SUMMARY
Polycomb proteins play essential roles in stem cell renewal and human disease. Recent studies of HOX genes and X-inactivation have provided evidence for RNA cofactors in Polycomb repressive complex 2 (PRC2). Here, we develop a RIP-seq method to capture the PRC2 transcriptome and identify a genome-wide pool of >9,000 PRC2-interacting RNAs in embryonic stem cells. The transcriptome includes antisense, intergenic, and promoter-associated transcripts, as well as many unannotated RNAs. A large number of transcripts occur within imprinted regions, oncogene and tumor suppressor loci, and stem-cell-related bivalent domains. We provide evidence for direct RNA-protein interactions, most likely via the Ezh2 subunit. We also identify Gtl2 RNA as a PRC2 cofactor that directs PRC2 to the reciprocally imprinted Dlk1 coding gene. Thus, Polycomb proteins interact with a genome-wide family of RNAs, some of which may be used as biomarkers and therapeutic targets for human disease.
INTRODUCTION
Transcriptome analyses have suggested that, although only 1–2% of the mammalian genome is protein-coding, 70–90% is transcriptionally active (Carninci et al., 2005; Kapranov et al., 2007; Mercer et al., 2009). Ranging from 100 nt to >100 kb, these transcripts are largely unknown in function, may originate within or between genes, and may be conserved and developmentally regulated (Kapranov et al., 2007; Guttman et al., 2009). Recent discoveries argue that a subset of these transcripts play crucial roles in epigenetic regulation. For example, genes in the human HOX-D locus are regulated in trans by HOTAIR RNA, produced by the unlinked HOX-C locus (Rinn et al., 2007), and during X-chromosome inactivation, Tsix, RepA, and Xist RNAs target a chromatin modifier in cis to control chromosome-wide silencing (Zhao et al., 2008). Interestingly, all four RNAs bind and regulate Polycomb Repressive Complex 2 (PRC2), the complex that catalyzes trimethylation of histone H3-lysine27 (H3-K27me3)(Schwartz and Pirrotta, 2008). These observations support the idea that long ncRNAs are ideal for targeting chromatin modifiers to specific alleles or unique locations in the genome (Lee, 2009) (Lee, 2010).
RNA-mediated recruitment is especially attractive for Polycomb proteins. First identified in Drosophila as homeotic regulators, Polycomb proteins are conserved from flies to mammals and control many aspects of development (Ringrose and Paro, 2004; Boyer et al., 2006; Lee et al., 2006; Schuettengruber et al., 2007; Pietersen and van Lohuizen, 2008; Schwartz and Pirrotta, 2008). Mammalian PRC2 contains four core subunits, Eed, Suz12, RbAp48, and the catalytic Ezh2. In humans, aberrant PRC2 expression is linked to cancer and disease (Sparmann and van Lohuizen, 2006; Bernardi and Pandolfi, 2007; Miremadi et al., 2007; Rajasekhar and Begemann, 2007; Simon and Lange, 2008). Despite growing recognition of Polycomb’s role in health, little is known about their regulation in vivo. In flies, Polycomb complexes may contain sequence-specific DNA-binding factors, such as Zeste, Pipsqueak (PSQ), or Pho, to help bind Polycomb-response elements (PRE) (Ringrose and Paro, 2004; Schwartz and Pirrotta, 2008). By contrast, mammalian Polycomb complexes are not thought to contain such subunits. Therefore, their mechanism of recruitment to thousands of genomic locations remains poorly understood, though PRE-like elements (Sing et al., 2009; Woo et al., 2010) and Jarid2 may facilitate binding (Li et al.; Pasini et al.; Peng et al., 2009; Shen et al., 2009). Interestingly, several PRC2 subunits have potential RNA-binding motifs (Denisenko et al., 1998; Bernstein and Allis, 2005; Bernstein et al., 2006b) – a possibility borne out by functional interactions between Tsix/RepA/Xist RNA and PRC2 for X-inactivation (Zhao et al., 2008) and by HOTAIR and PRC2 for HOX regulation (Rinn et al., 2007). Recent work also identified several short RNAs of 50–200 nt as candidate PRC2 regulators (Kanhere et al., 2010). Control of Polycomb repressive complex 1 (PRC1) may also involved RNA (Yap et al., 2010). Given these intriguing examples, here we investigate whether Polycomb complexes may associate with RNA on a genome-wide scale. We develop the RIP-seq method to capture PRC2-bound transcripts in murine ES cells and identify thousands of RNAs that specifically associate with PRC2.
RESULTS
Capturing the PRC2 transcriptome by RIP-seq
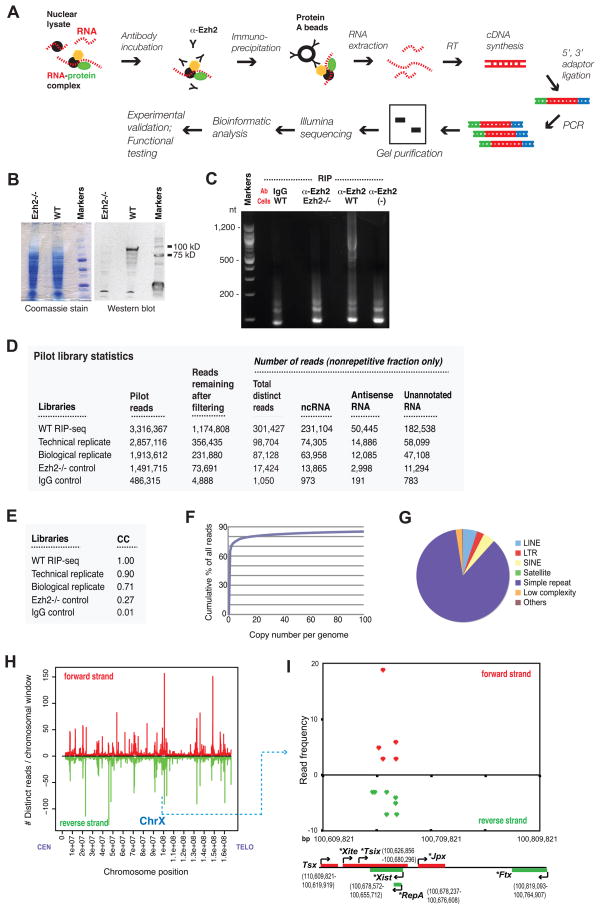
Native RNA immunoprecipitations (RIP) previously identified RepA, Xist, and Tsix as PRC2-interacting RNAs (Zhao et al., 2008). Here, we developed a method of capturing the genome-wide pool bound to PRC2 by combining native RIP (Zhao et al., 2008) and RNA-seq (Cloonan et al., 2008)(RIP-seq; Fig. 1A). Nuclear RNAs immunoprecipitated by α-Ezh2 antibodies were isolated from mouse ES cells (Lee and Lu, 1999) and an Ezh2−/− control (Shen et al., 2008) (Fig. 1B), cDNAs created using strand-specific adaptors, and those from 200–1,200 nt were purified and subjected to Illumina sequencing (Fig. 1C).
Figure 1. The RIP-seq technique and analysis of pilot libraries.
A. RIP-seq schematic.
B. Western blot analysis (right panel) of Ezh2 protein in wildtype (WT) and Ezh2−/− ES cells. Coomassie staining (left panel) shows equal loading.
C. Preparatory agarose gel for RIP product size selection.
D. Pilot library statistics for WT and control libraries for an equivalent number of cells (column 2), reads after filtering using criteria shown in Fig. S1 (column 3), and distinct reads after removing duplicates and repetitive elements (column 4).
E. CCs of indicated libraries in pairwise comparisons against the original WT library.
F. The cumulative frequency of WT reads mapping to elements with indicated genome copy numbers.
G. Relative frequencies of various repeats in the WT library. Elements repeated >10 times per genome accounted for <20% of all reads. Simple repeats accounted for 85.714% and LINEs, SINEs, LTRs, low-complexity repeats, and satellites represented 4.881%, 4.130%, 2.636%, 2.487%, and 0.002%, respectively.
H. Alignments of distinct WT pilot reads to the mouse X-chromosome. The number of reads per 100-kb window for both unique and repetitive elements are plotted from centromere (CEN) to distal telomere (TELO). 100-kilobase windows are nonoverlapping and consecutive. Reads were normalized such that those mapping to ‘n’ locations were counted as 1/nth of a read at each location. Chr, chromosome. Red, forward strand; green, reverse strand.
I. Zoom-in of the X-inactivation center showing pilot WT reads. The Ezh2−/− library is depleted of these reads. Freq ≥3 reads shown. *, ncRNA.
