Abstract
Gene therapy research has expanded from its original concept of replacing absent or defective DNA with functional DNA for transcription. Genetic material may be delivered via multiple vectors, including naked plasmid DNA, viruses and even cells with the goal of increasing gene expression; and the targeting of specific tissues or cell types is aimed at decreasing risks of systemic or side effects. As with the development of any drug, there is an amount of empiricism in the choice of gene target, route of administration, dosing and in particular the scaling-up from pre-clinical models to clinical trials. Systems Biology, whose arsenal includes high-throughput experimental and computational studies that account for the complexities of host-disease-therapy interactions, holds significant promise in aiding the development and optimization of gene therapies, including personalized therapies and the identification of biomarkers for success of these strategies. In this review we describe some of the obstacles and successes in gene therapy, using the specific example of growth factor gene delivery to promote angiogenesis and blood vessel remodeling in ischemic diseases; we also make references to anti-angiogenic gene therapy in cancer. The opportunities for Systems Biology and in silico modeling to improve on current outcomes are highlighted.
Keywords: Mathematical model, computational model, bioinformatics, angiogenesis, gene delivery, coronary artery disease, peripheral artery disease
Introduction
As the complexity of therapy-host interactions becomes increasingly apparent, the ability to predict the outcomes of interventions relies on a comprehensive dual approach of systematic measurement and integrative assembly of predictive computational models [1]. This is reflective of a shift from a one-gene-per-disease mindset to understanding of multi-gene complex disease processes, where the outcome is dependent on a synthesis of the behavior of interrelated networks and systems as a whole [2]. While systems biology has recently been applied to the study of diseases [3] and specific molecules or pathways [4], the scope of this review is to outline the potential for the application of systems biology practices to therapeutic design, and specifically to gene delivery. In particular we point out the advantages of this approach as regards personalized medicine. Systems biology principles have been applied only in part in the gene delivery arena so far, and by describing the approaches and opportunities, it is hoped that this review can spur further activity in this area.
Because the ‘systems’ designation is reflective of the complexity being studied rather than a specific limited set of tools, strict definitions are elusive; a recent analysis refers to Systems Biology studies as incorporating at least two of the following three characteristic components: High-throughput experiments; Computational models; and Bioinformatics [5]. Systems Medicine is a subset of Systems Biology related to human therapeutics. Systems Biology studies are: (i) Quantitative. Going beyond on/off, up/down, inhibit/repress representations of biological interactions [4]. (ii) Integrative. Compiling and analyzing data for multiple elements within the network and system, rather than a single readout [6,7]. (iii) Spanning multiple scales. Incorporating gene, message, protein and cell-level measurements and predictions, and possibly extended to tissue, organ and organism levels [8,9]. (iv) Predictive. Covering a sufficient fraction of the system to allow prediction of system behavior in untested states, and the ability to predict interventions to alter those states. Thus, systems biology approaches enable us to measure and predict multiple parameters simultaneously and under many conditions, giving a better picture of the state and dynamics of the system as a whole, rather than one specific element.
Gene therapy as it is currently practiced has failed to take advantage of the approaches of systems biology or systems medicine. Typically, patients receive treatments that are not based on their genomic or proteomic characteristics but rather on the up-regulation of the amount of a particular protein. As with other therapeutic approaches, empirically-derived maximum tolerated dose for a patient population, rather than maximally efficacious dose for the individual patient is used, which may not be the same, and thus the translation from pre-clinical animal model to human trial and to clinic is not optimal. Moreover, analysis of toxicity needs to consider both the effects of changes in protein concentration and the effects linked to the vector itself. Indeed, for plasmid DNA approaches the amount of DNA that can be delivered is often limited by the amount/cost of the vector, more so than the limiting effects of toxicity of the protein product.
Pre-clinical models strive to achieve homogeneity and reproducibility (e.g. using inbred strains of mice), while disease heterogeneity among patients that may manifest in heterogeneous activation of different genes, and in differential expression of multiple ligands, receptors and regulatory molecules may render certain therapies unsuccessful for some patients but effective for others. Taking advantage of systems biology can help to increase the success rate of translation. Possible reasons for failure of clinical trials include inappropriate selection of pre-clinical animal models, rendering the scale-up less accurate [10]; systems biology can help to identify the key drivers of therapeutic success, indicating which correlations between animal and human are most relevant. In target selection, systems biology can help identify targets (e.g. ligands, receptors or transcription factors) which are most likely to be effective for the broadest number of patients or patient subpopulations [11,12]. A systems approach also allows for comparison among multiple therapies targeting the same pathways, whether through delivery of genes, siRNA, shRNA or miRNA. Developing predictive and quantitative biomarkers for gene therapy would allow determination of patient subpopulations that are most likely to benefit from a given therapy, allowing the clinical trial to be more selective and increase the significance of successful outcomes and optimize dosing and timing of the gene therapy regimen [13].
