sterilely protected against challenge (CSP, SSP2/TRAP, AMA1, MSP1, MAL7P1.32, PFI0580c, PF10_0138, PF14_0315), whereas only three antigens were recognized prior to challenge in the unprotected subjects (CSP, AMA1, MAL7P1.32) (Figures 3 and 4). The three novel antigens recognized by protected but not unprotected volunteers (PFI0580c, PF10_0138, PF14_0315) may be of interest for vaccine development. Notably, the four most immunoreactive proteins recognized by sporozoite immune sera were previously characterized Pf antigens known to be expressed in the sporozoite and/or liver stage of the parasite life cycle: CSP [47], SSP2/TRAP [48], AMA1 [49] and MSP1 [49]. The sporozoite surface protein STARP [50, 51] was also recognized. These data confirm that proteins expressed during the sporozoite and/or liver stage but not the blood stage are recognized and that the protein microarray approach can effectively identify antigens expressed in the pre-erythrocytic as well as erythrocytic stages of the Pf parasite life cycle.
Importantly, the antigen repertoire in the protected sporozoite immunized group was unchanged after challenge. In contrast, immunized volunteers who developed clinical malaria following sporozoite challenge (experimentally infected) developed an additional subset of antibodies after challenge; some (n=21) were similar to the naturally exposed profile consistent with the development of patent blood-stage parasitemia, but many were unique to the experimentally infected group (n=38, none of which have been previously characterized). Six of the seven most immunodominant antigens recognized by the naturally exposed group, including known blood stage antigens MSP1, MSP2, MSP4, and Exp1, were recognized by the experimentally infected individuals. However antibodies against LSA3 (the most immunoreactive antigen recognized by the naturally exposed group) or against the blood stage specific antigens MSP6, MSP7, EBA175 or S-antigen did not develop post-challenge. The most dominant responses in the experimentally infected group were to MSP2, AMA1, MAL7P1.32 (hypothetical protein), PFI0580c (hypothetical protein), PFA0410w (hypothetical protein), and PFL0625c (translation initiation factor).
Profile of antigen recognition within a donor group
The profiles of antigen immunoreactivity were remarkably consistent among individuals within a given donor group, although individual specificities were apparent as expected from an outbred human population. The antigen-specific reactivity profile for each of the 12 naturally exposed subjects is presented in Figure 3. The signal intensities varied by about 10-fold between subjects. The profile of a pool of hyperimmune serum from 192 subjects resident in the same region and assayed in parallel appeared representative of an average of all individual responses, and a scatterplot of the pooled sera against the average of the individual sera gave a line with an R2 value of 0.83. Antibody profiles from individuals experimentally immunized with radiation-attenuated Pf sporozoites were also consistent among individuals, although some inter-individual variation was apparent as expected given the genetic heterogeneity of the human population (Figure 3). As noted above, these sera reacted with a distinct set of antigens as compared with sera from naturally exposed individuals.
Proteomic features of the immunodominant antigens
The data in Table 3 and Supplementary Table S5 summarize proteomic features that are enriched in the immunodominant antigen set relative to the whole Pf proteome. The average isoelectric point of all the proteins in the Pf proteome is 8.0, but the average isoelectric point of the top 48 immunodominant ORFs is 6.3, reflecting a propensity for more acidic structures or peptide fragments to be recognized by the host immune system; in the Systemic Lupus model, isoelectric point was identified as an excellent predictor of the antigenicity of spliceosomal autoantigens [52]. The average molecular weight of all the Pf proteins in the proteome is 87 kDa, but the 48 immunodominant antigens average nearly two times larger, presumably reflecting the presence of additional B cell epitopes in the larger sequences. A predicted signal peptide sequence is present in 42% of the immunodominant hits as compared to 15% of the whole Pf proteome, consistent with secretion of the protein product into the cytoplasm for recognition by the host immune system. Fifty percent of the proteins on the top 48 immunodominant hit list were detected by mass spectrometry analysis (MudPIT) of different stages of the Plasmodium parasite life cycle as compared with 18% of the whole Pf proteome (1000 proteins) (www.PlasmodDB.org), presumably reflecting the relative abundance of the immunodominant proteins. Notably, PfEMP1 was absent from this list. Combining the signal peptide and mass spectroscopy evidence produces a set of 1712 proteins predicted to contain ~70% of the top immunodominant hits (Table 3 and Supplementary Table S5). In summary, low isoelectric point, high molecular weight, a signal peptide, and evidence of expression by mass spectrometry are proteomic features associated with enhanced recognition by the humoral immune system.
