Abstract
Sarcomas are a group of heterogeneous tumours with varying genetic basis. Cytogenetic abnormalities range from distinct genomic rearrangements such as pathognomonic translocation events and common chromosomal amplification or loss, to more complex rearrangements involving multiple chromosomes. The different subtypes of liposarcoma are spread across this spectrum and constitute an interesting tumour type for molecular review. This paper will outline molecular pathogenesis of the three main subtypes of liposarcoma: well-differentiated/dedifferentiated, myxoid/round cell, and pleomorphic liposarcoma. Both the molecular basis and future avenues for therapeutic intervention will be discussed.
1. Introduction
An estimated 13,000 people were diagnosed with soft tissue and bone sarcoma in 2009 in America, of which liposarcomas constitute 20% [1, 2]. Despite their rarity these tumours have substantial morbidity and mortality, depending on histological subtype, tumour location, and volume with retroperitoneal sarcomas having particularly poor prognosis [3–9]. Liposarcomas may be classified morphologically into 3 main subtypes consisting of: well-differentiated liposarcoma/de-differentiated liposarcoma (WD/DDLPS), myxoid/round cell liposarcoma (MLPS) and pleomorphic liposarcoma (PLPS) [10]. The morphological diversity of liposarcoma reflects the great variation in biological behaviour ranging from tumours with low metastatic potential, that is, WDLPS, to tumours with high propensity to metastasise, that is, the round cell (RC) variant of MLPS or PLPS [11]. In addition to histological characteristics, anatomical location impacts upon prognosis, given that local control is a prime concern for curative intent.
Treatment is multimodal with surgical removal and radiotherapy used as cornerstones for local control, along with chemotherapy for systemic disease. Few therapeutic options are available for aggressive local or metastatic disease. Chemotherapy sensitivity varies considerably between subtypes with higher response rates in MLPS compared with WD/DDLPS (48% versus 11%) [12]. MLPS tumours are also highly radiosensitive [13, 14]. Given the small subgroup that is chemo-sensitive, and the overriding lack of chemo-curative disease there are avenues and a need for novel molecular therapies.
A recent histological and molecular review of 163 liposarcoma and lipomas at the Netherlands Cancer Institute resulted in 23% of tumours being reclassified based on cytogenetic information. This highlights the importance of molecular classification in these tumours and genetic alterations now considered an integral part of the WHO classification [15]. It is hoped that further insight into the molecular characteristics of liposarcomas will allow for accurate subclassification, whilst providing a platform for molecular therapies to be included in the current treatment approach. This paper will outline the current molecular basis of liposarcoma and potential strategies for therapeutic intervention.
2. Well- and De-differentiated Liposarcoma
WDLPS represents 40%–45% of all diagnosis of liposarcoma [16]. It is classified as a low-grade neoplasm; it is rarely metastatic and has a low recurrence rate (10%) occurring most often in the retroperitoneum and limbs. The World Health Organization (WHO) classifies WDLPS into three main subtypes: adipocytic, sclerosing, and inflammatory. Adipocytic (lipoma-like) liposarcoma is composed of mature adipocytes, which exhibit variation in cell size and focal nuclear atypia and hyperchromasia [16]. The sclerosing subtype shows scattered distinctive bizarre stromal cells associated with rare multivacuolated lipoblasts set in a fibrillary collagenous background [16]. Finally, the inflammatory subtype shows polyphenotypic lymphoplasmacytic infiltrate, with a B-cell predominance. Less is known about this rare subtype [16–18].
DDLPS represents progression from low grade to high-grade nonlipogenic morphology within a WDLPS. DDLPS is more aggressive and exhibits an increased rapidity of disease in contrast to WDLPS, with a metastatic rate of 10%–20% and overall mortality of 50%–75% [4, 7, 19]. In respect to tumour site, retroperitoneal tumours appear to have a worse prognosis [19]. Histologically, DDLPS consists of a WDPLS with a nonlipogenic component, either high-grade, most often resembling malignant fibrous histiocytoma (MFH), or low-grade resembling fibromatosis or low-grade myxofibrosarcoma. The presence of transition from WDLPS to DDLPS is used to differentiate between DDLPS and these other lesions [4, 7, 11, 19–21].
2.1. Molecular Genetics
A characteristic feature of WD/DDLPS is the presence of supernumerary ring and/or giant rod chromosomes [22]. These chromosomes contain amplified segments from the 12q13–15 region that can be identified with fluorescence in situ hybridization (FISH) and comparative genomic hybridization (CGH) [23]. Intensive research has identified several oncogenes residing in this region including MDM2, CDK4, HMGA2, TSPAN31, OS1, OS9, CHOP and GLI1 [11, 23–25]. The most compelling evidence to date demonstrates an oncogenic role in WD/DDLPS for MDM2, CDK4, HMGA2 and TSPAN31. Additional amplification events may also play a role in liposarcoma genesis, for example, c-Jun in the de-differentiation process [26].
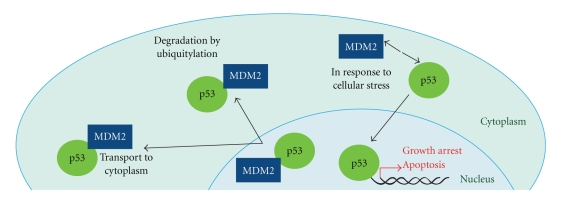
MDM2 amplification is a key feature of WD/DDLPS and is amplified and overexpressed in a number of other cancers, highlighting its importance in tumorigenesis (as reviewed [27]). MDM2 encodes a negative regulator of the tumour suppressor, p53. MDM2 binds to the transcription activation domain of p53, within an N-terminal hydrophobic pocket [28], blocking p53-dependent transcription [29–33] and recruitment of transcription coactivators [28]. MDM2 also acts as a ubiquitin ligase targeting p53 for proteasomal degradation through both cytoplasmic and nuclear proteasomes [34–36]. MDM2 is involved in its own auto-degradation to prevent MDM2 activity inhibiting p53 during times of cellular stress [37]. Thus MDM2 maintains tight control on cellular p53 levels through multiple mechanisms (see Figure 1) [38, 39]. Therapeutically this is important, as MDM2 inhibitors aim to reactivate p53 and thus allow it to actively induce cell death in response to appropriate stressors [40]. In addition to a functional downstream p53 signalling pathway, MDM2 amplification is a predictor of sensitivity to current MDM2 antagonists [40]. Amplification of MDM2 and mutation of p53 appear to be mutually exclusive events in WDLPS, but have been reported in DDLPS [41, 42]. p53 mutations have been associated with the de-differentiation process from WDLPS to DDLPS [41]. Pilotti et al. reported upon a subgroup of WD/DDLPS tumours. Retroperitoneal WD/DDLPS demonstrate mutual exclusivity between MDM2 amplification and p53 mutation. In non-retroperitoneal DDLPS, p53 mutations occur in the absence of MDM2 amplification suggesting involvement in the de-differentiation process [41].
Figure 1.
MDM2 binds to the transcriptional activation domain of p53, blocking transcription. MDM2 functions as a ubiquitin ligase, facilitating proteasomal degradation of p53. MDM2 releases p53 in response to cellular stress and p53 translocates to the nucleus where it acts as a transcription factor to enable growth arrest and apoptosis.
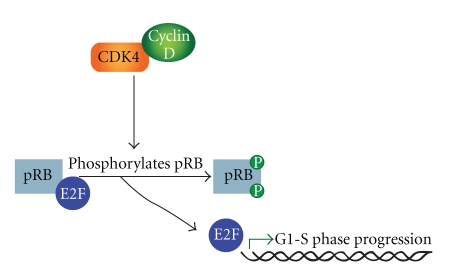
MDM2 is the most frequent amplification in WD/DDLPS (close to 100%) however CDK4 is shown to be amplified in over 90% of cases [16, 43, 44]. Given its role in the cell cycle and the frequency of amplification, CDK4 has been well researched in WD/DDLPS. The CDK4 gene encodes a 33-kD protein that forms complexes with the cyclin D family, to enable G1-S transition [45]. These CDK4/Cyclin D complexes phosphorylate pRb (encoded by RB1), with resultant activation of E2F target genes including E-type cyclins (see Figure 2) [46–48]. It has been suggested that CDK4 provides a selection advantage in WD/DDLPS and may contribute to transformation as CDK4 negative WDLPS exhibit more favorable prognostic features [46]. Coamplification of MDM2 and CDK4 is a common feature of WD/DDLPS and may result in proliferation through combined effects upon p53 and the cell cycle [49, 50]. Interestingly, the rearrangements of chromosome 12 on the giant rod chromosome are discontinuous and MDM2 and CDK4 may belong to different amplicons [51, 52]. Several studies [43, 53, 54] have suggested that immunohistochemical staining for both CDK4 and MDM2 may provide a useful diagnostic marker, although FISH and quantitative polymerase chain reaction (qPCR) are more effective. Although MDM2 and CDK4 are useful markers to aid in diagnosis, overexpression of these markers is not unique to WD/DDLPS [43, 54]. Further, the amplification and over-expression of CDK4 and MDM2 does not distinguish WDLPS from DDLPS [16, 23, 41].
Figure 2.
Cyclin dependent kinase CDK4 binds with cyclin D to form active complexes. This results in phosphorylation of Rb and dissociates pRb from the pRb-E2F complex. E2F binds DNA to upregulate transcription of genes required to progress to S phase.