Bioinformatics
Like computational modeling that we will review below, bioinformatics is an enabling methodology in the arsenal of systems biology. Modern bioinformatics generally includes application of computational and statistical methods to the analysis of genomic, proteomic, transcriptomic and metabolomic data (Omics), gene expression, cell signaling and metabolic pathways, protein-protein interactions, genome-wide association studies, and structure-based drug discovery. Bioinformatics can be regarded as a means to generate biological hypotheses and derive scientific knowledge from computer analysis of complex experimental data. Large-scale bioinformatic analysis of vector integration sites for gene therapy has led to new insights into vector–host cell interactions [14,15]. Genomic and proteomic approaches have been used to identify new targets for gene therapy in cardiovascular applications [16] and cancer [17]. However, the systematic use of this approach is still in its infancy relative to its potential. Bioinformatic approaches can be used to identify novel targets for both pro- and anti-angiogenic gene therapy strategies. A variety of systems biology approaches have been introduced in a study to engineer microvascular networks for ischemic diseases and also in application to tissue engineering; those include genomics, proteomics, transcriptomics, and pathway analysis integrated with high-throughput experimental methods, and computational modeling [4]
Mathematical and Computational Modeling of Gene Therapy
While the goal of gene therapy is the expression of the protein encoded by the delivered gene, it is not sufficient that the gene product is expressed. The downstream impact of this protein, expressed for a sufficient duration and magnitude, must exert a beneficial effect on the protein network, whether this “effect” is short or long term. Currently, gene expression studies often examine changes in target protein concentration over a period of time as opposed to the effects of these changes in protein expression on the overall pathways involved (Figure 2). The goal of systems biology is to overcome this by incorporating information on the expressed protein and the pathways in which it is involved, allowing us to optimize gene therapy approaches. This includes systematic use of mathematical models and in silico simulations of gene therapy. We can consider the following sequential steps each as candidates for systems biology studies: (1) target selection; (2) therapy design (e.g. promoters, enhancers, vector); (3) delivery (systemic, targeted); (4) uptake by cells; (5) expression of gene product; (6) impact on target and therapeutic outcome. In certain cases, even this last step alone can reveal considerable information on the design of gene therapy; for example, hypothetical fitness advantages conferred by delivered genes on a subset of cells – and the longevity of those advantages – are predicted by Markov models to have great impact on the prevalence of specific cell lineages in hematopoiesis [18,19]; these predictions guide the design of specific gene therapies that can provide these advantages. In other cases, several of the steps are simulated together, and mathematical and computational models can be used to optimize or identify markers of success and failure at each of the steps in gene therapy. A variety of model types and modeling methodologies can be used to quantify these steps. For example, models can be classified as deterministic vs stochastic or hybrid, continuum vs discrete, spatial (1-, 2-, or 3-dimensional) vs compartment, single- vs multi-scale. As far as modeling methodologies, models can be expressed in terms of algebraic equations, ordinary or partial differential equations (ODEs or PDEs), representation of stochastic processes using probability distributions, agent-based models (ABMs). Once the model is formulated in mathematical terms using a single or combination of methodologies, numerical methods are used to make the problem amenable for computer simulations, i.e. a computer algorithm. The problem is then solved on the computer (depending on the complexity of the problem, using a single processor or tens to thousands of processors) to generate predictions. Models often contain multiple parameters (e.g., kinetic coefficients, receptor expression, rates of degradation) whose values are not accurately known; this necessitates a sensitivity analysis where parameter values are varied within wide ranges to assess the sensitivity of the results to these variations. Many of the mathematical and computational models in the area of gene therapy have been reviewed in [20].
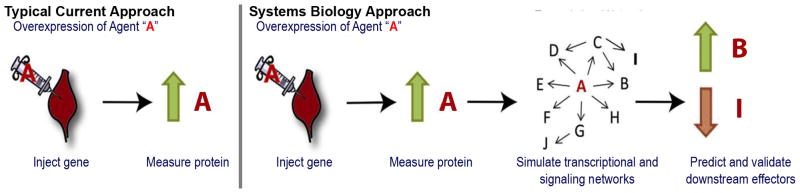
Figure 2. Evaluating success of gene delivery.
A vector (plasmid, virus or cell) carrying the gene for agent A injected causes overexpression of A. Traditionally, gene delivery studies have used elevated measurements of A (typically in blood) as confirmation that the treatment is working. However, the expression of A is only the start of the process of the tissue responding to the intervention. Agent A will interact with endogenous molecules and cause multifocal effects. It is the impact of these effects on the desired therapeutic outcome that defines the efficacy of the treatment, but these can be difficult to predict without the use of systems biology approaches and models.
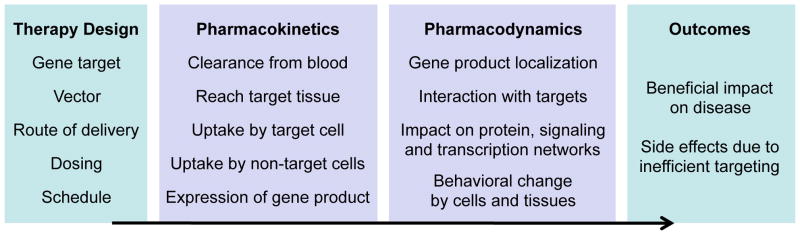
Effective therapeutic models would study both the pharmacokinetics (i.e. the fate of the gene vector in the body) and the pharmacodynamics (i.e. the ability of the vector to produce an effective gene product) (Figure 3), but many studies focus primarily on one or the other. So far no one modeling approach has integrated these together.
Figure 3. Systems Biology provides a predictive bridge between therapeutic design and outcomes.
Barriers between the gene delivery and the therapeutic outcome can be analyzed and simulated by applying existing frameworks for pharmacokinetics and pharmacodynamics. Systems biology allows the exploration of the potential space for therapy design, and of the space of possible interindividual variation, particularly in how the pharmacokinetics and pharmacodynamics would be altered. This aids in determination of the best therapy for each individual and the best therapies for groups of individuals.