Table 3.
Proteomic features associated with the subset of proteins identified as immunodominant
| Signal Peptide | EST Evidence | DNA Microarray |
Mass Spec. |
Signal Pep. & Mass Spec. | |||
|---|---|---|---|---|---|---|---|
| Sexual Stage | Asexual Stage | Sexual Stage | Asexual Stage | All Stages | |||
| Total ORFs in Category | 835 | 1244 | 2557 | 3039 | 3814 | 1000 | 1712 |
| % of Total Genome in Category | 15% | 23% | 46% | 55% | 69% | 18% | 33% |
| # of Immunodominant Hits in Category | 20 | 13 | 35 | 39 | 43 | 24 | 33 |
| % of Total Immunodominant Hits | 42% | 27% | 73% | 81% | 90% | 50% | 69% |
| Fold enrichment Immunodominant Hits | 2.8 | 1.2 | 1.6 | 1.5 | 1.3 | 2.8 | 2.1 |
Different features were identified that classify the genes in the Pf genome that are present in the antigen set identified here as immunodominant in the context of host immunity.
At least 12 of the immunodominant antigens contain primary amino acid sequence repeats; five of these antigens are already known and well-described antigens (S-antigen, LSA1, LSA3, CSP, STARP) (Supplementary Table S6). Of the hypothetical antigens identified as immunoreactive here, five antigens contain arginine rich domains, and one antigen has a high serine content (28%) but is not a member of the previously annotated SERA family [53]. One protein (PFL0625c) has a more complex repeat domain. Overall, data are consistent with the presence of immunodominant B cell epitopes in repeat regions.
DISCUSSION
To date, efforts to express Pf proteins on a large-scale have been largely unsuccessful [42–44, 54]. Even on a small scale, Pf proteins have proven particularly difficult to express in bacteria, yeast or insect cells using conventional methodologies as compared with other organisms, possibly due to their high A+T content (~82%, the highest A+T content of any known organism [1] ) and rare codon usage [55]. Herein, we have used a high throughput PCR recombination cloning and E. coli based cell-free in vitro transcription/translation system to express 250 putative Pf proteins. We report ≥ 93% expression efficiency, contrasting with rates reported in other large-scale Pf studies in E. Coli (7–16% [42]; 21% [44]; 6% [43]) or the wheat germ cell-free system (75% [54]). The actual frequency of expression in our E. coli-based system may be greater than 93% since in some cases the HA- or HIS-tags used to confirm expression may be present but conformationally obscured and not accessible to antibody on dot blots. These expression percentages agree well with other proteomes that we have investigated including vaccinia virus [21], F. tularensis [38], and M. tuberculosis (unpublished results). The high success rate seen with the cell-free transcription/translation system may be attributed, at least in part, to the fact that the system is supplemented with rare t-RNAs to help translate A+T-rich genes, that there is no cell to be killed by a potentially lethal expressed product, and that the proteins do not have to be purified prior to printing. It is understood that antibodies recognize their targets specifically even in the presence of large amounts of nonspecific proteins. The high efficiency of protein expression is maintained on the microarray by virtue of printing the expression reactions without further protein purification, since manipulation and handling cause significant loss. Furthermore, the microarrays are printed within a few hours after the expression reaction is complete reducing protein instability problems. Once the protein microarrays are printed, they are stored dry at room temperature and are stable under these conditions for many months (unpublished results).