HMGA2 is similarly located on 12q and frequently amplified in WD/DDLPS. This is a member of the high-mobility group of proteins [55, 56]. Previously referred to as HMGIC, it encodes an architectural transcription factor capable of remodeling DNA [57–59]. A direct role for HMGA2 in cellular transformation is demonstrated by NIH3T3 neoplastic transformation with the overexpression of HMGA2 [60]. In human sarcomas during chromosomal rearrangement, HMGA2 is fused to distant sequences, commonly occurring on other chromosomes and loses its 3′ translated end that also contains sites for Let-7 microRNAS [57]. Further support for HMGA2 involvement in adipogenic neoplasm development includes the xenograft model by Arlotta et al. [55] that showed mice expressing C-terminal truncated HMGA2 developed lipomas. Interestingly HMGA2 is frequently coamplified with MDM2 in human malignant tumours [57, 61], particularly WDLPS and DDLPS [52]. This raises the possibility that HMGA2 and MDM2 have a cooperating role in WD/DDLPS. Also included within the chromosome 12 q13–15 region is the transmembrane superfamily gene, sarcoma amplified sequence (SAS or TSPAN31) gene [62, 63]. TSPAN31 was originally identified and cloned from an amplified sequence in a malignant fibrous histiocytoma [63]. It has been identified in other subtypes of sarcoma, particularly de-differentiated liposarcoma [64, 65], although its precise role in the de-differentiation process is not well delineated. Forus et al. [66] showed TSPAN31 was as frequently amplified as MDM2 in 98 sarcomas. Both TSPAN31 and MDM2 were amplified in 8 of 11 liposarcoma samples, with MDM2 amplified alone in one additional tumour. WDLPS and DDLPS have shown co-amplification of 1q21-q22 and/or 12q21-q22 [11, 16, 23], along with amplification of chromosome 1(1q21-q23). Chromosome 1 amplified sequences include COAS1, COAS2 and COAS3 [67]. Nilsson et al. showed co-amplification of both COAS and MDM2 in 12/18 lipomatous tumours [68]. The biological function of the COAS genes remains a subject for study.
Recent studies into the WDLPS de-differentiation process have suggested a role for the c-Jun N-terminal kinase (JNK) pathway. Co-amplification of 1p32 and 6q23, that contain c-Jun, and Apoptosis Signaling Kinase 1 (ASK1), are seen in DDLPS but not WDLPS [69]. The proto-oncogene c-Jun encodes part of the activator protein transcription factor (AP-1) complex involved in cell proliferation, transformation and apoptosis [70]. ASK1 activates JNK [71, 72] ultimately leading to c-Jun activation and PPARγ inactivation. PPARγ is involved in the adipocytic differentiation process and its inhibition may result in de-differentiation. A further role for c-Jun in the de-differentiation process is demonstrated by overexpression in a 3T3-L1 adipocytic tumour xenograft model. Transfection of c-Jun into 3T3-L1 cells in vitro delays adipocytic differentiation [26].
3. Myxoid Liposarcoma
MLPS is the second most common subtype of liposarcoma and accounts for more than one third of liposarcomas and 10% of all adult soft tissue sarcomas. MLPS is characterized by the presence of spindle or ovoid cells set in a myxoid stroma with signet ring lipoblasts and a distinctive chicken-wire pattern vasculature. The presence of areas with greater cellularity, known as round cell (RC) de-differentiation, is associated with a worse prognosis [73]. Unusual sites of metastasis are common in MLPS with a propensity to metastasize to soft tissue and bone rather than lung [74, 75]. Thirty-one percent of MLPS patients develop metastasis with bone metastases constituting 56% of these [74]. MLPS exhibits inferior survival compared to other low-grade sarcoma subtypes with a 5-year disease survival rate of 85% [76, 77]. MLPS without RC is particularly radiosensitive with good local control rates with patients treated with adjuvant or neoadjuvant radiotherapy approaching 98% 5-year local control [13, 78].
3.1. Molecular Genetics
MLPS is characterised by the recurrent translocation t(12;16)(q13;p11) that results in the FUS-CHOP gene fusion that is present in over 95% of cases [79, 80]. In most cases, the amino terminal domain of FUS (also known as TLS) is fused to C/EBP homologous protein (CHOP, also known as DDIT3 or GADD153). In rare cases, an alternative translocation event is found t(12;22)(q13;q12) that results in formation of the novel fusion oncogene where EWS takes the place of FUS [81, 82]. There is strong evidence for these translocations to be the primary oncogenic event in MLPS as these tumours have a relatively normal karyotype, the exception being a few recurrent cases of trisomy 8 [83]. In addition, several growth factor pathways have been implicated in MLPS pathogenesis [84–86].
There are currently 11 different FUS-CHOP chimeras and 4 different known EWS-CHOP fusion genes. In the most common variants, a portion of the amino terminus of FUS is fused to the entire coding region of CHOP. The FUS-CHOP transcript type does not appear to have a significant impact upon clinical outcome, and RC content, necrosis and p53 expression remain stronger predictors of clinical outcome [79, 87]. There is evidence that the fusion transcript type may influence response to therapy although the studies are hindered by sample size [88–90]. Understanding how the FUS-CHOP fusion causes MLPS and uncovering any further molecular abnormalities in the disease will aid in development of novel targeted therapies.
FUS belongs to the FET family of RNA-binding proteins that consists of FUS, EWS, and TAF15 as well as the closely homologous, Drosophila SARFH (Cab) [80, 91, 92]. These structurally and functionally related RNA-binding proteins are composed of an SYGQ-rich amino terminus, an RNA recognition motif, a zinc finger motif, and at least one RGG rich repeat region [93, 94]. FET proteins are expressed in most human tissues and appear to be regulated following differentiation in neuroblastoma cells and spontaneously differentiating human embryonic stem cells [95].
Both FUS and EWS have been shown to localize to the nucleus and the cytoplasm, bind RNA, and are also involved in nucleo-cytoplasmic shuttling [96–98]. The FET family associate with various complexes involved in the induction of transcription, including RNA polymerase II (RNAPII), which regulates transcription and TFIID complexes, that binds DNA as part of the transcriptional machinery [91], implicating both FUS and EWS in transcriptional control. In addition, FUS has recently been shown to repress transcription of RNA polymerase III (RNAPIII), suggesting a broader role in regulation through multiple different mechanisms [99]. Noncoding RNAs are capable of allosterically modifying FUS in response to DNA damage to inhibit the transcription factor CREB-binding protein (CBP) and p300 histone acetyltransferase activity, resulting in transcriptional inhibition at the cyclin D1 promoter in cell lines and shows a further role for FUS in transcriptional control [100]. FUS has also been implicated in the DNA damage response as a downstream target of ATM, which can detect and coordinate DNA repair [101].
CHOP is induced in response to endoplasmic reticular stress and is involved in mediating cell death in response to such stress stimuli [102]. CHOP also plays a role in regulating differentiation in adipocytes by interfering with the process in response to metabolic stress [103]. Adipocytic differentiation is dependent on the coordinated expression of a group of transcription factors, the CCAAT/enhancer-binding protein (C/EBP) family of proteins [104]. The C/EBP family consists of six members from C/EBPα to ζ, and they require dimerisation to bind DNA and can form homodimers or heterodimers. CHOP is capable of binding to the C/EBP family members through their highly conserved leucine zipper domain and inhibiting their function. The leucine zipper dimerization domain and the adjacent basic region in CHOP are required for NIH-3T3 transformation with FUS-CHOP, highlighting the requirement for functional DNA binding and dimerization for FUS-CHOP induced oncogenesis [105].
As C/EBPα and C/EBPβ play an important role in the adipogenic differentiation and are regulated by CHOP, it is possible FUS-CHOP may interfere in cellular differentiation. In support, various studies suggest that FUS-CHOP functions by inhibiting adipogenesis and maintaining immature adipocytes in a continuous cycle of proliferation without differentiation [106–108]. Introduction of FUS-CHOP into mice, where expression of the transgene is driven by the ubiquitously expressed elongation factor 1α (EF1α) promoter, results specifically in liposarcomas with inherent induction of adipocyte specific genes such as PPARγ [109]. Further evidence of adipogenic differentiation block resulting from FUS-CHOP expression was shown in vitro where mice expressing FUS-CHOP under the control of the aP2 promoter, which is a downstream target of PPARγ expressed in immature adipocytes, failed to develop liposarcomas, indicating interference between PPARγ and aP2 activation [107].
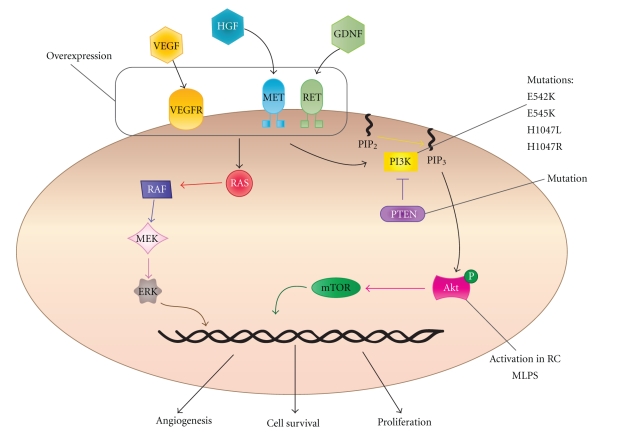
An emerging clinically relevant targetable pathway in MLPS involves the receptor tyrosine kinases (RTKs) MET, RET, and the PI3K signaling cascade (see Figure 3). RET is overexpressed in MLPS compared to normal fat [84] and high expression has been correlated with poor metastasis free survival in MLPS [108]. RET, IGF1R and IGF2 are highly expressed in MLPS and promote cell survival through both the PI3K/Akt and Ras-Raf-ERK/MAPK pathways [85, 86]. A panel of tyrosine kinases including PDGFRB, EGFR, MET, RET, and VEGFR2 are activated in both treated (with chemotherapy/radiotherapy or Trabectedin) and untreated cases of human MLPS [110]. In addition to activation of MET in clinical MLPS specimens, MET and the ligand HGF are potentially regulated by FUS-CHOP. Both MET and HGF are highly expressed in mesenchymal progenitor cells transfected with FUS-CHOP in a disease mimicking allograft mouse model [111]. In a small clinical cohort, specific Akt phosphorylation was observed in the RC variant and 2 treated cases that harboured PTEN mutations, implicating RTK pathways signaling through Akt in MLPS [110]. FLT1 (that encodes the VEGFR1 protein) is expressed as an indirect downstream effect of FUS-CHOP expression in both FUS-CHOP transfected HT1080 (fibrosarcoma) and MLPS cell lines however, VEGFR tyrosine kinase inhibitors did not have a notable impact on proliferation in MLPS cell lines indicating a separate role in these cells [112, 113].
