Abstract
Promoter region hyermethylation and transcriptional silencing is a frequent cause of tumour suppressor gene (TSG) inactivation in many types of human cancers. Functional epigenetic studies, in which gene expression is induced by treatment with demethylating agents, may identify novel genes with tumour-specific methylation. We used high-density gene expression microarrays in a functional epigenetic study of 11 renal cell carcinoma (RCC) cell lines. Twenty eight genes were then selected for analysis of promoter methylation status in cell lines and primary RCC. Eight genes (BNC1, PDLIM4, RPRM, CST6, SFRP1, GREM1, COL14A1 and COL15A1) demonstrated frequent (>30% of RCC tested) tumour-specific promoter region methylation. Hypermethylation was associated with transcriptional silencing. Re-expression of BNC1, CST6, RPRM, and SFRP1 suppressed the growth of RCC cell lines and RNAi knock-down of BNC1, SFRP1 and COL14A1 increased the growth of RCC cell lines. Methylation of BNC1 or COL14A1 was associated with a poorer prognosis independent of tumour size, stage or grade. The identification of these epigenetically inactivated candidate RCC tumour suppressor genes can provide insights into renal tumourigenesis and a basis for developing novel therapies and biomarkers for prognosis and detection.
Keywords: Renal cell carcinoma, methylation, epigenetics
INTRODUCTION
Kidney cancers account for about 2% of all cancers and more than 200,000 new cases of kidney cancer are diagnosed in the world each year (Ferlay et al., 2007). The most common form of kidney cancer in adults is renal cell carcinoma (RCC). Most RCC (~75%) are classified as clear cell (conventional) and the next most frequent subtype is papillary RCC (~15% of all cases) (Mancini et al., 2008). The prognosis of advanced RCC is poor, although newer treatments, based on knowledge of the molecular pathology of RCC but characterisation of the molecular pathology of RCC may provide a basis for developing novel approaches to therapy. Thus the most frequent genetic event in the evolution of sporadic clear cell RCC is inactivation of the VHL tumour suppressor gene (TSG) (Clifford et al., 1998; Foster et al., 1994; Herman et al., 1994; Latif et al., 1993). VHL inactivation leads to stabilisation of HIF-1 and HIF-2 transcription factors and activation of a wide repertoire of hypoxic response genes (Maxwell et al., 1999). HIF-mediated RCC growth may be antagonised by multi tyrosine kinas inhibitors such as sunitinib and sorafenib (Chowdhury et al., 2008). Hence identification of mechanisms of tumourigenesis in RCC can provide a basis for therapeutic intervention. Although large scale mutation analysis studies of RCC are in progress (see http://www.sanger.ac.uk/genetics/CGP/cosmic/), with the exception of VHL, none of the thousands of genes tested to date are mutated in >15% of tumours. Epigenetic inactivation of TSGs by methylation promoter region of CpG dinucleotides has also been implicated in the pathogenesis of RCC and some important TSGs are frequently inactivated by promoter hypermethylation but rarely mutated (e.g. RASSF1A) (Dallol et al., 2002; Morris et al., 2003; Morrissey et al., 2001). Hence, strategies to identify epigenetically inactivated TSGs are an important approach to elucidating the molecular pathogenesis of RCC. Previously, in order to investigate the utility of a functional epigenomic approach to identify candidate novel epigenetically inactivated RCC TSGs, we performed a pilot study using gene expression profiling of four RCC cell lines treated with the demethylated agent 5-Aza-2′-deoxycytidine (Morris et al., 2008; Morris et al., 2005). This led us to identify HAI-2/SPINT2 as a novel epigenetically inactivated RCC TSG (Morris et al., 2005). We now report the results of a large functional epigenetic screen of RCC in which 11 RCC cell lines were analysed using a high density gene expression microarray platform.
METHODS
Patients and samples
DNA from up to 61 primary RCCs (~80% clear cell and 20% non-clear cell) and matched adjacent macroscopically normal renal tissue and normal renal tissue (not required for surgical pathology) from 6 patients undergoing non-renal cancer surgery (mean age 57 years, range from 23-79 years) were analysed. Local research ethics committees approved the collection of samples and informed consent was obtained from each patient. This study was conducted according to the principles expressed in the Declaration of Helsinki.
Cell lines, 5-Aza-2′-deoxycytidine treatment and microarray analysis
RCC cell lines KTCL 26, RCC4, UMRC2, UMRC3, SKRC18, SKRC39, SKRC45, SKRC47, SKRC54, 786-0 and Caki-1 were routinely maintained in DMEM (Invitrogen, San Diego, CA) supplemented with 10% FCS at 37°C, 5% CO2. The demethylating agent 5-Aza-2′-deoxycytidine (Sigma) was freshly prepared in dd H2O and filter sterilized. Cell lines were plated in 75-cm2 flasks in DMEM supplemented with 10% FCS at differing densities, depending upon their replication factor, to ensure that both control and 5-Aza-2′-deoxycytidine treated lines reached approximately 75% confluency at the point of RNA extraction. Twenty-four hrs later, cells were treated with 5 μM 5-Aza-2′-deoxycytidine. The medium was changed 24 hrs after treatment and then changed again after 72hrs. RNA was prepared 5 days after treatment using RNABee (AMS Biotechnology). Total RNA from all 11 cell lines +/−5-Aza-2′-deoxycytidine was isolated using RNA-Bee reagent following manufacturer's instructions (AMS Bio) followed by purification using RNeasy Mini-columns (Qiagen). cRNA probes were prepared using the Affymetrix protocol and hybridized to HG-U133 plus2 GeneChip oligonucleotide arrays (Affymetrix). Array hybridisation and data production was done by the CRUK Paterson Institute Microarray Service (http://bioinformatics.picr.man.ac.uk/mbcf/).
RT PCR conditions
PCR cycling conditions consisted of 5 min at 95°C followed by 30 cycles of 45 sec of denaturation at 95°C, 45sec of annealing at 55-60°C and 45sec of extension at 72°C. Semi-quantitative analysis of expression was done using LabWorks software (Ultraviolet products, California). (RTPCR primers and conditions upon request).
Bisulfite Modification and Methylation Analysis
Bisulfite DNA sequencing was performed as described previously (Morris et al., 2008; Morris et al., 2005). Briefly, 0.5–1.0 μg of genomic DNA was denatured in 0.3 M NaOH for 15 min at 37°C, and then unmethylated cytosine residues were sulfonated by incubation in 3.12 M sodium bisulfite (pH 5.0; Sigma)/5 mM hydroquinone (Sigma) in a thermocycler (Hybaid) for 20 cycles of 30 s at 99°C and 15 min at 50°C. The sulfonated DNA was recovered using the Wizard DNA cleanup system (Promega) in accordance with the manufacturer's instructions. The conversion reaction was completed by desulfonating in 0.3 M NaOH for 10 min at room temperature. The DNA was ethanol-precipitated and resuspended in water.
Promoter Methylation Analysis
CpG islands were identified on the human genome browser and putative promoter regions were predicted by Promoter Inspector software (Genomatix). Details of bisulphite sequencing primers and COL15A1 MSP primers are provided in supplementary table 1. Examples of direct tumour-DNA sequencing traces are shown in supplementary figure 1. Methylation specific PCR analysis for the following genes was carried out using previously described MSP primers: BNC1 (Shames et al., 2006), SFRP1 (Nomoto et al., 2007), REPRIMO (Sato et al., 2006).