The bacterial cell-free expression system has two main disadvantages. Firstly, there is no glycosylation of proteins. In vaccinia immunized humans and animals we have observed reactivity to vaccinia extracellular enveloped virion proteins that are known to be glycosylated, even though the E. coli expression system does not generate glycosylated products [56]. Presumably the polyclonal response generated during infection of eukaryotic cells produces antibodies against the non-glycosylated regions of these proteins. However, it is possible that some Pf glycoproteins have not been recognized in this study. Secondly, some protein products may not be folded correctly due to the lack of a proper redox environment in which to form disulfide bonds [56], and so immune responses against other important conformational epitopes may also have been missed.
Data from other studies show that protein microarray chip signal intensity is proportional both to the titer of antibody in the sample [22] and the amount of antigen spotted on the chip (unpublished results). Since the amount of antigen per spot varies between different antigens, and the absolute amount of antigen per spot is not accurately known, one cannot directly compare signal intensities between different antigens. However, one can legitimately compare signal intensities of a given antigen between different sera samples. Moreover antibody titers against different antigens, obtained by plotting signal intensities for a given antigen against the serial dilution of sera to produces a sigmoid ‘titration curve’, can be compared between antigens. These considerations are similar for the development of traditional ELISA assays.
Here, using protein microarrays, we have screened ~5% of the entire Pf genome (250 putative proteins) for its capacity to be recognized by antibodies from individuals naturally or experimentally exposed to Pf, in order to identify immunodominant antigens and define immunoreactivity profiles amongst distinct donor groups, including individuals who are demonstrably protected from malaria. Within this subset was a panel of 21 known and well characterized Pf pre-erythrocytic and erythrocytic stage antigens. Inclusion of those antigens allowed us to verify that the protein microarray approach can effectively identify antigens known to be recognized by immune sera, that antigens expressed in stages other than the asexual erythrocyte stage can be recognized, and that the results of protein chip based screening correlate with results from a conventional ELISA assay.
A total of 72 highly immunoreactive proteins were identified (Supplementary Table S4). Almost all of the well-characterized antigens that are under clinical evaluation (with orthologs demonstrated to be protective in animal models of malaria) were identified by this analysis, and 16 of these ranked within the top 23 most immunoreactive antigens. In addition, 56 previously uncharacterized antigens were identified as serodominant. Of particular interest as high priority antigens for further evaluation are five hypothetical proteins on the list of the top 23 most immunoreactive antigens: MAL7P1.32, PFI0580c, PF10_0138, PFE0060w, and PFL2410w. Also of interest are two antigens previously shown to be recognized by T cells from irradiated sporozoite immunized volunteers: PF11_0226 (high T cell reactivity) and PF10_0179 (intermediate T cell reactivity) [46]. An additional subset of potential interest are those hypothetical proteins that did not meet our conservative criteria of positivity [(i) normalized signal intensity > 4, (ii) p < 0.05, and (iii) number of positive responses within a particular group ≥ 2] but that were highly significant on a statistical basis for recognition by protected but not unprotected irradiated sporozoite immunized volunteers (MAL7P1.29, PFL1130c, PF14_0470, PF11_0358, PF13_0350, PF11_0226); most of these were also recognized as significant by naturally exposed individuals consistent with boosting of the antigen-specific immune response in the field by sporozoite exposure. Overall, from a panel of 250 putative Pf proteins screened by protein microarrays, we identified 14 novel antigens which warrant additional consideration as potential target antigens for a subunit malaria vaccine.
Notably, the well characterized pre-erythrocytic stage antigens CSP, SSP2/TRAP, and AMA1 were recognized by both the protected and unprotected subgroups of volunteers immunized by irradiated sporozoites. The reactivity profiles did not reflect whether or not the volunteers were protected against subsequent sporozoite challenge, suggesting that other antigens yet to be defined may be targeted by the protective immune responses induced by immunization with radiation-attenuated Pf sporozoites. Those novel antigens that are not recognized in susceptible individuals but are reactive in individuals with sterile protective immunity are potentially more promising as targets for vaccine development than antigens currently in clinical development. CSP was the only antigen recognized by all irradiated sporozoite immunized volunteers, regardless of protection status, consistent with this protein being the dominant coat protein on the sporozoite.