Figure 3.
The PI3K pathway is highly active in MLPS, and this is potentiated at least in part by overexpression, and/or activation through RTKs such as MET, RET and VEGFRs. Upon ligand binding, RTKs activate downstream activation of genes involved in multiple cell processes such as cell survival, proliferation, and angiogenesis. These signals are mediated through the PI3K/Akt pathway and also through RAS. PIK3CA and PTEN mutations and Akt activation have also been documented in MLPS.
Akt activation, particularly in the RC variant, suggests a role for phosphoinositide 3-kinases (PI3K) [110]. PI3Ks are activated upon phosphorylation of membrane bound receptor tyrosine kinases. PI3K can activate many proteins including the protein serine-threonine kinase Akt, which when phosphorylated causes downstream activation and ultimately cell growth, cell cycle entry, and subsequently survival. The PI3K holoenzyme complex is composed of both a catalytic and regulatory subunit. The catalytic subunit, PIK3CA, encodes the p110α isoform and is commonly mutated in various cancer types including breast, colon, brain and gastric malignancies [114, 115]. A recent study showed 18% of MLPS patients (n = 71) had PIK3CA mutations in either the helical (E542K and E545K) or kinase (H1047L and H1047R) domain. The presence of a PIK3CA mutation was associated with a shortened disease specific survival [116]. Barretina et al. also showed one tumour with a homozygous PTEN mutation. PTEN is a tumour suppressor that dephosphorylates phosphoinositide substrates to negatively regulate the Akt signaling pathway [117], demonstrating more mechanisms for perturbation of the pathway.
4. Pleomorphic Liposarcoma
PLPS accounts for only 5% of liposarcomas and occurs mainly within the 55–65 year-old group [8, 118, 119]. PLPS mortality is 40% with no current clinical or pathological predictors of outcome [8, 120]. Histologically PLPS are similar to MFH with the addition of lipoblasts. Histology reveals a disorderly growth pattern, extreme cellularity, and cellular pleomorphism including bizarre giant cells [121]. Lesional cells are polygonal with pale eosinophilic cytoplasm and poorly demarcated boundaries. These lesional cells are interspersed with giant lipoblasts containing enlarged hyperchromatic, angular or globular nuclei [121, 122].
4.1. Molecular Genetics
Molecular studies of PLPS are limited by the scarcity of this disease. Tumours tend to show complex arrangements including gains: 1p, 1q21-q32,2q, 3p, 3q, 5p12-p15, 5q, 6p21, 7p, 7q22 (see reviews) [118, 123, 124]. reported literature shows losses i of 1q, 2q, 3p, 4q, 10q, 11q, 12p13, 13q14, 13q21-qter, 13q23-24, (see reviews) [123–125], Taylor et al. described that 60% of PLPS have a deletion of 13q14.2-q14.3, a region that includes the tumour suppressor RB1 [123]. Also amplified in PLPS, the mitotic arrest deficient (MAD2) may also play a critical role [126, 127]. As reported by Singer et al. [126], MAD2 was found to be over-expressed 13 fold in comparison to normal fat, although small sample size (n = 6) must be appreciated. As reported by Taylor et al. [123] additional deletions in PLPS include 17p13 and 17q11.2, where p53 and the sarcoma associated tumour suppressor gene, neurofibromatosis type 1 (NF-1) are located. Consistent with these observations, Barretina et al. [116] showed 16.7% of PLPS cases had mutations identified in p53, which are rarely seen in MLPS and WD/DDLPS.
5. Therapeutic Implications in Liposarcoma
The current modalities available (chemotherapy, surgery and radiotherapy) for the treatment of liposarcoma are limited, creating a need to identify novel therapeutics.
5.1. MDM2 Antagonists
Given MDM2 is consistently amplified in WD/DDLPS, and sensitivity to MDM2 antagonists (such as Nutlin-3a) is predicted by MDM2 amplification and an intact wild-type p53, it is an appealing therapeutic target [40]. First generation MDM2 inhibitors work via blocking the p53/MDM2 interaction. Nutlin-3a was heralded as one of the most promising MDM2 antagonists when it was shown to activate wild type p53 and induce cell cycle arrest and apoptosis in cancer cell lines [40]. These cell lines included osteosarcoma with amplified MDM2 [40, 128]. Nutlins require wild-type p53 and a functional downstream p53 pathway to be effective [128]. Müller et al. [40] showed downstream p53 dependent transcription and apoptosis in liposarcoma cell lines treated with Nutlin-3a [40].
Translation from in vitro to attractive in vivo therapeutic intervention requires that drugs pass Phase I requirements. Shangary et al. [129] designed spiro-oxindoles as a new class of inhibitors of the MDM2-p53 complex. Spiro-oxindoles bind to MDM2 with high affinity and activates the p53 pathway, inhibiting the growth of neoplastic cell lines with wild-type p53 [129, 130]. MI-219, the lead compound in this class, demonstrates greater potency along with a superior pharmacokinetic profile than Nutlin-3a [129, 131]. MI-219 has been shown to stimulate rapid p53 activation in tumour xenograft tissues with resultant inhibition of cell proliferation [131]. Studies using both Nutlin-3a and MI-219 show a p53 and p21 dependent cell cycle arrest in normal cells, along with p53 dependent cell death specifically in tumour cells [128, 129, 131, 132]. The ability of Nutlin-3a to induce apoptosis in tumours is variable, and osteosarcoma cell lines lacking MDM2 amplification are resistant to apoptosis [131]. Importantly, Nutlin-3a and MI-219 do not cause visible toxicity to animals, as assessed at necropsy [128, 129, 133].
Two oral MDM2 inhibitors have recently entered the clinical setting [134], JNJ-26854165 (Ortho Biotech; Johnson & Johnson) [135] and R7112 (Hoffmann-La Roche) [136]. Both agents are available in advanced stage or refractory solid tumours Phase I trials [134]. In addition, AT-219 (a derivative of MI-219) is in preclinical studies with phase I trials planned [134]. Of relevant interest, an MDM2 antagonist RO5045337 is about to recruit for a Phase I trial in liposarcoma patients [137].
5.2. CDK4 Antagonists
Targeting CDK4 is an attractive therapeutic strategy given its frequent overexpression in WD/DDLPS [138]. A number of CDK4 inhibitors are in the early pre-clinical development or Phase I and II trials [139]. First generation pan-CDK inhibitors include Flavopiridol and Seleciclib (R-Roscovitine), inhibiting CDK1, CDK2, CDK4, CDK6, CDK7, and CDK1, CDK2, CDK7 and CDK9 respectively [140]. Flavopiridol causes arrest in G1 and G2 phases in a range of solid tumour cell lines [139, 141, 142]. Flavopiridol is more potent if tumour cells are in S phase. Matranga and Shapiro [143] demonstrated recruitment to S phase using hydroxyurea, gemcitabine and cisplatin, followed by flavopiridol resulting in sequence-dependent cytotoxic synergy [143–145]. Flavopiridol and Seliciclib have been investigated in Phase I/II trials for haematological and solid tumours including sarcomas. Trials include Flavopiridol as a single agent and in combination with taxanes where synergism has been noted [141]. Both Flavopiridol and Seleciclib have shown disappointing results relating to clinical outcome and intolerable side effects [146, 147].
Newer generation CDK inhibitors include PD0332991, P27600, ZK 304709, R 547 and P1446A05. All are available in Phase I and II solid tumour trials [146]. PD0332991 is one of two more selective CDK inhibitors specific for CDK4 and CDK6. Preclinical data showed inhibition of cell growth through G1 arrest in pRb-positive tumour cell lines and antitumorigenic effects in xenograft models of colon carcinoma [148]. PD0332991 is available in Phase I and Phase II trials for solid and haematological malignancy. Finally, P1446A05 is the only single CDK4 selective inhibitor available [146]. No pre-clinical data is publicly available for this compound; however, it has been released as a Phase I drug for refractory solid tumour and haematological malignancies [146].
5.3. PPARγ Ligand Agonists
A critical regulator of terminal differentiation for the adipocytic lineage is a nuclear receptor peroxisome proliferator-activated receptor γ [149–151]. PPARγ is an attractive target in undifferentiated lipomatous tumours such as DDLPS and MLPS. PPARγ forms a heterodimeric complex with the retinoid X receptor (RXR). This complex regulates transcription of adipocyte-specific genes by binding sites on DNA. Agonist ligands for the PPARγ receptor have been shown to induce terminal differentiation of normal preadipocytes in human liposarcoma cells in vitro [149].
A Dana-Farber Cancer Institute Phase II clinical trial used Troglitazone, a synthetic PPARγ ligand, in patients with high-grade liposarcoma. This trial enrolled three patients. All patients showed histologic and biochemical differentiation in vivo, with reduction in immunohistochemical expression of proliferation marker Ki-67 [149]. A more recent study with 12 patients with Rosiglitazone, belonging to the same class of drugs (thiazolidinediones) as Troglitazone, was not as promising, with median progression free survival of 5.5 months. Treatment did not produce any convincing adipocytic differentiation with no correlation between the high expression of differentiation genes that was found in two patients, and clinical response [152].