Pharmacokinetic models
To better compare multiple possible therapeutic strategies, the pharmacokinetics of gene delivery are required. Recent mathematical studies have permitted the identification of the rate-limiting steps for retroviral delivery, focusing on extracellular and intracellular viral trafficking and integration [21]. The problem was formulated to simulate an experiment with mammalian cells at the bottom of a culture dish and retrovirus introduced to the medium. Mathematically, the vector distribution is described by a time-dependent one-dimensional diffusion equation with a decay term, and the concentration of target cells which carry viruses inside their cytoplasm is governed by an ordinary differential equation with respect to time. However, some of the terms are evaluated at time t-τ, where τ is the mean trafficking time of a virus in the cell cytoplasm which includes the times for reverse transcription and transport to the nucleus resulting in a delay differential equation. The distribution of virus-carrying cells containing k vectors is approximated by the Poisson probability density. This description is also related to stochastic models describing intracellular virus dynamics [22]. Conceptually similar models have been formulated for non-viral delivery described by a combination of kinetic equations and the distribution of plasmids in the cells described stochastically using Poisson and Bernoulli processes [23]. The results of simulations yielding probability distributions for the number of plasmids are validated against in vitro cell transfection experiments. A biophysically-detailed non-viral gene delivery model based on stochastic description was developed in [24]. Therefore, modeling methodology for viral and non-viral gene delivery has been developed and validated against in vitro data. Gene products that are extracellular, such as growth factors, add an additional level of complexity, as the transport of the protein throughout the target tissue, and potentially through the blood to other tissues, must also be included [25].
Pharmacodynamic models
Describing pharmacodynamics pertinent to gene therapy in biophysical and biochemical detail is a difficult task and only a few studies have been undertaken. Note that this problem is intimately related to models of gene expression and gene networks regulation [26]. It is also related to the intrinsic cell-to-cell variability of gene expression [27].
The simplest incorporation of gene product overexpression into a mathematical model is an additional production term, as in a recent model of endostatin delivery to tumor cells [28]. The major considerations are: (i) which cell types experience overexpression? This could depend on the ability of the cells to uptake the vector, as well as by design using a cell-specific promoter; (ii) what is the timeline of overexpression? Whether transient or stable, there is a characteristic time of upregulation and/or downregulation [29]; (iii) what is the peak overexpression? This level of expression of the gene product is likely to be the key to effective therapy; (iv) what is the variability in expression level from cell to cell? [30] There are several stochastic processes that can result in a range of copy numbers and thus expression differences between cells; (v) once expressed, what is the localization of the gene product? Growth factors with longer retention times (e.g. heparin-binding growth factors such as HGF) have shown some promise [31] in comparison to diffusible factors [32].
Effective pharmacodynamic models also include the host target of the gene product, e.g. VEGFR2 in the above endostatin model [28], or other components of the VEGF/VEGFR pathway in the case of VEGF gene delivery [30]. This has the advantage of detecting the typically nonlinear or even biphasic relationships between expression of the gene product and therapeutic efficacy. For example, VEGF delivery can induce endogenous production of additional factors that promote stabilization of newly formed blood vessels [33].
In the area of angiogenesis and vascular remodeling a variety of models have been developed, including models of pro- and anti-angiogenic therapeutic interventions; general recent reviews are available [4,9,34,35] Pro-angiogenic therapies in peripheral arterial disease are analyzed in [7,25].
The study of viral and nonviral gene delivery pharmacokinetics has so far been distinct from the study of endogenous signaling networks targeted for intervention. There are some technical challenges in integrating the two types of models together; for example, the pharmacodynamic models are frequently multicellular while the pharmackinetic models have to date focused on only the primary target cells, validating in vitro.
Gene therapy for neovascularization: the story so far
Both naked plasmid and adenoviral gene therapy of VEGF and other growth factors have been successful in multiple animal models of ischemic diseases such as peripheral and coronary artery disease [36–38]. Translation to human clinical trials, however, has not yet been successful despite a dozen clinical trials in those diseases delivering genes expressing VEGF or a VEGF-inducing transcription factor [7,36,39]). Delivery of other pro-angiogenic growth factors such as FGF in humans has also failed, though a meta-analysis suggests significant improvement when all the pro-angiogenic studies are pooled [40]. Indeed, no human gene therapy is yet FDA-approved. Finding an answer to why these clinical trials have failed – including testing hypotheses such as insufficient dosing [39] and insufficient inflammatory response [38] – requires integration of theoretical and experimental studies, scaling animal models up to human tissues. The specific disease present may play a role – while clinical trials of growth factor delivery in coronary artery disease and claudication in peripheral artery disease have had no major successes, there have been at least two promising recent Phase II trials in critical limb ischemia, delivering HGF [31] or FGF [41]. Interestingly, in both cases, plasmids were the vehicle and multiple injections were used (three and four doses, respectively, approximately two weeks apart). Anti-angiogenic approaches for use in cancer have also been studied, including delivery of natural angiogenesis inhibitors and cytokines that downregulate endogenous angiogenesis promoters [42].
Gene therapy for neovascularization: opportunities
Computational models can be used to predict the impact on efficacy of many parameters beyond target selection, and exploration of this space may lead to both improved and individualized therapies.