In pilot experiments, we performed RIP on 107 ES cells and included several control RIPs to assess the specificity of α-Ezh2 pulldowns. In the wildtype pulldown and its technical and biological replicates, α-Ezh2 antibodies precipitated 70–170 ng of RNA from 107 ES cells and yielded a cDNA smear of >200 nt (Fig. 1C, Fig. S1A). Treatment with RNAses eliminated products in this size range (Fig. S1B) and −RT samples yielded no products, suggesting that the immunoprecipitated material was indeed RNA. There was ~10-fold less RNA in the Ezh2−/− pulldown (~14 ng) and when wildtype cells were immunoprecipitated by IgG (~24 ng). A 500-fold enrichment over a mock RIP control (no cells) was also observed. In the >200 nt size range, control RIPs (null cells, IgG pulldowns, mock) were even further depleted of RNA, as these samples were dominated by adaptor and primer dimers. We computationally filtered out adaptor/primer dimers, rRNA, mitochondrial RNA, reads with <18 nt or indeterminate nucleotides, and homopolymer runs in excess of 15 bases (Fig. S1). From an equivalent number of cells, control RIPs were significantly depleted of reads (Fig. 1D). In wildtype libraries, 231,880–1.2 million reads remained after filtering. By contrast, only 4,888 to 73,691 reads remained in controls (Fig. 1D, columns 2 and 3). The overwhelming majority of transcripts in the controls were of spurious nature (adaptor/primer dimers, homopolymers, etc.). Therefore, wildtype RIPs exhibited substantial RNA enrichment and greater degrees of RNA complexity in comparison to control RIPs.
Approximately half of all reads in the wildtype libraries was represented three times or more. Even after removing duplicates to avoid potential PCR artifacts, the wildtype library contained 301,427 distinct reads (technical and biological replicates with 98,704 and 87,128, respectively), whereas control samples yielded only 1,050 (IgG) and 17,424 (null)(Fig. 1D). The wildtype libraries were highly similar among each other, with correlation coefficients (CC) of 0.71–0.90, as compared to 0.27–0.01 when compared against Ezh2−/− and IgG controls, respectively (Fig. 1E). Reads mapping to repetitive elements of >10 copies/genome accounted for <20% of total wildtype reads (Fig. 1F), with simple repeats being most common and accounting for 85.714%, whereas LINEs, SINEs, and LTRs were relatively under-represented (Fig. 1G). Because reads with ≤10 alignments have greatest representation, we hereafter focus analysis on these reads (a cutoff of ≤10 retains genes with low-copy genomic duplications).
We next examined their genome distribution by plotting distinct reads as a function of chromosome position (Fig. S2–S6). The alignments showed that PRC2-associated RNAs occurred on every chromosome in the wildtype libraries. Alignments for IgG and Ezh2−/− controls demonstrated few and sporadic reads. Therefore, our RIP-seq produced a specific and reproducible profile for the PRC2 transcriptome. A large number of wildtype reads hits the X-chromosome (Fig. 1H), and a zoom of the X-inactivation center showed that our positive controls – Tsix, RepA, and Xist RNAs – were each represented dozens of times (Fig. 1I). The high sensitivity of our RIP-seq detection was suggested by representation of RepA and Xist, which are in aggregate expressed at <10 copies/ES cell (Zhao et al., 2008). On the other hand, no hits occurred within other noncoding RNAs of the X-inactivation center. Thus, the RIP-seq technique was both sensitive and specific.
The PRC2 transcriptome
To obtain saturating coverage, we scaled up sequencing and obtained 31.9 million reads for the original wildtype sample and 36.4 million for its biological replicate. After removing duplicates and filtering as shown in Fig. S1-A, 1,030,708 and 852,635 distinct reads of alignment ≤10 remained for each library, respectively. These reads were then combined with pilot wildtype reads for subsequent analyses (henceforth, WT library) and all analyses were performed using the Ezh2−/− library as control.
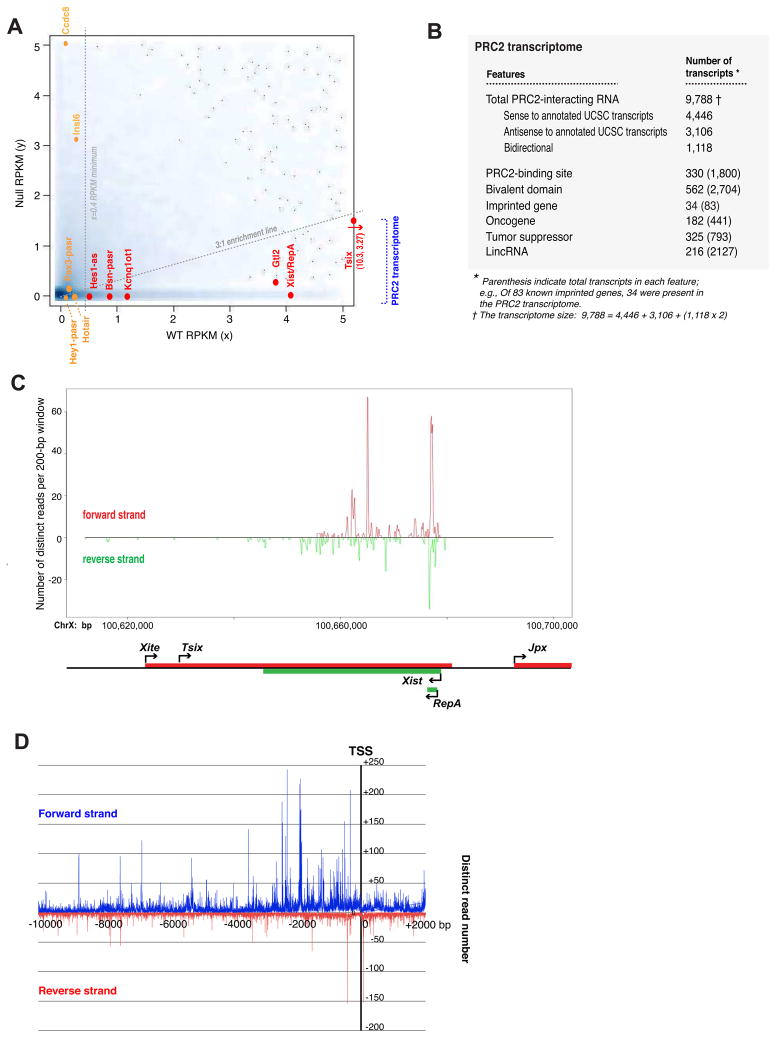
To determine a threshold for calling transcripts a member of the “PRC2 transcriptome”, we designed a strategy based on (i) the number of distinct reads per transcript, on the principle that bona fide PRC2-interacting transcripts should have higher read densities than background, and (ii) the relative representation in the WT versus null libraries, reasoning that bona fide positives should be enriched in the WT. We calculated genic representations using “reads per kilobase per million reads” (RPKM) as a means of normalizing for gene length and depth of sequencing (Mortazavi et al., 2008), and then mapped all 39,003 transcripts in the UCSC joined transcriptome to a scatterplot by their WT RPKM (x-axis) and their null RPKM (y-axis) values (Fig. 2A). Transcripts with zero or near-zero representation in both libraries accounted for the vast majority of datapoints [blue cloud at (0,0)]. Transcripts with nonzero x-values and a zero y-value indicated a population represented only in WT pulldowns (Fig. 2A, y=0 line).
Figure 2. Larger-scale sequencing to capture the PRC2 transcriptome.
A. The scatterplot maps 39,003 transcripts from the UCSC joined transcriptome database by their RPKM values in the wildtype library (x-axis) and the null library (y-axis). A UCSC transcript that is neither represented in the WT or null library is plotted at (0,0). Smoothing was performed by the function, smoothScatter, in R. Darker shades correspond to a greater density of genes at a given point on the graph. The 3:1 WT/null enrichment line and the x=0.4 threshold are shown as dotted grey lines. Transcripts meeting the criteria of ≥3:1 RPKM enrichment and WT RPKM≥0.4 are deemed strong positives and are shown in red, in a pool marked “PRC2 transcriptome”. Transcripts that fall below the cut-offs are considered background and are shown in orange. Tsix is off-chart (arrow) with (x,y) coordinates indicated.
B. Characteristics of the PRC2 transcriptome. Numbers in parentheses indicate the total number of genes in each category (e.g., Of 793 tumor suppressors, 325 are found in the PRC2 transcriptome).
