Abstract
The UBA domain is a conserved sequence motif among polyubiquitin binding proteins. For the first time, we demonstrate a systematic, high throughput approach to identification of UBA domain-interacting proteins from a proteome-wide perspective. Using the rabbit reticulocyte lysate in vitro expression cloning system, we have successfully identified eleven proteins that interact with p62’s UBA domain, and the majority of the eleven proteins are associated with neurodegenerative disorders, such as Alzheimer’s disease. Therefore, p62 may play a novel regulatory role through its UBA domain. Our approach provides an easy route to the characterization of UBA domain interacting proteins and its application will unfold the important roles that the UBA domain plays.
Keywords: Ubiquitin, Alzheimer Disease
Introduction
p62 is a novel cellular protein which was initially identified in humans as a phosphotyrosine independent ligand of the src homology 2 (SH2) domain of p56 lck (1, 2). p56lck is a member of the c-src family of cytoplasmic tyrosine kinases that is found predominantly in cells of lymphoid origin (3, 4). In addition to the interaction with p56 lck , p62 also associates with the Ser/Thr kinase (1, 2), atypical protein kinase C (5, 6), and ubiquitin (7). In addition to the SH2 domain, p62 possesses several structural motifs, including a ubiquitin associated (UBA) domain that is capable of binding ubiquitin nonconvalently (8, 9).
Ubiquitin (Ub) is a small polypeptide of 76 amino acids that can be convalently attached to other proteins at specific lysine residues through chains composed of one (mono) or several ubiquitin moieties (poly). In addition to its classical role in protein degradation, ubiquitin is emerging as a signal for protein transport and processing (10-12). Conjugation of ubiquitin to substrate proteins requires three enzymes: a ubiquitin activating enzyme E1, a ubiquitin-conjugating enzyme E2, and a ubiquitin ligase E3. Initially, E1 activates ubiquitin by forming a high energy thioester intermediate with the C-terminal glycine using ATP. The activated ubiquitin is sequentially transferred to E2, then to E3 which catalyzes isopeptide bond formation between the activated C-terminal glycine of ubiquitin and e-amino group of a lysine residue of the substrate. Following the linkage of the first ubiquitin chain, additional molecules of ubiquitin are attached to lysine side chains of the previously conjugated moiety to form branched polyubiquitin chains. The fate of ubiquitinated substrates depends on the number of ubiquitin moieties conjugated, as well as, the lysine linkage of Ub-Ub conjugation. The conjugation of ubiquitin to eukaryotic intracellular proteins is one way in which those proteins are targeted to the proteasome for subsequent rapid degradation. This mechanism is particularly important for short-lived regulatory proteins such as cyclins, cyclin-dependent protein kinase-inhibitors, p53, the nuclear factor kappa B precursor, and IkB (13). The ubiquitin-proteasome system consists of two steps: 1) the target protein is conjugated with polyubiquitin molecules, which mark the substrate for degradation; 2) the target protein is transferred to the 26S proteasome, unfolded and degraded.
The UBA domain is a conserved sequence motif among proteins that can bind polyubiquitin. It is comprised of ~45 amino acids (13). The amino acids 386-434 of p62, which bind polyubiquitin, has been shown to possess homology to other recently described UBA domains (9). Interestingly, proteins with UBA domains are more likely to bind polyubiquitin chains over monoubiquitin, such as the yeast UBA protein Rad23, a highly conserved protein involved in nucleotide excision repair (13). Recently, it has been shown that yeast cells lacking two UBA proteins (Dsk2 and Rad23) are deficient in protein degradation and that the UBA motif is essential for their function in proteolysis (14).
In addition to the important role in recycling of amino acids from damaged or misfolded proteins, ubiquitin-protein conjugation also has functions unrelated to proteasomal targeting. For example, polyubiquitination is required for the internalization of several yeast and mammalian cell surface proteins into the endocytic pathway (15, 16). Interestingly, p62 appears to sequester ubiquitinated substrates into a cytoplasmic structure referred to as a sequestosome, into which excess ubiquitinated proteins are segregated (17). In addition, p62 is an immediate early response gene product for a variety of signals (18). Thus, p62 appears to play a novel regulatory role for polyubiquitinated proteins and may have an essential function in cell proliferation and differentiation. We have developed a method that will enable identification of protein(s) that interact with p62’s UBA domain.
Materials and Methods
Materials
Human adult brain library 10×96 well plates with 100 cDNAs per well and Gold TNT SP6 Express 96 system plate were purchased from Promega Corporation (Madison, WI). [35S] methionine was from ICN Biomedicals, Inc. (Irvine, CA). Ubiquitin was from Sigma-Aldrich Corporation (St. Louis, MO). Polyubiquitin K48 chains (Ub2-7 K48) were from Affiniti Research Products Limited (Exeter, UK). Autoradiography enhancer En3HANCE was from Perkin Elmer, Inc. (Wellesley, MA). JM109 competent cells were from Promega Corporation (Madison, WI). Ubiquitin activating enzyme E1 was from Calbiochem (La Jolla, CA). Ubiquitin conjugating enzyme E2 (UbcH7) was from Affiniti Research Products Limited (Exeter, UK). The rabbit polyclonal anti-myc antibody, mouse monoclonal anti-myc and anti-ubiquitin antibodies were from Santa Cruz Biotechnology, Inc (Santa Cruz, CA). Ubiquitin K63 only (Ub K63, ubiquitin with all lysines mutated to arginines except K63), Ub K29, Ub K48 were from Boston Biochem Inc. (Cambridge, MA). Mammalian Cell Transfection Kit was from Specialty Media (Phillipsburg, NJ). Agarose-immobilised p62 UBA peptide corresponding to the amino acid sequence of the ubiquitin-binding protein p62 residues 387-436 (PPEADPRLIESLSQmlsmgfsdeggwltrllqtkny digaaldtiqyskh) was purchased from Affiniti Research Products Limited (Exeter, UK).