5.4. Trabectedin (ET-743)
Trabectedin (also known as Ecteinascidin or ET-743) is an antitumor drug isolated from the Caribbean marine tunicate, Ecteinascidia turbinata [153]. Trabectedin is an approved second-line agent for advanced soft tissue sarcoma and has been shown to be exquisitely sensitive to Trabectedin in Phase II clinical trials [154, 155]. The drug is a tetrahydroisoquinoline alkaloid whose main mechanism of action is through binding to the DNA minor groove with promoter and sequence specificity; however, it has also been shown to have effects on promoters that are regulated by major groove binding transcription factors [156–158]. Trabectedin does not appear to effect transcription of FUS-CHOP, but has been shown to dissociate the aberrant transcription factor from promoters of its target genes resulting in removal of the differentiation block by activating a differentiation cascade through the C/EBPs [88].
Trabectedin relies on intact nucleotide excision repair (NER) machinery and induces lethal DNA strand breaks in a transcription-couple NER dependant manner [159–161]. It has been suggested that these breaks are repaired by homologous recombination (HR), as HR-deficient cells, such as BRCA2 mutants, are 100 fold more sensitive to Trabectedin [162]. This effect is specific to HR- mediated double strand break repair as defects in the alternative pathway using nonhomologous end joining do not result in the same degree of Trabectedin sensitivity [161, 162].
FUS-CHOP modulates immune genes by activating NF-κB controlled cytokines IL-6 and IL-8 in a C/EBPβ- dependent manner [163, 164]. Proinflammatory cytokines and growth factors such as CCL2, CXCL8, IL-6, VEGF and PTX3 are highly expressed in both xenograft MLPS models and patient tumours. Trabectedin has been shown to reduce expression and production of these immune modulators, potentially altering the tumour microenvironment in a favorable way [165]. Thus, Trabectedin appears to affect the biological activity of FUS-CHOP and so far shows promise as a therapeutic in MLPS.
5.5. Receptor Tyrosine Kinase Pathway Inhibitors
The high frequency of PIK3CA and PTEN mutations suggests a role for PI3K inhibitors in MLPS. The nonisoform-specific PI3K inhibitors Wortmannin, and LY294002 have been widely used in biological research but are not particularly suited to clinical work due to their lack of specificity, Wortmannin's instability and LY294002's low potency (as reviewed [166]). GDC-0941 and PX-866 are promising PI3K inhibitors currently in clinical trials that have low nanomolar potency against class I isoforms of PI3K [167–169]. In lung cancer cell lines and xenograft models, PIK3CA mutants are more sensitive to GDC-0941 [170]. Similarly, PIK3CA mutant and PTEN-null tumours were sensitive to PX-866 in xenograft models, and phase I clinical trials for solid tumours are currently underway [169]. The Rapamycin derivate Everolimus inhibits the mTOR complex-1 (mTORC1), which is a downstream effector of PI3K. Both H1047R and E545K PI3K mutant cells are sensitive to Everolimus [171]. PIK3CA mutated MLPS represents an ideal candidate for PI3K inhibition.
As MET is activated in MLPS and there are many MET pathway inhibitors currently in development and in clinical trials (as reviewed in [172], MLPS may be a good candidate for MET inhibition. For example, the novel and promising inhibitor Foretinib (XL880) inhibits multiple kinases including both MET and VEGFR2 and exhibits extensive biological activity and clinical efficacy in an early Phase I clinical trial in metastatic or unresectable solid tumours [173].
6. Conclusion
Molecular-based therapeutics are not routinely used in liposarcoma, where surgery, radiotherapy, and chemotherapy remain the mainstay of treatment. Translation of targeted molecular therapeutics in sarcoma has been successfully demonstrated with Imatinib mesylate therapy in c-Kit positive gastrointestinal stromal tumour (GIST) [174]. A major challenge with the use of molecularly targeted therapeutics is to translate disease control into disease eradication. One strategy to achieve this goal is to combine two or more independent molecularly targeted agents in a disease where all of the targets are relevant. The dependence of WD/DDLPS on amplification of both MDM2 and CDK4 means that this disease represents an important candidate for combination therapy. Recent studies point towards RTK involvement in MLPS oncogenesis, particularly signaling through the PI3K/Akt pathway. This provides an important avenue for new research due to the large number of clinical trials currently underway that target this pathway. Although not considered a molecularly targeted therapeutic, treatment of MLPS with Trabectedin is currently in late stage clinical trials with promising results.
It is hoped that emerging technologies, such as next-generation sequencing, will be fundamental in revealing new molecular targets in liposarcoma. Similarly, advances in drug development should enable improvement of molecular therapies with greater sensitivity, specificity, potency, and limited toxicity. Combining technologies in both areas will allow for efficient clinical translation.
Conflicts of Interest
No potential conflict of interest is disclosed.
Acknowledgments
The authors acknowledge Dr. Maya Kansara and Dale Garsed for reviewing the manuscript and Dr. Catherine Mitchell for reviewing the histopathology. D. M. Thomas is a recipient of a Victorian Cancer Agency clinician scientist fellowship. S. Young is a recipient of an Australian Postgraduate Award. R. Conyers is a recipient of an NHMRC Postgraduate Scholarship. R. Conyers and S. Young contributed equally to this work.
References
- 1. N. C. Institute, A Snaphsot of Sarcoma, 2010, http://www.cancer.gov/cancertopics/types/soft-tissue-sarcoma.
- 2.Mack TM. Sarcomas and other malignancies of soft tissue, retroperitoneum, peritoneum, pleura, heart, mediastinum, and spleen. Cancer. 1995;75(1):211–244. doi: 10.1002/1097-0142(19950101)75:1+<211::aid-cncr2820751309>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer Journal for Clinicians. 2006;56(2):106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 4.Henricks WH, Chu YC, Goldblum JR, Weiss SW. Dedifferentiated liposarcoma: a clinicopathological analysis of 155 cases with a proposal for an expanded definition of dedifferentiation. American Journal of Surgical Pathology. 1997;21(3):271–281. doi: 10.1097/00000478-199703000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Linehan DC, Lewis JJ, Leung D, Brennan MF. Influence of biologic factors and anatomic site in completely resected liposarcoma. Journal of Clinical Oncology. 2000;18(8):1637–1643. doi: 10.1200/JCO.2000.18.8.1637. [DOI] [PubMed] [Google Scholar]
- 6.Lewis JJ, Leung D, Woodruff JM, Brennan MF. Retroperitoneal soft-tissue sarcoma: analysis of 500 patients treated and followed at a single institution. Annals of Surgery. 1998;228(3):355–365. doi: 10.1097/00000658-199809000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCormick D, Mentzel T, Beham A, Fletcher CDM. Dedifferentiated liposarcoma: clinicopathologic analysis of 32 cases suggesting a better prognostic subgroup among pleomorphic sarcomas. American Journal of Surgical Pathology. 1994;18(12):1213–1223. doi: 10.1097/00000478-199412000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Gebhard S, Coindre J-M, Michels J-J, et al. Pleomorphic liposarcoma: clinicopathologic, immunohistochemical, and follow-up analysis of 63 cases: a study from the French Federation of Cancer Centers Sarcoma Group. American Journal of Surgical Pathology. 2002;26(5):601–616. doi: 10.1097/00000478-200205000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Singer S, Antonescu CR, Riedel E, Brennan MF, Pollock RE. Histologic subtype and margin of resection predict pattern of recurrence and survival for retroperitoneal liposarcoma. Annals of Surgery. 2003;238(3):358–371. doi: 10.1097/01.sla.0000086542.11899.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kindblom L-G. Lipomatous tumors—how we have reached our present views, what controversies remain and why we still face diagnostic problems: a tribute to Dr Franz Enzinger. Advances in Anatomic Pathology. 2006;13(6):279–285. doi: 10.1097/01.pap.0000213053.00060.5a. [DOI] [PubMed] [Google Scholar]
- 11.Rieker RJ, Weitz J, Lehner B, et al. Genomic profiling reveals subsets of dedifferentiated liposarcoma to follow separate molecular pathways. Virchows Archiv. 2010;456(3):277–285. doi: 10.1007/s00428-009-0869-9. [DOI] [PubMed] [Google Scholar]
