Abstract
Purpose
Tubby-like proteins (TULPs) are a family of four proteins, two of which have been linked to neurosensory disease phenotypes. TULP1 is a photoreceptor-specific protein that is mutated in retinitis pigmentosa, an inherited retinal disease characterized by the degeneration of rod and cone photoreceptor cells. To investigate the function of TULP1 in maintaining the health of photoreceptors, the authors sought the identification of interacting proteins.
Methods
Immunoprecipitation from retinal lysates, followed by liquid chromatography tandem mass spectrometry and in vitro binding assays, were used to identify TULP1 binding partners. RT-PCR was performed on total RNA from wild-type mouse retina to identify the Dynamin-1 isoform expressed in the retina. Immunocytochemistry was used to determine the localization of TULP1 and Dynamin-1 in photoreceptor cells. Electroretinography (ERG) and light microscopy were used to phenotype tulp1–/– mice at a young age.
Results
Immunoprecipitation from retinal lysate identified Dynamin-1 as a possible TULP1 binding partner. GST pull-down assays further supported an interaction between TULP1 and Dynamin-1. In photoreceptor cells, Dynamin-1 and TULP1 colocalized primarily to the outer plexiform layer, where photoreceptor terminals synapse on second-order neurons and, to a lesser extent, to the inner segments, where polarized protein translocation occurs. ERG analyses in young tulp1–/– mice indicated a decreased b-wave at ages when the retina retained a full complement of photoreceptor cells.
Conclusions
These data indicated that TULP1 interacts with Dynamin-1 and suggested that TULP1 is involved in the vesicular trafficking of photoreceptor proteins, both at the nerve terminal during synaptic transmission and at the inner segment during protein translocation to the outer segment. These results also raised the possibility that normal synaptic function requires TULP1, and they motivate a closer look at synaptic architecture in the developing tulp1–/– retina.
The Tubby-like protein (TULP) family is found in multicellular organisms of both the plant and the animal kingdoms. In vertebrates, there are four members of the family: TUB, TULP1, TULP2, and TULP3.1 TULPs are localized primarily to neuronal tissues. TUB and TULP3 are widely distributed throughout the central nervous system, and TULP1 and TULP2 are restricted largely to the photoreceptors of the retina and testis, respectively.1–6 Though not well understood, TULPs appear to play an important role in neuronal development and function. In fact, two family members have been linked to neurosensory disease phenotypes, including photoreceptor degeneration. Mice homozygous for a mutation in the tub gene develop retinal and cochlear degeneration and adult-onset obesity associated with insulin resistance.4,7,8 Mutations in human TULP1 cause retinitis pigmentosa, a heterogeneous group of inherited retinal diseases in which the rod and cone photoreceptor cells degenerate, leading to blindness.9–12 Mice lacking TULP1 also develop early-onset, progressive, pan-retinal degeneration involving rod and cone photoreceptor cells.13,14 TULP3 appears to be required for normal neural development because tulp3–/– mice die at embryonic day 14.15 Embryos have numerous nervous system defects, including failure in neural tube development and increased neuronal apoptosis. Taken together, results from the analyses of TULP genes suggest that their primary site of action is in neuronal cells.
The role of TULPs in the nervous system remains unclear, though a variety of functions have been postulated for TUB, including vesicular trafficking,16 mediation of insulin signaling,17 gene transcription,18 G-protein signaling,19 and ribosomal RNA synthesis.5 TULP1 is located exclusively in photoreceptors, localizing in the inner segments and connecting cilium, perikarya, and outer plexiform layer (OPL).13,14,20 TULP1 is a cytoplasmic protein that associates with cellular membranes and the actin cytoskeleton.21 These results, and observations that rod and cone opsins are mislocalized in tulp1–/– photoreceptors, which also develop an abnormal accumulation of rhodopsin-bearing extracellular vesicles surrounding the ellipsoid region of the inner segments, have led us to hypothesize that TULP1 may be involved in actin cytoskeletal functions such as protein trafficking that take place at or near the plasma membrane from the inner segment through the connecting cilium into the outer segment of photoreceptor cells.13,20,21
A critical step toward understanding the physiology of TULPs and how TULP1 maintains the health of the photoreceptor cells would be the identification of TULP1 binding partners. In the present study, we show that TULP1 binds the neuronal-specific protein Dynamin-1 and that it colocalizes with Dynamin-1 at the OPL and inner segments of photoreceptors. Dynamin-1 is a GTPase involved in vesicular formation, trafficking, and recycling. Studies indicate that Dynamin-1 has roles in endocytosis, vesicle formation, and movement at the trans-Golgi network (TGN) and plasma membrane and in vesicle recycling at neuronal synapses. Consistent with a role for TULP1 at the photoreceptor-to-bipolar cell synapse, our current data show that the bipolar cell contributions to the electroretinogram (ERG) are markedly reduced in young tulp1–/– mice that retain a full complement of photoreceptor cells. The current data provide the first indication of a molecular pathway in which TULP1 may be involved, and they are consistent with the hypothesis that TULP1 is a member of the molecular machinery required for the polarized vesicular translocation of proteins, within photoreceptor outer segments and terminals.
Methods
Mice
The generation of tulp1–/– mice has been described previously.13 Homozygote tulp1–/– mice and wild-type littermate controls were derived from tulp1+/– heterozygous mating and were identified by PCR amplification of genomic DNA, as previously described.13 All experiments on animals were approved by the Institutional Animal Care and Use Committee of the Cleveland Clinic Foundation and were performed in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Immunoprecipitation and Protein Identification
Immunoprecipitation (IP) experiments were performed as previously reported.21,22 In brief, two bovine retinas were homogenized and lysed in 5 mL NP-40 lysis buffer supplemented with protease inhibitors. The suspension was centrifuged, and the supernatant was removed from the pellet. The retinal lysate was used for IP analysis with the polyclonal antibody M-tulp1N.20 Total retinal lysate protein (1 mg), determined by Bradford method, was added to 50 μL M-tulp1N coupled to the protein A beads. The binding reaction occurred overnight at 4°C. As a negative control, protein A beads alone were allowed to bind to retinal lysate. Reactions were centrifuged, supernatants were removed, and beads were washed and resuspended in lysis buffer. Immunoprecipitated antigens were fractionated by SDS-PAGE on a 10% gel. The gel was stained with colloidal Coomassie blue (Gel Code Blue; Pierce, Rockford, IL), and the bands were excised for in situ tryptic digestion and mass spectrometric analysis. Identification of proteins by liquid chromatography tandem mass spectrometry (LC MS/MS) was performed with a quadrupole time-of-flight (QTOF) instrument equipped for exact mass, high-sensitivity LC MS/MS analysis (CapLC System; Waters Corporation, Milford, MA), as described previously.21–24 Protein identifications from MS/MS data were made with search engines (ProteinLynx Global Server [Waters Corporation]; Mascot [Matrix Science, Boston, MA]) and the Swiss-Protein and NCBI protein sequence databases.