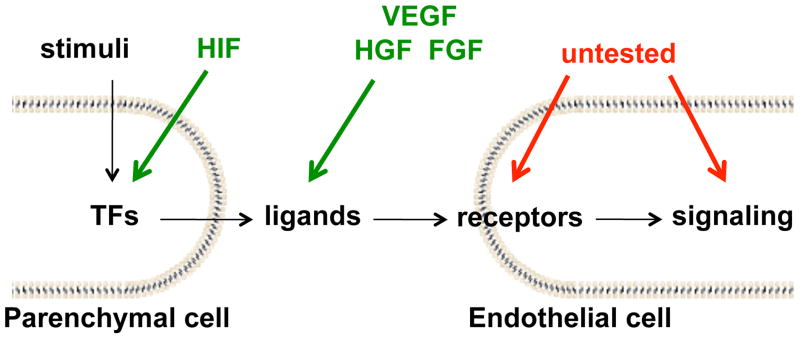
The failed clinical trials of gene delivery targeting VEGF performed to date should not in totality exclude the potential of gene delivery to target the vasculature. There are many additional available targets (Figure 1), including the VEGF-inducing transcription factor PGC1α [43,44], alternate ligands such as Placental Growth Factor PlGF, signaling pathway members, and the VEGF receptors themselves. The receptors are particularly intriguing as they have been observed to be elevated in exercise [45]. Perhaps the peak and duration of altered protein expression were insufficient.
Figure 1. Current and potential targets for pro-angiogenic gene therapy.
Although gene delivery of the pro-angiogenic cytokines VEGF, FGF and HGF, plus that of the transcription factor (TF) HIF, have so far not resulted in successful phase III trials, there are many approaches for targeting the same system, including alternate ligands, receptors, and downstream signaling targets. In particular, combination therapies have yet to be tried. Computational and systems biology are uniquely placed to compare these approaches using a consistent model system based on previous trials and preclinical experiments. Green arrows, tested in clinical trials; red arrows, not tested.
Most of the pro-angiogenic therapy studies have focused on increasing the expression of pro-angiogenic molecules. However a similar methodology would permit the delivery of siRNA, shRNA or miRNA to modulate expression of endogenous anti-angiogenic molecules, of which there are many. This would be particularly relevant in certain disease backgrounds where anti-angiogenic molecules are upregulated [46,47]. Negative regulation of VEGF family members such as soluble VEGF receptor 1 or other components alone or in combination could improve outcomes [25,48].
Rather than delivering naked DNA or viral vectors, the injection of cells that have been treated and transduced has the potential for well-targeted and well-tolerated delivery of the gene product [49,50]. This method has the advantage of establishing confirmed stable expression ex vivo before reimplantation [51,52], and may be particularly useful for highly localized induction [30]; focal induction of collaterals may be particularly effective in ischemic diseases as only a small number of collaterals are necessary for efficient restoration of flow [53]. Stable expression of gene products is still a major challenge of gene therapy. In many cases, expression is transient; this can result in a temporally-restricted response that may or may not be beneficial [54]. Gene therapy has typically been formulated a single-injection, but transient expression suggests that multiple dosing could be indicated. The pharmacokinetics of multiple dosing will be particularly important to study, as the transduction efficiency may decrease with each round of gene delivery (a problem that extends also to simultaneous delivery of multiple genes).
There are many issues associated with scaling up of doses from animal models to humans. The fraction of target cells successfully transduced is vastly lower in humans than in small animals; it has also been noted that gene doses and concomitant inflammation may also be lower in humans [38], resulting in less effective gene product production. Systems biological models can be formulated for both small animals and humans that take into account the impact of geometry, microanatomy and other differences in microenvironment. In addition, it may be advantageous to add larger animal models, such as swine to the studies of gene therapy targeting vasculature [47,55].
A particular strength of systems biology studies is the ability to predict effects on parts of the system that are not the direct or intended target of the therapy. In the case of gene therapy, the delivery of the vector to a multicellular tissue can result in ectopic expression, or an extracellular gene product may interact with other cells. Systemic delivery carries particular risks in this regard. Interfacing with studies of other major processes such as inflammation may aid in both therapy design and side effect reduction. Side effects and interactions of gene therapy with other drugs [56,57] would also be identifiable and this enables us to design around them. Synergy (or at least lack of interference) with other new promising therapies such as injected circulating endothelial progenitor cells (EPCs) could also be tested [58,59]. Conceptually, the idea that activity within a pathway is activated is probably more important than merely altering expression of a specific protein.
Mathematical models and bioinformatic studies designed to compare single therapies can also be used to test combinations of therapies. Gene therapy is increasingly being tested in combination with other modes of therapy [60]. In addition, some therapeutic processes (e.g. angiogenesis or arteriogenesis) appear to be best induced by multiple (two or three) complementary gene products [61–64].
Conclusion
Systems biology – the combination of experiment, bioinformatics and computational biology – can shed significant insight on both the challenges and opportunities for gene delivery. Uniquely it can synthesize many sources of information to provide detailed and testable predictions for therapy design, biomarker prediction, and side effects. It can be used to compare existing and novel therapeutic designs and to predict the behavior of multimodal or multigene therapy. While individual components of systems biology – e.g. computational modeling – have been applied to gene therapy studies as described here, a fully integrated systems approach has yet to be attempted in this area. As we look forward to the next decade of gene therapy studies, a one-size-fits-all approach to medicine is likely to lose ground in favor of a systems approach to the prediction of outcomes of, and biomarkers for, personalized medicine. Emphasis is increasing on personalized therapies, and systems biology – particularly bioinformatics and computational modeling – are extremely well suited to contribute in this area. Just as there is cell-to-cell variability in expression and behavior [23], there is also considerable inter-individual variation, possibly due to modulation by SNPs [65,66]. For each of the above opportunities in gene therapy design, there will be individuals who will respond better or worse than average, or not at all, particularly in the presence of disease backgrounds [48,67]. Thus we need biomarkers for the success of therapies. Typically biomarkers are identified in a post-hoc manner – as metrics that distinguished responders from non-responders. Bringing the tools of systems biology to bear allows us to predict biomarkers that can aid in therapy design and selection, as well as post-therapeutic monitoring.