Plasmid Constructs and Colony Formation Assay
The CST6, SFRP1 and REPRIMO expression constructs were made by cloning the full-length human coding regions amplified from kidney cell lines into the EcoR1-BamHII sites of pCDNA3.1 (Invitrogen) or pFLAG-CMV4 (Sigma-Aldrich) vectors. BNC1 was amplified from the I.M.A.G.E clone 40080551. Plasmid constructs were verified by sequencing. Six micrograms of empty vector and an equal Molar amount of expression vector were transfected, using Fugene (Roch), following the manufacturer's instructions, into 5 × 105 target cells (SKRC39, RCC4, 786-0, SKRC47 or HEK-293). Forty-eight hours after transfection, cells were seeded in a serial dilution and maintained in DMEM and 10% foetal bovine serum supplemented with 1 mg/mL G418 (Life Technologies). Surviving colonies were stained with 0.4% crystal violet (Sigma) in 50% methanol, 21 days after initial seeding, and counted. Each transfection was carried out in triplicate. Additionally, replicate experiments were carried out to obtain further clones for expression analysis. Expression was confirmed by RT-PCR and Western blot analysis.
Anchorage Independent Growth Assay
RCC clones stably expressing BNC1, CST6, REPRIMO or the empty vector control were suspended in 2ml DMEM 10% FCS, 3% agar. Cells were maintained by addition of 200 μl of DMEM 10% FCS, supplemented with 1mg/ml G418, weekly. After 5 weeks of growth a final count of colonies was performed. RNAi “silencer select” oligos against BNC1 (s2012), COL14A1 (s14677) and SFRP1 (s12713) or “Silencer select” control oligo no.1 (Ambion) were transfected into HEK-293 or KTCL26 cells using Interferin reagent (Polyplus) following the manufacture's instructions. After 24h incubation cells were seeded into 2ml DMEM 10% FCS, 3% agar. Cells were maintained by addition of 200 μl of DMEM 10% FCS weekly. After 3 weeks of growth a final count of colonies was performed. Cells not seeded into agar were incubated for a further 24h before efficiency of knock-down was assessed by RT-PCR.
Statistical analysis was performed as indicated with a significance level of 5%.
RESULTS
Identification of candidate silenced genes involved in RCC
Eleven RCC-derived cell lines were treated with the demethylating agent 5-Aza-2′-deoxycytidine (5 μM) for 5 days to re-activate epigenetically silenced/down-regulated genes and changes in gene expression were measured by Affymetrix U133 plus-2 microarrays that contain probes for >47000 unique transcripts. In order to prioritise genes for methylation analysis, we initially focussed on genes which showed a ten-fold increase in expression following de-methylation in three or more cell lines (RT-PCR analyses demonstrated a good correlation with microarray based estimates of changes in gene expression (see Supplementary Fig 1, Fig 1A and Fig 2A)), Transcripts that represented hypothetical proteins were also removed. 405 unique genes were left following this filter. Next we excluded genes that did not have a predicted promoter specific CpG island (as predicted by the human genome browser (www.genome.ucsc.edu) and Genomatix promoter inspector (www.genomatix.de)), 201 genes were left following this filter step. Genes known to be imprinted and genes that are not expressed in renal tissue (Array express (www.ebi.ac.uk/arrayexpress/)) were also removed, leaving 155 genes. Of these, 19 genes have previously been assessed for promoter methylation in RCC including KRT19, COL1A1 and IGFBP1 that are frequently methylated in RCC (banez de Caceres et al., 2006; Morris et al., 2008) (Supplementary table 2 lists the genes identified by this filtering process in full).
Figure 1.
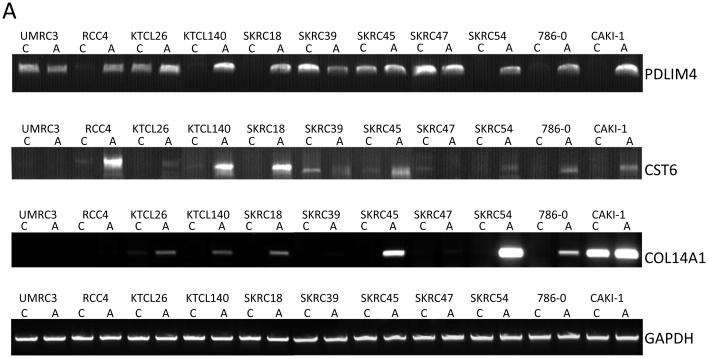
A, RT-PCR analysis of PDLIM4, CST6 and COL14A expression. PDLIM4 expression was restored (SKRC18, SKRC54, CAKI-1)/up-regulated (RCC4, KTCL140, 786-0) in 6 of 11 cell lines following treatment with the demethylating agent, 5-Aza-2′-deoxycytidine. CST6 expression was restored (KTCL26, SKRC18, SKRC54, 786-0, CAKI-1)/up-regulated (RCC4, KTCL140, SKRC45) in 8/11 cell lines. Global demethylation restored expression of COL14A in 5/11 cell lines (KTCL140, SKRC18, SKRC45, SKRC54, 786-0).
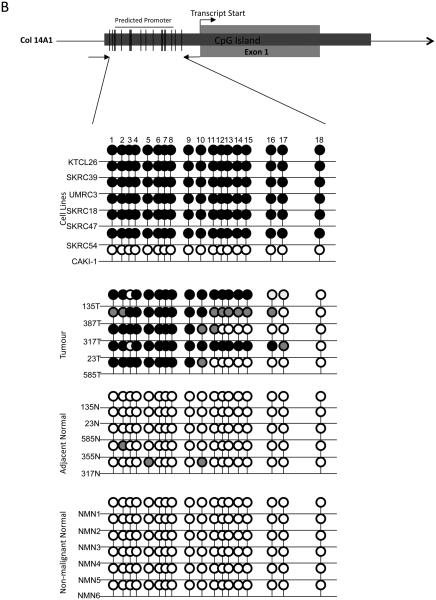
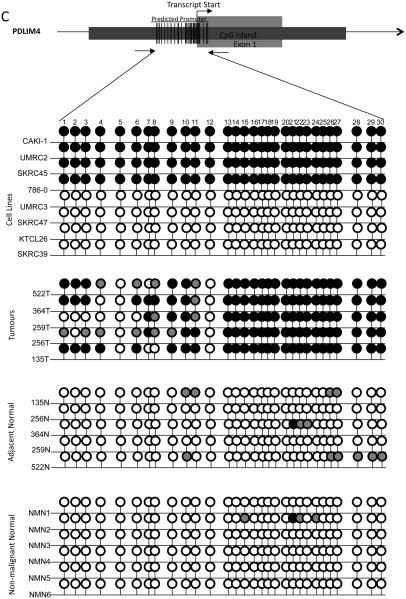
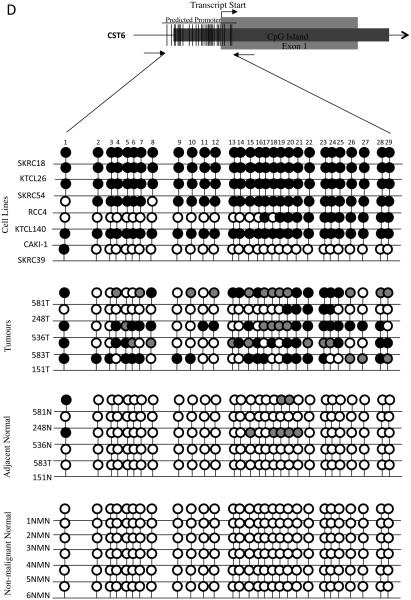
B-D, Bisulphite sequencing analysis of PDLIM4, CST6 and COL14A1 5′ CpG island regions. Primers were designed to amplify the predicted promoter region of the 5′ CpG island of PDLIM4, CST6 and COL14A. The schematic represents the region sequenced; vertical lines indicate individual CpGs. Sequencing indicated methylation in RCC cell lines correlated with gene silencing/ down-regulation: black circle; complete methylation; grey circle; partial methylation; empty circle; no methylation. Bisulfite sequencing of RCC tumours and adjacent normal tissue showed frequent methylation in tumours and infrequent methylation in normal tissue (see results for details). Analysis of normal tissue derived from non-malignant kidneys indicated no CpG island methylation, with the exception of PDLIM4 in which 3 CpGs were partially and 1 CpG was fully methylated in 1 non-malignant sample (NMN2). T=Tumour, N= normal tissue adjacent to tumour, NMN= non-malignant normal tissue. Representative sequencing traces are shown in supplementary fig. 1.