Isolation of RNA and Generation of Dynamin-1 cDNA
Total RNA from wild-type mouse retina was isolated (Trizol; Gibco, Grand Island, NY) according to the manufacturer's instructions. Total RNA was used for reverse transcription (RT) in a 25-μL reaction using a primer specific to the 3′ untranslated region of mouse Dynamin-1 (accession no. NM010065; 5′ ttagaggtcgaaggggggcct 3′). First-strand cDNA synthesis was performed (AMV First-Strand cDNA Synthesis Kit; Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The RT sample (2 μL) was used to PCR amplify full-length mouse Dynamin-1 cDNA using the GC-RICH PCR System (Roche, Indianapolis, IN) in a 50-μL reaction. The 5′ PCR primer was complementary to the initiation sequence, and the 3′ PCR primer included the corresponding stop codon of all Dynamin-1 isoforms (sense primer, 5′catgggcaaccgcggcatggaagac 3′; antisense primer, 5′ tcagagcctggctttgccagcgcg 3′). Automated DNA sequencing of the PCR product was performed to confirm that the complete and correct Dynamin-1 cDNA was isolated.
Western Blot
Western blot analysis was performed as previously described.20–22 Briefly, proteins were separated on SDS-polyacrylamide gels and electroblotted to polyvinylidene difluoride (PVDF) membranes. Membranes were incubated with primary antibodies, followed by peroxidase-conjugated secondary antibodies, and were detected by chemiluminescence.
Construction of Plasmids
The full-length mouse Dynamin-1 cDNA isolated from retina (amino acids 1–814) was subcloned into the SmaI site of the pGEX-2TK vector (designated GST-Dyn1), and the correct fusion of the construct was verified by automated sequencing. The full-length Dynamin-1 construct was used as a template to generate three different Dynamin-1 domain constructs. Primers 5′ catgggcaaccgcggcatggaagac 3′ and 5′ ttagaccagaatctcatcctggttccc 3′ were used to amplify the N-terminal domain consisting of the GTPase domain (amino acids 1–520, designated GST-Dyn1-N). Primers 5′ cgtcagcacgcccatgcccccgccc 3′ and 5′ tcagagcctggctttgccagcgcg 3′ were used to amplify the proline-rich domain (PRD) (amino acids 750 – 814, designated GST-Dyn1-PRD). Primers 5′ cattcgaaaggggtggttgaccatc 3′ and 5′ tcagagcctggctttgccagcgcg 3′ were used to amplify the C-terminal domain consisting of the pleckstrin homology (PH) domain, the GTPase effector domain (GED), and the PRD domain (amino acids 521–814, designated GST-Dyn1-C). DNA constructs were verified by automated sequence analysis.
Glutathione S-Transferase Pull-down Assay
Wild-type and domain specific glutathione S-transferase (GST)-Dynamin-1 fusion constructs were expressed as proteins in the Escherichia coli BL21 strain. Cells were sonicated in buffer (PBS, 1% Triton X-100, and proteinase inhibitors) and centrifuged (16,000g for 15 minutes), and the clarified cell lysate was incubated with 50 μL glutathione-Sepharose 4B beads at room temperature for 1 hour, washed 3 times with PBS, and resuspended in 50 μL buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 15 mM EDTA, 0.1% Triton X-100, 1 mM dithiothreitol [DTT], and proteinase inhibitors). The integrity and purity of each GST fusion protein was analyzed by SDS-PAGE and Coomassie staining. Each GST fusion protein bound to glutathione-Sepharose beads (approximately 10 μg) was incubated with bovine retinal lysate (approximately 1 mg total protein in 300 μL buffer) at 4°C overnight with gentle rotating. Approximately equal amounts of each GST fusion protein were used. Beads were then pelleted, washed 3 times in PBS, resuspended in 50 μL SDS sample buffer, and analyzed by SDS-PAGE on 10% gel, and immunoblotted with TULP1 primary antibodies.
Immunocytochemistry
Mouse eyes were fixed in 4% paraformaldehyde in PBS for 30 minutes. After removal of the cornea and lens, the posterior pole was immersed through a graded series of sucrose solutions as follows: 10% for 1 hour, 20% for 1 hour, and 30% overnight. The posterior pole was embedded in OCT freezing medium, frozen, and sectioned at 10-μm thickness with a cryostat (Leica, Wetzlar, Germany). Retinal sections were blocked in 5% goat serum before incubation with antibodies. TULP1 was stained using polyclonal M-tulp1N antibody at 4°C overnight; this was followed by incubation with goat anti–rabbit IgG (Alexa Fluor 488; Molecular Probes, Eugene, OR) for 1 hour. Dynamin-1 was stained using a monoclonal antibody (Hudy 1; Upstate Biotechnology, Lake Placid, NY) at 4°C overnight, followed by incubation with goat anti–mouse IgG (Alexa Fluor 532; Molecular Probes) for 1 hour. Sections were then rinsed 3 times with PBS and coverslipped with mounting medium with DAPI (Vectashield; Vector Laboratories, Burlingame, CA). Sections were examined with a fluorescent microscope (Microphot-2; Nikon, Tokyo, Japan) equipped with a CCD camera (SPOT 2; Diagnostic Instruments, Sterling Heights, MI).
Electroretinography
ERGs were recorded from tulp1–/– and control (tulp1+/– or +/+) littermates at postnatal day (P)12 and P15. After overnight dark adaptation, mice were anesthetized with ketamine (80 mg/kg) and xylazine (16 mg/kg). Eye drops were used to anesthetize the cornea (1% proparacaine HCl) and to dilate the pupil (1% tropicamide, 2.5% phenylephrine HCl, 1% cyclopentolate HCl). Mice were placed on a temperature-regulated heating pad throughout the recording session. ERGs were recorded using a stainless steel electrode that made contact with the corneal surface through a thin layer of 0.7% methylcellulose. Needle electrodes placed in the cheek and the tail served as reference and ground leads, respectively. Responses were differentially amplified (0.3–1500 Hz), averaged, and stored using a signal averaging system (UTAS E-3000; LKC Technologies, Gaithersburg, MD). ERGs were recorded to flash stimuli presented in a ganzfeld (LKC Technologies).