Acknowledgments
Supported by NIH grants R00 HL093219 (FMG), R01 HL101200 (ASP/BHA), and R01 CA138264 (ASP).
Contributor Information
Feilim Mac Gabhann, Email: feilim@jhu.edu.
Brian H. Annex, Email: annex@virginia.edu.
Aleksander S. Popel, Email: apopel@jhu.edu.
References
- 1.Auffray C, Chen Z, Hood L. Systems medicine: The future of medical genomics and healthcare. Genome medicine. 2009;1(1):2. doi: 10.1186/gm2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn AC, Tewari M, Poon CS, Phillips RS. The clinical applications of a systems approach. PLoS medicine. 2006;3(7):e209. doi: 10.1371/journal.pmed.0030209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramsey SA, Gold ES, Aderem A. A systems biology approach to understanding atherosclerosis. EMBO molecular medicine. 2010;2(3):79–89. doi: 10.1002/emmm.201000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Sefcik LS, Wilson JL, Papin JA, Botchwey EA. Harnessing systems biology approaches to engineer functional microvascular networks. Tissue engineering. 2010;16(3):361–370. doi: 10.1089/ten.teb.2009.0611. Excellent review of experimental datasets and predictive analysis in systems biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popel AS, Hunter PJ. Systems biology and physiome projects. WIREs Systems Biology and Medicine. 2009;1(2):153–158. doi: 10.1002/wsbm.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mac Gabhann F, Popel AS. Systems biology of vascular endothelial growth factors. Microcirculation. 2008;15(8):715–738. doi: 10.1080/10739680802095964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mac Gabhann F, Qutub AA, Annex BH, Popel AS. Systems biology of pro-angiogenic therapies targeting the vegf system. WIREs Systems Biology and Medicine. 2010 doi: 10.1002/wsbm.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qutub AA, Liu G, Vempati P, Popel AS. Integration of angiogenesis modules at multiple scales: From molecular to tissue. Pacific Symposium on Biocomputing. 2009:316–327. [PMC free article] [PubMed] [Google Scholar]
- 9.Qutub AA, Mac Gabhann F, Karagiannis ED, Vempati P, Popel AS. Multiscale models of angiogenesis. IEEE Eng Med Biol Mag. 2009;28(2):14–31. doi: 10.1109/MEMB.2009.931791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10**.Lowenstein PR, Castro MG. Uncertainty in the translation of preclinical experiments to clinical trials. Why do most phase iii clinical trials fail? Current gene therapy. 2009;9(5):368–374. doi: 10.2174/156652309789753392. This analysis of the failure of Phase III clinical trials describes the value of computational models and systems biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoenig MR, Bianchi C, Rosenzweig A, Sellke FW. Decreased vascular repair and neovascularization with ageing: Mechanisms and clinical relevance with an emphasis on hypoxia-inducible factor-1. Current molecular medicine. 2008;8(8):754–767. doi: 10.2174/156652408786733685. [DOI] [PubMed] [Google Scholar]
- 12.Hoenig MR, Bianchi C, Sellke FW. Hypoxia inducible factor-1 alpha, endothelial progenitor cells, monocytes, cardiovascular risk, wound healing, cobalt and hydralazine: A unifying hypothesis. Current drug targets. 2008;9(5):422–435. doi: 10.2174/138945008784221215. [DOI] [PubMed] [Google Scholar]
- 13.Castro MG, Lowenstein PR. Gene therapy continues to make progress: Clinical and regulatory perspectives. Current gene therapy. 2009;9(5):327–328. doi: 10.2174/156652309789753374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Gabriel R, Eckenberg R, Paruzynski A, Bartholomae CC, Nowrouzi A, Arens A, Howe SJ, Recchia A, Cattoglio C, Wang W, Faber K, et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nature medicine. 2009;15(12):1431–1436. doi: 10.1038/nm.2057. Describes a novel method for better identifying viral integration sites in the human genome following retroviral gene delivery. [DOI] [PubMed] [Google Scholar]
- 15*.Felice B, Cattoglio C, Cittaro D, Testa A, Miccio A, Ferrari G, Luzi L, Recchia A, Mavilio F. Transcription factor binding sites are genetic determinants of retroviral integration in the human genome. PloS one. 2009;4(2):e4571. doi: 10.1371/journal.pone.0004571. A bioinformatic analysis that reveals motifs, particularly transcription factor binding sites, for predicting retroviral integration sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira AJ, Raizada MK. Genomic and proteomic approaches for targeting of angiotensin-converting enzyme2 for cardiovascular diseases. Current opinion in cardiology. 2008;23(4):364–369. doi: 10.1097/HCO.0b013e328303b79b. [DOI] [PubMed] [Google Scholar]
- 17.Al-Humadi H, Zarros A, Al-Saigh R, Liapi C. Genetic basis and gene therapy trials for thyroid cancer. Cancer genomics & proteomics. 7(1):31–49. [PubMed] [Google Scholar]
- 18.Abkowitz JL, Catlin SN, Guttorp P. Strategies for hematopoietic stem cell gene therapy: Insights from computer simulation studies. Blood. 1997;89(9):3192–3198. [PubMed] [Google Scholar]