Figure 2.
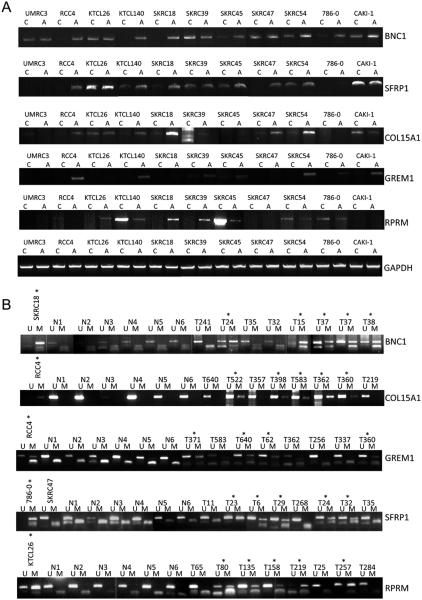
A, RT-PCR analysis of BNC1, SFRP1 and COL15A1, GREM1, and RPRM expression. BNC1 expression was restored (RCC4, KTCL140, SKRC18)/up-regulated (SKRC45, 786-0) in 5 of 11 cell lines following treatment with the demethylating agent, 5-Aza-2′-deoxycytidine. SFRP1 expression was restored (RCC4, SKRC47)/up-regulated (KTCL140, SKRC18, SKRC45) in 5/11 cell lines. COL15A1 expression was restored (RCC4)/up-regulated (KTCL140, SKRC18, SKRC47, SKRC54) in 5/11 cell lines. GREM1 expression was restored (RCC4, SKRC140, SKRC54, CAKI-1)/up-regulated (SKRC39) in 5/11 cell lines following treatment with the demethylating agent, 5-Aza-2′-deoxycytidine. Global demethylation restored expression of RPRM in 3/11 cell lines (KTCL26, SKRC18, SKRC39).
B, Methylation-specific PCR analysis of BNC1, SFRP1 and COL15A, GREM1, and RPRM1. MSP analysis showed frequent (34-53%) promoter methylation in tumours, infrequent or absent methylation in adjacent tissue and no methylation in samples derived from non-malignant normal tissue (see results for details). RCC tumour cell lines were used for each MSP assay as positive controls for methylated and unmethylated DNA. Representative samples are shown. M=product derived from methylation-specific primers; U= products derived from unmethylated specific primers, T= tumour sample, N= non-malignant sample, *=methylated tumours. MSP and USP product are of a similar size. Smaller bands are primer dimmers.
To evaluate the microarray fold-change filtering selection criteria we analysed the methylation status of 3 genes (LRCH1, KLF2 FZD8) that had expression fold-changes of <10 (range 2 to 7-fold). All three genes were unmethylated in the cell lines (supplementary table 1).
24 genes from our shortlist were analysed for promoter methylation in RCC cell lines and primary tumours. In addition we analysed promoter methylation status of RPRM which, although not meeting the selection criteria had previously been shown to be inactivated by methylation in a number of other tumour types (Bernal et al., 2008; Sato et al., 2006; Takahashi et al., 2005). One of the 25 genes (NOG) did not demonstrate detectable promoter region hypermethylation. Six genes (DMRTB1, SCYA20, CLDN6, TPM2L1, LOXL and ZFP42) were methylated in RCC cell lines and tumours but also in non-malignant renal tissue and so were not investigated further. A further 10 genes demonstrated both promoter region methylation in RCC cell lines but no (FOXG1B, RARRES1, DKK1 and UQCRH) or infrequent (RGPD5 (7%), ITGBL (9%), KLF4 (10%), TLL1 (10%), PTGIS (13%) and EMX2 (18%)) promoter methylation in primary RCC tumours.
Eight of 28 (29%) genes (BNC1, COL14A1, COL15A1 CST6, SFRP1, GREM1, RPRM and PDLIM4) demonstrated frequent (methylation frequency >30%, range 35-53%) methylation in primary RCC, no methylation in normal kidney from non-RCC patients and upregulation of gene expression following demethylation of methylated RCC cell lines (Table 1, Fig 1A and 2A). Five of 8 genes were methylated in renal tumours but not (or rarely) in matched normal renal tissue (BNC1, COL14A1, CST6, PDLIM4 and SFRP1).
Table 1.
A summary of genes identified in this study and their frequency of promoter methylation in RCC-derived cell lines, sporadic tumours, kidney tissue resected from an area adjacent to the tumour and kidney tissue from patients with no kidney cancer.
| Gene Symbol |
Accession no. |
Gene Name | Loci | Cell line methylation frequency |
Tumour methylation frequency |
Adjacent kidney methylation frequency |
Non-malignant kidney methylation frequency |
|---|---|---|---|---|---|---|---|
| COL15A1 | NM_001855 | Collagen, type Xv, alpha 1 | 9q22 | 7/9 | 35/65 (53%) | 9/30 (30%) | 0/6 |
| GREM1 | NM_204978 | Gremlin-1 | 15q13 | 7/11 | 11/27(41%) | 7/29 (24%) | 0/6 |
| COL14A1 | NM_021110 | Collagen, type XIV, alpha 1 | 8q24 | 7/11 | 18/41 (44%) | 1/20 (5%) | 0/6 |
| PDLIM4 | NM_003687 | PDZ and LIM domain 4 | 5q31 | 5/10 | 13/30 (43%) | 0/22 (0%) | 1/6 |
| RPRM | NM_019845 | Reprimo, TP53 dependant G2 arrest mediator | 2q23 | 4/9 | 23/52 (44%) | 8/44 (18%) | 0/6 |
| SFRP1 | NM_003012 | Secreted frizzled-related protein 1 | 8p11 | 5/10 | 20/58 (34%) | 0/20 (0%) | 0/6 |
| CST6 | NM_001323 | Cystatin E/M | 11q13 | 8/11 | 28/61 (46%) | 4/35 (11%) | 0/6 |
| BNC1 | NM_001717 | Basonuclin 1 | 15q25 | 5/11 | 27/59 (46%) | 1/20 (5%) | 0/6 |
The promoter region of the 5′ CpG island of BNC1 was methylated in 46% (27/59) of primary RCC (Fig 2B). One of 20 (5%) adjacent normal samples (from a patient with a methylated tumour) also demonstrated promoter methylation. Methylation of the COL14A1 promoter was detected in 44% (18/41) primary RCC tumours analysed (1/20 adjacent normal kidney samples (from a patient with a methylated tumour) and 0/6 samples from patients without kidney cancer) (Fig 1B). Methylation of PDLIM4 promoter was detected in 43% (13/30) primary RCC but not in any normal renal tissue samples (n=22) (Fig 1C). SFRP1 promoter methylation was present in 34% (20/58) sporadic primary RCC but not in adjacent normal tissue samples (Fig 2B). The CST6 predicted promoter was methylated in 46% (28/61) primary RCC. A small number of normal kidney samples adjacent to RCC (11%, 4/35) demonstrated a low level of promoter methylation (all of which showed methylation in the matched tumour tissue) (Fig 1D).