Light Microscopy
At the completion of the ERG session, mice were euthanatized by cervical dislocation and the eyes were fixed in 2% paraformaldehyde, 2% glutaraldehyde. After overnight fixation, the anterior segment was removed, and the retina was rinsed 3 times with 0.1 M PO4 buffer, followed by postfixation in 1% OsO4 for 30 minutes. The retina was dehydrated in a gradient series of ethanol, rinsed 3 times with propylene oxide, and infiltrated overnight in a 1:1 mixture of propylene oxide and Epon. The next day the tissue was embedded in Epon.
Results
Identification of Dynamin-1 as a TULP1 Binding Partner
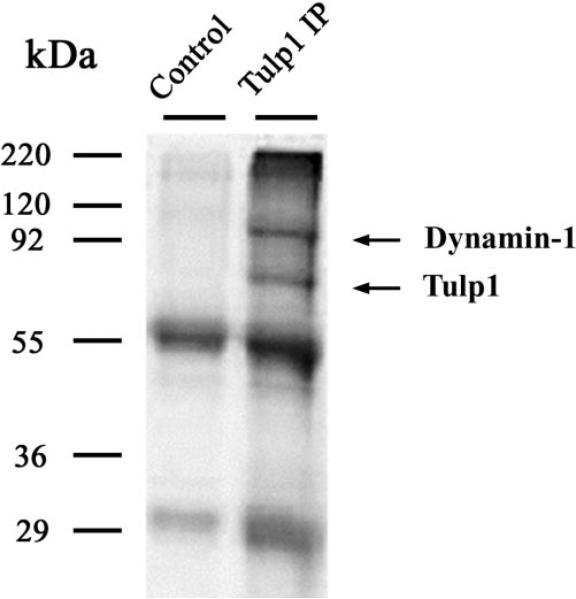
To identify retinal proteins that interact with TULP1, TULP1 was immunoprecipitated from bovine retinal homogenates using M-tulp1N antibodies and coprecipitating proteins identified by mass spectrometry.22 Four independent IP experiments were performed. Figure 1 shows a representative SDS-PAGE result. Bands from the IP lane were excised, and proteins were identified by LC MS/MS sequence analysis. The major band in the TULP1 immunoprecipitates at approximately 70 kDa was identified in all four independent IP experiments as TULP1 by MS/MS sequence analysis, yielding six tryptic peptides covering 13.5% of the protein. The major band present in the control lane at approximately55 kDa was identified as tubulin, whereas no peptide match was identified for the band at approximately 30 kDa.
Figure 1.
Immunoprecipitation of a TULP1 containing complex from retina. Gel Code blue stain of an SDS-PAGE gel separating retinal lysates incubated with TULP1 antibodies. Lane 1: immunoprecipitated product after binding with protein A beads without TULP1 antibodies. Lane 2: immunoprecipitated product incubated with TULP1 antibodies coupled to protein A beads and demonstrates a pattern of bands absent in the control lane. The bands were excised and digested with trypsin, and the peptides were identified by LC MS/MS. The major band at approximately 70 kDa was identified as TULP1. The major band at approximately 95 kDa was identified as Dynamin-1.
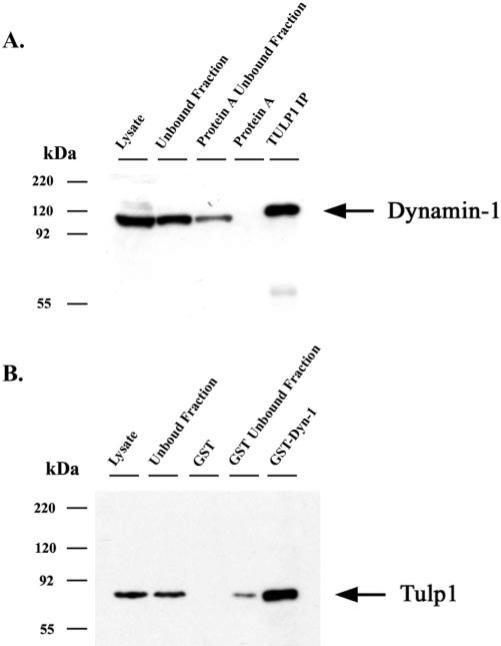
Figure 1 shows that additional bands were detected in the TULP1 protein complex. The major band at approximately 95 kDa was identified in all four independent IP experiments as Dynamin-1 by MS/MS sequence analysis of up to 16 tryptic peptides covering 23.7% of the protein. Given that Dynamin-1 was identified in multiple IP experiments, we further pursued this interaction by performing pull-down assays. Reciprocal experiments attempting to immunoprecipitate TULP1 from retinal lysate with Dynamin-1 antibodies were unsuccessful. The inability of Dynamin-1 antibodies to immunoprecipitate TULP1 in our experiments could be attributed to several factors. First, Dynamin-1 might have had a stronger affinity for other proteins than TULP1. Second, the Dynamin-1 antibodies were not appropriate for IP studies (e.g., the epitope was masked or structurally modified by protein interactions). Third, the experimental conditions were not optimized to detect an interaction. In Figure 2A, retinal lysates were incubated with TULP1 antibodies coupled to protein A beads, and Dynamin-1 was detected by Western blot of the immunoprecipitated products. Dynamin-1 was not detected in the control lane isolating the immunoprecipitated products from protein A beads alone, demonstrating little, if any, nonspecific interaction between Dynamin-1 and the protein A beads. In Figure 2B, bovine retinal lysates were incubated with a fusion protein of GST and Dynamin-1 (GST-Dyn1) or with GST alone. GST-Dyn1 or GST and their associated proteins were isolated from the lysates with glutathione-Sepharose and were then detected with TULP1 antibodies. TULP1 immunoreactivity was present only in the sample incubated with GST-Dyn1 and not in the sample incubated with GST alone. These data provide further supporting evidence that TULP1 and Dynamin-1 interact.
Figure 2.