- 19.Abkowitz JL, Golinelli D, Guttorp P. Strategies to expand transduced hematopoietic stem cells in vivo. Mol Ther. 2004;9(4):566–576. doi: 10.1016/j.ymthe.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 20**.Parra-Guillen ZP, Gonzalez-Aseguinolaza G, Berraondo P, Troconiz IF. Gene therapy: A pharmacokinetic/pharmacodynamic modelling overview. Pharmaceutical research. 2010 doi: 10.1007/s11095-010-0136-4. in press. Excellent review of both pharmacokinetic and pharmacodynamic models for simulating gene therapy. [DOI] [PubMed] [Google Scholar]
- 21**.Tayi VS, Bowen BD, Piret JM. Mathematical model of the rate-limiting steps for retrovirus-mediated gene transfer into mammalian cells. Biotechnology and bioengineering. 2010;105(1):195–209. doi: 10.1002/bit.22515. Computational model of multiple steps in retroviral delivery, including cell surface binding, entry, reverse transcription, nuclear import and integration. [DOI] [PubMed] [Google Scholar]
- 22.Lagache T, Dauty E, Holcman D. Physical principles and models describing intracellular virus particle dynamics. Current opinion in microbiology. 2009;12 (4):439–445. doi: 10.1016/j.mib.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 23**.Schwake G, Youssef S, Kuhr JT, Gude S, David MP, Mendoza E, Frey E, Radler JO. Predictive modeling of non-viral gene transfer. Biotechnology and bioengineering. 2010;105(4):805–813. doi: 10.1002/bit.22604. A combined computational-experimental approach, which shows that a two-step stochastic model describes well the transfection and co-transfection rates of plasmids. [DOI] [PubMed] [Google Scholar]
- 24*.Dinh AT, Pangarkar C, Theofanous T, Mitragotri S. Understanding intracellular transport processes pertinent to synthetic gene delivery via stochastic simulations and sensitivity analyses. Biophysical journal. 2007;92(3):831–846. doi: 10.1529/biophysj.106.095521. A computational model of gene delivery using polymeric approaches, accounting for cell geometry and endosomal trafficking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu FT, Stefanini MO, Mac Gabhann F, Kontos CD, Annex BH, Popel AS. Vegf and soluble vegf receptor-1 (sflt-1) distributions in peripheral arterial disease: An in silico model. American journal of physiology. 2010;298(6):H2174–H2191. doi: 10.1152/ajpheart.00365.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooke EJ, Savage RS, Wild DL. Computational approaches to the integration of gene expression, chip-chip and sequence data in the inference of gene regulatory networks. Seminars in cell & developmental biology. 2009;20(7):863–868. doi: 10.1016/j.semcdb.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Niepel M, Spencer SL, Sorger PK. Non-genetic cell-to-cell variability and the consequences for pharmacology. Current opinion in chemical biology. 2009;13(5–6):556–561. doi: 10.1016/j.cbpa.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Billy F, Ribba B, Saut O, Morre-Trouilhet H, Colin T, Bresch D, Boissel JP, Grenier E, Flandrois JP. A pharmacologically based multiscale mathematical model of angiogenesis and its use in investigating the efficacy of a new cancer treatment strategy. Journal of theoretical biology. 2009;260(4):545–562. doi: 10.1016/j.jtbi.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 29*.Berraondo P, Gonzalez-Aseguinolaza G, Troconiz IF. Semi-mechanistic pharmacodynamic modelling of gene expression and silencing processes. Eur J Pharm Sci. 2009;37(3–4):418–426. doi: 10.1016/j.ejps.2009.03.013. Co-delivering reporter and expression plasmids, gene silencing was well described by a mathematical model of the gene products. [DOI] [PubMed] [Google Scholar]
- 30.Mac Gabhann F, Ji JW, Popel AS. Multi-scale computational models of pro-angiogenic treatments in peripheral arterial disease. Annals of biomedical engineering. 2007;35(6):982–994. doi: 10.1007/s10439-007-9303-0. [DOI] [PubMed] [Google Scholar]
- 31.Powell RJ, Simons M, Mendelsohn FO, Daniel G, Henry TD, Koga M, Morishita R, Annex BH. Results of a double-blind, placebo-controlled study to assess the safety of intramuscular injection of hepatocyte growth factor plasmid to improve limb perfusion in patients with critical limb ischemia. Circulation. 2008;118(1):58–65. doi: 10.1161/CIRCULATIONAHA.107.727347. [DOI] [PubMed] [Google Scholar]
- 32.Rajagopalan S, Mohler ER, 3rd, Lederman RJ, Mendelsohn FO, Saucedo JF, Goldman CK, Blebea J, Macko J, Kessler PD, Rasmussen HS, Annex BH. Regional angiogenesis with vascular endothelial growth factor in peripheral arterial disease: A phase ii randomized, double-blind, controlled study of adenoviral delivery of vascular endothelial growth factor 121 in patients with disabling intermittent claudication. Circulation. 2003;108(16):1933–1938. doi: 10.1161/01.CIR.0000093398.16124.29. [DOI] [PubMed] [Google Scholar]
- 33.Korpisalo P, Yla-Herttuala S. Stimulation of functional vessel growth by gene therapy. Integr Biol (Camb) 2010;2(2–3):102–112. doi: 10.1039/b921869f. [DOI] [PubMed] [Google Scholar]