In vitro expression cloning
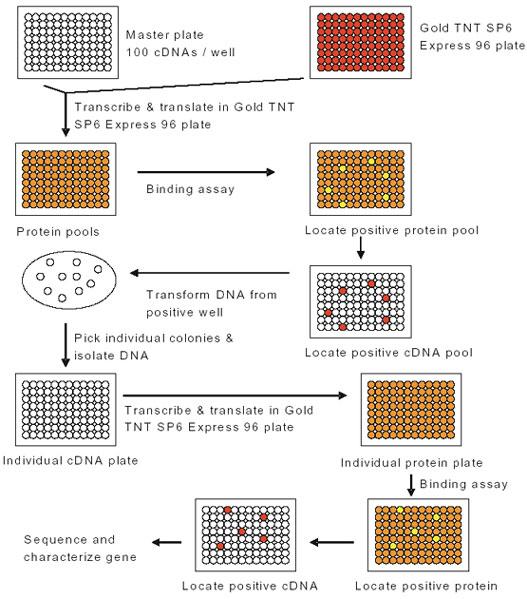
To search for novel proteins that bind to the UBA domain of p62, we performed in vitro expression cloning (IVEC) using the ProteoLink IVEC system. The human adult brain library 96 well plates with 100 cDNAs per well was transcribed and translated employing the Gold TNT SP6 Express 96 plate and [35S] methionine. The TNT Quick-coupled transcription-translation system contained a rabbit reticulocyte lysate pre-mixed with most of the reaction components necessary to carry out transcription/translation in the lysate, including all of the amino acids except methionine. [35S] Methionine was used to label newly synthesized proteins. The reactions were set up according to the manufacturer’s instructions. Rabbit reticulocyte lysate has been shown to be capable of carrying out ubiquitination of proteins that were translated in such an in vitro translation system (19, 20). The reactions mixtures also contained ubiquitin so that the newly synthesized proteins could be ubiquitinated. The reactions were incubated at 30°C for 2 hours. The resulting proteins were assayed to determine their binding ability with p62’s UBA domain. Potential positive “hits” were further subdivided and re-assayed to link individual clones to the protein of interest (Fig. 1).
Fig. 1. Schematic description of the in vitro expression cloning (IVEC) system for the primary proteome-wide screen and subsequent isolation of cDNAs encoding the proteins of interest (from IVEC manual).
p62 UBA pull down assays
Each translated pool was resuspended in binding buffer (25 mM Tris pH 7.5, 125 mM NaCl, 0.1% NP-40) and used as a source of protein in p62 UBA pull down assays. Proteins that specifically interact with the UBA domain of p62 were isolated by interaction with agarose-immobilised p62-UBA peptide (amino acid 387-436 of p62) (5 mg) for 2 hours at 4ºC, then washed three times in washing buffer (25 mM Tris pH 7.6, 100 mM NaCl, 1% NP-40). Bound proteins were released by addition of SDS-sample buffer and separated by SDS-PAGE. The SDS-PAGE gels were fixed in 50% methanol, 10% acetic acid for 30 min, stained in 0.2% Commassie Brilliant Blue R-250, 45% methanol, 10% acetic acid for 15 min, destained in 10% acetic acid, 50% methanol overnight, and enhanced in autoradiography enhancer En3HANCE for 1 hr and exposed to X-Ray film.
Library Screening
By combining 4 pools as one mixed pool, 96 protein pools were divided into 24 mixed protein pools for use in p62 UBA pull down assays. Positive mixed protein pools were selected and individual pools were retested for its ability to bind p62’s UBA domain. The individual cDNA pool from which the positive protein pool was generated was transformed into JM109 competent cells and plated on LB ampicillin plate. Individual colonies were chosen to grow overnight in 1 ml of LB media plus ampicillin. Plasmid DNA was purified from the cell culture and used for TNT Quick coupled in vitro transcription/ translation. The individual protein synthesized from each plasmid DNA chosen was screened for its ability to bind p62’s UBA domain. To confirm the interaction with p62’s UBA domain, the final resulting individual proteins were used in the coupled TNT/p62 UBA pull down assays. The cDNA inserts were sequenced in the Genomics Core Facility at Auburn University and the sequences were compared with known sequences in NCBI database by BLAST analysis.
Transfection and in vitro ubiquitination
Human embryonic kidney 293 (HEK 293) cells were cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM) containing 10% heat-inactivated fetal calf serum and transfected with myc-tagged HSP70 plasmid using the Mammalian Cell Transfection Kit. Cells were harvested and lysed in 1 ml of SDS lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 10 mM NaF, 0.5% TX-100, 1 mM Na3VO4, 2 mg/ml aprotinin, 2 mg/ml leupeptin, 1 mM PMSF, 1% SDS) for 30 min on ice, followed by centrifugation at 14000 rpm for 15 min at 4°C to remove the insoluble fraction. The protein concentration of the supernatant was determined using the Bio-Rad DC protein assay reagent with bovine serum albumin (BSA) as standard. Equal amount of protein (750 mg) was immunoprecipitated with anti-myc and collected with agarose-coupled secondary antibody. To the agarose beads containing the immunoprecipitated HSP70, 50 ml of reaction buffer (50 mM Tris-HCl pH 7.5, 2.5 mM MgCl2, 2 mM DTT, 2 mM ATP) was added containing 100 ng E1, 200 ng E2 (UbcH7), and 100 mg of E3 (Flag-tagged TRAF6) along with 5 mg GST-WT-Ub, GST-K29R Ub, GST-K48R Ub, GST-K63R Ub, or K63 Ub. Control samples without HSP70, E1, E2, E3, or GST-WT-Ub were also included. Reactions were carried out by continuous shaking at 37°C for 2 hours and then washed three times with reaction buffer. The proteins were released by boiling for 2 min in SDS-PAGE sample buffer, separated on 7.5% SDS-PAGE and Western blotted for anti-ubiquitin.
Results and Discussion
In vitro transcription/translation
To search for novel proteins that bind to the UBA domain of p62, we performed in vitro expression cloning (IVEC) using the ProteoLink IVEC system from Promega (Cat. No. L6500). The human adult brain library 96 well plates with 100 cDNAs per well were transcribed and translated employing the Gold TNT SP6 Express 96 plate in the presence of [35S] methionine and ubiquitin (25 mg/ml, Sigma). By combining 4 protein pools as one mixed pool, 96 protein pools were divided into 24 mixed pools (Fig. 2A, 2B).
Fig. 2. Protein synthesized by the ProteoLink IVEC system in the presence of [35S] methionine.