An important finding from this research is the demonstration that protein microarrays can identify, as antibody targets, proteins expressed only in the pre-erythrocytic stage of the Plasmodium parasite (e.g., CSP, SSP2/TRAP), including liver-stage specific antigens such as LSA1 which was dominant in naturally exposed subjects. T cell responses rather than antibody responses are considered the important immune responses against antigens expressed in these stages. This finding is supported by the identification here of two hypothetical antigens known to be recognized by T cells from irradiated sporozoite immunized volunteers (PF11_0226 and PF10_0179) [46]. Previously, although antigens known to elicit T cell responses are likely to be recognized by serum antibodies, it had been considered that a protein-antibody based approach might preferentially identify blood stage proteins that are targets of antibody responses. It should be noted, however, that demonstration that a given protein is recognized by antibodies does not constitute evidence of B-cell mediated protective responses. Antibodies that recognize Pf antigens may be elicited against proteins released from dead parasites or as a result of schizogony. There is therefore no direct link between antibody recognition and protection in the absence of further functional information. Confirmation that the novel antigens identified by protein microarrays elicit protective immune responses in humans will take additional research.
It should be also noted that the protein microarray approach is particularly suited for screening large numbers of sera. We have seen in other pathogens that antibody profiles generated by humans are similar although the specificities vary between individuals [21, 23]. We term those proteins that are most frequently recognized in a population as "serodominant" (in contrast to "immunodominant" which usually refers to antigens recognized in the context of a single individual). Serodominant proteins may be important targets for development vaccines and diagnostics. In other studies, we are screening T cell and antibody responses at the level of the whole vaccinia proteome in subjects immunized with the licensed vaccinia-virus based smallpox vaccine, Dryvax (Wyeth). We have observed that there is overlap in the antibody profiles revealed by protein microarray and the immunodominant antigens recognized by proliferative (CD4+) T cells (Jing et al, in press), as would be predicted for cognate T-B cell collaboration.
In addition to identifying serodominant antigens which may represent promising targets for vaccine development, we show that protein microarrays identify characteristic immunoreactive antigen profiles recognized by serum antibodies from distinct donor groups of differing malaria immune status. Within a donor group, there is individual-to-individual variation in the strength and breadth of the profiled immune responses, as expected, and it is encouraging that the method can distinguish these differences between individuals. From a vaccine perspective, comparing the profiles between protected and unprotected subjects in vaccine studies using whole organisms (eg, immunization with radiation attenuated Pf sporozoites, genetically attenuated parasites [57–59] or parasitized erythrocytes [60]) may lead to the identification of antigens associated with protection that may be useful in subunit vaccines. The identification of unique immunoreactivity profiles among donor groups of differing immune status, and the relationship between these antigen(s) and protective immunity, may also be useful for dissecting the molecular basis of immunity and for identifying immune correlates of protection. Data will also promote understanding of host-pathogen immunity.
In summary, herein we define the antibody profiles that develop in humans after exposure to a complex parasite that poses an enormous public health threat worldwide. We show that we can discriminate between individuals with sterile immunity against experimental malaria challenge following vaccination with radiation-attenuated Pf sporozoites, individuals with partial immunity acquired by natural exposure to malaria, individuals who were not protected against experimental malaria challenge following vaccination with radiation-attenuated Pf sporozoites, and healthy controls with no history of malaria exposure. We have further identified 72 highly immunoreactive Pf antigens which may represent promising targets for vaccine development. Our results have basic science implications by contributing to a better understanding of host-pathogen immunity, as well as practical implications for vaccine development.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Denis Heck, UCI Microarray Facility, for array printing. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the U.S. Government.