GST pull-down assay indicating that TULP1 and Dynamin-1 interact. (A) Western blot probed with Dynamin-1 antibodies. Lane 1: retinal lysate. Lane 2: unbound fraction after incubation of lysate with Protein A beads. Lane 3: unbound fraction after incubation of lysate with TULP1 antibodies coupled to Protein A beads. Lane 4: immunoprecipitated products from Protein A beads alone. Lane 5: immunoprecipitated products from TULP1 antibodies coupled to Protein A beads. (B) Western blot probed with TULP1 antibodies. Lane 1: retinal lysate. Lane 2: unbound fraction after incubation of lysate with GST-bound glutathione beads. Lane 3: pull-down products from GST bound glutathione beads. Lane 4: unbound fraction after incubation of lysate with GST-Dynamin-1– bound glutathione beads. Lane 5: pull-down products from GST-Dynamin-1– bound glutathione beads.
Retina-Specific Dynamin-1 Isoform
Dynamin-1 is predicted to have four known domains, each with different functions and binding partners.25 We sought to determine whether a specific domain is responsible for the interaction between TULP1 and Dynamin-1. Before this experiment could be performed, it was necessary to identify the Dynamin-1 isoform expressed in the retina given that a total of eight different splice variants have been identified.26,27 To this end, the full-length cDNA corresponding to the retina-specific Dynamin-1 isoform was amplified by RT-PCR from retinal cDNA by oligonucleotide primers complementary to all Dynamin-1 splice variants. Sequencing results of the cDNA clones revealed that only one Dynamin-1 isoform is expressed in the mouse retina. There are two reported splicing regions for Dynamin-1.26 The first region, corresponding to amino acids 399 to 444, contains one of two sequences of identical size but different nucleotide sequences, designated as the a or the b form. The second splicing region is in the C-terminal region making four distinct tails, designated as a, b, c, or d. Our sequencing results indicate that the retina-specific isoform corresponds to the splice variant designated Dynamin-1 (ac).26 This isoform has a 26-nucleotide deletion causing a frameshift mutation and therefore a premature stop codon truncating the protein at amino acid 814. Figure 3 shows the predicted amino acid sequence of this isoform. The two splicing regions are in bold letters, and the known domains are highlighted in different colors.
Figure 3.
Predicted amino acid sequence and protein domains of the retina-specific Dynamin-1 isoform. Retina-specific Dynamin-1 isoform consists of 814 amino acids. Two splicing regions are delineated in bold. Five known domains are indicated as follows: blue, GTPase domain; red, middle domain of unknown function; black, PH domain; green, GED; and purple, PRD.
Dynamin-1 and TULP1 Domain Interactions
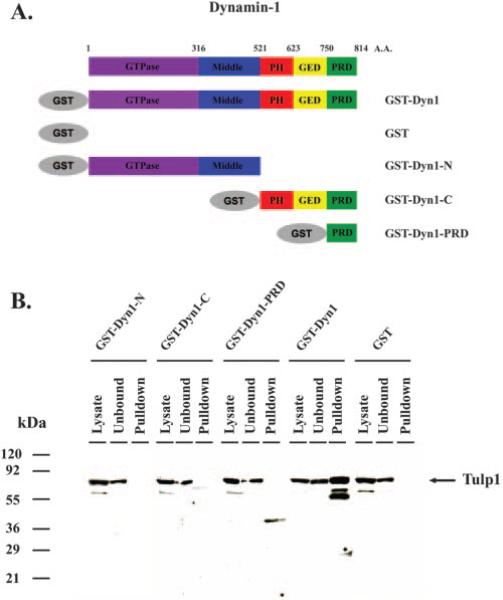
Having cloned the Dynamin-1 isoform expressed in mouse retina, we next sought to determine whether a specific domain of Dynamin-1 is responsible for the interaction with TULP1 in the retina. A series of four GST-Dynamin-1 fusion proteins corresponding to the full-length protein (GST-Dyn1), the N-terminal region (GST-Dyn1-N) consisting of the GTPase domain, the C-terminal region (GST-Dyn1-C) consisting of the PH, GED and PRD domains, and the PRD domain alone (GST-Dyn1-PRD) were generated and used in GST pull-down assays. Figure 4A shows a schematic representation of the truncation series of GST Dynamin-1 fusion proteins. In the pull-down assays, bovine retinal lysates were incubated independently with each of the four GST fusion proteins or with GST alone, and associated proteins were isolated from the lysates with glutathione-Sepharose and screened with TULP1 antibodies. Figure 4B shows that binding of TULP1 was detected only with the intact GST-Dyn1 pull-down sample and not with the pull-down samples from lysates incubated with GST-Dyn1-N, GST-Dyn1-C, GST-Dyn1-PRD, or GST alone. Combined, these data suggest that TULP1 interacts with Dynamin-1 and imply that the interaction is dependent on the conformation of the full-length protein and not an isolated region of the protein.
Figure 4.
GST pull-down assay with different domains of Dynamin-1. (A) Schematic representation of the Dynamin-1 protein and the truncation series of Dynamin-1 GST fusion proteins generated for the binding assays. (B) GST pull-down products were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with TULP1 antibodies. Lane 1: bovine retinal lysate. Lanes 2, 3: unbound fraction and the pull-down products using the N-terminal domain of Dynamin-1 (GST-Dyn1-N). Lanes 5, 6: unbound fraction and the pull-down products using the C-terminal domain of Dynamin-1 (GST-Dyn1-C). Lanes 8, 9: unbound fraction and the pull-down products using the PRD domain of Dynamin-1 (GST-Dyn1-PRD). Lanes 11, 12: unbound fraction and the pull-down products using full-length Dynamin-1 (GST-Dyn1). Lanes 14, 15: unbound fraction and pull-down products using GST alone. Positions of the molecular mass standards are indicated in kDa. Binding of TULP1 was detected only with the GST-Dyn1 sample and not in the samples from lysates incubated with GST-Dyn1-N, GST-Dyn1-C, GST-Dyn1-PRD, or GST alone.