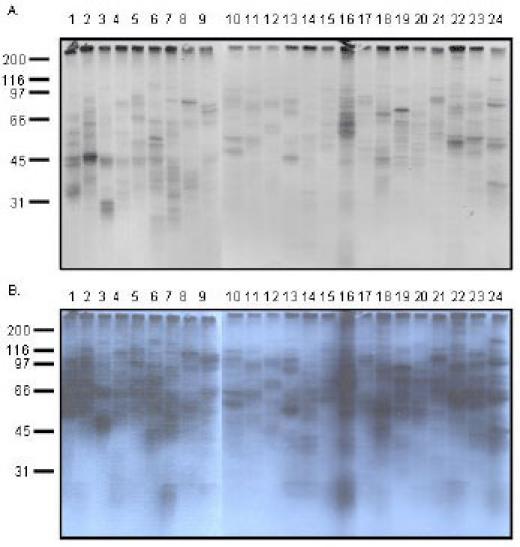
96 protein pools were generated by employing TNT Quick Coupled in vitro transcription/translation system. The 96 protein pools were then divided into 24 mixed protein pools by combining four protein pools as one mixed protein pool. A: 1 day exposure; B: 3 days exposure.
Each lane contained more than 100 proteins (theoretically 400) with different molecular weight. Therefore, each lane appeared as a smear, indicating that the in vitro transcription/translation system from Promega worked successfully. In order to examine whether proteins synthesized in the IVEC system are also ubiquitinated, Western blot analysis was performed by blotting the newly synthesized proteins (in the presence of cold methionine instead of 35S methionine) with ubiquitin monoclonal antibody. In the mixed protein pools, each of the 24 lanes appeared as a smear, indicating that proteins synthesized by the IVEC system are also ubiquitinated (Fig. 3A). Furthermore, the rabbit reticulocyte lysate in the IVEC system can utilize different lysine linkages of ubiquitin (i.e., Ub K29, Ub K48, and Ub K63) for ubiquitination (Fig. 3B). In order to investigate whether the agarose-immobilised p62 UBA peptide has binding specificity, a mixed protein pool synthesized by IVEC system was tested in a pull down assay in the presence of agarose beads alone or in the presence of p62 UBA agarose beads (Fig. 3C). Our results revealed that proteins that bound to p62’s UBA domain could not be pulled down by agarose beads alone, indicating that the agarose-immobilised p62 UBA peptide had binding specificity.
Fig. 3. Western blot analysis of the proteins synthesized by the ProteoLink IVEC system with ubiquitin antibody and specificity of UBA binding.
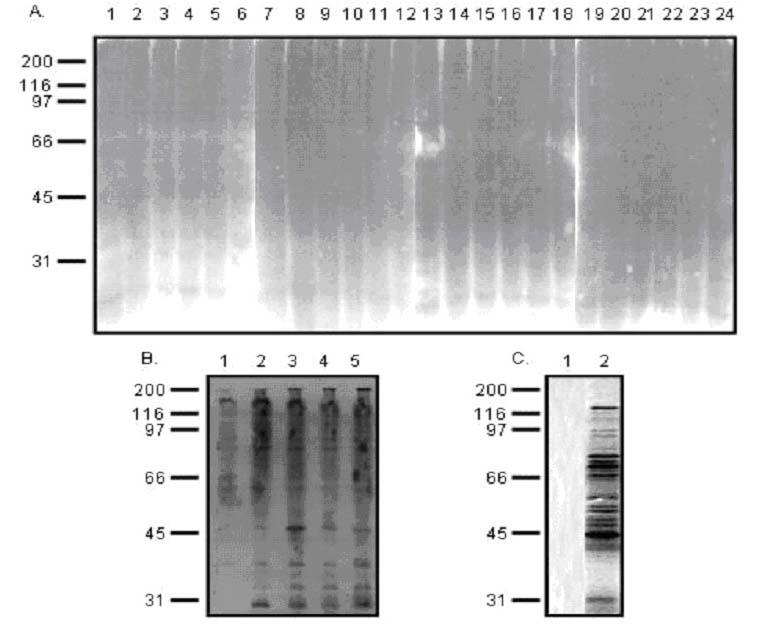
A: Mixed protein pools. 96 protein pools were generated by employing TNT Quick Coupled in vitro transcription/translation system. The 96 protein pools were divided into 24 mixed protein pools and separated by SDS-PAGE, followed by immunoblotting with ubiquitin monoclonal antibody. B: Proteins synthesized in the IVEC system in the absence of ubiquitin (lane 1) or presence of wild type ubiquitin (lane 2), ubiquitin K29 (lane 3), ubiquitin K48 (lane 4), and ubiquitin K63 (lane 5). C: Proteins synthesized in the IVEC system were labeled by 35S methionine and used for pull down assays. Lane 1: proteins recovered with agarose beads alone; Lane 2: proteins recovered with p62 UBA agarose beads.
p62 UBA pull down and library screen
In order to identify proteins that bind to p62’s UBA domain, p62 UBA pull down assays were performed. Out of the 24 mixed protein pools, several pools contained [35S] methionine-labeled bands in the primary p62 UBA pull down assays (Fig. 4). We chose 6 pools (pool # 2, 4, 8, 14, 20, 21) because of their stronger signal to specifically identify which individual protein pool in the mixed pools has the ability to bind to p62’s UBA domain. Therefore, a secondary screen was conducted on the 6 positive individual mixed pools (representing 24 individual protein pools) which bound with p62’s UBA domain (Fig. 5). Mixed protein pool # 2 generated a positive protein with molecular weight of 51 KDa (Fig. 4), and only individual protein pool “c” out of the four protein pools (a, b, c, d) that comprised protein pool #2 had a protein with the same molecular weight (Fig. 5). Depending on the size of the protein pulled down in the secondary screen compared to the primary screen (Fig. 4), individual protein pools “c”, “h”, “i”, “o”, “t”, and “v” were identified (Fig. 5).
Fig. 4. Pull down assay of p62’s UBA domain with proteins (mixed protein pool) synthesized by IVEC system in the presence of [35S] methionine.

The 24 mixed proteins pools were incubated with agarose-immobilised p62 UBA beads for 2 hours at 4ºC and washed three times in washing buffer. Bound proteins were released by boiling for 2 min in SDS-PAGE sample buffer and separated on 10% SDS-PAGE. Positive “hits” were marked by arrows and the positive protein pools were selected for subsequent deconvulation.
Fig. 5. Pull down assay of p62’s UBA domain with proteins (individual protein pool) synthesized by IVEC system in the presence of [35S] methionine.