C. Higher resolution analysis of the X-inactivation center. Distinct reads were smoothed with a sliding 200-bp window on the x-axis and their representations plotted on the y-axis.
D. Metagene analysis: distinct reads from the PRC2 transcriptome are plotted as a function of distance from TSS.
We established a density minimum by using control transcripts as calibration points. Xist/RepA scored an RPKM of 4.19, implying 126 distinct reads per million. Tsix scored 10.35, and Bsn-pasr (~300-nt Bsn-promoter associated transcript (Kanhere et al., 2010)) scored 0.95. The imprinted antisense transcript, Kcnq1ot1, has been proposed to interact with PRC2, though whether it does so directly is not known (Pandey et al., 2008). Kcnq1ot1 scored 1.17. For negative controls, we used transcripts that a priori should not be in the WT library. For example, Hotair is expressed later in development only in caudal tissues (Rinn et al., 2007). It scored 0.25, implying only a single representation per million. Two other promoter-associated RNAs, Hey1-pasr and Pax3-pasr (Kanhere et al., 2010), are <200 nt and fell outside of our size-selection scheme. They scored 0.28 and 0.11, respectively, suggesting ≪1 distinct reads per million. Cytoplasmically localized protein-coding mRNAs that are not expected to be PRC2-interacting also showed low RPKM [Insl6 0.27, Ccdc8 0.22]. We consider these low representations background. On the basis of the calibration points, we set the RPKM minimum at x=0.40, which falls between values for positive and negative controls.
To determine an appropriate enrichment threshold, we examined WT/null RPKM ratios for the same calibrators. Xist/RepA scored 4.18/0, implying hundreds to thousands of representations in the WT library but none in the null. Tsix scored 10.35/3.27, Bsn-pasr 0.95/0, and Kcnq1ot1 1.17/0. The negative controls scored low ratios, with Pax3-pasr at 0.11/0.26, Hey1-pasr 0.28/0, Hotair 0.25/0, Insl6 0.27/3.09, and Ccdc8 0.22/5.04. On this basis, we set the enrichment cutoff at 3:1. The combined criteria for transcript inclusion *RPKM(WT)≥0.4, RPKM(WT)/RPKM(null)≥3.0+ are expected to eliminate false positives and subtract background based on direct comparisons between WT and null libraries using an established set of controls.
By these criteria, we estimate the PRC2 transcriptome at 9,788 RNAs (Table S-I). Some 4,446 transcripts in the joined UCSC transcriptome (39,003 transcripts) were included in our PRC2 transcriptome (Fig. 2B). Another 3,106 UCSC transcripts were hit but only on the reverse strand, implying the existence of 3,106 previously unannotated antisense RNAs. Some 1,118 UCSC transcripts were hit in both directions, implying the existence of 2,236 additional distinct transcripts. 19% of reads did not have a hit in the UCSC database. These “orphan reads” suggest that the transcriptome may include other novel transcripts. Therefore, 9,788 represents a lower bound on the actual PRC2 transcriptome in ES cells. Because the total mouse transcriptome is believed to be anywhere from 40,000 to 200,000, the PRC2 transcriptome comprises 5–25% of all mouse transcripts, depending on the actual size of the total transcriptome.
Epigenetic features
We examined specific epigenetic features (Fig. 2B, Tables I, S-II to S-VI). Interestingly, the RepA region within Xist and the 3′ end of Tsix were represented many times (Fig. 2C), a region consistent with the proposed Ezh2 footprint (Zhao et al., 2008). In a metagene analysis, we queried the relationship of transcripts to transcription start sites (TSS) by plotting read numbers as a function of distance (Fig. 2D). On the forward strand, enrichment was observed at −2.0 to +0.001 kb; on the reverse strand, peaks were discernible at −0.5 to +0.1 kb. The enrichment occurred above background (null, IgG controls)(Fig. S1C). TSS association is notable given the existence of short transcripts at promoters (Kapranov et al., 2007; Core et al., 2008; Seila et al., 2008; Taft et al., 2009), PRC2’s preferred occupancy near promoters (Boyer et al., 2006; Lee et al., 2006; Schwartz et al., 2006; Ku et al., 2008), and identification of several TSS-associated RNAs which bind PRC2 (Kanhere et al., 2010).
Table I. Imprinted regions hit by the PRC2 transcriptome.
Intersection of the PRC2 transcriptome with imprinted gene coordinates (http://www.geneimprint.com/). The imprinted gene’s chromosome localization and nucleotide coordinates in mm9 are shown in column 2. ‘Chr strand’ (column 3) refers to the orientation of the gene. A hit in the ‘plus’ strand (column 4) indicates that PRC2-interacting transcript is on the top strand of the chromosome, whereas a hit in the ‘minus’ strand (column 5) indicates that the PRC2-interacting transcript is on the bottom strand. A hit to the opposite strand of the reference transcript implies that the PRC2-interacting RNA is complementary to reference gene. Hits to both strands imply that both strands of the reference transcript interact with PRC2. Note that the PRC2-interacting transcript need not be the reference gene itself, but a distinct transcript that overlaps in position.
| Imprinted refGene hit by transcriptome | chr location and coordinate (mm9) | chr strand | plus strand hit | minus strand hit |
|---|---|---|---|---|
| Nespas | chr2:174096737-174130936 | − | Y | Y |
| Gnas | chr2:174099821-174182244 | + | Y | Y |
| Ngfb | chr3:101938712-102009789 | + | Y | Y |
| Trp73 | chr4:153422952-153524316 | − | Y | Y |
| Sgce | chr6:4614349-4707098 | − | Y | N |
| Peg10 | chr6:4687379-4720475 | + | Y | N |
| Pon3 | chr6:5160851-5216232 | − | N | Y |
| Dlx5 | chr6:6817804-6842067 | − | Y | Y |
| Atp10a | chr7:65903701-66094160 | + | N | Y |
| Ube3a | chr7:66474119-66572096 | + | Y | Y |
| Ndn | chr7:69483233-69504813 | + | Y | N |
| H19 | chr7:149751436-149774050 | − | Y | Y |
| Igf2 | chr7:149826672-149855393 | − | Y | Y |
| Ascl2 | chr7:150142730-150165131 | − | Y | N |
| Cd81 | chr7:150228699-150263828 | + | Y | Y |
| Tssc4 | chr7:150245272-150266991 | + | Y | Y |
| Trpm5 | chr7:150247696-150285512 | − | Y | Y |
| Kcnq1 | chr7:150283158-150622946 | + | Y | Y |
| Cdkn1c | chr7:150634244-150656900 | − | Y | N |
| Nap1l4 | chr7:150689483-150744994 | − | Y | N |
| Tnfrsf23 | chr7:150841711-150881776 | − | Y | N |
| Osbpl5 | chr7:150864666-150937867 | − | Y | N |
| Sdhd | chr9:50394450-50421921 | − | N | Y |
| Plagl1 | chr10:12800714-12859693 | + | N | Y |
| Dcn | chr10:96935000-96990784 | + | Y | N |
| Ddc | chr11:11704105-11800403 | − | N | Y |
| Gtl2 | chr12:110773827-110809921 | + | Y | N |
| Rtl1 | chr12:110818378-110843612 | − | Y | Y |
| Kcnk9 | chr15:72335722-72389882 | − | N | Y |
| Igf2r | chr17:12865278-12972529 | − | N | Y |
| Air | chr17:12931160-12954858 | + | N | Y |
| Impact | chr18:13120760-13161456 | + | N | Y |
| Tsix | chrX:100616855-100690295 | + | Y | Y |
| Xist | chrX:100645711-100688571 | − | Y | Y |
We next asked how much of the PRC2 transcriptome intersects PRC2-binding sites (Boyer et al., 2006; Lee et al., 2006) and bivalent domains in ES cells (Bernstein et al., 2006a; Mikkelsen et al., 2007; Ku et al., 2008). Notably, 562 of 2,704 bivalent domains (21%) and 330 of 1,800 Suz12-binding sites (18%) were hit by at least one RNA (Fig. 2B, Table S-II, S-III), raising the possibility that RNA may be involved in recruiting or retaining Polycomb complexes in a subset of binding sites and control stem cell fate. Sites which do not intersect our transcriptome may recruit PRC2 using other mechanisms.