- 34.Peirce SM. Computational and mathematical modeling of angiogenesis. Microcirculation. 2008;15(8):739–751. doi: 10.1080/10739680802220331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byrne HM. Dissecting cancer through mathematics: From the cell to the animal model. Nature reviews. 2010;10(3):221–230. doi: 10.1038/nrc2808. [DOI] [PubMed] [Google Scholar]
- 36**.Gupta R, Tongers J, Losordo DW. Human studies of angiogenic gene therapy. Circulation research. 2009;105(8):724–736. doi: 10.1161/CIRCRESAHA.109.200386. A comprehensive review of applications of pro-angiogenic gene therapy in ischemic diseases, e.g. coronary artery disease and peripheral artery disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrovic D. The role of vascular endothelial growth factor gene as the genetic marker of atherothrombotic disorders and in the gene therapy of coronary artery disease. Cardiovascular & hematological agents in medicinal chemistry. 2010;8(1):47–54. doi: 10.2174/187152510790796183. [DOI] [PubMed] [Google Scholar]
- 38.Yla-Herttuala S. Gene therapy with vascular endothelial growth factors. Biochemical Society transactions. 2009;37(Pt 6):1198–1200. doi: 10.1042/BST0371198. [DOI] [PubMed] [Google Scholar]
- 39.Karvinen H, Yla-Herttuala S. New aspects in vascular gene therapy. Current opinion in pharmacology. 2010;10(2):208–211. doi: 10.1016/j.coph.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 40.De Haro J, Acin F, Lopez-Quintana A, Florez A, Martinez-Aguilar E, Varela C. Meta-analysis of randomized, controlled clinical trials in angiogenesis: Gene and cell therapy in peripheral arterial disease. Heart and vessels. 2009;24(5):321–328. doi: 10.1007/s00380-008-1140-z. [DOI] [PubMed] [Google Scholar]
- 41.Nikol S, Baumgartner I, Van Belle E, Diehm C, Visona A, Capogrossi MC, Ferreira-Maldent N, Gallino A, Wyatt MG, Wijesinghe LD, Fusari M, et al. Therapeutic angiogenesis with intramuscular nv1fgf improves amputation-free survival in patients with critical limb ischemia. Mol Ther. 2008;16(5):972–978. doi: 10.1038/mt.2008.33. [DOI] [PubMed] [Google Scholar]
- 42**.Samaranayake H, Maatta AM, Pikkarainen J, Yla-Herttuala S. Future prospects and challenges of antiangiogenic cancer gene therapy. Human gene therapy. 2010;21(4):381–396. doi: 10.1089/hum.2010.017. A comprehensive review of applications of anti-angiogenic gene therapy for cancer. [DOI] [PubMed] [Google Scholar]
- 43.Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, Cooper M, Laznik D, Chinsomboon J, Rangwala SM, Baek KH, et al. Hif-independent regulation of vegf and angiogenesis by the transcriptional coactivator pgc-1alpha. Nature. 2008;451(7181):1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 44.Chinsomboon J, Ruas J, Gupta RK, Thom R, Shoag J, Rowe GC, Sawada N, Raghuram S, Arany Z. The transcriptional coactivator pgc-1alpha mediates exercise-induced angiogenesis in skeletal muscle. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(50):21401–21406. doi: 10.1073/pnas.0909131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji JW, Mac Gabhann F, Popel AS. Skeletal muscle vegf gradients in peripheral arterial disease: Simulations of rest and exercise. American journal of physiology. 2007;293(6):H3740–3749. doi: 10.1152/ajpheart.00009.2007. [DOI] [PubMed] [Google Scholar]
- 46.Sodha NR, Clements RT, Boodhwani M, Xu SH, Laham RJ, Bianchi C, Sellke FW. Endostatin and angiostatin are increased in diabetic patients with coronary artery disease and associated with impaired coronary collateral formation. American journal of physiology. 2009;296(2):H428–434. doi: 10.1152/ajpheart.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sodha NR, Boodhwani M, Clements RT, Xu SH, Khabbaz KR, Sellke FW. Increased antiangiogenic protein expression in the skeletal muscle of diabetic swine and patients. Arch Surg. 2008;143(5):463–470. doi: 10.1001/archsurg.143.5.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hazarika S, Dokun AO, Li Y, Popel AS, Kontos CD, Annex BH. Impaired angiogenesis after hindlimb ischemia in type 2 diabetes mellitus: Differential regulation of vascular endothelial growth factor receptor 1 and soluble vascular endothelial growth factor receptor 1. Circulation research. 2007;101(9):948–956. doi: 10.1161/CIRCRESAHA.107.160630. [DOI] [PubMed] [Google Scholar]
- 49.Roncalli J, Tongers J, Renault MA, Losordo DW. Biological approaches to ischemic tissue repair: Gene- and cell-based strategies. Expert review of cardiovascular therapy. 2008;6(5):653–668. doi: 10.1586/14779072.6.5.653. [DOI] [PubMed] [Google Scholar]
- 50.Tongers J, Roncalli JG, Losordo DW. Therapeutic angiogenesis for critical limb ischemia: Microvascular therapies coming of age. Circulation. 2008;118(1):9–16. doi: 10.1161/CIRCULATIONAHA.108.784371. [DOI] [PubMed] [Google Scholar]
- 51.Misteli H, Wolff T, Fuglistaler P, Gianni-Barrera R, Gurke L, Heberer M, Banfi A. High-throughput flow cytometry purification of transduced progenitors expressing defined levels of vascular endothelial growth factor induces controlled angiogenesis in vivo. Stem cells (Dayton, Ohio) 2010;28(3):611–619. doi: 10.1002/stem.291. [DOI] [PubMed] [Google Scholar]