Colocalization of TULP1 and Dynamin-1 in the Retina
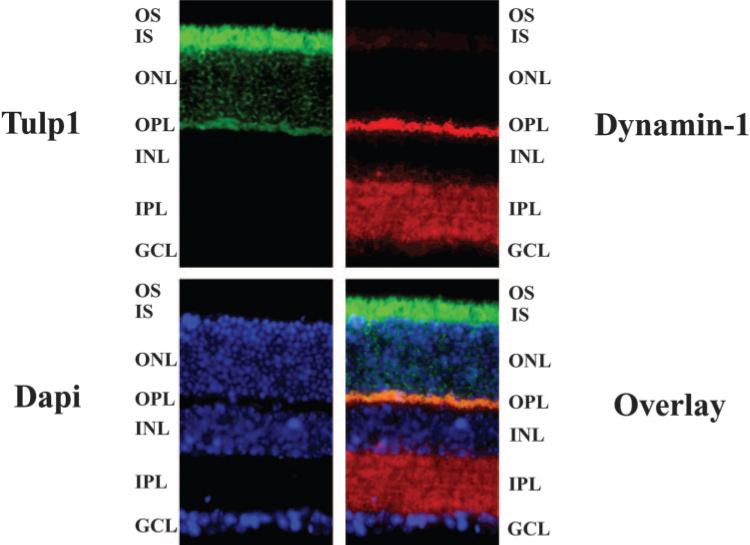
Given our IP and pull-down results that indicate Dynamin-1 and TULP1 interact, we sought evidence of an association in a cellular context by localizing the two proteins in the retina. Figure 5 shows 10-week-old wild-type mouse retinal sections stained with antibodies to TULP1 and Dynamin-1. Immunohistochemical analysis shows that TULP1 is localized primarily to the photoreceptor inner segments and the OPL, consistent with our previous reports.13,20 Dynamin-1 is localized primarily to the OPL and the inner plexiform layer (IPL). There is, however, a reproducible but low-level Dynamin-1 signal in the photoreceptor inner segments. No signal was detected for TULP1 or Dynamin-1 when wild-type retinas were incubated with secondary antibodies alone (data not shown). Our data indicate that TULP1 and Dynamin-1 colocalize primarily to the OPL containing the photoreceptor synaptic terminals and, to a lesser extent, to the inner segments of photoreceptors, providing additional evidence of a possible physiological interaction.
Figure 5.
TULP1 colocalizes with Dynamin-1 in wild-type mouse retinal sections. Immunofluorescence localization of Tulp1 (green) and Dynamin-1 (red) in 10-week-old wild-type mouse retinal sections. Nuclei are labeled with DAPI (blue). Tulp1 and Dynamin-1 colocalize to the synaptic region and, to a lesser extent, the inner segment of photoreceptor cells. OS, outer segment; IS, inner segment; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer.
Synaptic Defect in tulp1–/– Retina
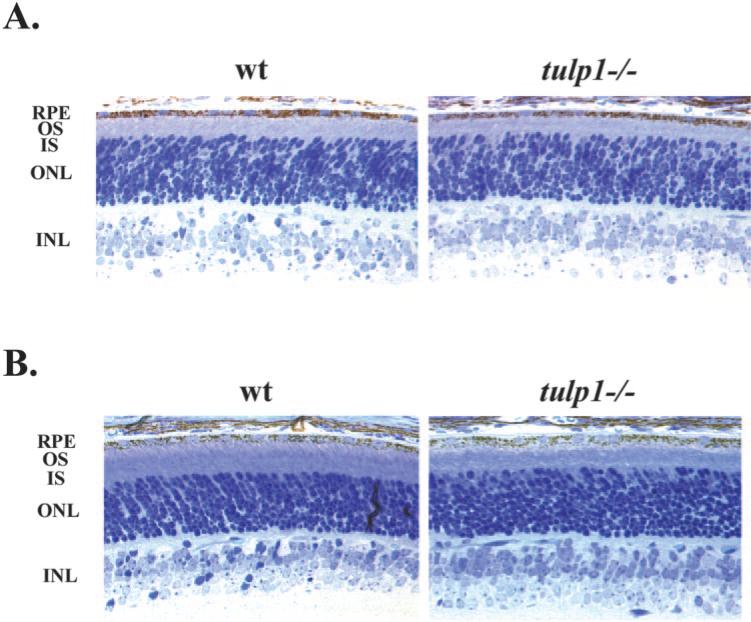
Our data indicating that TULP1 and Dynamin-1 colocalize to the OPL suggest that TULP1 may play an important role in establishing or maintaining photoreceptor synaptic terminals. To evaluate this, we examined retinal function in tulp1–/– mice. In view of the rapid photoreceptor degeneration in tulp1–/– mice, we focused our analysis on two early time points, P12 and P15. As shown in Figure 6, tulp1–/– mice retain a normal complement of photoreceptors at P12 (Fig. 6A) and P15 (Fig. 6B). At both ages, inner and outer segments are clearly seen, though these are slightly shorter and disorganized in tulp1–/– mice at P15 than in wild-type control littermates.
Figure 6.
At young ages, tulp1–/– retinas maintain a full complement of photoreceptor cells. (A) At P12, the ONL of tulp1–/– retinas was normal in thickness, and the OS and IS appeared nearly the same length as in wild-type (wt) littermate controls. The INL was also normal in thickness. (B) At P15, the ONL of tulp1–/– retinas was normal in thickness yet less organized, and the OS appeared slightly shorter and more disorganized than in wild-type littermate controls. Small gaps near the outer limiting membrane were also seen in the ONL of tulp1–/– retinas that were not detected in the wild-type retinas. The INL was of normal thickness. OS, outer segment; IS, inner segment; OPL, outer plexiform layer.
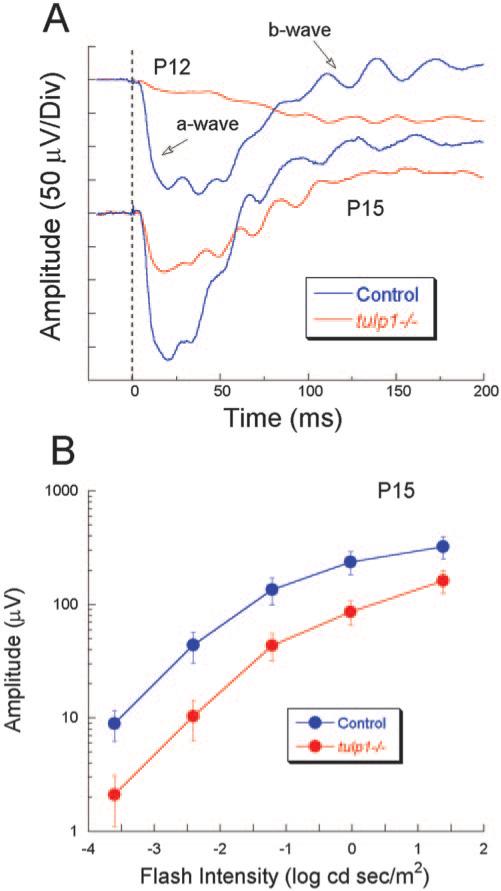
Figure 7A compares ERGs obtained in response to a high-intensity stimulus from tulp1–/– mice and control littermates (tulp1+/– and tulp1+/+) at P12 and P15. In agreement with previous studies, control mice have clear a- and b-waves at P12.28 These results indicate the presence of functional invaginating synapses between photoreceptors and depolarizing bipolar cells. In P12 tulp1–/– mice, the a-wave is markedly reduced in amplitude. Moreover, a b-wave never rises above the a-wave minimum. By P15, the control response has grown substantially, as the retina matures. At this age in tulp1–/– mice, responses have also matured, and a distinct b-wave component is now observed. To more closely examine responses at P15, Figure 7B presents intensity-response functions for control and tulp1–/– mice. Compared with control, the amplitude of the b-wave is reduced in tulp1–/– throughout the intensity range by approximately the same proportion.