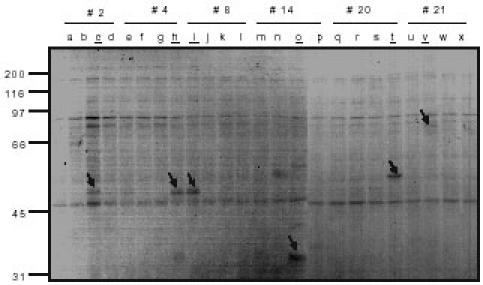
The 6 positive mix protein pools (# 2, 4, 8, 14, 20, 21 from Fig. 4) representing 24 individual protein pools were incubated with agarose-immobilised p62 UBA beads for 2 hours at 4ºC and the beads were washed three times in washing buffer. Bound proteins were released by boiling for 2 min in SDS-PAGE sample buffer and separated on 10% SDS-PAGE. Positive “hits” were marked by arrows and the positive individual protein pools were underlined.
To specifically identify which protein in the individual protein pool has the ability to bind p62’s UBA domain, the cDNAs from the positive individual protein pools were then transformed into JM109 competent cells and plated out on LB ampicillin plates. Individual colonies were chosen to grow overnight in 1 ml of LB media plus ampicillin. Plasmid DNAs were purified from the cell culture and used for TNT Quick coupled in vitro transcription/translation. The individual protein synthesized from each plasmid DNA was retested for its ability to bind p62’s UBA domain. By synthesizing individual protein from individual plasmid using the Gold TNT Quick coupled in vitro transcription/translation system and subjecting them to p62 UBA pull-down assays, 11 positive clones were isolated from the 6 positive individual pools. It is not surprising that 5 more clones showed binding ability with p62’s UBA domain since there are 100 cDNAs in each positive individual pool and some of them could have lower binding ability and therefore showed weak signal in the mixed protein pool. It is also possible that they are not as efficiently synthesized in the mixed TNT reaction as in the individual TNT reaction in which only one cDNA was used as template. The 11 positive plasmids were sequenced and compared with known cDNA sequences in NCBI database using BLAST analysis with results shown in Table 1.
Table 1.
Sequencing result of individual cDNA/Protein that binds to p62’s UBA domain
| Well # | Plasmid | cDNA Homology and resulting protein | Putative function | |
| A7 | #19 | 98% | Myelin basic protein | Decreased in Alzheimer’s Disease (AD) |
| A7 | #23 | 98% | 14-3-3 protein zeta isoform | Located in neurofibrillary tangles of AD |
| B4 | #18 | 96% | Syntaxin binding protein (munc18) 1 | Interact with X11 and reduce b-amyloid |
| B4 | #24 | 99% | FK506 binding protein (FKBP) 14 | Ligands as neurological disorders therapeutics |
| C5 | #19 | 99% | Homeobox protein Meis2 | Brain development |
| E7 | #9 | 97% | Transketolase | Decreased activity in dementia of AD |
| E7 | #15 | 100% | Heat shock cognate hsp70 | Protect cell death |
| F3 | #10 | 97% | Reelin isoform b | Suppress tau Phosphorylation |
| G8 | #16 | 94% | Calcium/calmodulin kinase II | May phosphate tau, leading to AD |
| G8 | #24 | 99% | Unc51 like kinase II | Plays role in axonal elongation |
| G10 | #13 | 98% | Nuclear receptor co-repressor 1 | Link with Huntington’s disease |
Interestingly, the proteins identified in the screen fall into three distinct categories. One set are proteins that are associated with Alzheimer’s disease, including myelin basic protein, 14-3-3 protein, syntaxin binding protein munc18, transketolase, heat shock protein HSP70, reelin, and calcium/calmodulin kinase II (Table 1). Significant decrease in the amount of myelin basic protein has been reported in the white matter of Alzheimer’s disease patients, accompanied by increased quantities of b-amyloid peptides (21). The presence of b-amyloid peptides containing senile plaques and neurofibrillary tangles are the two major pathological features in the brain of patients with Alzheimer’s disease (22). Interestingly, 14-3-3 proteins have also been demonstrated to be components of neurofibrillary tangles of Alzheimer’s disease brains (23). Syntaxin binding protein munc18 can powerfully regulate amyloid precursor protein metabolism and b-amyloid secretion through direct and indirect interactions with X11 proteins (24). The activity of transketolase has been reported to be reduced in dementia of Alzheimer’s type brain (25). Heat shock protein HSP70 expression is significantly increased in the temporal cortex of patients with Alzheimer’s disease (26). Besides HSP70, other heat shock proteins are also linked with Alzheimer’s disease. For example, increased synthesis of HSP27 has been suggested to play a role in preventing neuronal injury in AD (27), and alpha-crystallin heat shock protein has a close relationship with neurofibrillary tangles of AD brains (28). Reelin is a large secreted protein that controls cortical layering by signaling through the very low density lipoprotein receptor and apolipoprotein E receptor 2, thereby inducing tyrosine phosphorylation of the adaptor protein Disabled-1 (Dab1) and suppressing tau phosphorylation (29). Neurofibrillary tangles comprised of highly phosphorylated tau proteins are a key component of Alzheimer’s disease (30). Enhanced activity of calcium/calmodulin kinase II has been suggested to contribute to phosphorylation of tau protein and lead to neurofibrillary tangle deposition and neuronal death in Alzheimer’s disease (31). Although the relationship between p62 and neurofibrillary tangles or neuritic plaques is unclear, both neurofibrillary tangles and dystrophic neuritis of neuritic plaques are associated with ubiquitin (32), suggesting that dysfunction in ubiquitin-mediated proteolysis and the resulting accumulation of ubiquitin-conjugated proteins may contribute to the origination of dystrophic neuritis and neurofibrillary tangles. Furthermore, p62 has been recently reported to accumulate early in neurofibrillary tangles in Alzheimer’s disease (33), suggesting that p62 may play an important role in Alzheimer’s disease by interacting with those proteins through its UBA domain.
A second set of proteins identified in the screen that bind to p62’s UBA domain are associated with brain development, including homeobox protein Meis2 and unc51 like kinase II (Table 1). Although Meis proteins are not extensively studied in humans, these proteins have been shown to be required for hindbrain development in the zebrafish (34). Unc51 like kinase II has been demonstrated to play a role in axonal elongation (35, 36), which is needed for the formation of complicated neuronal networks. The third set of proteins that exhibit ability to bind p62’s UBA domain are proteins that are linked with other neurodegenerative diseases, including FK506 binding proteins and nuclear receptor corepressor I (Table 1). FK506 (tacrolimus) is a potent immunosuppressive drug used in the treatment of patients after organ transplantation and in selected autoimmune disorders (37). FK506 is activated upon binding to members of the immunophilin family of proteins, which were designated as FK506 binding proteins (38). Immunophilins are chaperone proteins and FK506 binding proteins have been suggested as therapeutics for neurological disorders (39, 40). Nuclear receptor corepressor I has been suggested to play a role in Huntington’s disease because it is able to interact with huntingtin (41). The proteins identified here suggest that p62’s UBA domain has the ability to interact with multiple proteins that play important roles in neurodegenerative diseases. Further screening from the whole genome-wide perspective will be necessary to define the important role that p62’s UBA domain plays.