Three genes, COL15A1, RPRM and GREM1 demonstrated frequent promoter methylation in primary RCC (53% (34/65), 44% (23/52) and 41% (11/27) respectively) (Fig 2B), but methylation was also detected relatively frequently in macroscopically normal renal tissue from patients with RCC (30% (9/30), 18% (8/44), 24% (7/29) respectively (P=0.034, P= 0.009 and P=0.025 respectively compared to primary RCC). However there was a strong correlation between the presence of methylation in normal renal tissue and in tumour material. Thus 8/9 normal renal tissue with COL15A1 methylation in normal tissue also displayed methylation in the corresponding tumour and the corresponding figures for RPRM and GREM1 were 7/8 and 5/7.
Functional Analysis of the tumour suppressor activity of epigenetically inactivated genes
The effect of re-expressing four of the epigenetically silenced RCC genes (SFRP1, RPRM, CST6, and BNC1) on in vitro growth characteristics of RCC cell lines was assessed using colony formation and anchorage independent growth assays.
Colony formation
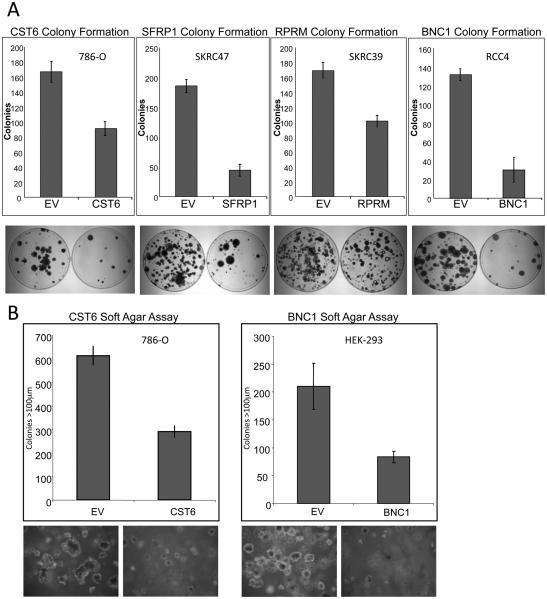
The tumour suppressor activity of SFRP1, RPRM, CST6, and BNC1 was assessed by in vitro colony formation assays. Following transfection of wild-type BNC1, CST6, RPRM and SFRP1 expression plasmids into non-expressing RCC cell lines (RCC4, 786-0, SKRC39 and SKRC47 respectively) there was a significantly reduced number of G418 resistant colonies compared to cell lines transfected with an empty vector control for 3 independent experiments (Fig 3A).
Figure 3.
A, Re-expression of CST6, SFRP1, RPRM and BNC1 in RCC cells results in growth suppression. Exogenous re-expression of selected genes in RCC cell lines that do not express their respective genes resulted in a significant reduction of in vitro colony formation compared to RCC lines transfected with an empty vector (EV). Equal amounts of empty vector or pCDNA3.1-wtCST6, or pCDNA3.1-wtSFRP1, or pCDNA3.1-wtRPRM, or pCDNA3.1-wtBNC1 were transfected into 786-O (CST6), SKRC47 (SFRP1), RCC4 (BNC1) or SKRC39 (RPRM) cells. Each experiment was done in triplicate. There was a statistically significant reduction of colonies in each of the re-expression experiments (p=0.003, p<0.0001, p<0.0001 and p=0.001 respectively). Shown below each graph are representative plates showing reduction of colonies following gene re-expression.
B, Re-expression of CST6 or over-expression of BNC1 inhibits anchorage-independent growth. A, clones of 786-O-pCDNA3.1 and 786-0O-pCDNA3.1-wtCST6 or HEK293-pCDNA3.1 and HEK293-pCDNA3.1-wtBNC1were seeded at the same density into soft agar and incubated for 5 weeks. 786-O clones not expressing CST6 (pCDNA3.1) produced many large (>100 μm) colonies. In contrast, 786-O clones expressing exogenous wtCST6 did not grow as robustly. Three independent experiments showed a 53% reduction of large colonies (p=0.0001) after 5 weeks of incubation. Similarly exogenous over-expression of BNC1 resulted in 54% fewer large colonies (p=0.001). Below each graph is shown a representative image of clones after 5 weeks of incubation (X100 magnification).
The number of BNC1 expressing RCC4 clones was reduced by 77% compared to those transfected with an empty plasmid (SD=10%, t=12.43, P<0.0001). Re-expression of CST6 in 786-0 cells reduced their colony forming ability by 47% (SD=6%, t=6.629, P=0.003). RPRM expressing SKRC39 clone numbers were reduced by 40% (SD=5%, t=8.328, P=0.001). The number of SFRP1 expressing SKRC47 clones was reduced by 76% compared to SKRC47 clones containing the empty vector (SD=5%, t=16.644, P<0.0001) (Fig. 3A).
Anchorage independent growth
The effect of re-expression of, RPRM and CST6 on anchorage-independent growth in a soft agar colony formation assay was assessed in stably transfected RCC cell line clones. Colonies were counted after initial seeding and incubation in soft agar for 5 weeks. Each experiment was done in triplicate with 3 independent clones. Cells transfected with empty vector showed robust colony growth, whereas, colony growth was greatly reduced when CST6 was re-expressed in 786-0 cells, both the number and size of colonies was statistically significantly reduced The number of colonies ≥100μm was reduced by 53% (SD=4%, t=16.15, P<0.0001) in clones expressing CST6 when compared to the control clones (Fig. 3B).
Re-expression of RPRM in SKRC39 cells did not significantly alter the anchorage independent growth capabilities of SKRC39 cells. Both clones re-expressing RPRM and those with RPRM silenced grew robustly when suspended in soft agar.
As the BNC1 silenced RCC4 cell line did not grow well in an anchorage independent manner, to investigate the effect of alterations of BNC1 expression on cell growth we produced BNC1 over-expressing clones of HEK293s. Whereas HEK293 cells transfected with an empty vector grew robustly, BNC1 overexpressing clones produced statistically significantly fewer colonies ≥100μm (mean (±SD) reduction 54% (±5%), t=5.92 P=0.001) (Fig 3B).
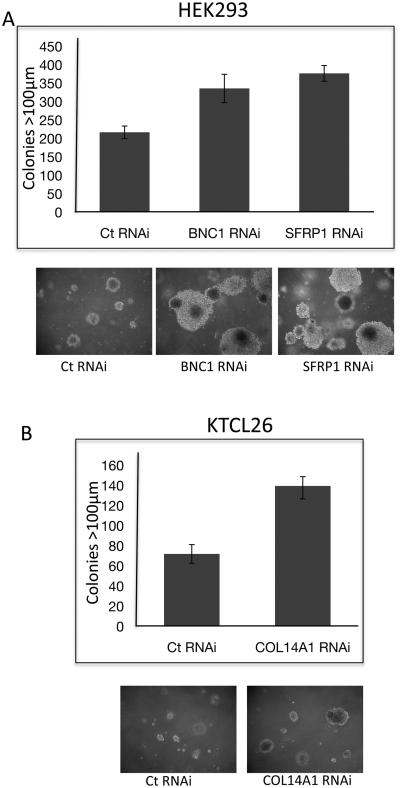
To investigate the possible tumourigenic advantage of loss of expression of BNC1, SFRP1 and COL14A we utilised RNAi methodology to knock down BNC1 and SFRP1 expression in HEK293s and COL14A expression in the KTCL26 RCC cell line (sufficient knockdown of COL14A expression was obtained in HEK293 cells). Twenty-four hours after RNAi transfection cells were seeded into 3% agar, colonies >100μm were counted 21 days later. 55% more colonies >100μm were produced by HEK293 cells with reduced BNC1 (SD=11%, t=−6.9, p<0.0001) expression compared to cells transfected with a control RNAi oligo (Fig 4A). Reduced expression of SFRP1 resulted in 74% more colonies >100μm (SD=6%, t=−14.3, p<0.0001) than control cells (Fig 4A). The growth suppressing activity of re-expression of SFRP1 in RCC cell lines has been previously demonstrated (Gumz et al., 2007). However, to our knowledge, this is the first time that an increase in growth potential has been associated with reduced SFRP1 expression in RCC. The colony forming capability of the RCC cell line KTCL26 was less than that of HEK293s. However, this was increased by 93% in those cells where COL14A expression was reduced by RNAi (SD=9%, t=−10.44, p<0.0001) (Fig 4B).