Figure 7.
Electroretinographic analyses of tulp1–/– mice indicate a synaptic defect. (A) ERGs obtained to a high-intensity stimulus presented to the dark-adapted eye. Tracings indicate the average response obtained from all mice tested at P12 (5 tulp1–/–; 6 control) or P15 (11 tulp1–/–; 6 control). At both ages, there was a clear reduction in overall amplitude, and the b-wave component was essentially missing at P12. (B) Intensity-response functions for the ERG b-wave at P15. When plotted on a log amplitude axis, the amplitude of the b-wave is reduced proportionately across flash intensity.
Discussion
TULPs play a vital role in maintaining normal cellular function in the neurons in which they are expressed. TULP1 is expressed specifically in the photoreceptor cells of the retina. Mutations in TULP1 cause retinitis pigmentosa in patients and photoreceptor degeneration in mice.9–14 Our initial studies on TULP1 suggested that the protein might play a role in the trafficking of photoreceptor outer segment proteins.13,20 To pursue this hypothesis, we reasoned that identifying TULP1 binding partners would be a critical step toward understanding the function of TULP1 in maintaining the health photoreceptors and its participation in the disease process of photoreceptor degeneration. In this study, we isolated a TULP1-containing complex from the retina and identified a neuronal-specific protein, Dynamin-1, that interacts with TULP1. GST pull-down assays supported the binding as specific rather than the result of contamination or background.
Dynamin-1 is an essential component of vesicle formation in receptor-mediated endocytosis, synaptic vesicle recycling, and vesicle trafficking in and out of the TGN.25,27 Dynamin-1 is thought to act as a polymeric contractile scaffold at the interface between membranes and actin to generate vesicles at different cellular sites.29,30 The protein contains five distinct protein domains: an N-terminal GTPase domain, a middle domain of unknown function, a pleckstrin-homology (PH) domain, a domain with a propensity to form coiled coils (also named the GTPase effector domain [GED]), and a C-terminal proline-rich domain (PRD). Several in vitro and in vivo studies have demonstrated that Dynamin-1 binds to phosphoinositides of membranes through its PH domain and to multiple effector molecules through its PRD domain.27 The GTPase domain contains the GTP-binding motifs required for guanine-nucleotide binding and hydrolysis. These observations have shown that these distinct domains work synergistically toward vesicle formation and point to a mechanochemical function for Dynamin-1.
Numerous studies have focused on identifying proteins that bind to the different domains of Dynamin-1 and suggest functional and structural roles for each domain. To probe whether a known functional Dynamin-1 domain binds TULP1, we cloned the retina-specific Dynamin-1 isoform and performed in vitro binding assays with a series of Dynamin-1 GST truncation mutants. Distinct Dynamin-1 isoforms may provide functional differences through interacting with various binding proteins and targeting to different intracellular locations, including the plasma membrane and the TGN, where they function in distinct types of vesicle formation that may include polarized neurons.25,31 Dynamin-1 isoform sequence differences might specify the binding partners of Dynamin-1 and alter the specificity of targeting through unique interactions with other molecules. The isoform identified in retina has previously been detected in neurons of the rat brain and cultured PC-12 cells.26 The protein has a substantial alteration in the sequence near the C terminus compared with the classical Dynamin-1 and is predicted to be an 814-amino acid protein.26 The sequence missing in this isoform is essential for clathrin association, involved in vesicle formation at several stages of the secretory and endocytic pathways.32 In photoreceptors, post-Golgi transport vesicles do not contain clathrin, suggesting that the expression of this isoform by the retina is plausible.33 After cloning the Dynamin-1 isoform expressed in retina, we made a series of four truncation mutants fused to GST to perform pull-down assays. Under our assay conditions, TULP1 interacts only with the full-length Dynamin-1 fusion protein and not with the isolated N-terminal, C-terminal, or PRD domains, suggesting a conformation-dependent interaction dictated by the intact Dynamin-1 protein structure.
Given our IP and pull-down results indicating that Dynamin-1 and TULP1 interact, we next sought evidence of an association in a cellular context by localizing the two proteins in the retina. Immunohistochemical experiments performed on wild-type mouse retina indicate that Dynamin-1 is expressed primarily in the OPL, IPL, and, to a lesser extent, the inner segments of photoreceptor cells. TULP1 is also localized to the inner segments and photoreceptor synapse in the OPL, in addition to the connecting cilium and perikarya.13,20 Our data indicate that TULP1 and Dynamin-1 colocalize in photoreceptor cells, providing additional support for a physiological interaction. In addition, the colocalization observed in the OPL encourages closer examination of the synaptic complex using cell-specific, presynaptic, and postsynaptic markers.