- 12.Jones RL, Fisher C, Al-Muderis O, Judson IR. Differential sensitivity of liposarcoma subtypes to chemotherapy. European Journal of Cancer. 2005;41(18):2853–2860. doi: 10.1016/j.ejca.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 13.de Vreeze RSA, de Jong D, Haas RL, Stewart F, van Coevorden F. Effectiveness of radiotherapy in myxoid sarcomas is associated with a dense vascular pattern. International Journal of Radiation Oncology Biology Physics. 2008;72(5):1480–1487. doi: 10.1016/j.ijrobp.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Pitson G, Robinson P, Wilke D, et al. Radiation response: an additional unique signature of myxoid liposarcoma. International Journal of Radiation Oncology Biology Physics. 2004;60(2):522–526. doi: 10.1016/j.ijrobp.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 15.de Vreeze RSA, de Jong D, Nederlof PM, et al. Added value of molecular biological analysis in diagnosis and clinical management of liposarcoma: a 30-year single-institution experience. Annals of Surgical Oncology. 2009;14(3):686–693. doi: 10.1245/s10434-009-0806-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dei Tos AP, Doglioni C, Piccinin S, et al. Coordinated expression and amplification of the MDM2, CDK4, and HMGI-C genes in atypical lipomatous tumours. Journal of Pathology. 2000;190(5):531–536. doi: 10.1002/(SICI)1096-9896(200004)190:5<531::AID-PATH579>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 17.Kraus MD, Guillou L, Fletcher CDM. Well-differentiated inflammatory liposarcoma: an uncommon and easily overlooked variant of a common sarcoma. American Journal of Surgical Pathology. 1997;21(5):518–527. doi: 10.1097/00000478-199705000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Argani P, Facchetti F, Inghirami G, Rosai J. Lymphocyte-rich well-differentiated liposarcoma: report of nine cases. American Journal of Surgical Pathology. 1997;21(8):884–895. doi: 10.1097/00000478-199708000-00002. [DOI] [PubMed] [Google Scholar]
- 19.Hasegawa T, Seki K, Hasegawa F, et al. Dedifferentiated liposarcoma of retroperitoneum and mesentery: varied growth patterns and histological grades—a clinicopathologic study of 32 cases. Human Pathology. 2000;31(6):717–727. doi: 10.1053/hupa.2000.8222. [DOI] [PubMed] [Google Scholar]
- 20.Tontonoz P, Singer S, Forman BM, et al. Terminal differentiation of human liposarcoma cells induced by ligands for peroxisome proliferator-activated receptor γ and the retinoid X receptor. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(1):237–241. doi: 10.1073/pnas.94.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss SW, Rao VK. Well-differentiated liposarcoma (atypical lipoma) of deep soft tissue of the extremities, retroperitoneum, and miscellaneous sites: a follow-up study of 92 cases with analysis of the incidence of “dedifferentiation”. American Journal of Surgical Pathology. 1992;16(11):1051–1058. doi: 10.1097/00000478-199211000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Rosai J, Akerman M, Dal Cin P, et al. Combined morphologic and karyotypic study of 59 atypical lipomatous tumors: evaluation of their relationship and differential diagnosis with other adipose tissue tumors. (A report of the CHAMP Study Group) American Journal of Surgical Pathology. 1996;20(10):1182–1189. doi: 10.1097/00000478-199610000-00002. [DOI] [PubMed] [Google Scholar]
- 23.Pedeutour F, Forus A, Coindre J-M, et al. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes and Cancer. 1999;24(1):30–41. [PubMed] [Google Scholar]
- 24.Forus A, Weghuis DO, Smeets D, Fodstad O, Myklebost O, Van Kessel AG. Comparative genomic hybridization analysis of human sarcomas: I. Occurrence of genomic imbalances and identification of a novel major amplicon at 1q21-q22 in soft tissue sarcomas. Genes Chromosomes and Cancer. 1995;14(1):8–14. doi: 10.1002/gcc.2870140103. [DOI] [PubMed] [Google Scholar]
- 25.Szymanska J, Virolainen M, Tarkkanen M, et al. Overrepresentation of 1q21-23 and 12q13-21 in lipoma-like liposarcomas but not in benign lipomas: a comparative genomic hybridization study. Cancer Genetics and Cytogenetics. 1997;99(1):14–18. doi: 10.1016/s0165-4608(96)00436-0. [DOI] [PubMed] [Google Scholar]
- 26.Mariani O, Brennetot C, Coindre J-M, et al. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell. 2007;11(4):361–374. doi: 10.1016/j.ccr.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nature Reviews Cancer. 2006;6(12):909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 28.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends in Cell Biology. 2010;20(5):299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendrysa SM, McElwee MK, Michalowski J, O’Leary KA, Young KM, Perry ME. mdm2 is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Molecular and Cellular Biology. 2003;23(2):462–473. doi: 10.1128/MCB.23.2.462-473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubbutat MHG, Ludwig RL, Levine AJ, Vousden KH. Analysis of the degradation function of Mdm2. Cell Growth and Differentiation. 1999;10(2):87–92. [PubMed] [Google Scholar]
- 31.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69(7):1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 32.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362(6423):857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 33.Thut CJ, Goodrich JA, Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes and Development. 1997;11(15):1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 35.Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 36.Hu C-Q, Hu Y-Z. Small molecule inhibitors of the p53-MDM2. Current Medicinal Chemistry. 2008;15(17):1720–1730. doi: 10.2174/092986708784872375. [DOI] [PubMed] [Google Scholar]
- 37.Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. The EMBO Journal. 2004;23(7):1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Molecular Cell. 2006;21(3):307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Seminars in Cancer Biology. 2003;13(1):49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 40.Müller CR, Paulsen EB, Noordhuis P, Pedeutour F, Sæter G, Myklebost O. Potential for treatment of liposarcomas with the MDM2 antagonist Nutlin-3A. International Journal of Cancer. 2007;121(1):199–205. doi: 10.1002/ijc.22643. [DOI] [PubMed] [Google Scholar]
- 41.Pilotti S, Della Torre G, Lavarino C, et al. Distinct mdm2/p53 expression in patterns in liposarcoma subgroups: implications for different pathogenetic mechanisms. Journal of Pathology. 1997;181(1):14–24. doi: 10.1002/(SICI)1096-9896(199701)181:1<14::AID-PATH730>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 42.Schneider-Stock R, Walter H, Radig K, et al. MDM2 amplification and loss of heterozygosity at Rb and p53 genes: no simultaneous alterations in the oncogenesis of liposarcomas. Journal of Cancer Research and Clinical Oncology. 1998;124(10):532–540. doi: 10.1007/s004320050211. [DOI] [PubMed] [Google Scholar]
- 43.Binh MBN, Sastre-Garau X, Guillou L, et al. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: a comparative analysis of 559 soft tissue neoplasms with genetic data. American Journal of Surgical Pathology. 2005;29(10):1340–1347. doi: 10.1097/01.pas.0000170343.09562.39. [DOI] [PubMed] [Google Scholar]
- 44.Nakayama T, Toguchida J, Wadayama B-I, Kanoe H, Kotoura Y, Sasaki MS. MDM2 gene amplification in bone and soft-tissue tumors: association with tumor progression in differentiated adipose-tissue tumors. International Journal of Cancer. 1995;64(5):342–346. doi: 10.1002/ijc.2910640511. [DOI] [PubMed] [Google Scholar]
- 45.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochimica et Biophysica Acta. 2002;1602(1):73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 46.Italiano A, Bianchini L, Gjernes E, et al. Clinical and biological significance of CDK4 amplification in well-differentiated and dedifferentiated liposarcomas. Clinical Cancer Research. 2009;15(18):5696–5703. doi: 10.1158/1078-0432.CCR-08-3185. [DOI] [PubMed] [Google Scholar]
- 47.Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98(6):859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 48.Day PJ, Cleasby A, Tickle IJ, et al. Crystal structure of human CDK4 in complex with a D-type cyclin. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(11):4166–4170. doi: 10.1073/pnas.0809645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dei Tos AP, Doglioni C, Piccinin S, et al. Molecular abnormalities of the p53 pathway in dedifferentiated liposarcoma. Journal of Pathology. 1997;181(1):8–13. doi: 10.1002/(SICI)1096-9896(199701)181:1<8::AID-PATH700>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 50.Hisaoka M, Tsuji S, Morimitsu Y, et al. Detection of TLS/FUS-CHOP fusion transcripts in myxoid and round cell liposarcomas by nested reverse transcription-polymerase chain reaction using archival paraffin-embedded tissues. Diagnostic Molecular Pathology. 1998;7(2):96–101. doi: 10.1097/00019606-199804000-00006. [DOI] [PubMed] [Google Scholar]
- 51.Kanoe H, Nakayama T, Murakami H, et al. Amplification of the CDK4 gene in sarcomas: tumor specificity and relationship with the RB gene mutation. Anticancer Research. 1998;18(4 A):2317–2321. [PubMed] [Google Scholar]
- 52.Italiano A, Bianchini L, Keslair F, et al. HMGA2 is the partner of MDM2 in well-differentiated and dedifferentiated liposarcomas whereas CDK4 belongs to a distinct inconsistent amplicon. International Journal of Cancer. 2008;122(10):2233–2241. doi: 10.1002/ijc.23380. [DOI] [PubMed] [Google Scholar]
- 53.Sirvent N, Coindre J-M, Maire G, et al. Detection of MDM2-CDK4 amplification by fluorescence in situ hybridization in 200 paraffin-embedded tumor samples: utility in diagnosing adipocytic lesions and comparison with immunohistochemistry and real-time PCR. American Journal of Surgical Pathology. 2007;31(10):1476–1489. doi: 10.1097/PAS.0b013e3180581fff. [DOI] [PubMed] [Google Scholar]
- 54.Aleixo PB, Hartmann AA, Menezes IC, Meurer RT, Oliveira AM. Can MDM2 and CDK4 make the diagnosis of well differentiated/ dedifferentiated liposarcoma? An immunohistochemical study on 129 soft tissue tumours. Journal of Clinical Pathology. 2009;62(12):1127–1135. doi: 10.1136/jcp.2009.070201. [DOI] [PubMed] [Google Scholar]