Figure 4.
Knock-down of expression of BNC1, SFRP1 or COL14A1 increases anchorage-independent growth potential. A. RNAi-induced reduced expression of BNC1 or SFRP1 in HEK-293 cells resulted in the growth of significantly more colonies >100μm in diameter compared to cells transfected with a control RNAi oligo when seeded at the same density into soft agar (p<0.0001 in both cases). B. RNAi-induced reduced expression of COL14A1 in KTCL26 cells resulted in the growth of significantly more colonies >100μm in diameter compared to cells transfected with a control RNAi oligo when seeded at the same density into soft agar (p<0.0001).
Analysis of patient survival and gene methylation
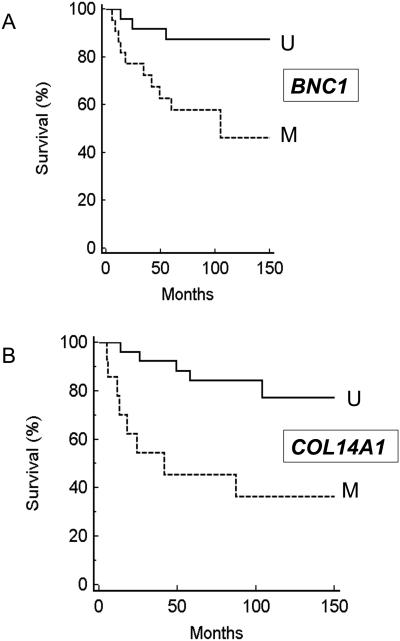
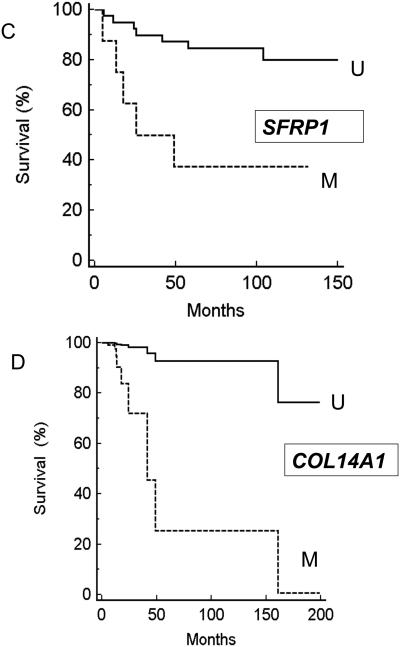
We initially investigated whether the presence of tumour methylation for BNC1, PDLIM4, RPRM, CST6, SFRP1, GREM1, COL14A1 and COL15A1) was associated with changes in patient survival (time to cancer death) or recurrence/cancer death. No significant association was detected for CST6 (P=0.38 and P=0.76 respectively by Kaplan-Meier analysis), COL15A1 (P=0.25 and P=0.16), GREM1 (0.07 and 0.08), RPRM (P=0.83 and 0.94), or PDLIM4 (P=0.38 and 0.32). However methylation of BNC1 (P=0.017 (Hazard ratio (HR)= 3.69 (95%CI= 1.27-10.93) and P=0.018 (HR=3.07 (95%CI= 1.22-8.16)), COL14A1 (P=0.005 (HR= 4.07 (95%CI= 1.73-20.3) and P=0.007 (HR=3.49 (95%CI= 1.55-16.1)) or SFRP1 (P=0.002 (HR=4.88 (95%CI= 2.76-87.95)) and P=0.003 (HR=4.44 (95%CI= 2.75-86.2)) was associated with a significantly poorer prognosis (see Figure 4A-C). We then investigated the relationship between survival, tumour grade, size and stage and methylation status of BNC1, COL14A1 and SFRP1 using Cox proportional hazard analysis. Analysis for SFRP1 methylation, grade, size and stage gave a strong association with survival (P=0.0002) but the effect of SFRP1 methylation status was not statistically significant (P=0.4). However for a similar analysis for BNC1, tumour stage (P=0.019; HR=2.4536 (95% confidence interval (95%CI)= 1.16-5.19)), BNC1 methylation (P=0.033; HR=4.87 (95%CI 1.14- 20.888) and tumour size (p=0.04; HR=1.23 (95%CI= 1.009-1.49) were each significantly associated with prognosis, and under a similar analysis for COL14A1, methylation of COL14A1 (P=0.0067) (Hazard ratio (HR)= 6.56 (95% CI 1.69 to 25.38) and tumour stage (p=0.0009) (HR=3.42 (95%CI 1.67 to 7.02) were identified as significant prognostic factors. A combined analysis was then performed for survival and BNC1 and COL14A1 methylation status, tumour grade, size and stage. Methylation of COL14A1 (P=0.0067; HR=18.37 (95%CI=2.27-148.8)) was the most significant predictor of survival, followed by stage (P=0.015; HR=3.5 (95%CI=1.28-9.57) and grade (P=0.08; HR=2.03 (95%CI=0.92-4.46) (BNC1 methylation status was not a significant prognostic factor (P=0.38) (Figure 4D).
There were no significant differences in BNC1, PDLIM4, RPRM, CST6, SFRP1, GREM1, COL14A1 and COL15A1 promoter methylation frequencies bwtween clear cell RCC and non-clear cell RCC) and no significant differences between methylation status of VHL mutated and VHL wild-type clear cell RCC.
DISCUSSION
Cancer specific genetic and epigenetic alterations resulting in tumour suppressor gene (TSG) inactivation are frequent events in RCC. However, whereas the mutation spectrum causing TSG inactivation is usually diverse (so limiting the utility for tumour screening programmes of detecting an individual TSG mutation), TSG inactivation by promoter hypermethylation provides a more homogeneous target for molecular screening strategies. Furthermore, large scale gene sequencing studies of RCC and other cancers have revealed that relatively few genes are mutated frequently. Thus, apart from VHL, there are no reports of a TSG that is frequently mutated in RCC (http://www.sanger.ac.uk/genetics/CGP/cosmic/). In contrast, at least 10 genes have been reported to be hypermethylated >20% of RCC (13 and references within) and we have now reported a further 8 genes that demonstrate promoter methylation in >30% of RCC.
Functional epigenomic screens provide a useful strategy to identify novel TSGs and have been employed to identify epigenetically inactivated TSGs in a number of different tumour types (Lodygin et al., 2005; Sato et al., 2003; Shames et al., 2006; Yamashita et al., 2002), including two studies of RCC (Ibanez de Caceres et al., 2006; Morris et al., 2008; Morris et al., 2005). Previously we identified SPINT2 as a novel RCC TSG (Morris et al., 2005), but many candidate TSGs upregulated after treatment of RCC cell lines with 5-Aza-2′-deoxycytidine do not show tumour-specific promoter methylation (Morris et al., 2008). Therefore in order to maximise the potential for identifying additional genes epigenetically silenced in RCC we extended our previous study (Morris et al., 2008; Morris et al., 2005) to include more cell lines (11 versus 4) and higher density microarrays (47000 transcripts versus 11000 genes). Seven of the 8 genes we report in this study (BNC1, RPRM, CST6, PDLIM4, GREM1, COL14A1 and COL15A1) have not previously been reported to be methylated in RCC, and, to our knowledge, COL14A1 and COL15A1 have not previously been reported to be methylated in neoplasia. While this study was in progress SFRP1 was reported to be methylated in RCC (Awakura et al., 2008; Dahl et al., 2007; Gumz et al., 2007)
SFRP1 encodes a negative regulator the Wnt/beta-catenin pathway. Thus SFRP1 molecules bind to and sequester Wnt molecules away from their cognate receptors, the Frizzled family. Activation of the Wnt/beta-catenin pathway is associated with upregulation of a range of oncogenic targets (e.g. Cyclin D1, VEGF, cMYC and cMET) implicated in the pathogenesis of cancer. We detected SFRP1 promoter methylation in 34% of RCC compared to 68% in the study of Dahl et al (Dahl et al., 2007). Re-expression of SFRP1 in a null RCC-cell line has previously been shown to associate with reduced tumour cell growth (Gumz et al., 2007). We confirmed this and also demonstrated that reduced expression of endogenous SFRP1 increases the tumourogenic potential of an RCC-cell line.