In previous studies, we provided evidence that TULP1 may function in the transport of rhodopsin from its site of synthesis in the inner segment through the connecting cilium to its final destination in the outer segment of the photoreceptor cell.13,20 This is based on the photoreceptor distribution of TULP1 and the photoreceptor disease phenotype in tulp1–/– mice. As described, TULP1 is localized to specialized compartments of the photoreceptor involved in protein transport. In addition, tulp1–/– mice develop early-onset, progressive photoreceptor degeneration that involve both rods and cones. Before retinal degeneration, rod and cone opsins are mislocalized, and an abnormal accumulation of rhodopsin-bearing extracellular vesicles is found surrounding the ellipsoid region of the inner segments, indicating a misrouting of rhodopsin transport carriers. These results indicate that TULP1 might function in trafficking proteins from the inner segment through the connecting cilium to the outer segment. We recently provided evidence that TULP1 interacts and colocalizes with actin.21 The actin cytoskeleton is widely believed to play an important role in intracellular protein transport.34,35 Numerous studies implicate actin in several processes associated with protein movement, including vesicle assembly and polarized transport. In fact, recently the light-driven translocation of the visual proteins arrestin and transducin have been shown to partially rely on the active motor-driven elements of the actin cytoskeleton.36 Interestingly, interactions between Dynamin-1 and the actin cytoskeleton have also been reported.29,30,37 Dynamin-1 interacts with several actin-binding proteins and is thought to be intimately involved in actin-membrane dynamics. Because actin and Dynamin-1 have been implicated in the formation and movement of many types of vesicles, interactions between actin, Dynamin-1, and TULP1 have broad implications in protein trafficking specific to the photoreceptor cells. Photoreceptor cells are known to have an active intracellular membrane and protein trafficking system to support their continuous renewal of outer segments and the proteins localized to the outer segments. Here we provide evidence that TULP1 binds and colocalizes with Dynamin-1 in photoreceptor cells, suggesting that TULP1 may be a part of a protein complex involved in the molecular pathway of vesicular protein transport occurring from the inner segment to the outer segment and the synapse.
To further evaluate the photoreceptor synapse in tulp1–/– retinas, we examined retinal function in young tulp1–/– mice before the onset of photoreceptor cell loss. At P12, the amplitude of the a-wave was reduced, indicative of an abnormality in the phototransduction process. In addition, the b-wave never rose above the baseline at P12. These abnormalities occurred before degeneration, indicating that TULP1 plays an important role developmentally in establishing outer segment function and the photoreceptor-to-bipolar cell synapse. ERG b-wave reductions are noted in a wide range of mutants involving the photoreceptor output synapse.38 These abnormalities are unlikely to be secondary to an outer segment defect inducing a reduction in photoreceptor light sensitivity. A reduction in the effective stimulus intensity would be expected to cause the relative amplitude of the b-wave to increase, similar to what is seen in wild-type mice when stimulus intensity is decreased. In addition, in other models of photoreceptor degeneration, the b-wave is relatively spared, presumably because of remodeling of the photoreceptor output synapse.39 These functional studies suggest that it would be worthwhile to examine more closely the synaptic architecture in the developing tulp1–/– retina before significant photoreceptor degeneration occurs.
The localization and interaction studies presented here support a physical interaction between TULP1 and Dynamin-1; however, a functional interaction has yet to be unequivocally established. Our results also support a role for TULP1 in protein trafficking in the photoreceptor cells of the retina and provide evidence that defective protein transport pathways may be a pathologic mechanism responsible for retinal degeneration. Future experiments aimed at identifying the role that TULP1 may play in a transport pathway will help to clarify a poorly understood aspect of photoreceptor biology relevant to human retinal disease.
Acknowledgments
The authors thank Sanjoy Bhattacharya for assistance with immunoprecipitations, Jian Sun for bioinformatics, Xiarong Gu for mass spectrometry analysis, and Jiang Wu for electroretinographic analysis.
Supported by National Institutes of Health Grants EY16072, EY06603, EY14239, and EY15638, Foundation Fighting Blindness, and a Research to Prevent Blindness Challenge Grant.
Footnotes
Disclosure: Q. Xi, None; G.J.T. Pauer, None; S.L. Ball, None; M. Rayborn, None; J.G. Hollyfield, None; N.S. Peachey, None; J.W. Crabb, None; S.A. Hagstrom, None
References
- 1.North MA, Naggert JK, Yan Y, Noben-Trauth K, Nishina PM. Molecular characterization of TUB, TULP1, and TULP2, members of the novel tubby gene family and their possible relation to ocular diseases. Proc Natl Acad Sci USA. 1997;94:3128–3133. doi: 10.1073/pnas.94.7.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishina PM, North MA, Ikeda A, Yan Y, Naggert JK. Molecular characterization of a novel tubby gene family member, TULP3, in mouse and humans. Genomics. 1998;54:215–220. doi: 10.1006/geno.1998.5567. [DOI] [PubMed] [Google Scholar]
- 3.Sahly I, Gogat K, Kobetz A, et al. Prominent neuronal-specific tub gene expression in cellular targets of tubby mice mutation. Hum Mol Genet. 1998;7:1437–1447. doi: 10.1093/hmg/7.9.1437. [DOI] [PubMed] [Google Scholar]
- 4.Kleyn PW, Fan W, Kovats SG, et al. Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell. 1996;85:281–290. doi: 10.1016/s0092-8674(00)81104-6. [DOI] [PubMed] [Google Scholar]
- 5.He W, Ikeda S, Bronson RT, et al. GFP-tagged expression and immunohistochemical studies to determine the subcellular localization of the tubby gene family members. Brain Res Mol Brain Res. 2000;81:109–117. doi: 10.1016/s0169-328x(00)00164-9. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda S, He W, Ikeda A, Naggert JK, North MA, Nishina PM. Cell-specific expression of tubby gene family members (tub, Tulp1, 2, and 3) in the retina. Invest Ophthalmol Vis Sci. 1999;40:2706–2712. [PubMed] [Google Scholar]
- 7.Ohlemiller KK, Hughes RM, Mosinger-Ogilvie J, Speck JD, Grosof DH, Silverman MS. Cochlear and retinal degeneration in the tubby mouse. Neuroreport. 1995;6:845–849. doi: 10.1097/00001756-199504190-00005. [DOI] [PubMed] [Google Scholar]