We also queried the extent of overlap with a group of intergenic ncRNA dubbed “lincRNA” (Guttman et al., 2009). Intersecting 2,127 mouse lincRNA with our 9,788 transcripts revealed an overlap of 216 (Fig. 2B, Table S-IV), indicating that lincRNA account for ~2% of the PRC2 transcriptome. Of human lincRNA, 260 may have potential to associate with PRC2 (Khalil et al., 2009). To ask whether the 260 human lincRNA overlap with the 216 mouse lincRNA in our PRC2 transcriptome, we mapped syntenic coordinates in the mouse by LiftOver (http://genome.ucsc.edu/cgi-bin/hgLiftOver) but found no recognizable homology between the two subsets. Thus, our transcriptome represents a large and distinct set of PRC2-interacting RNAs.
Because misregulation of Polycomb proteins is often associated with cancer, we intersected PRC2-interacting RNAs with oncogene and tumor suppressor loci (Sparmann and van Lohuizen, 2006; Bernardi and Pandolfi, 2007; Miremadi et al., 2007; Rajasekhar and Begemann, 2007; Simon and Lange, 2008). Intriguingly, of 441 oncogenes and 793 tumor suppressors (http://cbio.mskcc.org/CancerGenes), 182 (41%) and 325 (41%) respectively have at least one PRC2-interacting transcript of either orientation (Fig. 2B, Tables S-V, S-VI), suggesting that RNA may play a role in misregulating Polycomb recruitment in cancer. Notable examples include c-Myc, Brca1, Klf4, and Dnmt1.
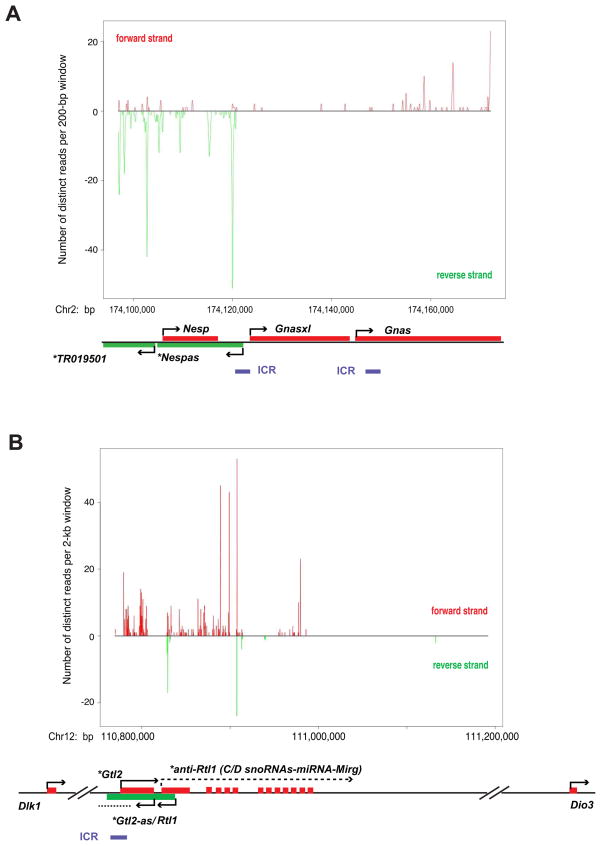
Finally, like X-chromosome inactivation, genomic imprinting must be regulated in cis. Imprinted genes are controlled by a cis-acting ‘imprinting control region’ (ICR) that dictates parent-specific expression (Edwards and Ferguson-Smith, 2007; Thorvaldsen and Bartolomei, 2007). Interestingly, ICRs are generally associated with long transcripts (Williamson et al., 2006; Pandey et al., 2008; Wan and Bartolomei, 2008), many of which were found in the PRC2 transcriptome (Fig. 2B, Table I). They include H19, Gtl2, Kcnq1ot1, and Nespas. Multiple hits occurred in Nespas RNA/TR019501 (Fig. 3A), an antisense RNA from the primary ICR thought to regulate the Nesp/Gnas cluster (Coombes et al., 2003; Williamson et al., 2006). Also hit repeatedly was Gtl2 (Fig. 3B), the locus believed to control Dlk1 imprinting (Edwards et al., 2008), along with anti-Rtl1 and an antisense counterpart of Gtl2 (here dubbed Gtl2-as). Hits within ICR-associated long transcripts hint that RNA may regulate imprinted clusters by targeting PRC2.
Figure 3. Hits to select imprinted loci.
A,B. Read density plots for Nesp/Gnas (A) and Dlk1/Gtl2 (B) imprinted clusters. Distinct reads are smoothed with sliding consecutive 200-bp or 2-kb windows on the x-axis and their representations plotted on the y-axis. *, ncRNA. Chr, chromosome. Red, forward strand; green, reverse strand. The Ezh2−/− library is depleted of these reads.
Validation of RNA-PRC2 interactions
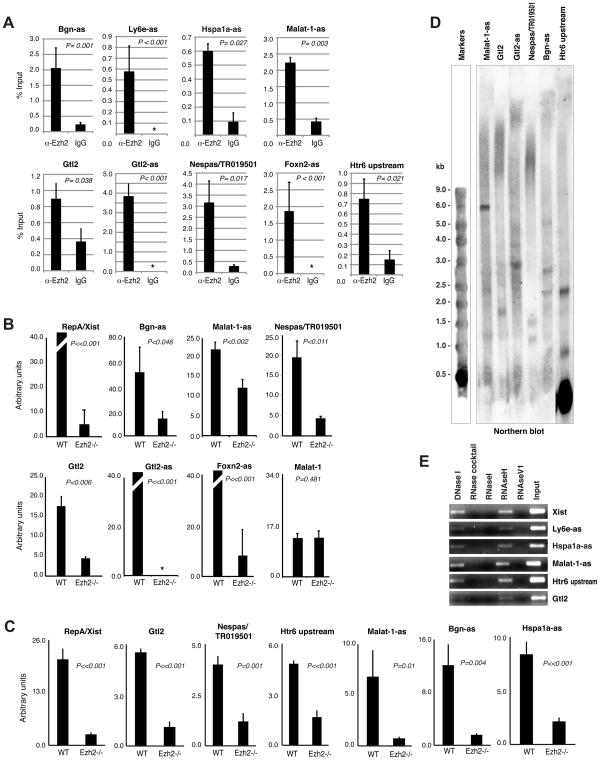
We next validated RNA-protein interactions by several approaches. First, we performed RIP-qPCR and found that candidate RNAs had significant enrichment in the α-Ezh2 relative to IgG pulldowns (Fig. 4A). Strong positive pulldowns were observed for the imprinted Gtl2, its antisense partner Gtl2-as/Rtl1, and Nespas/TR019501. A number of previously unknown antisense transcripts or RNAs linked to disease loci was also enriched, including Hspa1a-as (antisense to Hsp70), Malat-1-as (antisense to Malat-1), Bgn-as (antisense to Bgn), Ly6e-as (antisense to lymphocyte antigen 6 complex locus E), Foxn2-as (antisense to Foxn2), and an RNA upstream of Htr6 serotonin receptor. Second, we compared the amount of RNA pulled down by α-Ezh2 in WT versus Ezh2−/− ES cells (Fig. 4B). In every case, the RNA was significantly more enriched in WT. By contrast, the negative control Malat-1 sense transcript showed no enrichment. Third, we performed UV-crosslink RIP, an alternative method of testing RNA-protein interactions in vivo based on the ability of UV to covalently link RNA to protein at near-zero Angstroms (Ule et al., 2005). Because crosslinking occurs only at short range and complexes are isolated with disruptive sonication and high-salt washes, this method better detects direct RNA-protein interactions and may avoid reassociation artifacts during RNA isolation. Enrichment of candidate RNAs was similarly observed using this method (Fig. 4C). Combined, these data support the specificity of RIP-seq and suggest direct interactions between RNA and Ezh2.
Figure 4. Confirmation by native RIP/qRT-PCR and UV-crosslinked RIP.
A. qRT-PCR to compare α-Ezh2 and IgG pulldowns. The experiments were performed 2–3 times in triplicate. Error bar = 1 standard deviation (SD). P was calculated using the two-tailed student t-test. Asterisks, undetectable levels.
B. qRT-PCR after native α-Ezh2 RIP of wildtype and null ES cells, each normalized to IgG RIP values. Values for Xist, Gtl2-as, and Foxn2-as were off-chart. Experiments were performed 2–4 times in triplicate. 1 SD shown. P is calculated using the paired, two-tailed student t-test. Asterisks, undetectable RNA levels.
C. Confirmation of native RIP by UV-crosslinked RIP. Each experiment was performed 2–4 times in triplicate, normalized to IgG pulldowns, and compared to that of Ezh2−/− controls using the t-test (P). 1 SD shown.
