Abstract
Protists account for the bulk of eukaryotic diversity. Through studies of gene and especially genome sequences the molecular basis for this diversity can be determined. Evident from genome sequencing are examples of versatile metabolism that go far beyond the canonical pathways described for eukaryotes in textbooks. In the last 2–3 years, genome sequencing and transcript profiling has unveiled several examples of heterotrophic and phototrophic protists that are unexpectedly well-equipped for ATP production using a facultative anaerobic metabolism, including some protists that can (Chlamydomonas reinhardtii) or are predicted (Naegleria gruberi, Acanthamoeba castellanii, Amoebidium parasiticum) to produce H2 in their metabolism. It is possible that some enzymes of anaerobic metabolism were acquired and distributed among eukaryotes by lateral transfer, but it is also likely that the common ancestor of eukaryotes already had far more metabolic versatility than was widely thought a few years ago. The discussion of core energy metabolism in unicellular eukaryotes is the subject of this review. Since genomic sequencing has so far only touched the surface of protist diversity, it is anticipated that sequences of additional protists may reveal an even wider range of metabolic capabilities, while simultaneously enriching our understanding of the early evolution of eukaryotes.
Keywords: amoebae, anaerobic metabolism, ecology, eukaryotic evolution, lateral gene transfer, mitochondria
Introduction
Protists are a polyphyletic assortment of mostly unicellular organisms that account for the bulk of eukaryotic diversity. Much of this diversity is unseen, evident only from small subunit ribosomal RNA gene sequences extracted from environmental samples. Furthermore, novel species and unexpected diversity from every major protist group continue to be identified from diverse environments, including oxic and sub-oxic sediments from marine and freshwater study sites, the open ocean, and freshwater locations (Amaral Zettler et al. 2002; Dawson and Pace 2002; Kolisko et al. 2010; Moreira and Lopez-Garcia 2002; Slapeta et al. 2005; Stoeck et al. 2009). In current views of eukaryotic phylogeny, these protist groups are combined, together with metazoans and fungi, into six major eukaryotic clades1 that are thought to be descended from an ancestral diversification and radiation of the earliest eukaryotic organism(s) around 1–1.5 Ga years ago (Burki et al. 2008; Cavalier-Smith and Chao 2010; Embley and Martin 2006; Hampl et al. 2009; Kim et al. 2006; Koonin 2010; Richards and Cavalier-Smith 2005; Roger and Simpson 2009; Simpson and Roger 2004; Stechmann and Cavalier-Smith 2003; Fig. 1).
Figure 1.
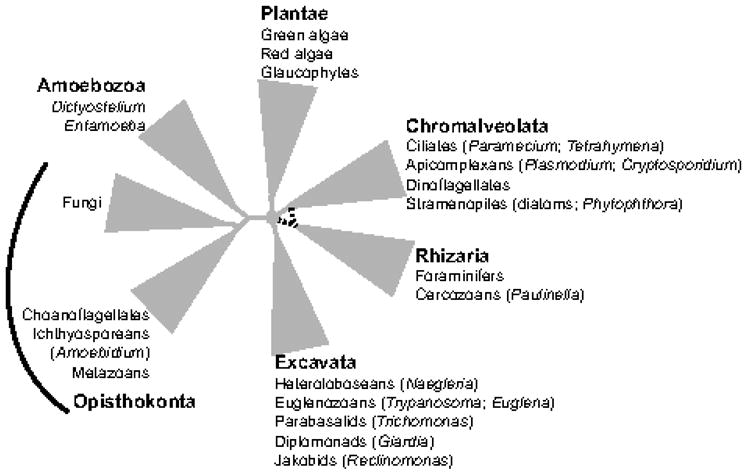
The organisation of eukaryotes into six major groups, each of which contains protists. Identities of the six major supergroups are highlighted by the bold font. Dotted lines reflect the uncertainty regarding the relationship between Chromalveolata and Rhizaria (Burki et al. 2008; Hampl et al. 2009). The position of the root for eukaryotic evolution is not known.