FUNDING
This work was supported by National Institute of Allergy and Infectious Diseases Grants U01AI056464 and 1U01AI061363, and by funds allocated to the Naval Medical Research Center by the US Army Medical Research Materiel Command (work unit 6000.RAD1.F.A0309). The bioinformatics and primer design in this work was supported by National Institutes of Health Biomedical Informatics Training Program Grant 5T15LM007743 and National Science Foundation Grant MRI EIA-0321390 to Pierre Baldi and the Institute for Genomics and Bioinformatics.
Abbreviations
- EST
expressed sequence tag
- LCM
laser capture microdissection
- ORF
open reading frame
- MudPIT
multidimensional protein identification technology
- Pf
Plasmodium falciparum
- SI
signal intensity
Footnotes
AUTHOR CONTRIBUTIONS
DLD and PLF conceived and designed the study, assisted in data analysis and interpretation, and wrote the manuscript. PLF and DHD contributed to the supervision and execution of the research. YM, BU, CV executed the research and assisted in data analysis and preparation of the manuscript figures. DM and XL provided the protein microarrays. SS, SH, AR and PB were responsible for the bioinformatic and statistical analysis. PLB and JCA assisted in the initial selected of open reading frames for analysis. DAF was responsible for the studies with irradiated sporozoite immunized volunteers that provided key specimens for analysis. JAO was responsible for the field studies with Kenyan volunteers that provided key specimens for analysis.
COMPETING INTERESTS
The authors declare that no competing interests exist.
REFERENCES
- 1.Gardner MJ, Hall N, Fung E, White O, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Florens L, Washburn MP, Raine JD, Anthony RM, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 3.Lasonder E, Ishihama Y, Andersen JS, Vermunt AM, et al. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419:537–542. doi: 10.1038/nature01111. [DOI] [PubMed] [Google Scholar]
- 4.Bozdech Z, Llinas M, Pulliam BL, Wong ED, et al. The Transcriptome of the Intraerythrocytic Developmental Cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Roch KG, Zhou Y, Blair PL, Grainger M, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 6.Le Roch KG, Johnson JR, Florens L, Zhou Y, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall N, Karras M, Raine JD, Carlton JM, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 8.Kawakami Y, Rosenberg SA. Human tumor antigens recognized by T-cells. Immunol Res. 1997;16:313–339. doi: 10.1007/BF02786397. [DOI] [PubMed] [Google Scholar]
- 9.Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255:1261–1266. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 10.Rotzschke O, Falk K, Deres K, Schild H, et al. Isolation and analysis of naturally processed peptides as recognized by cytotoxic T cells. Nature. 1990;348:252–254. doi: 10.1038/348252a0. [DOI] [PubMed] [Google Scholar]
- 11.Van Bleek GM, Nathenson SG. Isolation of an endogenously processed immunodominant viral peptide from the class I H-2Kb molecule. Nature. 1990;348:213–216. doi: 10.1038/348213a0. [DOI] [PubMed] [Google Scholar]
- 12.Currier JR, deSouza M, Chanbancherd P, Bernstein W, et al. Comprehensive screening for human immunodeficiency virus type 1 subtype-specific CD8 cytotoxic T lymphocytes and definition of degenerate epitopes restricted by HLA-A0207 and -C(W)0304 alleles. J Virol. 2002;76:4971–4986. doi: 10.1128/JVI.76.10.4971-4986.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauer GM, Ouchi K, Chung RT, Nguyen TN, et al. Comprehensive analysis of CD8(+)-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. J Virol. 2002;76:6104–6113. doi: 10.1128/JVI.76.12.6104-6113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maecker HT, Dunn HS, Suni MA, Khatamzas E, et al. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol Methods. 2001;255:27–40. doi: 10.1016/s0022-1759(01)00416-1. [DOI] [PubMed] [Google Scholar]