In vitro ubiquitination
It has been reported that polyubiquitin chains assembled through lysine 48 of ubiquitin act as a signal for substrate proteolysis by the 26S proteasome (42-44). In order to understand whether the proteins identified in our screen bind to the p62’s UBA domain through lysine 48 (K48), polyubiquitin K48 chains were added to the p62 UBA pull down assay (Fig. 6). Inclusion of polyubiquitin K48 chains in the assay should compete for the binding of substrate to the p62’s UBA domain and reduce the interaction of those proteins with the p62’s UBA domain if those proteins are assembled through K48 chains. An alternative interpretation for polyubiquitin K48 chain competition is that the ubiquitin chains are competing for the same binding site as the binding partners which are either ubiquitinated or non-ubiquitinated. We randomly chose five proteins out of the 11 binding partners for the competition pull down (Fig. 6). Out of the five proteins, four proteins (# 2, 3, 4, and 5) showed reduced binding ability with p62’s UBA domain when polyubiquitin K48 chains were included (Fig. 6A, 6B). However, K48 chains failed to compete with HSP70, suggesting that p62’s UBA domain binds to HSP70 through a ubiquitin lysine linkage other than K48. Interestingly, it has been reported that heat shock protein 70 cognate (HSP70) is ubiquitinated by CHIP (carboxyl terminus of Hsc70-interacting protein) via ubiquitin chain synthesis that uses either K29 or K63 (45). In order to examine which lysine linkage utilized by HSP70 binds to p62’s UBA domain, in vitro ubiquitination assay was performed by incubating lysates from HEK cells expressing HSP70 with E1, E2, and E3 in reaction buffer (50 mM Tris-HCl pH 7.5, 2.5 mM MgCl2, 2 mM DTT, 2 mM ATP). As control, the ubiquitination of HSP70 utilizing the rabbit reticulocyte lysate was also investigated by Western blot analysis. Our results revealed that HSP70 was ubiquitinated in the IVEC system (Fig. 7A, 7B), and the rabbit reticulocyte lysate contained enzymes such as TRAF6 (E3) and UbcH7 (E2) for in vitro ubiquitination (Fig. 7C).
Fig. 6. Polyubiquitin K48 chain (Ub2-7 K48) competition pull down assay.
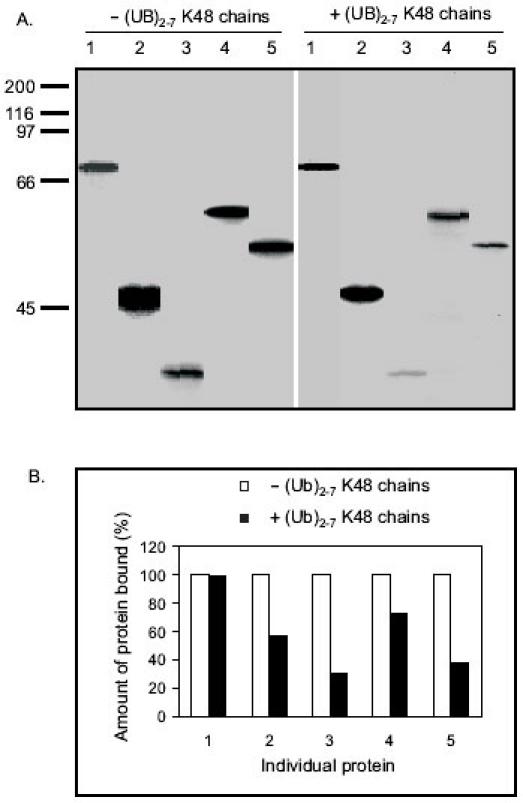
A: The individual proteins were synthesized by employing TNT Quick Coupled in vitro transcription/translation system and incubated with 5 mg agarose-immobilised p62 UBA beads +/- polyubiquitin K48 chains for 2 hours at 4ºC. The beads were washed three times in washing buffer and bound proteins were released by boiling for 2 min in SDS-PAGE sample buffer and separated on 10% SDS-PAGE. B: The autoradiogram was scanned and the relative amount of protein (%) bound to p62’s UBA domain +/- polyubiquitin K48 chains was graphed. The amount of protein bound to p62’s UBA domain without addition of polyubiquitin K48 chains was considered 100%. 1: HSP70; 2: Meis2; 3: 14-3-3; 4: Reelin; 5: MBP.
Fig. 7. In vitro ubiquitination using the rabbit reticulocyte lysate.
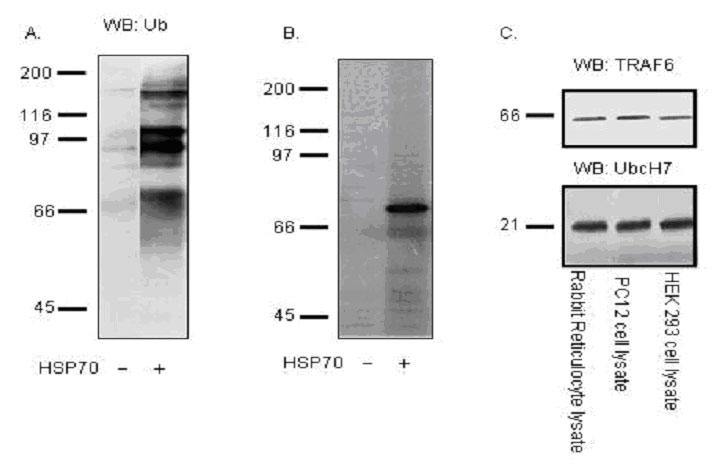
A: HSP70 Protein was synthesized employing TNT Quick Coupled in vitro transcription/translation system in the presence of ubiquitin, resolved on 10% SDS-PAGE gels, transferred to nitrocellulose membrane and western blotted with ubiquitin monoclonal antibody. B: HSP70 Protein was synthesized employing TNT Quick Coupled in vitro transcription/translation system in the presence of ubiquitin and 35S-methionine, resolved on 10% SDS-PAGE and exposed to X-ray film. C: Western blot of rabbit reticulocyte lysate with TRAF6 (E3) and UbcH7 (E2).