RPRM (Reprimo, TP53 dependent G2 arrest mediator candidate) is a downstream mediator of p53-induced G2 cell cycle arrest (Ohki et al., 2000). Reprimo induced cell cycle arrest is thought to be mediated by an indirect inhibition of Cdc2-CyclinB1 complex translocation to the nucleus (Ohki et al., 2000). Epigenetic silencing of RPRM has previously been reported in a variety of cancers including gastric, gall bladder, colorectal, oesophageal, breast and prostate (Bernal et al., 2008; Sato et al., 2006; Takahashi et al., 2005). We have now demonstrated that it is also frequently methylated in RCC and that re-expression RPRM reduces the tumour forming properties of RCC-derived cell lines.
CST6 is a type 2 secreted cystatin. It inhibits Cathepsin B, L, H and V, members of a family of lysosomal proteases. Cathepsins are involved in multiple biological processes such as apoptosis, intracellular protein catabolism and pericellular matrix re-modelling (Bromme & Kaleta, 2002). It has been suggested that an imbalance of cathepsins and their respective inhibitors may promote tumour cell invasion (Bellail et al., 2004) and so loss of CST6 expression by epigenetic silencing might be predicted to contribute to the malignant phenotype. Consistent with this we found that re-expression of CST6 reduced the ability of RCC cells to form colonies in an anchorage-independent manner. CST6 has also been shown to be epigenetically silenced and reduced in vitro tumourigenicity in breast cancer (Ai et al., 2006; Shridhar et al., 2004) and gliomas (Qiu et al., 2008).
PDLIM4 encodes a LIM domain candidate tumour gene mapping to 5q31.1. Although methylation of PDLIM4 has not been reported previously in RCC tumours, Boumber et al (Boumber et al., 2007) reported frequent PDLIM4 promoter methylation in acute myelogenous leukaemia and colon cancer. Gremlin (GREM1) is an inhibitor of TGFbeta signalling that has previously been reported to be frequently methylated in lung, breast and bladder cancers (Suzuki et al., 2005). We found frequent methylation of GREM1 suggesting that this may contribute to the perturbed TGFbeta signalling that is detected in many human cancers, including RCC. GREM1 methylation was detected in some samples of adjacent normal renal tissue from RCC patients with GREM1 tumour methylation. Similar findings were obtained for RPRM and COL15A1, these results would be consistent with the hypothesis that GREM1, RPRM and COL15A1 methylation might occur as part of a premalignant field effect, as has been described in bronchial epithelium (Wistuba, 2007).
Basonuclin (BNC1) is a zinc finger transcription factor that interacts with the promoters of both RNA polymerases I and II. In silico analysis suggests that basonuclin target genes may be implicated in chromatin structure, transcription/DNA-binding, adhesion/cell-cell junction, signal transduction, and intracellular transport (Wang et al., 2006). Shames et al (Shames et al., 2006) reported that BNC1 was methylated in breast, lung, prostate and colon cancers, suggesting that BNC1 methylation might be of relevant to the diagnosis and management of a range of human cancers.
Ibanez et al. have reported epigenetic silencing of COL1A1 in RCC (Ibanez de Caceres et al., 2006). In this study we identified frequent methylation of COL15A1 and COL14A1 in RCC. COL15A1 encodes a nonfibrillary proteoglycan which is found in many tissue types forming an integral unit of the collagenous network subjacent to the basement membrane (Amenta et al., 2005). The COOH-terminal end of COL15A1 which has homology to endostatin has been shown to have anti-angiogenic properties and is capable of inhibiting tumour growth in a xenograph renal model (Ramchandran et al., 1999). Moreover, recently COL15A1 was identified in a fibroblast-tumour cell hybrid screen as a potent suppressor of tumour growth, and re-expression of COL15A1 in a HeLa derivative cell line completely suppressed tumour formation in nude mice (Harris et al., 2007). We have identified frequent COL15A1 methylation in RCC (53%). Collagen XIVA1 (COL14A1) is a large extracellular matrix glycoprotein that associate with mature collagen fibrils (Schuppan et al., 1990). Although methylation of COL14A1 has not previously been reported in human cancer, Schuppan et al (1990) noted frequent absence of COL14A1 in the vicinity of invading tumours such as Kaposi sarcoma and oral squamous cell carcinoma. In addition we note data from the Sanger cancer genome re-sequencing project (http://www.sanger.ac.uk/genetics/CGP/Studies/) that identifies COL14A1 mutations in ~1% (n=4) of RCC sequenced (n=412). COL14A1 interacts with Decorin (Ehnis et al., 1997) which regulates fibrillogenesis and downregulates activity of a number of receptors (EGFR, IGF-IR, LRP) implicated in cell growth and survival (Brandan et al., 2006; Santra et al., 2002; Schaefer et al., 2007).
Loss of expression of epigenetically silenced TSGs can affect a wide variety of cellular processes, including those which directly affect cell growth and proliferation. We investigated whether re-expressing SFRP1, RPRM, CST6 and BNC1 in RCC cell lines would influence in vitro analyses of tumourigenesis. BNC1 has not previously been demonstrated to affect tumour cell growth, but we found that re-expression of BNC1 suppressed tumour cell growth. Similar results were found for RPRM, CST6 and SFRP1. Although RPRM and CST6 have not previously been studied in RCC these findings are consistent with those reported in cervical and breast cancer cell lines (Ohki et al., 2000; Shridhar et al., 2004). We have also shown that reducing the expression of BNC1, SFRP1 and COL14A1 in an embryonic kidney cell line or an RCC-cell line increased tumourogenic potential as assessed by soft agar colony formation assays.
The methylated genes identified in this and previous studies of RCC (Ibanez de Caceres et al., 2006; Morris et al., 2008; Morris et al., 2005; Morris et al., 2003), represent a variety of functions. However we note that many of these genes can be linked to pathways associated with the VHL TSG that has a gatekeeper function for RCC (analogous to that of APC in colorectal cancer). The VHL gene product has a key role in regulating the hypoxia inducible transcription factors HIF-1 and HIF-2 and, in turn, these influence a wide repertoire of target genes. Although dysregulation of hypoxia-inducible genes (particularly those regulated by HIF-2) is strongly linked to risk of RCC with VHL inactivation, HIF-dysregulation appears to be necessary but insufficient for VHL-related RCC tumourigenesis (Clifford et al., 2001; Raval et al., 2005). A number of HIF-independent functions have been ascribed to pVHL. In the context of COL14A1 and COL15A1 methylation it is interesting to note that analysis of Caenorhabditis elegans worms mutant for vhl-1 demonstrated a clear HIF-independent link between pVHL and extracellular matrix function (Bishop et al., 2004). Recently, pVHL has been reported to regulate the p53 pathway (relevant to RPRM) and the Wnt/beta catenin pathway (downstream of SFRP1) by targeting beta catenin for proteasomal degradation (Chitalia et al., 2008).