- 15.Davenport MP, Hill AV. Reverse immunogenetics: from HLA-disease associations to vaccine candidates. Molecular Medicine Today. 1996;2:38–45. doi: 10.1016/1357-4310(96)88757-0. [DOI] [PubMed] [Google Scholar]
- 16.Hill AV, Elvin J, Willis AC, Aidoo M, et al. Molecular analysis of the association of HLA-B53 and resistance to severe malaria. Nature. 1992;360:434–439. doi: 10.1038/360434a0. [DOI] [PubMed] [Google Scholar]
- 17.Kung LA, Snyder M. Proteome chips for whole-organism assays. Nat Rev Mol Cell Biol. 2006;7:617–622. doi: 10.1038/nrm1941. [DOI] [PubMed] [Google Scholar]
- 18.Zhu H, Bilgin M, Bangham R, Hall D, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 19.Ramachandran N, Hainsworth E, Demirkan G, LaBaer J. On-chip protein synthesis for making microarrays. Methods Mol Biol. 2006;328:1–14. doi: 10.1385/1-59745-026-X:1. [DOI] [PubMed] [Google Scholar]
- 20.Bahl A, Brunk B, Coppel RL, Crabtree J, et al. PlasmoDB: the Plasmodium genome resource. An integrated database providing tools for accessing, analyzing and mapping expression and sequence data (both finished and unfinished) Nucleic Acids Res. 2002;30:87–90. doi: 10.1093/nar/30.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies DH, Liang X, Hernandez JE, Randall A, et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies DH, Wyatt LS, Newman FK, Earl PL, et al. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J Virol. 2008;82:652–663. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benhnia MR, McCausland MM, Su H-P, Singh K, et al. Redundancy and plasticity or neutralizing antibody responses are cornerstone attributes of the human immune response to the smallpox vaccine. J. Virol. 2008;82:3751–3768. doi: 10.1128/JVI.02244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charoenvit Y, Leef ML, Yuan LF, Sedegah M, Beaudoin RL. Characterization of Plasmodium yoelii monoclonal antibodies directed against stage-specific sporozoite antigens. Infect. Immun. 1987;55:604–608. doi: 10.1128/iai.55.3.604-608.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charoenvit Y, Mellouk S, Cole C, Bechara R, et al. Monoclonal, but not polyclonal, antibodies protect against Plasmodium yoelii sporozoites. J Immunol. 1991;146:1020–1025. [PubMed] [Google Scholar]
- 26.Egan JE, Hoffman SL, Haynes JD, Sadoff JC, et al. Humoral immune responses in volunteers immunized with irradiated Plasmodium falciparum sporozoites. Am J Trop Med Hyg. 1993;49:166–173. doi: 10.4269/ajtmh.1993.49.166. [DOI] [PubMed] [Google Scholar]
- 27.Beier JC, Oster CN, Onyango FK, Bales JD, et al. Plasmodium falciparum incidence relative to entomologic inoculation rates at a site proposed for testing malaria vaccines in western Kenya. Am J Trop Med Hyg. 1994;50:529–536. doi: 10.4269/ajtmh.1994.50.529. [DOI] [PubMed] [Google Scholar]
- 28.McElroy PD, Beier JC, Oster CN, Beadle C, et al. Predicting outcome in malaria: correlation between rate of exposure to infected mosquitoes and level of Plasmodium falciparum parasitemia. Am. J. Trop. Med. Hyg. 1994;51:523–532. [PubMed] [Google Scholar]
- 29.Beadle C, McElroy PD, Oster CN, Beier JC, et al. Impact of transmission intensity and age on Plasmodium falciparum density and associated fever: implications for malaria vaccine trial design. J Infect Dis. 1995;172:1047–1054. doi: 10.1093/infdis/172.4.1047. [DOI] [PubMed] [Google Scholar]