TRAF6 was chosen as an E3 in this in vitro ubiquitination assay due to its RING domain, a common feature of E3 ligases, and the observation that p62 is a scaffold for TRAF6 interaction (46). Therefore, in vitro ubiquitination assays using the E1-E2-E3 system were performed in the presence of either ubiquitin wild type or ubiquitin mutants (K29R, K48R, and K63R). If one lysine mutant blocks the ubiquitination of HSP70, it would suggest that the ubiquitination of HSP70 utilizes that specific lysine linkage. Our results revealed that HSP70 utilizes K63 linkage to assemble polyubiquitin chains to bind to p62’s UBA domain since only the K63R ubiquitin mutant blocked the ubiquitination of HSP70 (Fig. 8A). A similar result was also observed when reactions were conducted with wild type ubiquitin or mutant ubiquitin with all lysines mutated to arginines except K63 and the ubiquitination of HSP70 occurred only in the reaction that has either intact K63 ubiquitin or wild type ubiquitin (Fig. 8B). This finding is consistent with previous reports (45), demonstrating that HSP70 is K63-polyubiquitinated. Furthermore, the in vivo interaction of HSP70 and p62 was confirmed by transfecting myc-tagged HSP70 into HEK 293 cells in the presence of the proteasome inhibitor MG132 and subjecting cell lysates to p62 immunoprecipitation and Western blot with anti-myc antibody (Fig. 8C). The interaction between HSP70 and p62 in vivo took place only when MG132 was included, suggesting that the interaction in vivo is dependent upon the ubiquitination of HSP70. The specific type of polyubiquitin chain recognized by p62’s UBA domain is not yet known and studies are underway lab to determine p62’s interaction with specific polyubiquitin chains, however, our preliminary studies suggest that p62’s UBA domain may recognize K63 linked polyubiquitin chains.
Fig. 8. In vitro ubiquitination of HSP70 using E1-E2-E3 system and in vivo interaction of p62 with HSP70.
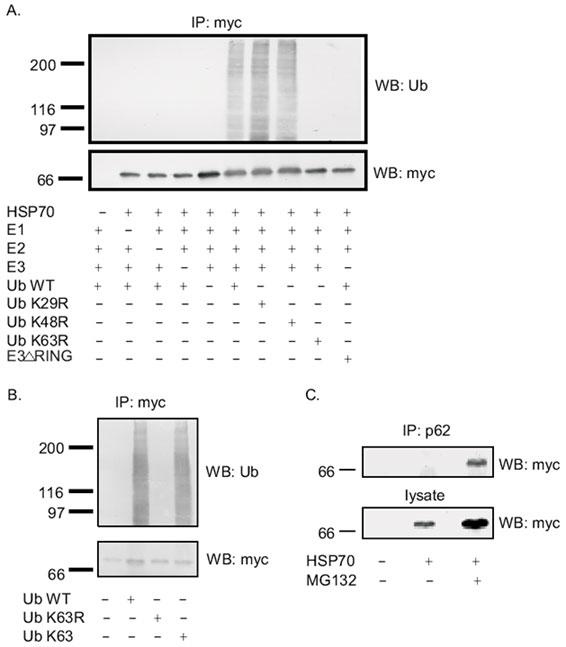
A: myc-tagged HSP70 protein expressed in HEK cells was immunoprecipitated by myc-polyclonal antibody and used as a source of substrate for in vitro ubiquitination +/- E1, E2, E3, WT-Ub, Ub K29R, Ub K48R, Ub K63R. The reactions were separated on 10% SDS-PAGE and Western blotted with myc monoclonal antibody (bottom panel) to detect HSP70 or with ubiquitin monoclonal antibody (top panel) to detect ubiquitination. B: myc-tagged HSP70 protein expressed in HEK cells was immunoprecipitated by myc-polyclonal antibody and used as a source of substrate for in vitro ubiquitination in the absence or presence of wild type ubiquitin or Ub K63R or Ub K63. The reactions were separated on 10% SDS-PAGE and Western blotted with myc monoclonal antibody (bottom panel) to detect HSP70 or with ubiquitin monoclonal antibody (top panel) to detect ubiquitination. C: In vivo interaction of HSP70 and p62. Transfection of myc-tagged HSP70 into HEK293 cells was performed and the cell lysates were subjected to immunoprecipitation with p62 polyclonal antibody, followed by Western blot with anti-myc monoclonal antibody.
In summary, for the first time, we demonstrate a systematic approach to identify UBA domain binding proteins from a proteome wide perspective. This approach could be readily adapted to high throughput screening. Using the rabbit reticulocyte lysate in vitro expression cloning system, we have successfully identified eleven proteins in the human adult brain that interact with the UBA domain of p62, and the majority of the eleven proteins are associated with neurodegenerative disorders, such as Alzheimer’s disease. This is a very interesting finding since 9600 cDNAs have been screened and only 11 of them showed binding specificity with p62’s UBA domain. Studies are underway to unfold the functional roles of p62 in the ubiquitin system. Our approach provides an easy route to the characterization of UBA domain binding proteins at the level of the whole proteome, its application will unfold the important roles that p62’s UBA domain plays. This method could be easily adapted to identify proteins that interact with other UBA domains as well.
Acknowledgments
This research work was supported by National Institute of Health Grant NINDS 33661. We thank Dr. Cam Patterson (University of North Carolina at Chapel Hill) for providing the myc-tagged HSP70 construct.
Appendix
Protocols
In vitro transcription/translation
Remove one Gold TNT SP6 Express 96 plate from storage at -80°C and thaw on ice for 10 minutes. Briefly spin to pull material down to the bottom of the wells.
Remove one ProteoLink Human Adult Brain cDNA library plate from storage at -20°C and thaw. Briefly spin to pull material down to the bottom of the wells.
Carefully peel away and discard the seal. The seal of the library should be discarded to avoid cross contamination.
For each reaction, i.e., one 20 ml pre-dispended well of lysate, add 1 ml of [35S] methionine (1000Ci/mol at 10 mCi/ml), 2 ml of cDNA template (0.25 mg/ml), 1 ml of ubiquitin (25 mg/ml), 1 ml of nuclease-free water. Seal the plate containing the reactions with a new adhesive plate seal. Reseal the cDNA library plate with a new adhesive plate seal and store it at -20°C.