- 55.Arlotta P, Tai AK-F, Manfioletti G, Clifford C, Jay G, Ono SJ. Transgenic mice expressing a truncated form of the high mobility group I-C protein develop adiposity and an abnormally high prevalence of lipomas. The Journal of Biological Chemistry. 2000;275(19):14394–14400. doi: 10.1074/jbc.m000564200. [DOI] [PubMed] [Google Scholar]
- 56.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Progress in Nucleic Acid Research and Molecular Biology. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 57.Meza-Zepeda LA, Berner J-M, Henriksen J, et al. Ectopic sequences from truncated HMGIC in liposarcomas are derived from various amplified chromosomal regions. Genes Chromosomes and Cancer. 2001;31(3):264–273. doi: 10.1002/gcc.1143. [DOI] [PubMed] [Google Scholar]
- 58.Mantovani F, Covaceuszach S, Rustighi A, et al. NF-κB mediated transcriptional activation is enhanced by the architectural factor HMGI-C. Nucleic Acids Research. 1998;26(6):1433–1439. doi: 10.1093/nar/26.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwanbeck R, Manfioletti G, Wiśniewski JR. Architecture of high mobility group protein I-C·DNA complex and its perturbation upon phosphorylation by Cdc2 kinase. The Journal of Biological Chemistry. 2000;275(3):1793–1801. doi: 10.1074/jbc.275.3.1793. [DOI] [PubMed] [Google Scholar]
- 60.Fedele M, Berlingieri MT, Scala S, et al. Truncated and chimeric HMGI-C genes induce neoplastic transformation of NIH3T3 murine fibroblasts. Oncogene. 1998;17(4):413–418. doi: 10.1038/sj.onc.1201952. [DOI] [PubMed] [Google Scholar]
- 61.Berner J-M, Meza-Zepeda LA, Kools PFJ, et al. HMGIC, the gene for an architectural transcription factor, is amplified and rearranged in a subset of human sarcomas. Oncogene. 1997;14(24):2935–2941. doi: 10.1038/sj.onc.1201135. [DOI] [PubMed] [Google Scholar]
- 62.Gamberi G, Ragazzini P, Benassi MS, et al. Analysis of 12q13-15 genes in parosteal osteosarcoma. Clinical Orthopaedics and Related Research. 2000;(377):195–204. doi: 10.1097/00003086-200008000-00026. [DOI] [PubMed] [Google Scholar]
- 63.Meltzer PS, Jankowski SA, Dal Cin P, Sandberg AA, Paz IB, Coccia MA. Identification and cloning of a novel amplified DNA sequence in human malignant fibrous histiocytoma derived from a region of chromosome 12 frequently rearranged in soft tissue tumors. Cell Growth. 1991;2(10):495–501. [PubMed] [Google Scholar]
- 64.Florenes VA, Moelandsmo GM, Forus A, Andreassen A, Myklebost O, Fodstad O. MDM2 gene amplification and transcript levels in human sarcomas: relationship to TP53 gene status. Journal of the National Cancer Institute. 1994;86(17):1297–1302. doi: 10.1093/jnci/86.17.1297. [DOI] [PubMed] [Google Scholar]
- 65.Noble-Topham SE, Burrow SR, Eppert K, et al. SAS is amplified predominantly in surface osteosarcoma. Journal of Orthopaedic Research. 1996;14(5):700–705. doi: 10.1002/jor.1100140504. [DOI] [PubMed] [Google Scholar]
- 66.Forus A, Florenes VA, Maelandsmo GM, Meltzer PS, Fodstad O, Myklebost O. Mapping of amplification units in the q13-14 region of chromosome-12 in human sarcomas—some amplica do not include MDM2. Cell Growth and Differentiation. 1993;4(12):1065–1070. [PubMed] [Google Scholar]
- 67.Meza-Zepeda LA, Forus A, Lygren B, et al. Positional cloning identifies a novel cyclophilin as a candidate amplified oncogene in 1q21. Oncogene. 2002;21(14):2261–2269. doi: 10.1038/sj.onc.1205339. [DOI] [PubMed] [Google Scholar]
- 68.Nilsson M, Meza-Zepeda LA, Mertens F, Forus A, Myklebost O, Mandahl N. Amplification of chromosome 1 sequences in lipomatous tumors and other sarcomas. International Journal of Cancer. 2004;109(3):363–369. doi: 10.1002/ijc.11716. [DOI] [PubMed] [Google Scholar]
- 69.Chibon F, Mariani O, Derré J, et al. A subgroup of malignant fibrous histiocytomas is associated with genetic changes similar to those of well-differentiated liposarcomas. Cancer Genetics and Cytogenetics. 2002;139(1):24–29. doi: 10.1016/s0165-4608(02)00614-3. [DOI] [PubMed] [Google Scholar]
- 70.Vallone D, Battista S, Pierantoni GM, et al. Neoplastic transformation of rat thyroid cells requires the junB and fra-1 gene induction which is dependent on the HMGI-C gene product. The EMBO Journal. 1997;16(17):5310–5321. doi: 10.1093/emboj/16.17.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coindre J-M, Pédeutour F, Aurias A. Well-differentiated and dedifferentiated liposarcomas. Virchows Archiv. 2010;456(2):167–179. doi: 10.1007/s00428-009-0815-x. [DOI] [PubMed] [Google Scholar]
- 72.Chibon F, Mariani O, Derré J, et al. ASK1 (MAP3K5) as a potential therapeutic target in malignant fibrous histiocytomas with 12q14q-q15 and 6q23 amplifications. Genes Chromosomes and Cancer. 2004;40(1):32–37. doi: 10.1002/gcc.20012. [DOI] [PubMed] [Google Scholar]
- 73.Fletcher CKU, Krishnan K, Mertens F. Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon, France: IARC Press; 2002. [Google Scholar]
- 74.Schwab JH, Boland P, Guo T, et al. Skeletal metastases in myxoid liposarcoma: an unusual pattern of distant spread. Annals of Surgical Oncology. 2007;14(4):1507–1514. doi: 10.1245/s10434-006-9306-3. [DOI] [PubMed] [Google Scholar]
- 75.Sheah K, Ouellette HA, Torriani M, Nielsen GP, Kattapuram S, Bredella MA. Metastatic myxoid liposarcomas: imaging and histopathologic findings. Skeletal Radiology. 2008;37(3):251–258. doi: 10.1007/s00256-007-0424-1. [DOI] [PubMed] [Google Scholar]
- 76.ten Heuvel SE, Hoekstra HJ, Van Ginkel RJ, Bastiaannet E, Suurmeijer AJH. Clinicopathologic prognostic factors in myxoid liposarcoma: a retrospective study of 49 patients with long-term follow-up. Annals of Surgical Oncology. 2007;14(1):222–229. doi: 10.1245/s10434-006-9043-7. [DOI] [PubMed] [Google Scholar]
- 77.Canter RJ, Qin L-X, Ferrone CR, Maki RG, Singer S, Brennan MF. Why do patients with low-grade soft tissue sarcoma die? Annals of Surgical Oncology. 2008;15(12):3550–3560. doi: 10.1245/s10434-008-0163-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chung PWM, Deheshi BM, Ferguson PC, et al. Radiosensitivity translates into excellent local control in extremity myxoid liposarcoma: a comparison with other soft tissue sarcomas. Cancer. 2009;115(14):3254–3261. doi: 10.1002/cncr.24375. [DOI] [PubMed] [Google Scholar]
- 79.Antonescu CR, Tschernyavsky SJ, Decuseara R, et al. Prognostic impact of P53 status, TLS-CHOP fusion transcript structure, and histological grade in myxoid liposarcoma: a molecular and clinicopathologic study of 82 cases. Clinical Cancer Research. 2001;7(12):3977–3987. [PubMed] [Google Scholar]
- 80.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363(6430):640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 81.Panagopoulos I, Höglund M, Mertens F, Mandahl N, Mitelman F, Åman P. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene. 1996;12(3):489–494. [PubMed] [Google Scholar]
- 82.Dal Cin P, Sciot R, Panagopoulos I, et al. Additional evidence of a variant translocation t(12;22) with EWS/CHOP fusion in myxoid liposarcoma: clinicopathological features. Journal of Pathology. 1997;182(4):437–441. doi: 10.1002/(SICI)1096-9896(199708)182:4<437::AID-PATH882>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 83.Sreekantaiah C, Karakousis CP, Leong SPL, Sandberg AA. Trisomy 8 as a nonrandom secondary change in myxoid liposarcoma. Cancer Genetics and Cytogenetics. 1991;51(2):195–205. doi: 10.1016/0165-4608(91)90132-e. [DOI] [PubMed] [Google Scholar]
- 84.Thelin-Järnum S, Lassen C, Panagopoulos I, Mandahl N, Åman P. Identification of genes differentially expressed in TLS-CHOP carrying myxoid liposarcomas. International Journal of Cancer. 1999;83(1):30–33. doi: 10.1002/(sici)1097-0215(19990924)83:1<30::aid-ijc6>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 85.Cheng H, Dodge J, Mehl E, et al. Validation of immature adipogenic status and identification of prognostic biomarkers in myxoid liposarcoma using tissue microarrays. Human Pathology. 2009;40(9):1244–1251. doi: 10.1016/j.humpath.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 86.Tao Y, Pinzi V, Bourhis J, Deutsch E. Mechanisms of disease: signaling of the insulin-like growth factor 1 receptor pathway—therapeutic perspectives in cancer. Nature Clinical Practice Oncology. 2007;4(10):591–602. doi: 10.1038/ncponc0934. [DOI] [PubMed] [Google Scholar]
- 87.Bode-Lesniewska B, Frigerio S, Exner U, Abdou MT, Moch H, Zimmermann DR. Relevance of translocation type in myxoid liposarcoma and identification of a novel EWSR1-DDIT3 fusion. Genes Chromosomes and Cancer. 2007;46(11):961–971. doi: 10.1002/gcc.20478. [DOI] [PubMed] [Google Scholar]
- 88.Forni C, Minuzzo M, Virdis E, et al. Trabectedin (ET-743) promotes differentiation in myxoid liposarcoma tumors. Molecular Cancer Therapeutics. 2009;8(2):449–457. doi: 10.1158/1535-7163.MCT-08-0848. [DOI] [PubMed] [Google Scholar]
- 89.Grosso F, Jones RL, Demetri GD, et al. Efficacy of trabectedin (ecteinascidin-743) in advanced pretreated myxoid liposarcomas: a retrospective study. Lancet Oncology. 2007;8(7):595–602. doi: 10.1016/S1470-2045(07)70175-4. [DOI] [PubMed] [Google Scholar]
- 90.Frapolli R, Tamborini E, Virdis E, et al. Novel models of Myxoid Liposarcoma Xenografts mimicking the biological and pharmacological features of human tumors. Clinical Cancer Research. 2010;16(20):4958–4967. doi: 10.1158/1078-0432.CCR-10-0317. [DOI] [PubMed] [Google Scholar]