- 30.Doolan DL, Hoffman SL, Southwood S, Wentworth PA, et al. Degenerate cytotoxic T cell epitopes from P. falciparum restricted by multiple HLA-A and HLA-B supertype alleles. Immunity. 1997;7:97–112. doi: 10.1016/s1074-7613(00)80513-0. [DOI] [PubMed] [Google Scholar]
- 31.Sundaresh S, Doolan DL, Hirst S, Mu Y, et al. Identification of humoral immune responses in protein microarrays using DNA microarray data analysis techniques. Bioinformatics. 2006;22:1760–1766. doi: 10.1093/bioinformatics/btl162. [DOI] [PubMed] [Google Scholar]
- 32.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;(18 Suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 33.Kreil DP, Karp NA, Lilley KS. DNA microarray normalization methods can remove bias from differential protein expression analysis of 2D difference gel electrophoresis results. Bioinformatics. 2004;20:2026–2034. doi: 10.1093/bioinformatics/bth193. [DOI] [PubMed] [Google Scholar]
- 34.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 35.Hatfield GW, Hung SP, Baldi P. Differential analysis of DNA microarray gene expression data. Mol Microbiol. 2003;47:871–877. doi: 10.1046/j.1365-2958.2003.03298.x. [DOI] [PubMed] [Google Scholar]
- 36.Hung SP, Baldi P, Hatfield GW. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J Biol Chem. 2002;277:40309–40323. doi: 10.1074/jbc.M204044200. [DOI] [PubMed] [Google Scholar]
- 37.Long AD, Mangalam HJ, Chan BY, Tolleri L, et al. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem. 2001;276:19937–19944. doi: 10.1074/jbc.M010192200. [DOI] [PubMed] [Google Scholar]
- 38.Sundaresh S, Randall A, Unal B, Petersen JM, et al. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics. 2007;23:508–518. doi: 10.1093/bioinformatics/btm207. [DOI] [PubMed] [Google Scholar]
- 39.Choe SE, Boutros M, Michelson AM, Church GM, Halfon MS. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 2005;6:R16. doi: 10.1186/gb-2005-6-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storey JD. A direct approach to false discovery rates. J. Royal Stat Soc. 2002;64:479–498. [Google Scholar]
- 41.Allison DB, et al. A mixture model approach for the analysis of microarray gene expression data. Comput Stat Data Anal. 2002;39:1–20. [Google Scholar]
- 42.Aguiar JC, LaBaer J, Blair PL, Shamailova VY, et al. High-throughput generation of P. falciparum functional molecules by recombinational cloning. Genome Res. 2004;14:2076–2082. doi: 10.1101/gr.2416604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehlin C, Boni E, Buckner FS, Engel L, et al. Heterologous expression of proteins from Plasmodium falciparum: results from 1000 genes. Mol Biochem Parasitol. 2006;148:144–160. doi: 10.1016/j.molbiopara.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Vedadi M, Lew J, Artz J, Amani M, et al. Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol Biochem Parasitol. 2007;151:100–110. doi: 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Wu CC, MacCoss MJ, Mardones G, Finnigan C, et al. Organellar proteomics reveals Golgi arginine dimethylation. Mol Biol Cell. 2004;15:2907–2919. doi: 10.1091/mbc.E04-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doolan DL, Southwood S, Freilich DA, Sidney J, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci U S A. 2003;100:9952–9957. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dame JB, Williams JL, McCutchan TF, Webe JL, et al. Structure of the gene encoding the immunodominant surface antigen on the sporozoite of the human malaria parasite Plasmodium falciparum. Science. 1984;225:593–599. doi: 10.1126/science.6204383. [DOI] [PubMed] [Google Scholar]
- 48.Robson KJ, Hall JR, Jennings MW, Harris TJ, et al. A highly conserved amino-acid sequence in thrombospondin, properdin and in proteins from sporozoites and blood stages of a human malaria parasite. Nature. 1988;335:79–82. doi: 10.1038/335079a0. [DOI] [PubMed] [Google Scholar]