D. Northern blot analysis of indicated RNA species.
E. Native RIP with RNAse pretreatment, followed by qRT-PCR quantification.
Nearly half of the transcripts identified by RIP-seq were previously unannotated (Fig. 2B). To verify their existence, we performed Northern analysis and found discrete transcripts in ES cells (Fig. 4D). To confirm the nature of nucleic acids precipitated by α-Ezh2, we pretreated nuclear extracts with RNases of different substrate specificities. Digesting with single-stranded RNase (RNase I) and double-stranded RNase (RNAse V1) abolished RNA pulldown, whereas digesting with RNase H (which degrades the RNA strand in RNA:DNA hybrids) and DNase I had no effect (Fig. 4E). Thus, the RNAs in complex with PRC2 have single- and double-stranded character.
Direct binding of RNA to PRC2
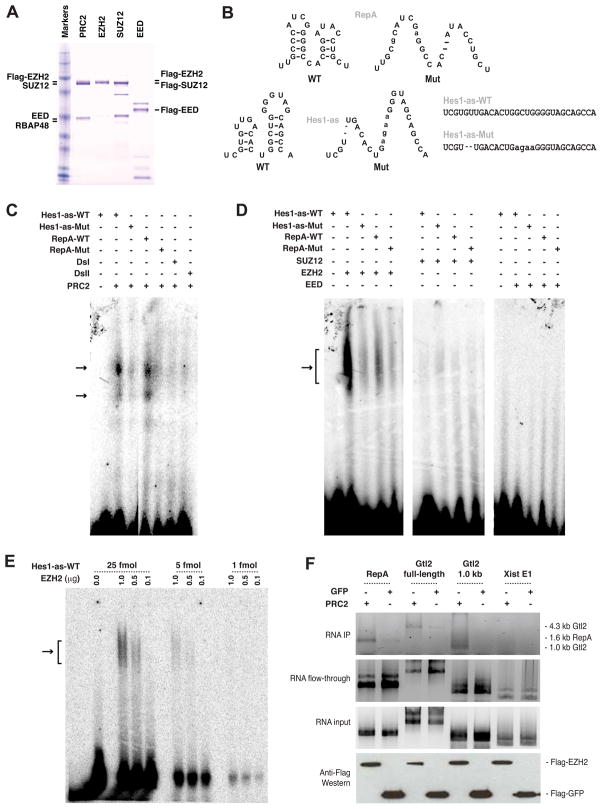
We next addressed whether RNA directly binds PRC2 by in vitro biochemical analyses using purified recombinant human PRC2 subunits, EED, EZH2, SUZ12, and RBAP48 (Fig. 5A). We find an antisense RNA for Hes1, a transcription factor in the Notch signaling pathway (Axelson, 2004). Hes1-as contains a double stem-loop structure, a motif also found in RepA (Zhao et al., 2008)(Fig. 5B). In an RNA electrophoretic mobility shift assay (EMSA), both the 28-nt RepA and 30-nt Hes1-as probes were shifted by PRC2, whereas RNAs derived from other regions of Xist (DsI, DsII) were not. Mutating the stem-loop structures reduced PRC2 binding. To determine which subunit of PRC2 binds Hes1-as, we performed EMSA using specific subunits (Fig. 5A,D,E). EZH2 strongly shifted wildtype but not mutated Hes1-as RNA, whereas neither SUZ12 nor EED shifted Hes1-as. The RNA-protein shift was always more discrete when whole PRC2 was used, suggesting that other subunits stabilize the interaction. These results show that Hes1-as RNA directly and specifically interacts with PRC2 and Ezh2 is the RNA-binding subunit.
Figure 5. Biochemical analysis shows direct interactions between RNA and PRC2.
A. Coomassie-stained gel of human PRC2 and subunits. Different migrations reflect Flag-tagged versus untagged versions of each protein.
B. WT and mutant (Mut) versions of RepA and Hes1-as double stem-loop structures.
C. RNA EMSA using purified PRC2 complex and end-labeled probes. Negative controls: DsI and DsII, RNA sequences from Xist outside of RepA. Double shifts indicate presence of multiple subcomplexes of PRC2.
D. RNA EMSA using purified PRC2 subunits. The lanes were run on the same gel but separated in the image because a lane was cut out between each panel.
E. Titration of 1–25 fmoles of Hes1-as RNA probe against 0.1–1.0 μg of EZH2.
F. RNA pulldown assays using purified PRC2 and indicated RNA probes loaded in equal moles. 25% of the IP fraction, 10% of flow-through, and 10% of RNA input are shown.
We also examined Gtl2 RNA. Because Gtl2 is 1.7–4.3 kb and too large to test by EMSA, we performed RNA pulldown assays (Fig. 5F). We in vitro-transcribed Gtl2, a truncated form (1.0-kb from the 5′ end), RepA, and Xist exon 1 (negative control), and tested equal molar amounts of each RNA in pulldown assays using Flag-PRC2 or Flag-GFP proteins. Both full-length and truncated Gtl2 RNAs were consistently enriched in PRC2 pulldowns. RepA RNA was also enriched, whereas Xist exon 1 was not. These results demonstrated that Gtl2 RNA – most likely its proximal 1.0 kb – directly and specifically binds PRC2.
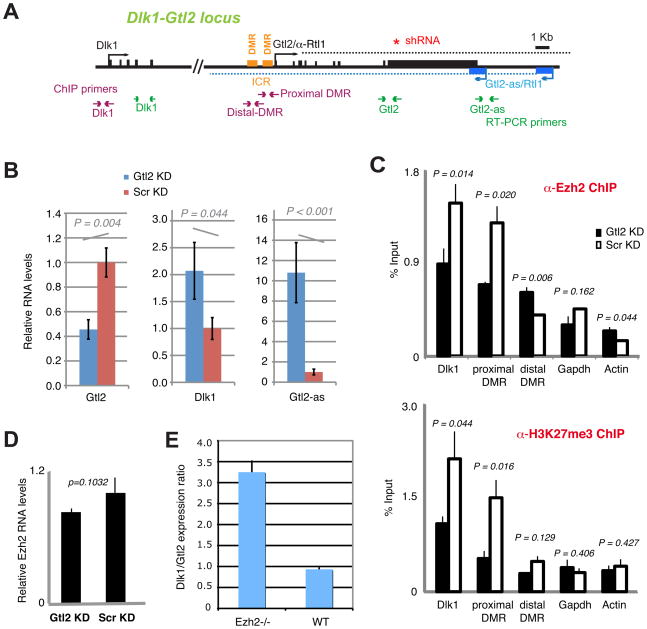
Gtl2-PRC2 interactions regulate gene expression at Dlk1-Gtl2
To investigate whether RIP-seq succeeded in discovering new functions, we focused on Gtl2-PRC2 interactions at Dlk1-Gtl2, the imprinted disease locus linked to the sheep Callipyge (gluteal hypertrophy), murine growth dysregulation, and human cancers (Edwards et al., 2008; Takahashi et al., 2009). The maternally expressed Gtl2 is associated with the ICR (Fig. 6A) and has been proposed to regulate paternally expressed Dlk1 (Lin et al., 2003; Takahashi et al., 2009), but the mechanism of action is currently unknown. To determine if the Gtl2 transcript per se regulates Dlk1, we knocked down Gtl2 in ES cells and observed a 2-fold upregulation of Dlk1, consistent with the idea that Dlk1 changed from mono- to bi-allelic expression (Fig. 6B). Gtl2-as was also upregulated. Because shRNAs target RNA for degradation post-transcriptionally, these experiments demonstrate that Gtl2 functions as RNA.
Figure 6. Gtl2 controls Dlk1 by targeting PRC2.
A. Map of Dlk1-Gtl2 and the positions of shRNAs and primer pairs used in RIP and ChIP. Dotted lines indicate that the transcripts may extend further.
B. qRT-PCR of Gtl2, Dlk1, and Gtl2-as RNA levels after Gtl2 knockdown (KD). Pools of knockdown cells are used. RNA levels are normalized to Gapdh levels and compared to levels in scrambled knockdown controls (Scr). Experiments were performed in triplicates two times. One SD shown. P is calculated using a two-tailed student t-test between Gtl2 versus Scr KDs.
C. qChIP of PRC2 association in KD cells. ChIP was carried out with α-Ezh2 and α-H3K27me3 antibodies, with normal rabbit IgG as control (not shown). qPCR levels are expressed as a percentage of input DNA. DMR, differentially methylated region. ICR, imprint control region. One SD shown. P, determined by two-tailed student t-tests of Gtl2 versus Scr KD.
D. qRT-PCR of Ezh2 mRNA levels in Gtl2- and Scr-KD clones. Averages and standard errors shown for two independent experiments.