Gently vortex the plate to mix.
Incubate the reaction at 30°C for 2 hours.
Use the translated proteins as source for p62 UBA pull down assays.
p62 UBA pull down assay
Wash p62 UBA agarose beads (Affiniti, UK) with binding buffer (25 mM Tris pH 7.5, 125 mM NaCl, 0.1% NP-40) three times.
Add equal volume to p62 UBA beads to make 50% slurry.
Add 5 ml of 50% p62 UBA beads to each TNT protein pool (mixed pools or individual pool).
Bring all reactions to a total of 50 ml with binding buffer.
Gently shake the reactions in a mixer at 4°C for 2 hours.
Wash the beads with 500 ml of washing buffer (25 mM Tris pH 7.6, 100 mM NaCl, 1% NP-40) three times to remove the unbound proteins.
Add 50 ml of SDS sample buffer, boiling for 2 minutes to release the bound proteins. Separate proteins on 10% SDS-PAGE.
Transfection and in vitro ubiquitination
Culture HEK 293 cells in high glucose Dulbecco’s modified Eagle’s medium (DMEM) containing 10% heat-inactivated fetal calf serum.
Transfect HEK 293 cells with 10 mg of myc-tagged HSP70 plasmid by calcium phosphate procedure.
After 48 hours transfection, lyse cells in 1 ml of triton lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 10 mM NaF, 0.5% TX-100, 1 mM Na3VO4, 2 mg/ml aprotinin, 2 mg/ml leupeptin, 1 mM PMSF) containing 1% SDS for 30 min on ice.
Centrifuge at 14000 rpm for 15 min at 4°C to remove the insoluble fraction.
Measure the protein concentration using the Bio-Rad DC protein assay reagent with bovine serum albumin as standard.
Combine equal amount of protein (750 mg) with 4 mg of anti-myc monoclonal antibody in SDS lysis buffer. The total volume of the reaction is 1 ml.
Rotate reactions gently at 4°C for 2 hours.
Add 50 ml of 50% anti-mouse IgG agarose beads in triton lysis buffer to each reaction. Rotate reactions gently at 4°C for 2 hours.
Wash agarose beads three times with triton lysis buffer to remove unbound protein.
Wash agarose beads once with reaction buffer (50 mM Tris-HCl, pH 7.5, 2.5 mM MgCl2, 2 mM DTT, 2 mM ATP).
To the agarose beads containing the immunoprecipitated HSP70, add 100 ng E1(Calbiochem), 200 ng E2 (UbcH7, Affiniti, UK), 100 mg of E3 (Flag-tagged TRAF6), 5 mg GST-WT-Ub, GST-K29R Ub, GST K48R Ub, or GST-K63R Ub in 50 ml of reaction buffer. Also include control samples without HSP70, E1, E2, E3, or GST-WT-Ub.
Shake the reactions continuously at 37°C for 2 hours.
Wash beads three times with reaction buffer.
Add 50 ml of SDS sample buffer, boiling for 2 minutes to release the bound proteins. Separate proteins on 10% SDS-PAGE.
References
- Park I, Chung J, Walsh CT, Yun Y, Strominger JL, Shin J. Phosphotyrosine-independent binding of a 62-kDa protein to the src homology 2 (SH2) domain of p56lck and its regulation by phosphorylation of Ser-59 in the lck unique N-terminal region. Proc Natl Acad Sci. 1995;92:12338–12342. doi: 10.1073/pnas.92.26.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung I, Strominger JL, Shin J. Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc Natl Acad Sci. 1996;93:5991–5995. doi: 10.1073/pnas.93.12.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Howell B. The when and how of Src regulation. Cell. 1993;73:1051–1054. doi: 10.1016/0092-8674(93)90634-3. [DOI] [PubMed] [Google Scholar]
- Weiss A, Littman DR. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- Puls A, Schmidt S, Grawe F, Stabel S. Interaction of protein kinase C zeta with ZIP, a novel protein kinase C-binding protein. Proc Natl Acad Sci. 1997;94:6191–6196. doi: 10.1073/pnas.94.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez P, De Carcer G, Sandoval IV, Moscat J, Diaz-Meco MT. Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol Cel Biol. 1998;18:3069–3080. doi: 10.1128/mcb.18.5.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamundi RK, Joung I, Strominger JL, Shin J. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding protein. J Biol Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- Geetha T, Wooten MW. Structure and functional properties of the ubiquitin-binding protein p62. FEBS Lett. 2001;512:19–24. doi: 10.1016/S0014-5793(02)02286-X. [DOI] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Bio. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- Hicke L. A new ticket for entry into budding vesicles-ubiquitin. Cell. 2001;106:527–530. doi: 10.1016/s0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. TIBS. 1996;21:172–173. [PubMed] [Google Scholar]
- Rao H, Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J Biol Chem. 2002;277:11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Hicke L. Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J. 1997;11:1215–1226. doi: 10.1096/fasebj.11.14.9409540. [DOI] [PubMed] [Google Scholar]
- Shin J. p62 and the sequestosome, a novel mechanism for protein metabolism. Arch Pharm Res. 1998;21:829–833. doi: 10.1007/BF02976748. [DOI] [PubMed] [Google Scholar]
- Lee YH, Ko J, Joung I, Shin J. Immediate early response of the p62 gene encoding a non-proteasomal multiubiquitin chain binding protein. FEBS Lett. 1998;438:297–300. doi: 10.1016/S0014-5793(98)01323-4. [DOI] [PubMed] [Google Scholar]
- Sato S, Ward CL, Kopito RR. Cotranslational ubiquitination of cystic fibrosis transmembrane conductance regulator in vitro. J Biol Chem. 1998;273:7189–7192. doi: 10.1074/jbc.273.13.7189. [DOI] [PubMed] [Google Scholar]
- Tibbles KW, Brierley I, Cavanagh D, Brown TD. A region of the coronavirus infectious bronchitis virus a1 polyprotein encoding the 3C-like protease domain is subject to rapid turnover when expressed in rabbit reticulocyte lysate. J Gen Virol. 1995;76:3059–3070. doi: 10.1099/0022-1317-76-12-3059. [DOI] [PubMed] [Google Scholar]