The identification of frequently methylated RCC TSGs can highlight critical pathways that might be targeted for novel therapeutic interventions. In addition, the detection of methylated DNA in urine or serum might be used as a biomarkers for the diagnosis, staging or risk stratification of RCC (Battagli et al., 2003; Hoque et al., 2004; Urakami et al., 2006). VHL inactivation occurs early in tumourigenesis but, to date, has not been demonstrated to be a significant prognostic indicator (Smits et al., 2008). Thus, given the infrequency of mutations in other TSGs in RCC, detection of TSG methylation would appear to presents a promising strategy for prognostic biomarkers in RCC. However, only a few potential RCC methylation prognostic biomarkers have been reported (Breault et al., 2005; Costa et al., 2007; Christoph et al., 2006; Gonzalgo et al., 2004; McRonald et al., 2009; Yamada et al., 2006). We found that methylation of BNC1, COL14A1 and SFRP1 was each associated with significantly poorer patient survival. However for SFRP1 this association was not independent of conventional prognostic factors (stage, size and grade). In multivariate analysis with conventional prognostic factors, methylation of COL14A1 and BNC1 were each significant predictors of poorer survival. Combining the COL14A1 and BNC1 methylation data identified COL14A1 methylation as an independent prognostic factor with higher statistical significance than stage, size and grade.
Supplementary Material
Figure 5.
Panel A: Kaplan Meier survival analysis for BNC1 methylation status and Survival (M=methylated U=Unmethylated); Panel B: Kaplan Meier survival analysis for COL14A1 methylation status and survival (M=methylated U=Unmethylated); panel C: Kaplan Meier survival analysis for BNC1 methylation status and survival (M=methylated U=Unmethylated). Panel D: Cox proportional hazard analysis for COL14A1 methylation status (analysis also incorporates tumour stage. Grade and size and BNC1 methylation status).
ACKNOWLEDGEMENTS
We thank Cancer Research UK for financial support.
References
- Ai L, Kim WJ, Kim TY, Fields CR, Massoll NA, Robertson KD, Brown KD. Cancer Res. 2006;66:7899–909. doi: 10.1158/0008-5472.CAN-06-0576. [DOI] [PubMed] [Google Scholar]
- Amenta PS, Scivoletti NA, Newman MD, Sciancalepore JP, Li D, Myers JC. J Histochem Cytochem. 2005;53:165–76. doi: 10.1369/jhc.4A6376.2005. [DOI] [PubMed] [Google Scholar]
- Awakura Y, Nakamura E, Ito N, Kamoto T, Ogawa O. Oncol Rep. 2007;20:1257–63. [PubMed] [Google Scholar]
- Battagli C, Uzzo RG, Dulaimi E, Ibanez de Caceres I, Krassenstein R, Al-Saleem T, Greenberg RE, Cairns P. Cancer Res. 2003;63:8695–9. [PubMed] [Google Scholar]
- Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG. Int J Biochem Cell Biol. 2004;36:1046–69. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Bernal C, Aguayo F, Villarroel C, Vargas M, Diaz I, Ossandon FJ, Santibanez E, Palma M, Aravena E, Barrientos C, Corvalan AH. Clin Cancer Res. 2008;14:6264–9. doi: 10.1158/1078-0432.CCR-07-4522. [DOI] [PubMed] [Google Scholar]
- Bishop T, Lau KW, Epstein AC, Kim SK, Jiang M, O'Rourke D, Pugh CW, Gleadle JM, Taylor MS, Hodgkin J, Ratcliffe PJ. PLoS Biol. 2004;2:e289. doi: 10.1371/journal.pbio.0020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumber YA, Kondo Y, Chen X, Shen L, Gharibyan V, Konishi K, Estey E, Kantarjian H, Garcia-Manero G, Issa JP. Cancer Res. 2007;67:1997–2005. doi: 10.1158/0008-5472.CAN-06-3093. [DOI] [PubMed] [Google Scholar]
- Brandan E, Retamal C, Cabello-Verrugio C, Marzolo MP. J Biol Chem. 2006;281:31562–71. doi: 10.1074/jbc.M602919200. [DOI] [PubMed] [Google Scholar]
- Breault JE, Shiina H, Igawa M, Ribeiro-Filho LA, Deguchi M, Enokida H, Urakami S, Terashima M, Nakagawa M, Kane CJ, Carroll PR, Dahiya R. Clin Cancer Res. 2005;11:557–64. [PubMed] [Google Scholar]
- Bromme D, Kaleta J. Curr Pharm Des. 2002;8:1639–58. doi: 10.2174/1381612023394179. [DOI] [PubMed] [Google Scholar]
- Chitalia VC, Foy RL, Bachschmid MM, Zeng L, Panchenko MV, Zhou MI, Bharti A, Seldin DC, Lecker SH, Dominguez I, Cohen HT. Nat Cell Biol. 2008;10:1208–16. doi: 10.1038/ncb1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Larkin JM, Gore ME. Eur J Cancer. 2008;44:2152–61. doi: 10.1016/j.ejca.2008.06.028. [DOI] [PubMed] [Google Scholar]
- Christoph F, Weikert S, Kempkensteffen C, Krause H, Schostak M, Kollermann J, Miller K, Schrader M. Clin Cancer Res. 2006;12:5040–6. doi: 10.1158/1078-0432.CCR-06-0144. [DOI] [PubMed] [Google Scholar]
- Clifford SC, Cockman ME, Smallwood AC, Mole DR, Woodward ER, Maxwell PH, Ratcliffe PJ, Maher ER. Hum Mol Genet. 2001;10:1029–38. doi: 10.1093/hmg/10.10.1029. [DOI] [PubMed] [Google Scholar]
- Clifford SC, Prowse AH, Affara NA, Buys CH, Maher ER. Genes Chromosomes Cancer. 1998;22:200–9. doi: 10.1002/(sici)1098-2264(199807)22:3<200::aid-gcc5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Costa VL, Henrique R, Ribeiro FR, Pinto M, Oliveira J, Lobo F, Teixeira MR, Jerónimo C. BMC Cancer. 2007;23:133. doi: 10.1186/1471-2407-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl E, Wiesmann F, Woenckhaus M, Stoehr R, Wild PJ, Veeck J, Knuchel R, Klopocki E, Sauter G, Simon R, Wieland WF, Walter B, Denzinger S, Hartmann A, Hammerschmied CG. Oncogene. 2007;26:5680–91. doi: 10.1038/sj.onc.1210345. [DOI] [PubMed] [Google Scholar]
- Dallol A, Forgacs E, Martinez A, Sekido Y, Walker R, Kishida T, Rabbitts P, Maher ER, Minna JD, Latif F. Oncogene. 2002;21:3020–8. doi: 10.1038/sj.onc.1205421. [DOI] [PubMed] [Google Scholar]
- Ehnis T, Dieterich W, Bauer M, Kresse H, Schuppan D. J Biol Chem. 1997;272:20414–9. doi: 10.1074/jbc.272.33.20414. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P. Ann Oncol. 2007;18:581–92. doi: 10.1093/annonc/mdl498. [DOI] [PubMed] [Google Scholar]
- Foster K, Prowse A, van den Berg A, Fleming S, Hulsbeek MM, Crossey PA, Richards FM, Cairns P, Affara NA, Ferguson-Smith MA, et al. Hum Mol Genet. 1994;3:2169–73. doi: 10.1093/hmg/3.12.2169. [DOI] [PubMed] [Google Scholar]
- Gonzalgo ML, Yegnasubramanian S, Yan G, Rogers CG, Nicol TL, Nelson WG, Pavlovich CP. Clin Cancer Res. 2004;10:7276–83. doi: 10.1158/1078-0432.CCR-03-0692. [DOI] [PubMed] [Google Scholar]
- Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, Sun LZ, Ahlquist DA, Wood CG, Copland JA. Clin Cancer Res. 2007;13:4740–9. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- Harris A, Harris H, Hollingsworth MA. Mol Cancer Res. 2007;5:1241–5. doi: 10.1158/1541-7786.MCR-07-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM, et al. Proc Natl Acad Sci U S A. 1994;91:9700–4. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque MO, Begum S, Topaloglu O, Jeronimo C, Mambo E, Westra WH, Califano JA, Sidransky D. Cancer Res. 2004;64:5511–7. doi: 10.1158/0008-5472.CAN-04-0799. [DOI] [PubMed] [Google Scholar]
- Ibanez de Caceres I, Dulaimi E, Hoffman AM, Al-Saleem T, Uzzo RG, Cairns P. Cancer Res. 2006;66:5021–8. doi: 10.1158/0008-5472.CAN-05-3365. [DOI] [PubMed] [Google Scholar]
- Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. Science. 1993;260:1317–20. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H. Cancer Res. 2005;65:4218–27. doi: 10.1158/0008-5472.CAN-04-4407. [DOI] [PubMed] [Google Scholar]
- Mancini V, Battaglia M, Ditonno P, Palazzo S, Lastilla G, Montironi R, Bettocchi C, Cavalcanti E, Ranieri E, Selvaggi FP. Urol Oncol. 2008;26:225–38. doi: 10.1016/j.urolonc.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- McRonald FE, Morris MR, Gentle D, Winchester L, Baban D, Ragoussis J, Clarke NW, Brown MD, Kishida T, Yao M, Latif F, Maher ER. Mol Cancer. 2009;8:31. doi: 10.1186/1476-4598-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MR, Gentle D, Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, Teh BT, Latif F, Maher ER. Br J Cancer. 2008;98:496–501. doi: 10.1038/sj.bjc.6604180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MR, Gentle D, Abdulrahman M, Maina EN, Gupta K, Banks RE, Wiesener MS, Kishida T, Yao M, Teh B, Latif F, Maher ER. Cancer Res. 2005;65:4598–606. doi: 10.1158/0008-5472.CAN-04-3371. [DOI] [PubMed] [Google Scholar]
- Morris MR, Hesson LB, Wagner KJ, Morgan NV, Astuti D, Lees RD, Cooper WN, Lee J, Gentle D, Macdonald F, Kishida T, Grundy R, Yao M, Latif F, Maher ER. Oncogene. 2003;22:6794–801. doi: 10.1038/sj.onc.1206914. [DOI] [PubMed] [Google Scholar]
- Morrissey C, Martinez A, Zatyka M, Agathanggelou A, Honorio S, Astuti D, Morgan NV, Moch H, Richards FM, Kishida T, Yao M, Schraml P, Latif F, Maher ER. Cancer Res. 2001;61:7277–81. [PubMed] [Google Scholar]
- Nomoto S, Kinoshita T, Kato K, Otani S, Kasuya H, Takeda S, Kanazumi N, Sugimoto H, Nakao A. Br J Cancer. 2007;97:1260–5. doi: 10.1038/sj.bjc.6604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki R, Nemoto J, Murasawa H, Oda E, Inazawa J, Tanaka N, Taniguchi T. J Biol Chem. 2000;275:22627–30. doi: 10.1074/jbc.C000235200. [DOI] [PubMed] [Google Scholar]
- Qiu J, Ai L, Ramachandran C, Yao B, Gopalakrishnan S, Fields CR, Delmas AL, Dyer LM, Melnick SJ, Yachnis AT, Schwartz PH, Fine HA, Brown KD, Robertson KD. Lab Invest. 2008;88:910–25. doi: 10.1038/labinvest.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchandran R, Dhanabal M, Volk R, Waterman MJ, Segal M, Lu H, Knebelmann B, Sukhatme VP. Biochem Biophys Res Commun. 1999;255:735–9. doi: 10.1006/bbrc.1999.0248. [DOI] [PubMed] [Google Scholar]
- Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Mol Cell Biol. 2005;25:5675–86. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra M, Reed CC, Iozzo RV. J Biol Chem. 2002;277:35671–81. doi: 10.1074/jbc.M205317200. [DOI] [PubMed] [Google Scholar]
- Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Cancer Res. 2003;63:3735–42. [PubMed] [Google Scholar]
- Sato N, Fukushima N, Matsubayashi H, Iacobuzio-Donahue CA, Yeo CJ, Goggins M. Cancer. 2006;107:251–7. doi: 10.1002/cncr.21977. [DOI] [PubMed] [Google Scholar]
- Schaefer L, Tsalastra W, Babelova A, Baliova M, Minnerup J, Sorokin L, Grone HJ, Reinhardt DP, Pfeilschifter J, Iozzo RV, Schaefer RM. Am J Pathol. 2007;170:301–15. doi: 10.2353/ajpath.2007.060497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppan D, Cantaluppi MC, Becker J, Veit A, Bunte T, Troyer D, Schuppan F, Schmid M, Ackermann R, Hahn EG. J Biol Chem. 1990;265:8823–32. [PubMed] [Google Scholar]
- Shames DS, Girard L, Gao B, Sato M, Lewis CM, Shivapurkar N, Jiang A, Perou CM, Kim YH, Pollack JR, Fong KM, Lam CL, Wong M, Shyr Y, Nanda R, Olopade OI, Gerald W, Euhus DM, Shay JW, Gazdar AF, Minna JD. PLoS Med. 2006;3:e486. doi: 10.1371/journal.pmed.0030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shridhar R, Zhang J, Song J, Booth BA, Kevil CG, Sotiropoulou G, Sloane BF, Keppler D. Oncogene. 2004;23:2206–15. doi: 10.1038/sj.onc.1207340. [DOI] [PubMed] [Google Scholar]
- Smits KM, Schouten LJ, van Dijk BA, Hulsbergen-van de Kaa CA, Wouters KA, Oosterwijk E, van Engeland M, van den Brandt PA. Clin Cancer Res. 2008;14:782–7. doi: 10.1158/1078-0432.CCR-07-1753. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Shigematsu H, Shames DS, Sunaga N, Takahashi T, Shivapurkar N, Iizasa T, Frenkel EP, Minna JD, Fujisawa T, Gazdar AF. Br J Cancer. 2005;93:1029–37. doi: 10.1038/sj.bjc.6602837. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Takahashi T, Suzuki M, Shigematsu H, Shivapurkar N, Echebiri C, Nomura M, Stastny V, Augustus M, Wu CW, Wistuba II, Meltzer SJ, Gazdar AF. Int J Cancer. 2005;115:503–10. doi: 10.1002/ijc.20910. [DOI] [PubMed] [Google Scholar]
- Urakami S, Shiina H, Enokida H, Hirata H, Kawamoto K, Kawakami T, Kikuno N, Tanaka Y, Majid S, Nakagawa M, Igawa M, Dahiya R. Clin Cancer Res. 2006;12:6989–97. doi: 10.1158/1078-0432.CCR-06-1194. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang S, Schultz RM, Tseng H. Biochem Biophys Res Commun. 2006;348:1261–71. doi: 10.1016/j.bbrc.2006.07.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistuba II. Curr Mol Med. 2007;7:3–14. doi: 10.2174/156652407779940468. [DOI] [PubMed] [Google Scholar]
- Yamada D, Kikuchi S, Williams YN, Sakurai-Yageta M, Masuda M, Maruyama T, Tomita K, Gutmann DH, Kakizoe T, Kitamura T, Kanai Y, Murakami Y. Int J Cancer. 2006;118:916–23. doi: 10.1002/ijc.21450. [DOI] [PubMed] [Google Scholar]
- Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, Sato F, Meltzer SJ, Sidransky D. Cancer Cell. 2002;2:485–95. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.