E. qRT-PCR of Dlk1 expression in Ezh2−/− versus WT cells relative to Gtl2 expression. One SD shown.
To address if the RNA operates by attracting PRC2 to Dlk1, we carried out quantitative chromatin immunoprecipitation (ChIP) using α-Ezh2 and α-H3-K27me3 antibodies. Indeed, when Gtl2 RNA was knocked down, we detected a two-fold decrease in Ezh2 recruitment to the Dlk1 promoter and a commensurate decrease in H3-K27 trimethylation in cis (Fig. 6C), consistent with increased Dlk1 expression (Fig. 6B). We also saw decreased Ezh2 recruitment and H3-K27 trimethylation at a differentially methylated region (DMR) of the ICR proximal to Gtl2, whereas lesser effects were seen at the distal DMR (Fig. 6C). Because the distal DMR is genetically upstream of Gtl2 (Lin et al., 2003; Takahashi et al., 2009), we did not expect regulation by Gtl2. Gapdh and Actin controls did not show significant decreases after Gtl2 knockdown, and decreased Ezh2 recruitment to Dlk1 was not the result of generally decreased Ezh2 levels in Gtl2-knockdown cells (Fig. 6D). These data argue that Gtl2 indeed functions by attracting PRC2 to Dlk1. In further support, abolishing Ezh2 phenocopied the Gtl2 knockdown, with a ~3-fold increase in Dlk1 expression relative to Gtl2 levels (Fig. 6E). Given direct Gtl2-PRC2 interactions (Fig. 5) and loss of Ezh2/H3-K27me3 at Dlk1 when Gtl2 is knocked down (Fig. 6), we conclude that Gtl2-PRC2 interactions regulate Dlk1 by targeting PRC2 to Dlk1 in cis.
DISCUSSION
RNA cofactors for Polycomb complexes such as RepA/Xist for X-inactivation (Zhao et al., 2008) and HOTAIR for HOX-D (Rinn et al., 2007) have implicated RNA in Polycomb control. Here, we have developed the RIP-seq technology to capture a genome-wide pool of long transcripts (>200 nt) associated with PRC2. The PRC2 transcriptome consists of ~10,000 RNAs in mouse ES cells, likely accounting for 5–25% of expressed sequences in mice, depending on the actual size of the total mouse transcriptome. Transcriptome characterization has identified classes of medically significant targets, including dozens of imprinted loci, hundreds of oncogene and tumor suppressor loci, and multiple stem-cell-related domains, some of which may be used as biomarkers and therapeutics targets in the future.
Our data demonstrate that at least a subset of RNAs directly interact with Polycomb proteins in vivo and that the most likely interacting subunit is Ezh2. A recent study indicates that Suz12 also interacts with RNA (Kanhere et al., 2010). Differences between bacterially- and baculovirus-produced subunits could result in varying post-translational modifications with effects on binding properties. However, it seems more attractive to posit that multiple subunits of PRC2 can be regulated by RNA, which could modulate binding between PRC2 subunits, binding affinities of PRC2 for chromatin, and/or Ezh2 catalytic rates. This scenario would amplify the number of potential mechanisms by which RNA regulates Polycomb. Our study suggests thousands of RNA cofactors for Ezh2, the bait used for RIP-seq, specifically as part of the PRC2 complex. To our knowledge, Ezh2 is only present in Polycomb complexes, as biochemical purification using tagged Ezh2 identifies only Polycomb-related peptides (Li et al., 2010) and knocking out other subunits of PRC2 results in rapid degradation of Ezh2 (Pasini et al., 2004; Montgomery et al., 2005; Schoeftner et al., 2006).
Both cis and trans mechanisms may be utilized by RNAs in the PRC2 transcriptome. While HOTAIR works in trans (Rinn et al., 2007; Gupta et al.), the large number of antisense transcripts in the transcriptome suggests that many, like Tsix, may function by directing PRC2 to overlapping or linked coding loci in cis. We have provided the example of a linked RNA, Gtl2, which binds and targets PRC2 to Dlk1 locus to direct H3K27 trimethylation in cis. Long ncRNAs present an attractive mechanism to target chromatin modifiers to specific locations, as they remain tethered to the site of transcription and can co-transcriptionally direct enzymatic activities to a unique region (Lee, 2009, 2010).
In conclusion, our study implies that RNA cofactors may be a general feature of Polycomb regulation. Regulation by RNA need not be specific to Polycomb proteins. RIP-seq technology can be utilized to identify RNA cofactors for other chromatin modifiers, and different cell types might have distinct transcriptomes consistent with their developmental profiles. Because chromatin modifiers such as PRC2 play a central role in maintaining stem cell pluripotency and in cancer, a genome-wide profile of regulatory RNAs will be a valuable resource in the quest to diagnose and treat disease.
EXPERIMENTAL PROCEDURES
RIP-seq
RNA immunoprecipitation was performed (Zhao et al., 2008) using 107 wildtype 16.7 (Lee and Lu, 1999) and Ezh2−/− (Shen et al., 2008) ES cells. To construct RIP-seq libraries, cell nuclei were isolated, nuclear lysates were prepared, treated with 400 U/ml DNAse, and incubated with α-Ezh2 antibodies (Active Motif) or control IgG (Cell Signaling Technology). RNA-protein complexes were immunoprecipitated with protein A agarose beads and RNA extracted using Trizol (Invitrogen). To preserve strand information, template switching was used for the library construction (Cloonan et al., 2008). 20–150 ng RNA and Adaptor1 (5′-CTTTCCCTACACGACGCTCTTCCGATCTNNNNNN-3′) were used for first-strand cDNA synthesis using Superscript II Reverse Transcription Kit (Invitrogen). Superscript II adds non-template CCC 3′ overhangs, which were used to hybridize to Adaptor2-GGG template-switch primer (5′-CAAGCAGAAGACGGCATACGAGCTCTTCCGATCTGGG-3′). During 1st-strand cDNA synthesis, samples were incubated with adaptor1 at 20 °C for 10 min, followed by 37 °C for 10 min and 42 °C for 45 min. Denatured template switch primer was then added and each tube incubated for 30 min at 42 °C, followed by 75 °C for 15 min. Resulting cDNAs were amplified by forward (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′) and reverse (5′-CAAGCAGAAGACGGCATACGAGCTCTTCCGATCT-3′) Illumina primers. PCR was performed by Phusion polymerase (BioRad) as follows: 98 °C for 30s, 20 – 24 cycles of [98°C 10s, 65°C 30s, 72°C 30s], and 72°C for 5 min. PCR products were loaded on 3% NuSieve gel for size-selection and 200–1,200 bp products were excised and extracted by QIAEX II Agarose Gel Extraction Kit (Qiagen). Minus-RT samples generally yielded no products. DNA concentrations were quantitated by PicoGreen. 5–10 μl of 2 – 20 nM cDNA samples were sequenced by the Sequencing Core Facility of the Dept. of Molecular Biology, MGH, on the Illumina GAII.
Bioinformatic analysis
Complete RIP-seq datasets can be accessed through GEO via series GSE17064. Except as noted below, all analyses were performed using custom C++ programs. Image processing and base calling were performed using the Illumina pipeline. 3′ adaptor sequences were detected by crossmatch and matches of ≥5 bases were trimmed, homopolymer reads filtered, and reads matching the mitochondrial genome and ribosomal RNAs excluded from all subsequent analyses. Remaining sequences were then aligned to the mm9 mouse reference genome using shortQueryLookup (Batzoglou et al., 2002). Alignments with ≤1 error were retained. Because library construction and sequencing generate sequence from the opposite strand of the PRC2-bound RNA, in all further analysis, we treated each read as if it were reverse-complemented. To determine the correlation coefficients comparing the original α-Ezh2 RIP-seq library to its technical and biological replicates and also to RIP-seq of the Ezh2−/− control line, we compared the number of reads per gene between two samples and, for each pair, we computed the Pearson correlation between the number of reads mapped to each refGene. That is, for each sample, we created a vector of counts of reads mapped to each refGene and computed the Pearson correlation between all pairs of vectors.