- 91.Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. The EMBO Journal. 1996;15(18):5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 92.Stolow DT, Haynes SR. Cabeza, a Drosophila gene encoding a novel RNA binding protein, shares homology with EWS and TLS, two genes involved in human sarcoma formation. Nucleic Acids Research. 1995;23(5):835–843. doi: 10.1093/nar/23.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morohoshi F, Arai K, Takahashi E-I, Tanigami A, Ohki M. Cloning and mapping of a human RBP56 gene encoding a putative RNA binding protein similar to FUS/TLS and EWS proteins. Genomics. 1996;38(1):51–57. doi: 10.1006/geno.1996.0591. [DOI] [PubMed] [Google Scholar]
- 94.Morohoshi F, Ootsuka Y, Arai K, et al. Genomic structure of the human RBP56/hTAF(II)68 and FUS/TLS genes. Gene. 1998;221(2):191–198. doi: 10.1016/s0378-1119(98)00463-6. [DOI] [PubMed] [Google Scholar]
- 95.Andersson MK, Ståhlberg A, Arvidsson Y, et al. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biology. 2008;9, article 37 doi: 10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zinszner H, Immanuel D, Yin Y, Liang F-X, Ron D. A topogenic role for the oncogenic N-terminus of TLS: nucleolar localization when transcription is inhibited. Oncogene. 1997;14(4):451–461. doi: 10.1038/sj.onc.1200854. [DOI] [PubMed] [Google Scholar]
- 97.Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. Journal of Cell Science. 1997;110(15):1741–1750. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]
- 98.Belyanskaya LL, Gehrig PM, Gehring H. Exposure on cell surface and extensive arginine methylation of ewing sarcoma (EWS) Protein. The Journal of Biological Chemistry. 2001;276(22):18681–18687. doi: 10.1074/jbc.M011446200. [DOI] [PubMed] [Google Scholar]
- 99.Tan AY, Manley JL. TLS inhibits RNA polymerase III transcription. Molecular and Cellular Biology. 2010;30(1):186–196. doi: 10.1128/MCB.00884-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454(7200):126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gardiner M, Toth R, Vandermoere F, Morrice NA, Rouse J. Identification and characterization of FUS/TLS as a new target of ATM. Biochemical Journal. 2008;415(2):297–307. doi: 10.1042/BJ20081135. [DOI] [PubMed] [Google Scholar]
- 102.Wang X-Z, Lawson B, Brewer JW, et al. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153) Molecular and Cellular Biology. 1996;16(8):4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Batchvarova N, Wang X-Z, Ron D. Inhibition of adipogenesis by the stress-induced protein CHOP (Gadd153) The EMBO Journal. 1995;14(19):4654–4661. doi: 10.1002/j.1460-2075.1995.tb00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes and Development. 1991;5(9):1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 105.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes and Development. 1994;8(21):2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- 106.Kuroda M, Ishida T, Takanashi M, Satoh M, Machinami R, Watanabe T. Oncogenic transformation and inhibition of adipocytic conversion of preadipocytes by TLS/FUS-CHOP type II chimeric protein. American Journal of Pathology. 1997;151(3):735–744. [PMC free article] [PubMed] [Google Scholar]
- 107.Pérez-Mancera PA, Vicente-Dueñas C, González-Herrero I, Sánchez-Martín M, Flores-Corral T, Sánchez-García I. Fat-specific FUS-DDIT3-transgenic mice establish PPARγ inactivation is required to liposarcoma development. Carcinogenesis. 2007;28(10):2069–2073. doi: 10.1093/carcin/bgm107. [DOI] [PubMed] [Google Scholar]
- 108.Cheng H, Dodge J, Mehl E, et al. Validation of immature adipogenic status and identification of prognostic biomarkers in myxoid liposarcoma using tissue microarrays. Human Pathology. 2009;40(9):1244–1251. doi: 10.1016/j.humpath.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 109.Pérez-Losada J, Pintado B, Gutiérrez-Adán A, et al. The chimeric FUS/TLS-CHOP fusion protein specifically induces liposarcomas in transgenic mice. Oncogene. 2000;19(20):2413–2422. doi: 10.1038/sj.onc.1203572. [DOI] [PubMed] [Google Scholar]
- 110.Negri T, Virdis E, Brich S, et al. Functional mapping of receptor tyrosine kinases in myxoid liposarcoma. Clinical Cancer Research. 2010;16(14):3581–3593. doi: 10.1158/1078-0432.CCR-09-2912. [DOI] [PubMed] [Google Scholar]
- 111.Riggi N, Cironi L, Provero P, et al. Expression of the FUS-CHOP fusion protein in primary mesenchymal progenitor cells gives rise to a model of myxoid liposarcoma. Cancer Research. 2006;66(14):7016–7023. doi: 10.1158/0008-5472.CAN-05-3979. [DOI] [PubMed] [Google Scholar]
- 112.Engström K, Willén H, Kåbjörn-Gustafsson C, et al. The myxoid/round cell liposarcoma fusion oncogene FUS-DDIT3 and the normal DDIT3 induce a liposarcoma phenotype in transfected human fibrosarcoma cells. American Journal of Pathology. 2006;168(5):1642–1653. doi: 10.2353/ajpath.2006.050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Andersson MK, Göransson M, Olofsson A, Andersson C, Åman P. Nuclear expression of FLT1 and its ligand PGF in FUS-DDIT3 carrying myxoid liposarcomas suggests the existence of an intracrine signaling loop. BMC Cancer. 2010;10, article 249 doi: 10.1186/1471-2407-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670, article 554) doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 115.Thomas RK, Baker AC, DeBiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nature Genetics. 2007;39(3):347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 116.Barretina J, Taylor BS, Banerji S, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nature Genetics. 2010;42(8):715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 118.Guillou L, Wadden C, Coindre J-M, Krausz T, Fletcher CDM. ’Proximal-type’ epithelioid sarcoma, a distinctive aggressive neoplasm showing rhabdoid features: clinicopathologic, immunohistochemical, and ultrastructural study of a series. American Journal of Surgical Pathology. 1997;21(2):130–146. doi: 10.1097/00000478-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 119.Miettinen M, Enzinger FM. Epithelioid variant of pleomorphic liposarcoma: a study of 12 cases of a distinctive variant of high-grade liposarcoma. Modern Pathology. 1999;12(7):722–728. [PubMed] [Google Scholar]
- 120.Downes KA, Goldblum JR, Montgomery EA, Fisher C. Pleomorphic liposarcoma: a clinicopathologic analysis of 19 cases. Modern Pathology. 2001;14(3):179–184. doi: 10.1038/modpathol.3880280. [DOI] [PubMed] [Google Scholar]
- 121.Stewart MG, Schwartz MR, Alford BR. Atypical and malignant lipomatous lesions of the head and neck. Archives of Otolaryngology—Head and Neck Surgery. 1994;120(10):1151–1155. doi: 10.1001/archotol.1994.01880340091016. [DOI] [PubMed] [Google Scholar]
- 122.Allen PW, Strungs I, MacCormac LB. Atypical subcutaneous fatty tumors: a review of 37 referred cases. Pathology. 1998;30(2):123–135. doi: 10.1080/00313029800169066. [DOI] [PubMed] [Google Scholar]
- 123.Taylor BS, Barretina J, Socci ND, et al. Functional copy-number alterations in cancer. PLoS ONE. 2008;3(9) doi: 10.1371/journal.pone.0003179. Article ID e3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Idbaih A, Coindre J-M, Derré J, et al. Myxoid malignant fibrous histiocytoma and pleomorphic liposarcoma share very similar genomic imbalances. Laboratory Investigation. 2005;85(2):176–181. doi: 10.1038/labinvest.3700202. [DOI] [PubMed] [Google Scholar]
- 125.Schmidt H, Bartel F, Kappler M, et al. Gains of 13q are correlated with a poor prognosis in liposarcoma. Modern Pathology. 2005;18(5):638–644. doi: 10.1038/modpathol.3800326. [DOI] [PubMed] [Google Scholar]
- 126.Singer S, Socci ND, Ambrosini G, et al. Gene expression profiling of liposarcoma identifies distinct biological types/subtypes and potential therapeutic targets in well-differentiated and dedifferentiated liposarcoma. Cancer Research. 2007;67(14):6626–6636. doi: 10.1158/0008-5472.CAN-07-0584. [DOI] [PubMed] [Google Scholar]
- 127.Dotiwala F, Harrison JC, Jain S, Sugawara N, Haber JE. Mad2 prolongs DNA damage checkpoint arrest caused by a double-strand break via a centromere-dependent mechanism. Current Biology. 2010;20(4):328–332. doi: 10.1016/j.cub.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 129.Shangary S, Qin D, McEachern D, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(10):3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ding K, Lu Y, Nikolovska-Coleska Z, et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. Journal of Medicinal Chemistry. 2006;49(12):3432–3435. doi: 10.1021/jm051122a. [DOI] [PubMed] [Google Scholar]
- 131.Shangary S, Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annual Review of Pharmacology and Toxicology. 2009;49:223–241. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ringshausen I, O’Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10(6):501–514. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 133.Tovar C, Rosinski J, Filipovic Z, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(6):1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dickens MP, Fitzgerald R, Fischer PM. Small-molecule inhibitors of MDM2 as new anticancer therapeutics. Seminars in Cancer Biology. 2009;20(1):10–18. doi: 10.1016/j.semcancer.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 135.Arts JPM, Valckx A, Blattner C, et al. JNJ-26854165- a novel hdm2 antagonist in clinical development showing broad-spectrum pre-clinical anti-tumour activity against solid malignancies. Proceedings of the American Association for Cancer Research. 2008;49 abstract no. 1592. [Google Scholar]
- 136.Pipeline RP. 2008 (http://www.ascenta.com/)
- 137. ClinicalTrials.gov—RO5045337, 2010, http://clinicaltrials.gov/ct2/results?term=RO5045337.