- Roher AE, Weiss N, Kokjohn TA, Kuo Y, Kalback W, Anthony J, Watson D, Leuhers DC, Walker D, Emmerling M, Goux W, Beach T. Increased Aβ peptide and reduced cholesterol and myelin proteins characterize white matter degeneration in Alzheimer’s Disease. Biochemistry. 2002;41:11080–11090. doi: 10.1021/bi026173d. [DOI] [PubMed] [Google Scholar]
- Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- Layfield R, Fergusson J, Aitken A, Lowe J, Landon M, Mayer RJ. Neurofibrillary tangles of Alzheimer's disease brains contain 14-3-3 proteins. Neurosci Lett. 1996;209:57–60. doi: 10.1016/0304-3940(96)12598-2. [DOI] [PubMed] [Google Scholar]
- Ho CS, Marinescu V, Steinhilb ML, Gaut JR, Turner RS, Stuenkel EL. Synergistic effects of Munc18a and X11 proteins on amyloid precursor protein metabolism. J Biol Chem. 2002;277:27021–27028. doi: 10.1074/jbc.M201823200. [DOI] [PubMed] [Google Scholar]
- Blass JP, Sheu KF, Cooper AJ, Jung EH, Gibson GE. Thiamin and Alzheimer's disease. J Nutr Sci Vitaminol. 1992;Spec No:401–404. doi: 10.3177/jnsv.38.special_401. [DOI] [PubMed] [Google Scholar]
- Yoo BC, Seidl R, Cairns N, Lubec G. Heat-shock protein 70 levels in brain of patients with Down syndrome and Alzheimer's disease. J Neural Transm. 1999;57:315–322. doi: 10.1007/978-3-7091-6380-1_22. [DOI] [PubMed] [Google Scholar]
- Renkawek K, Bosman GJ, Gaestel M. Increased expression of heat-shock protein 27KDa in Alzheimer disease: a preliminary study. Neuroreport. 1993;5:14–16. doi: 10.1097/00001756-199310000-00003. [DOI] [PubMed] [Google Scholar]
- Mao JJ, Katayama S, Watanabe C, Harada Y, Noda K, Yamamura Y, Nakamura S. The relationship between alpha-crystallin and neurofibrillary tangles in Alzheimer's disease. Neuropath Appl Neuro. 2001;27:180–188. doi: 10.1046/j.1365-2990.2001.00310.x. [DOI] [PubMed] [Google Scholar]
- Beffert U, Morfini G, Bock HH, Reyna H, Brady ST, Herz J. Reelin-mediated signaling locally regulates protein kinase B/Akt and glycogen synthase kinase 3β. J Biol Chem. 2002;277:49958–49964. doi: 10.1074/jbc.M209205200. [DOI] [PubMed] [Google Scholar]
- Ohkubo N, Lee YD, Morishima A, Terashima T, Kikkawa S, Tohyama M, Sakanaka M, Tanaka J, Maeda N, Vitek MP, Mitsuda N. Apolipoprotein E and Reelin ligands modulate tau phosphorylation through an apolipoprotein E receptor/disabled-1/glycogen synthase kinase-3β cascade. FASEB J. 2003;17:295–297. doi: 10.1096/fj.02-0434fje. [DOI] [PubMed] [Google Scholar]
- McKee AC, Kosik KS, Kennedy MB, Kowall NW. Hippocampal neurons predisposed to neurofibrillary tangle formation are enriched in type II calcium/calmodulin-dependent protein kinase. J Neuropath Exp Neur. 1990;49:49–63. doi: 10.1097/00005072-199001000-00006. [DOI] [PubMed] [Google Scholar]
- Perry G, Friedman R, Shaw G, Chau V. Ubiquitin is detected in neurofibrillary tangles and senile plaque neurites of Alzheimer disease brains. Proc Natl Acad Sci. 1987;84:3033–3036. doi: 10.1073/pnas.84.9.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusisto E, Salminen A, Alafuzoff I. Early accumulation of p62 in neurofibrillary tangles in Alzheimer's disease: possible role in tangle formation. Neuropath Appl Neuro. 2002;28:228–237. doi: 10.1046/j.1365-2990.2002.00394.x. [DOI] [PubMed] [Google Scholar]
- Choe SK, Vlachakis N, Sagerstrom CG. Meis family proteins are required for hindbrain development in the zebrafish. Development. 2002;129:585–595. doi: 10.1242/dev.129.3.585. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Bio. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- Okazaki N, Yan J, Yuasa S, Ueno T, Kominami E, Masuho Y, Koga H, Muramatsu M. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Mol Brain Res. 2000;85:1–12. doi: 10.1016/S0169-328X(00)00218-7. [DOI] [PubMed] [Google Scholar]
- Fung JJ, Starzl TE. FK506 in solid organ transplantation. Ther Drug Monit. 1995;17:592–595. doi: 10.1097/00007691-199512000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SL, Crabtree GR. The mechanism of action of cyclosporin A and FK506. Immunol Today. 1992;13:136–142. doi: 10.1016/0167-5699(92)90111-J. [DOI] [PubMed] [Google Scholar]
- Christner C, Herdegen T, Fischer G. FKBP ligands as novel therapeutics for neurological disorders. Mini-Reviews in Medicinal Chemistry. 2001;1:377–397. doi: 10.2174/1389557013406675. [DOI] [PubMed] [Google Scholar]
- Avramut M, Zeevi A, Achim CL. The immunosuppressant drug FK506 is a potent trophic agent for human fetal neurons. Dev Brain Res. 2001;132:151–157. doi: 10.1016/S0165-3806(01)00307-8. [DOI] [PubMed] [Google Scholar]
- Boutell JM, Thomas P, Neal JW, Weston VJ, Duce J, Harper PS, Jones AL. Aberrant interactions of transcriptional repressor proteins with the Huntington's disease gene product, huntingtin. Hum Mol Genet. 1999;8:1647–1655. doi: 10.1093/hmg/8.9.1647. [DOI] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Gregori L, Poosch MS, Cousins G, Chau V. A uniform isopeptide-linked multiubiquitin chain is sufficient to target substrate for degradation in ubiquitin-mediated proteolysis. J Biol Chem. 1990;265:8354–8357. [PubMed] [Google Scholar]
- Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- Wooten MW, Seibenhener ML, Mamidipudi V, Diaz-Meco MT, Barker PA, Moscat J. The atypical protein kinase C-interacting protein p62 is a scaffold for NF-kappaB activation by nerve growth factor. J Biol Chem. 2001;276:7709–7712. doi: 10.1074/jbc.C000869200. [DOI] [PubMed] [Google Scholar]