Locations of repetitive sequences in mm9 (RepeatMasker) were obtained from the UCSC Genome Browser database (http://hgdownload.cse.ucsc.edu/goldenPath/mm9/database). The overlap of PRC2 transcriptome reads with these repeats was obtained by intersecting coordinates of RepeatMasker data with coordinates of read alignments. The UCSC transcriptome was used as general reference (http://hgdownload.cse.ucsc.edu/goldenPath/mm9/database/transcriptome.txt.gz). To obtain a set of non-overlapping distinct transcribed regions, we sorted the UCSC transcriptome transcripts by start coordinate and merged overlapping transcripts on the same strand (joined UCSC transcriptome: 39,003 transcripts total). We then intersected read alignment coordinates with those of the merged UCSC transcripts to determine the number of UCSC transcripts present in the PRC2 transcriptome. Hits to the transcripts were converted to RPKM units, where the read count is 1/(n*K*M), and n is the number of alignments in the genome, K is the transcript length divided by 1,000, and M is the sequencing depth including only reads mapping to mm9 divided by 1,000,000 (Mortazavi et al., 2008). This normalization allows for comparisons between transcripts of differing lengths and between samples of differing sequencing depths. To generate promoter maps, promoter regions were defined as −10,000 to +2000 bases relative to TSS (obtained from refGene catalog, UCSC Genome Browse). We plotted read counts overlapping promoter regions, except that the limit of 10 alignments was relaxed. For the chromosomal alignments in Fig. 1H and Supp. Figures, read numbers were computed for all non-overlapping consecutive 100 kb windows on each chromosome. Reads were normalized such that those mapping to n locations were counted as 1/nth of a read at each location. Graphs were plotted using custom scripts written in R. To generate Tables S-II though S-VI, a list of all enriched transcripts were found by comparing the RPKM scores on each strand for all transcripts in the WT and Ezh2−/− samples. Then their coordinates were intersected with coordinates of the feature of interest. Features not in NCBI37/mm9 mouse assembly coordinates were converted to those coordinates using UCSC’s LiftOver utility (http://genome.ucsc.edu/cgi-bin/hgLiftOver). Only features whose coordinates were convertible are shown.
RIP/qRT-PCR
Validation RIPs were performed as described (Zhao et al., 2008) using 5 μl of rabbit anti-mouse-Ezh2 antibodies (Active Motif) or normal rabbit IgG (Millipore). RIP was followed by quantitative, strand-specific RT-PCR using the iCycler iQ™ Real-time detection system (BioRad). Gene-specific PCR primer pairs are:
| Malat-1: | Forward 5′-GCCTTTTGTCACCTCACT-3′ |
| Reverse 5′-CAAACTCACTGCAAGGTCTC-3′ | |
| Malat1-as: | Forward 5′-TACTGGGTCTGGATTCTCTG-3′ |
| Reverse 5′-CAGTTCCGTGGTCTTTAGTG-3′ | |
| Foxn2-as: | Forward5′-GGCTATGCTCATGCTGTAAC |
| Reverse 5′-GTTACTGGCATCTTTCTCACA-3′ | |
| Ly6e-as: | Forward 5′-CCACACCGAGATTGAGATTG-3′ |
| Reverse 5′-GCCAGGAGAAAGACCATTAC-3′ | |
| Bgn-as: | Forward 5′-TGTGAACCCTTTCCTGGA-3′ |
| Reverse 5′-CTTCACAGGTCTCTAGCCA-3′ | |
| Gtl2: | Forward 5′-CGAGGACTTCACGCACAAC -3′ |
| Reverse 5′-TTACAGTTGGAGGGTCCTGG -3′ | |
| Gtl2-as: | Forward 5′-CACCCTGAACATCCAACA-3′ |
| Reverse 5′-CATCTGCTTTTCCTACCTGG-3′ | |
| Hapa1-upstream: | Forward 5′-GGTCCAAAATCGGCAGT-3′ |
| Reverse 5′-GTCCTCAAATCCCTACCAGA-3′ | |
| Htr6-downstream: | Forward 5′-ACACGGTCGTGAAGCTAGGTA-3′ |
| Reverse 5′-CAGTTGGAGTAGGCCATTCCC-3′ | |
| Nespas/TR019501: | Forward 5′-AGATGAGTCCAGGTGCTT-3′ |
| Reverse 5′-CAAGTCCAGAGTAGCCAAC-3′ |
Xist-Forward 3F5 and -Reverse 2R primers have been described (Zhao et al., 2008). For strand-specific cDNA synthesis, the reverse primer was used, qPCR carried out with SYBR green (BioRad), and threshold crossings (Ct) recorded. Each value was normalized to input RNA levels.
Northern blot analysis
5 μg of poly(A+) RNA were isolated from 16.7 ES cells, separated by 0.8% agarose gel containing formaldehyde, blotted onto Hybond-XL (GE Healthcare), and hybridized to probe using ULTRAhyb (Ambion) at 42°C. Probes were generated using Strip-EZ PCR kit (Ambion) and amplified from genomic DNA with: Malat1-AS-F, 5′-TGGGCTATTTTTCCTTACTGG-3′; Malat1-AS-R, 5′-GAGTCCCTTTGCTGTGCTG-3′; (Gtl2) Meg3-F, 5′-GCGATAAAGGAAGACACATGC-3′; Meg3-R, 5′-CCACTCCTTACTGGCTGCTC-3′; Meg3 ds-F3, 5′-ATGAAGTCCATGGTGACAGAC-3′; Meg3 ds-R2, 5′-ACGCTCTCGCATACACAATG-3′; Rtl1-F, 5′-GTTGGGGATGAAGATGTCGT-3′; Rtl1-R, 5′-GAGGCACAAGGGAAAATGAC-3′; Nespas ds-F, 5′-TGGACTTGCTACCCAAAAGG-3′; Nespas ds-R, 5′-CGATGTTGCCCAGTTATCAG-3′; Bgn-AS-F, 5′-CAACTGACCTCATAAGCAGCAC-3′; Bgn-AS-R, 5′-AGGCTGCTTTCTGCTTCACA-3′; Htr6 up-F, 5′-ATACTGAAGTGCCCGGAGTG-3′; Htr6 up-R, 5′-CAGGGGACAGACATCAGTGAG-3′.
UV-crosslink RIP
UV-crosslink IP was performed as described (Ule et al., 2005), except that transcripts in the RNA-protein complexes were not trimmed by RNAse treatment prior to RNA isolation in order to preserve full-length RNA for RT-PCR. Mouse ES cells were UV-irradiated at 254 nm, 400 mJ/cm2 (Stratagene Stratalinker), cell nuclei were lysed in RSB-Triton buffer (10mM Tris-HCl, 100mM NaCl, 2.5 mM MgCl2, 35 μg/mL digitonin, 0.5% triton X-100) with disruptive sonication. Nuclear lysates were pre-cleared with salmon sperm DNA/protein agarose beads for 1 hr at 4°C and incubated with antibodies overnight. RNA/antibody complexes were then precipitated with Protein A Dynabeads (Invitrogen), washed first in a low-stringency buffer (1XPBS [150 mM NaCl], 0.1% SDS, 0.5% deoxycholate, 0.5% NP-40), then washed twice in a high-stringency, high-salt buffer (5XPBS [750 mM NaCl], 0.1% SDS, 0.5% deoxycholate, 0.5% NP-40), and treated with proteinase K. RNA was extracted using Trizol (Invitrogen) and RT-qPCR was performed as described above.
Expression and purification of human PRC2 components
For expression of human PRC2 subunits, N-terminal flagged-tagged EZH2 and SUZ12 in pFastBac1 were expressed in Sf9 cells (Francis et al., 2001). For expression of the whole PRC2 complex, flag-tagged EZH2 was coexpressed with untagged SUZ12, EED, and RBAP48. Extracts were made by four freeze-thaw cycles in BC300 buffer (20mM HEPES pH 7.9, 300mM KCl, 0.2mM EDTA, 10% glycerol, 1mM DTT, 0.2mM PMSF, and complete protease inhibitors (Roche)) and bound to M2 beads for 4 h and washed with BC2000 before eluting in BC300 with 0.4mg/ml flag peptide. EZH2 and PRC2 were adjusted to 100mM KCl and loaded onto a HiTrap Heparin FF 1ml column and eluted with a 100–1000mM KCl gradient. Peak fractions were concentrated using Amicon ultra 10kDa MWCO concentrators (Millipore) and loaded onto a Superose 6 column equilibrated with BC300. Peak fractions were collected and concentrated. For SUZ12, the flag elution was concentrated and loaded onto a Superdex 200 column equilibrated with BC300.
Electrophoretic mobility shifting assays (EMSA)
RNA-EMSA is performed as previously described (Zhao et al., 2008). The 30 nt Hes-1 probe (~270 bp downstream of TSS in an antisense direction) was used for gel shifts. RNA probes were radiolabeled with [γ-33p]ATP using T4 polynucleotide kinase (Ambion). Purified PRC2 proteins (1 μg) were incubated with labeled probe for 1hr at 4 C. RNA–protein complexes were separated on a 4% non-denaturing polyacrylamide gel in 0.5×TBE at 250 V at 4 °C for 1 h. Gels were dried and exposed to Kodak BioMax film.