- 49.Bodescot M, Silvie O, Siau A, Refour P, et al. Transcription status of vaccine candidate genes of Plasmodium falciparum during the hepatic phase of its life cycle. Parasitol Res. 2004;92:449–452. doi: 10.1007/s00436-003-1061-9. [DOI] [PubMed] [Google Scholar]
- 50.Fidock DA, Bottius E, Brahimi K, Moelans II, et al. Cloning and characterization of a novel Plasmodium falciparum sporozoite surface antigen, STARP. Mol.Biochem.Parasitol. 1994;64:219–232. doi: 10.1016/0166-6851(94)00012-3. [DOI] [PubMed] [Google Scholar]
- 51.Pasquetto V, Fidock DA, Gras H, Badell E, et al. Plasmodium falciparum sporozoite invasion is inhibited by naturally acquired or experimentally induced polyclonal antibodies to the STARP antigen. Eur. J. Immunol. 1997;27:2502–2513. doi: 10.1002/eji.1830271007. [DOI] [PubMed] [Google Scholar]
- 52.McClain MT, Ramsland PA, Kaufman KM, James JA. Anti-sm autoantibodies in systemic lupus target highly basic surface structures of complexed spliceosomal autoantigens. J Immunol. 2002;168:2054–2062. doi: 10.4049/jimmunol.168.4.2054. [DOI] [PubMed] [Google Scholar]
- 53.Miller SK, Good R, Drew DR, Delorenzi M, et al. A subset of plasmodium falciparum SERA genes are expressed and appear to play an important role in the erythrocytic cycle. J Biol Chem. 2002 doi: 10.1074/jbc.M206974200. [DOI] [PubMed] [Google Scholar]
- 54.Tsuboi T, Takeo S, Iriko H, Jin L, et al. Wheat germ cell-free system-based production of malaria proteins for discovery of novel vaccine candidates. Infect Immun. 2008;76:1702–1708. doi: 10.1128/IAI.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber JL. Molecular biology of malaria parasites. Exp.Parasitol. 1988;66:143–170. doi: 10.1016/0014-4894(88)90087-2. [DOI] [PubMed] [Google Scholar]
- 56.Yadava A, Ockenhouse CF. Effect of codon optimization on expression levels of a functionally folded malaria vaccine candidate in prokaryotic and eukaryotic expression systems. Infect Immun. 2003;71:4961–4969. doi: 10.1128/IAI.71.9.4961-4969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mueller AK, Camargo N, Kaiser K, Andorfer C, et al. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc Natl Acad Sci U S A. 2005;102:3022–3027. doi: 10.1073/pnas.0408442102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Dijk MR, Douradinha B, Franke-Fayard B, Heussler V, et al. Genetically attenuated, P36p-deficient malarial sporozoites induce protective immunity and apoptosis of infected liver cells. Proc Natl Acad Sci U S A. 2005;102:12194–12199. doi: 10.1073/pnas.0500925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mueller AK, Labaied M, Kappe SH, Matuschewski K. Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature. 2005;433:164–167. doi: 10.1038/nature03188. [DOI] [PubMed] [Google Scholar]
- 60.Wykes M, Good MF. A case for whole-parasite malaria vaccines. Int J Parasitol. 2007;37:705–712. doi: 10.1016/j.ijpara.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Sacci JB, Jr, Ribeiro JM, Huang F, Alam U, et al. Transcriptional analysis of in vivo Plasmodium yoelii liver stage gene expression. Mol Biochem Parasitol. 2005;142:177–183. doi: 10.1016/j.molbiopara.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 62.Florens L, Liu X, Wang Y, Yang S, et al. Proteomics approach reveals novel proteins on the surface of malaria-infected erythrocytes. Mol Biochem Parasitol. 2004;135:1–11. doi: 10.1016/j.molbiopara.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 63.Kappe SH, Gardner MJ, Brown SM, Ross J, et al. Exploring the transcriptome of the malaria sporozoite stage. Proc Natl Acad Sci U S A. 2001;98:9895–9900. doi: 10.1073/pnas.171185198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.