- 52.Ozawa CR, Banfi A, Glazer NL, Thurston G, Springer ML, Kraft PE, McDonald DM, Blau HM. Microenvironmental vegf concentration, not total dose, determines a threshold between normal and aberrant angiogenesis. The Journal of clinical investigation. 2004;113(4):516–527. doi: 10.1172/JCI18420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mac Gabhann F, Peirce SM. Collateral capillary arterialization following arteriolar ligation in murine skeletal muscle. Microcirculation. 2010 doi: 10.1111/j.1549-8719.2010.00034.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gounis MJ, Spiga MG, Graham RM, Wilson A, Haliko S, Lieber BB, Wakhloo AK, Webster KA. Angiogenesis is confined to the transient period of vegf expression that follows adenoviral gene delivery to ischemic muscle. Gene therapy. 2005;12(9):762–771. doi: 10.1038/sj.gt.3302481. [DOI] [PubMed] [Google Scholar]
- 55.Boodhwani M, Voisine P, Ruel M, Sodha NR, Feng J, Xu SH, Bianchi C, Sellke FW. Comparison of vascular endothelial growth factor and fibroblast growth factor-2 in a swine model of endothelial dysfunction. Eur J Cardiothorac Surg. 2008;33(4):645–650. doi: 10.1016/j.ejcts.2007.12.016. discussion 251–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boodhwani M, Mieno S, Feng J, Sodha NR, Clements RT, Xu SH, Sellke FW. Atorvastatin impairs the myocardial angiogenic response to chronic ischemia in normocholesterolemic swine. The Journal of thoracic and cardiovascular surgery. 2008;135(1):117–122. doi: 10.1016/j.jtcvs.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 57.Boodhwani M, Mieno S, Voisine P, Feng J, Sodha N, Li J, Sellke FW. High-dose atorvastatin is associated with impaired myocardial angiogenesis in response to vascular endothelial growth factor in hypercholesterolemic swine. The Journal of thoracic and cardiovascular surgery. 2006;132(6):1299–1306. doi: 10.1016/j.jtcvs.2006.05.060. [DOI] [PubMed] [Google Scholar]
- 58.Kaur S, Kumar TR, Uruno A, Sugawara A, Jayakumar K, Kartha CC. Genetic engineering with endothelial nitric oxide synthase improves functional properties of endothelial progenitor cells from patients with coronary artery disease: An in vitro study. Basic research in cardiology. 2009;104(6):739–749. doi: 10.1007/s00395-009-0039-x. [DOI] [PubMed] [Google Scholar]
- 59.Tongers J, Roncalli JG, Losordo DW. Role of endothelial progenitor cells during ischemia-induced vasculogenesis and collateral formation. Microvascular research. 2010 doi: 10.1016/j.mvr.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Candolfi M, Kroeger KM, Muhammad AK, Yagiz K, Farrokhi C, Pechnick RN, Lowenstein PR, Castro MG. Gene therapy for brain cancer: Combination therapies provide enhanced efficacy and safety. Current gene therapy. 2009;9(5):409–421. doi: 10.2174/156652309789753301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benest AV, Stone OA, Miller WH, Glover CP, Uney JB, Baker AH, Harper SJ, Bates DO. Arteriolar genesis and angiogenesis induced by endothelial nitric oxide synthase overexpression results in a mature vasculature. Arteriosclerosis, thrombosis, and vascular biology. 2008;28(8):1462–1468. doi: 10.1161/ATVBAHA.108.169375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J, Wei Y, Liu K, Yuan C, Tang Y, Quan Q, Chen P, Wang W, Hu H, Yang L. Synergistic effects of fgf-2 and pdgf-bb on angiogenesis and muscle regeneration in rabbit hindlimb ischemia model. Microvascular research. 2010 doi: 10.1016/j.mvr.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 63.Matyas L, Schulte KL, Dormandy JA, Norgren L, Sowade O, Grotzbach G, Palmer-Kazen U, Rubanyi GM, Wahlberg E. Arteriogenic gene therapy in patients with unreconstructable critical limb ischemia: A randomized, placebo-controlled clinical trial of adenovirus 5-delivered fibroblast growth factor-4. Human gene therapy. 2005;16(10):1202–1211. doi: 10.1089/hum.2005.16.1202. [DOI] [PubMed] [Google Scholar]
- 64.de Paula EV, Flores-Nascimento MC, Arruda VR, Garcia RA, Ramos CD, Guillaumon AT, Annichino-Bizzacchi JAM. Dual gene transfer of fibroblast growth factor-2 and platelet derived growth factor-bb using plasmid deoxyribonucleic acid promotes effective angiogenesis and arteriogenesis in a rodent model of hindlimb ischemia. Transl Res. 2009;153(5):232–239. doi: 10.1016/j.trsl.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 65.Kitsios G, Zintzaras E. Ace (i/d) polymorphism and response to treatment in coronary artery disease: A comprehensive database and meta-analysis involving study quality evaluation. BMC medical genetics. 2009;10(50) doi: 10.1186/1471-2350-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petrovic D. Vascular endothelial growth factor gene polymorphisms and myocardial infarction. Cardiology. 2009;114(1):8–10. doi: 10.1159/000210188. [DOI] [PubMed] [Google Scholar]
- 67.Boodhwani M, Sellke FW. Therapeutic angiogenesis in diabetes and hypercholesterolemia: Influence of oxidative stress. Antioxidants & redox signaling. 2009;11(8):1945–1959. doi: 10.1089/ars.2009.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]