RNA pulldown assays
We incorporated T7 promoter sequence into forward primers for PCR products of RepA, Xist exon 1, and truncated Gtl2. Full-length Gtl2 was cloned into pYX-ASC and XistE1 into pEF1/V5/HisB (Invitrogen). Specific primer sequences were:
RepA-F: TAATACGACTCACTATAGGGAGACCCATCGGGGCCACGGATACCTGTGTGTCC
RepA-R: TAATAGGTGAGGTTTCAATGATTTACATCG
Truncated-Gtl2-F: TAATACGACTCACTATAGGGAGATTCTGAGACACTGACCATGTGCCCAGTGCACC
Truncated-Gtl2-R: CGTCGTGGGTGGAGTCCTCGCGCTGGGCTTCC
Xist E1-F: ATGCTCTGTGTCCTCTATCAGA
Xist E1-R: GAAGTCAGTATGGAGGGGGT
RNAs were then transcribed using the Mega Script T7 (Ambion), purified using Trizol, and slow-cooled to facilitate secondary structure formation. For pulldown assays, 3μg of Flag-PRC2 or Flag-GFP and 5 pmol of RNA supplemented with 20U RNAsin were incubated for 30 min on ice. 10μl of flag beads were added and incubated on a rotating wheel at 4°C for 1 hr. Beads were washed 3 times with 200 μl buffer containing 150mM KCl, 25mM Tris pH 7.4, 5mM EDTA, 0.5mM DTT, 0.5% NP40 and 1mM PMSF. RNA-protein complexes were eluted from flag beads by addition of 35μl of 0.2M-glycine pH2.5. Eluates were neutralized by addition of 1/10th volume of 1M Tris pH 8.0 and analyzed by gel electrophoresis.
Knockdown analysis and qRT-PCR
shRNA oligos were cloned into MISSION pLKO.1-puro (Sigma-Aldrich) vector and transfected into wild-type mouse ES cells by Lipofectamine 2000 (Invitrogen). After 10 days of puromycin selection, cells were collected and qRT-PCR was performed to confirm RNA knockdown. The corresponding scrambled sequence (MISSION Non-target shRNA) was used as a control (Scr). The shRNA oligos for Gtl2: (Top strand) 5′ - CCG GGC AAG TGA GAG GAC ACA TAG GCT CGA GCC TAT GTG TCC TCT CAC TTG CTT TTT G - 3′; (Bottom strand) 5′ - AAT TCA AAA AGC AAG TGA GAG GAC ACA TAG GCT CGA GCC TAT GTG TCC TCT CAC TTG C - 3′. qPCR primers for Gtl2 and Gtl2-as RNAs are as described above. Primers for Dlk1 RNAs: (Forward) 5′ - ACG GGA AAT TCT GCG AAA TA -3′; (Reverse) 5′ - CTT TCC AGA GAA CCC AGG TG -3′. Another Gtl2 shRNA was purchased from Open Biosystems (V2MM_97929). Ezh2 levels after knockdown with this shRNA were tested by qPCR (Zhao et al., 2008). After testing multiple clones, we concluded that Gtl2 could be knocked down in early passage clones (50–70%), but knockdown clones were difficult to maintain in culture long-term.
DNA ChIP and real-time PCR
ChIP was performed as described (Zhao et al., 2008). 5 μl of α-Ezh2 antibodies (Active Motif 39103), normal rabbit IgG (Upstate 12–370), and α-H3K27me3 (Upstate) were used per IP. Real-time PCR for ChIP DNA was performed at the Gtl2-proximal DMR with prGtl2F/prGtl2R, at the Gtl2-distal DMR with DMR-F/DMR-R, at the Dlk1 promoter with prDlk1F/prDlk1R, and at the Gapdh promoter with prGAPDH-F/prGAPDH-R. Primer sequences are as follows:
| proximal-DMR | 5′ - CATTACCACAGGGACCCCATTTT |
| proximal-DMR | 5′ - GATACGGGGAATTTGGCATTGTT |
| prDlk1F | 5′ - CTGTCTGCATTTGACGGTGAAC |
| prDlk1R | 5′ - CTCCTCTCGCAGGTACCACAGT |
| distal-DMR-F | 5′ - GCCGTAAAGATGACCACA |
| distal-DMR-R | 5′ - GGAGAAACCCCTAAGCTGTA |
| prGAPDH-F | 5′ - AGCATCCCTAGACCCGTACAGT |
| prGAPDH-R | 5′ – GGGTTCCTATAAATACGGACTGC |
| prActin-F | 5′ – GCA GGC CTA GTA ACC GAG ACA |
| prActin-R | 5′ – AGT TTT GGC GAT GGG TGC T |
Supplementary Material
Acknowledgments
S. Orkin generously provided the Ezh2−/− cell line. We thank M. Anguera for pEF1/V5/HisB and Xist E1 primers, B. del Rosario for Flag-GFP, and all members of the laboratory for many valuable discussions. J.T.K. is supported by a Postgraduate Scholarship from the Natural Sciences and Engineering Research Council of Canada, R.E.K. by the NIH (GM43901), J.J.S by the Korean Ministry of Education, Science, and Technology, and J.T.L. by the NIH (RO1-GM090278). J.T.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Axelson H. The Notch signaling cascade in neuroblastoma: role of the basic helix-loop-helix proteins HASH-1 and HES-1. Cancer Lett. 2004;204:171–178. doi: 10.1016/S0304-3835(03)00453-1. [DOI] [PubMed] [Google Scholar]
- Batzoglou S, Jaffe DB, Stanley K, Butler J, Gnerre S, Mauceli E, Berger B, Mesirov JP, Lander ES. ARACHNE: a whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006a;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006b;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Coombes C, Arnaud P, Gordon E, Dean W, Coar EA, Williamson CM, Feil R, Peters J, Kelsey G. Epigenetic properties and identification of an imprint mark in the Nesp-Gnasxl domain of the mouse Gnas imprinted locus. Mol Cell Biol. 2003;23:5475–5488. doi: 10.1128/MCB.23.16.5475-5488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisenko O, Shnyreva M, Suzuki H, Bomsztyk K. Point mutations in the WD40 domain of Eed block its interaction with Ezh2. Mol Cell Biol. 1998;18:5634–5642. doi: 10.1128/mcb.18.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Edwards CA, Mungall AJ, Matthews L, Ryder E, Gray DJ, Pask AJ, Shaw G, Graves JA, Rogers J, Dunham I, et al. The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 2008;6:e135. doi: 10.1371/journal.pbio.0060135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009 doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs Are Transcribed from Repressed Polycomb Target Genes and Interact with Polycomb Repressive Complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. The X as model for RNA’s niche in epigenomic regulation. Cold Spring Harb Perspect Biol. 2010;2:a003749. doi: 10.1101/cshperspect.a003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miremadi A, Oestergaard MZ, Pharoah PD, Caldas C. Cancer genetics of epigenetic genes. Hum Mol Genet. 2007;16(Spec No 1):R28–49. doi: 10.1093/hmg/ddm021. [DOI] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji Coordinates Control of PRC2 Enzymatic Activity and Target Gene Occupancy in Pluripotent Cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Rajasekhar VK, Begemann M. Concise review: roles of polycomb group proteins in development and disease: a stem cell perspective. Stem Cells. 2007;25:2498–2510. doi: 10.1634/stemcells.2006-0608. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. Embo J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Woojin K, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji Modulates Polycomb Activity and Self-Renewal versus Differentiation of Stem Cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Okamoto A, Kobayashi R, Shirai M, Obata Y, Ogawa H, Sotomaru Y, Kono T. Deletion of Gtl2, imprinted non-coding RNA, with its differentially methylated region induces lethal parent-origin-dependent defects in mice. Hum Mol Genet. 2009;18:1879–1888. doi: 10.1093/hmg/ddp108. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Bartolomei MS. SnapShot: imprinted gene clusters. Cell. 2007;130:958. doi: 10.1016/j.cell.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Wan LB, Bartolomei MS. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]
- Williamson CM, Turner MD, Ball ST, Nottingham WT, Glenister P, Fray M, Tymowska-Lalanne Z, Plagge A, Powles-Glover N, Kelsey G, et al. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet. 2006;38:350–355. doi: 10.1038/ng1731. [DOI] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers Polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.