- 138.Chen Y-NP, Sharma SK, Ramsey TM, et al. Selective killing of transformed cells by cyclin/cyclin-dependent kinase 2 antagonists. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(8):4325–4329. doi: 10.1073/pnas.96.8.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. Journal of Clinical Oncology. 2006;24(11):1770–1783. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]
- 140.De Azevedo WF, Leclerc S, Meijer L, Havlicek L, Strnad M, Kim S-H. Inhibition of cyclin-dependent kinases by purine analogues. Crystal structure of human cdk2 complexed with roscovitine. European Journal of Biochemistry. 1997;243(1-2):518–526. doi: 10.1111/j.1432-1033.1997.0518a.x. [DOI] [PubMed] [Google Scholar]
- 141.Shapiro GI. Preclinical and clinical development of the cyclin-dependent kinase inhibitor flavopiridol. Clinical Cancer Research. 2004;10(12):4270s–4275s. doi: 10.1158/1078-0432.CCR-040020. [DOI] [PubMed] [Google Scholar]
- 142.Sedlacek HH. Mechanisms of action of flavopiridol. Critical Reviews in Oncology/Hematology. 2001;38(2):139–170. doi: 10.1016/s1040-8428(00)00124-4. [DOI] [PubMed] [Google Scholar]
- 143.Matranga CB, Shapiro GI. Selective sensitization of transformed cells to flavopiridol-induced apoptosis following recruitment to S-phase. Cancer Research. 2002;62(6):1707–1717. [PubMed] [Google Scholar]
- 144.Bible KC, Kaufmann SH. Cytotoxic synergy between flavopiridol (NSC 649890, L86-8275) and various antineoplastic agents: the importance of sequence of administration. Cancer Research. 1997;57(16):3375–3380. [PubMed] [Google Scholar]
- 145.Jung CP, Motwani MV, Schwartz GK. Flavopiridol increases sensitization to gemcitabine in human gastrointestinal cancer cell lines and correlates with down-regulation of ribonucleotide reductase M2 subunit. Clinical Cancer Research. 2001;7(8):2527–2536. [PubMed] [Google Scholar]
- 146.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nature Reviews Drug Discovery. 2009;8(7):547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 147.Benson C, White J, De Bono J, et al. A phase I trial of the selective oral cyclin-dependent kinase inhibitor seliciclib (CYC202; R-Roscovitine), administered twice daily for 7 days every 21 days. British Journal of Cancer. 2007;96(1):29–37. doi: 10.1038/sj.bjc.6603509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fry DW, Harvey PJ, Keller PR, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Molecular Cancer Therapeutics. 2004;3(11):1427–1437. [PubMed] [Google Scholar]
- 149.Demetri GD, Fletcher CDM, Mueller E, et al. Induction of solid tumor differentiation by the peroxisome proliferator-activated receptor-γ ligand troglitazone in patients with liposarcoma. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(7):3951–3956. doi: 10.1073/pnas.96.7.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPARγ2: tissue-specific regulator of an adipocyte enhancer. Genes and Development. 1994;8(10):1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 151.Sears IB, MacGinnitie MA, Kovacs LG, Graves RA. Differentiation-dependent expression of the brown adipocyte uncoupling protein gene: regulation by peroxisome proliferator-activated receptor γ . Molecular and Cellular Biology. 1996;16(7):3410–3419. doi: 10.1128/mcb.16.7.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Debrock G, Vanhentenrijk V, Sciot R, Debiec-Rychter M, Oyen R, Van Oosterom A. A phase II trial with rosiglitazone in liposarcoma patients. British Journal of Cancer. 2003;89(8):1409–1412. doi: 10.1038/sj.bjc.6601306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Takebayashi Y, Pourquier P, Yoshida A, Kohlhagen G, Pommier Y. Poisoning of human DNA topoisomerase I by ecteinascidin 743, an anticancer drug that selectively alkylates DNA in the minor groove. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(13):7196–7201. doi: 10.1073/pnas.96.13.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fayette J, Boyle H, Chabaud S, et al. Efficacy of trabectedin for advanced sarcomas in clinical trials versus compassionate use programs: analysis of 92 patients treated in a single institution. Anti-Cancer Drugs. 2010;21(1):113–119. doi: 10.1097/CAD.0b013e328333057b. [DOI] [PubMed] [Google Scholar]
- 155.Thornton KA. Trabectedin: the evidence for its place in therapy in the treatment of soft tissue sarcoma. Core Evidence. 2010;4:191–198. doi: 10.2147/ce.s5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pommier Y, Kohlhagen G, Bailly C, Waring M, Mazumder A, Kohn KW. DNA sequence- and structure-selective alkylation of guanine N2 in the DNA minor groove by ecteinascidin 743, a potent antitumor compound from the caribbean tunicate Ecteinascidia turbinata. Biochemistry. 1996;35(41):13303–13309. doi: 10.1021/bi960306b. [DOI] [PubMed] [Google Scholar]
- 157.Friedman D, Hu Z, Kolb EA, Gorfajn B, Scotto KW. Ecteinascidin-743 inhibits activated but not constitutive transcription. Cancer Research. 2002;62(12):3377–3381. [PubMed] [Google Scholar]
- 158.Minuzzo M, Marchini S, Broggini M, Faircloth G, D’Incalci M, Mantovani R. Interference of transcriptional activation by the antineoplastic drug ecteinascidin-743. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6780–6784. doi: 10.1073/pnas.97.12.6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Takebayashi Y, Pourquier P, Zimonjic DB, et al. Antiproliferative activity of ecteinascidin 743 is dependent upon transcription-coupled nucleotide-excision repair. Nature Medicine. 2001;7(8):961–966. doi: 10.1038/91008. [DOI] [PubMed] [Google Scholar]
- 160.Guirouilh-Barbat J, Redon C, Pommier Y. Transcription-coupled DNA double-strand breaks are mediated via the nucleotide excision repair and the Mre11-Rad50-Nbs1 complex. Molecular Biology of the Cell. 2008;19(9):3969–3981. doi: 10.1091/mbc.E08-02-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Soares DG, Escargueil AE, Poindessous V, et al. Replication and homologous recombination repair regulate DNA double-strand break formation by the antitumor alkylator ecteinascidin 743. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(32):13062–13067. doi: 10.1073/pnas.0609877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tavecchio M, Simone M, Erba E, et al. Role of homologous recombination in trabectedin-induced DNA damage. European Journal of Cancer. 2008;44(4):609–618. doi: 10.1016/j.ejca.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 163.Göransson M, Elias E, Ståhlberg A, Olofsson A, Andersson C, Åman P. Myxoid liposarcoma FUS-DDIT3 fusion oncogene induces C/EBP β-mediated interleukin 6 expression. International Journal of Cancer. 2005;115(4):556–560. doi: 10.1002/ijc.20893. [DOI] [PubMed] [Google Scholar]
- 164.Göransson M, Andersson MK, Forni C, et al. The myxoid liposarcoma FUS-DDIT3 fusion oncoprotein deregulates NF-κB target genes by interaction with NFKBIZ. Oncogene. 2009;28(2):270–278. doi: 10.1038/onc.2008.378. [DOI] [PubMed] [Google Scholar]
- 165.Germano G, Frapolli R, Simone M, et al. Antitumor and anti-inflammatory effects of trabectedin on human myxoid liposarcoma cells. Cancer Research. 2010;70(6):2235–2244. doi: 10.1158/0008-5472.CAN-09-2335. [DOI] [PubMed] [Google Scholar]
- 166.Workman P, Clarke PA, Raynaud FI, Van Montfort RLM. Drugging the PI3 kinome: from chemical tools to drugs in the clinic. Cancer Research. 2010;70(6):2146–2157. doi: 10.1158/0008-5472.CAN-09-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Folkes AJ, Ahmadi K, Alderton WK, et al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. Journal of Medicinal Chemistry. 2008;51(18):5522–5532. doi: 10.1021/jm800295d. [DOI] [PubMed] [Google Scholar]
- 168.Raynaud FI, Eccles SA, Patel S, et al. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Molecular Cancer Therapeutics. 2009;8(7):1725–1738. doi: 10.1158/1535-7163.MCT-08-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ihle NT, Lemos R, Jr., Wipf P, et al. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic ras is a dominant predictor for resistance. Cancer Research. 2009;69(1):143–150. doi: 10.1158/0008-5472.CAN-07-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.O'Brien C, Wallin JJ, Sampath D, et al. Predictive biomarkers of sensitivity to the phosphatidylinositol 3′ kinase inhibitor GDC-0941 in breast cancer preclinical models. Clinical Cancer Research. 2010;16(14):3670–3683. doi: 10.1158/1078-0432.CCR-09-2828. [DOI] [PubMed] [Google Scholar]
- 171.Di Nicolantonio F, Arena S, Tabernero J, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. The Journal of Clinical Investigation. 2010;120(8):2858–2866. doi: 10.1172/JCI37539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cecchi F, Rabe DC, Bottaro DP. Targeting the HGF/Met signalling pathway in cancer. European Journal of Cancer. 2010;46(7):1260–1270. doi: 10.1016/j.ejca.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Eder JP, Shapiro GI, Appleman LJ, et al. A phase I study of foretinib, a multi-targeted inhibitor of c-Met and vascular endothelial growth factor receptor 2. Clinical Cancer Research. 2010;16(13):3507–3516. doi: 10.1158/1078-0432.CCR-10-0574. [DOI] [PubMed] [Google Scholar]
- 174.Joensuu H, Fletcher C, Dimitrijevic S, Silberman S, Roberts P, Demetri G. Management of malignant gastrointestinal stromal tumours. Lancet Oncology. 2002;3(11):655–664. doi: 10.1016/s1470-2045(02)00899-9. [DOI] [PubMed] [Google Scholar]