- 8.Noben-Trauth K, Naggert JK, North MA, Nishina PM. A candidate gene for the mouse mutation tubby. Nature. 1996;380:534–538. doi: 10.1038/380534a0. [DOI] [PubMed] [Google Scholar]
- 9.Hagstrom SA, North MA, Nishina PM, Berson EL, Dryja TP. Recessive mutations in the gene encoding tubby-like protein TULP1 in patients with retinitis pigmentosa. Nat Genet. 1998;18:174–176. doi: 10.1038/ng0298-174. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee P, Kleyn PW, Knowles JA, et al. TULP1 mutation in two extended Dominican kindreds with autosomal recessive retinitis pigmentosa. Nat Genet. 1998;18:177–179. doi: 10.1038/ng0298-177. [DOI] [PubMed] [Google Scholar]
- 11.Paloma E, Hjelmqvist L, Bayes M, et al. Novel mutations in the TULP1 gene causing autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000;41:656–659. [PubMed] [Google Scholar]
- 12.Gu SM, Lennon A, Li Y, et al. Tubby-like protein-1 mutations in autosomal recessive retinitis pigmentosa. Lancet. 1998;351:1103–1104. doi: 10.1016/S0140-6736(05)79384-3. [DOI] [PubMed] [Google Scholar]
- 13.Hagstrom SA, Duyao M, North MA, Li T. Retinal degeneration in tulp1−/− mice: vesicular accumulation in the interphotoreceptor matrix. Invest Ophthalmol Vis Sci. 1999;40:2795–2802. [PubMed] [Google Scholar]
- 14.Ikeda S, Shiva N, Ikeda A, et al. Retinal degeneration but not obesity is observed in null mutants of the tubby-like protein 1 gene. Hum Mol Genet. 2000;9:155–163. doi: 10.1093/hmg/9.2.155. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda A, Ikeda S, Gridley T, Nishina PM, Naggert JK. Neural tube defects and neuroepithelial cell death in Tulp3 knockout mice. Hum Mol Genet. 2001;10:1325–1334. doi: 10.1093/hmg/10.12.1325. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda A, Zheng QY, Zuberi AR, Johnson KR, Naggert JK, Nishina PM. Microtubule-associated protein 1A is a modifier of tubby hearing (moth1). Nat Genet. 2002;30:401–405. doi: 10.1038/ng838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapeller R, Moriarty A, Strauss A, et al. Tyrosine phosphorylation of tub and its association with src homology 2 domain-containing proteins implicate tub in intracellular signaling by insulin. J Biol Chem. 1999;274:24980–24986. doi: 10.1074/jbc.274.35.24980. [DOI] [PubMed] [Google Scholar]
- 18.Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro L. Implication of tubby proteins as transcription factors by structure- based functional analysis. Science. 1999;286:2119–2125. doi: 10.1126/science.286.5447.2119. [DOI] [PubMed] [Google Scholar]
- 19.Santagata S, Boggon TJ, Baird CL, et al. G-protein signaling through tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 20.Hagstrom SA, Adamian M, Scimeca M, Pawlyk BS, Yue G, Li T. A role for the Tubby-like protein 1 in rhodopsin transport. Invest Ophthalmol Vis Sci. 2001;42:1955–1962. [PubMed] [Google Scholar]
- 21.Xi Q, Pauer GJ, Marmostein AD, Crabb JW, Hagstrom SA. Tubby-like protein 1 (TULP1) interacts with F-actin in photoreceptor cells. Invest Ophthalmol Vis Sci. 2005;46:4754–4761. doi: 10.1167/iovs.05-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xi Q, Pauer GJ, West KA, Crabb JW, Hagstrom SA. Retinal degeneration caused by mutations in TULP1. Adv Exp Med Biol. 2003;533:303–308. doi: 10.1007/978-1-4615-0067-4_37. [DOI] [PubMed] [Google Scholar]
- 23.Crabb JW, Miyagi M, Gu X, et al. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci USA. 2002;99:14682–14687. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.West KA, Yan L, Shadrach K, et al. Protein database: human retinal pigment epithelium. Mol Cell Proteomics. 2003;2:37–49. doi: 10.1074/mcp.d200001-mcp200. [DOI] [PubMed] [Google Scholar]
- 25.van der Bliek AM. Functional diversity in the dynamin family. Trends Cell Biol. 1999;9:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- 26.Cao H, Garcia F, McNiven MA. Differential distribution of dynamin isoforms in mammalian cells. Mol Biol Cell. 1998;9:2595–2609. doi: 10.1091/mbc.9.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNiven MA, Cao H, Pitts KR, Yoon Y. The dynamin family of mechanoenzymes: pinching in new places. Trends Biochem Sci. 2000;25:115–120. doi: 10.1016/s0968-0004(99)01538-8. [DOI] [PubMed] [Google Scholar]
- 28.Bakall B, Marmorstein LY, Hoppe G, Peachey NS, Wadelius C, Marmorstein AD. Expression and localization of bestrophin during normal mouse development. Invest Ophthalmol Vis Sci. 2003;44:3622–3628. doi: 10.1167/iovs.03-0030. [DOI] [PubMed] [Google Scholar]
- 29.Orth JD, McNiven MA. Dynamin at the actin-membrane interface. Curr Opin Cell Biol. 2003;15:31–39. doi: 10.1016/s0955-0674(02)00010-8. [DOI] [PubMed] [Google Scholar]
- 30.Schafer DA. Regulating actin dynamics at membranes: a focus on dynamin. Traffic. 2004;5:463–469. doi: 10.1111/j.1600-0854.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 31.Urrutia R, Henley JR, Cook T, McNiven MA. The dynamins: redundant or distinct functions for an expanding family of related GTPases? Proc Natl Acad Sci USA. 1997;94:377–384. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shpetner HS, Herskovits JS, Vallee RB. A binding site for SH3 domains targets dynamin to coated pits. J Biol Chem. 1996;71:13–16. doi: 10.1074/jbc.271.1.13. [DOI] [PubMed] [Google Scholar]
- 33.Deretic D, Papermaster DS. Polarized sorting of rhodopsin on post-Golgi membranes in frog retinal photoreceptor cells. J Cell Biol. 1991;13:1281–1293. doi: 10.1083/jcb.113.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 35.Stamnes M. Regulating the actin cytoskeleton during vesicular transport. Curr Opin Cell Biol. 2002;4:428–433. doi: 10.1016/s0955-0674(02)00349-6. [DOI] [PubMed] [Google Scholar]
- 36.Peterson JJ, Orisme W, Fellows J, et al. A role for cytoskeletal elements in the light-driven translocation of proteins in rod photoreceptors. Invest Ophthalmol Vis Sci. 2005;46:3988–3998. doi: 10.1167/iovs.05-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee E, De Camilli P. Dynamin at actin tails. Proc Natl Acad Sci USA. 2002;99:161–166. doi: 10.1073/pnas.012607799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang B, Heckenlively JR, Bayley PR, et al. The nob2 mouse, a null mutation in Cacna1f: anatomical and functional abnormalities in the outer retina and their consequences on ganglion cell visual responses. Vis Neurosci. 2006;23:11–24. doi: 10.1017/S095252380623102X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aleman TS, LaVail MM, Montemayor R, et al. Augmented rod bipolar cell function in partial receptor loss: an ERG study in P23H rhodopsin transgenic and aging normal rats. Vision Res. 2001;41:2779–2797. doi: 10.1016/s0042-6989(01)00157-2. [DOI] [PubMed] [Google Scholar]