The number of sequenced nuclear genomes of protists is small, even when considered against the small fraction of protist biodiversity that has been morphologically described. Yet, a sufficient variety of free-living and parasitic protists have been sampled from across the tree of eukaryotic evolution to confidently infer that the last common ancestor of extant eukaryotes was an unexpectedly complex organism, and already possessed almost all of the cytological and molecular elements that are considered to represent classic characteristics in eukaryotic cell biology – a nucleus, an actin-tubulin-based cytoskeleton and its associated molecular motors (dynein, myosin, kinesin), amoeboid- and flagellar-motility, an elaborate endomembrane system and organellar metabolism, including mitochondria and peroxisomes, eukaryotic-style signalling systems, and the involvement of small, non-coding RNA molecules in the regulation of gene expression (discussed generally in Fritz-Laylin et al. 2010; Koonin 2010, but see also Chapman and Carrington 2007; Embley 2006; Field and Dacks 2009; Gabaldon 2010; Richards and Cavalier-Smith 2005; Wickstead and Gull 2006). Thus, even though some relationships within and between the major eukaryotic clades, including the root position for eukaryotic evolution, are unresolved and often debated (Cavalier-Smith 2010; Embley and Martin 2006; Koonin 2010; Yoon et al. 2008), it is unlikely that the basic premise of ‘ancestral eukaryotic complexity’ will change substantially.
Changes in cellular metabolism are often associated with the radiation of organisms into novel niches. In the case of parasites, the moderation or loss of core metabolism is classically associated with the transition to obligate parasitism. For instance, loss of biosynthetic pathways – notably, purine biosynthesis or various pathways of amino acid and lipid biosynthesis – provide obvious examples of metabolic streamlining in both parasites and many free-living heterotrophs (e.g. el Kouni 2003; Fritz-Laylin et al. 2010; Gaulin et al. 2010; Kaneshiro 1987; Payne and Loomis 2006; Wood and Gottlieb 1978). Invariably, the loss of a biosynthetic pathway reflects the availability of the essential end-product in the environment, although there are also oxygen-dependent pathways, such as sterol biosynthesis (Fischer and Pearson 2007; Summons et al. 2006), that are not likely to be feasible under micro-oxic or anoxic conditions. Indeed, we are not aware of any obligate microaerophile or anaerobe in which sterol biosynthesis occurs. Another notable example of metabolic streamlining is the absence of mitochondrial energy metabolism from a variety of anaerobic protistan parasites and the Microsporidia. For many years, it was widely thought that these parasites were primitively without mitochondria, and were therefore extant descendents from the earliest diverging eukaryotic lineages (reviewed in Embley and Martin 2006). However, during the last fifteen years or so, phylogenetic studies and detailed cellular analyses of the supposedly ‘amitochondriate’ eukaryotes have revealed the presence of organelles of mitochondrial ancestry in all known extant eukaryotes. Therefore, the moderation and, in some organisms, outright loss of mitochondrial energy-generating pathways are also a consequence of niche adaptation (Hjort et al. 2010; Roger and Simpson 2009; Shiflett and Johnson 2010), at least in the instances identified thus far.
Biochemical insight into the streamlined core energy metabolism of anaerobic eukaryotes was classically led by studies with the parasitic protists Trichomonas vaginalis, Giardia lamblia, and Entamoeba histolytica. Each of these parasites ferments carbohydrates, and to a lesser extent also utilises some amino acids as energy sources (Edwards et al. 1992; Loftus et al. 2005; Yarlett et al. 1996). Significantly, the energy metabolism in these parasites, as well as other anaerobic protists and fungi, requires oxygen-labile enzymes rarely seen in other eukaryotes – e.g. pyruvate:ferredoxin oxidoreductase (PFO) and FeFe-hydrogenase. The publication of genome sequences for the fore-mentioned parasites (Carlton et al. 2007; Loftus et al. 2005; Morrison et al. 2007), corroborated the biochemical evidence for the loss of mitochondrial functions and, together with recent bioinformatics studies, biochemical analyses and expressed-sequence tag (EST) surveys of gene expression in unrelated anaerobic protists (Boxma et al. 2005; de Graaf et al. 2009; Gill et al. 2007; Mogi and Kita 2010; Stechmann et al. 2008), also help provide insight into the reductive processes that have repeatedly led to independent degeneracy of mitochondrial function during eukaryotic evolution (e.g. reviewed in Hjort et al. 2010).
Unexpectedly, an analysis of the Naegleria gruberi genome sequence, as well as EST profiles for several other heterotrophic protists suggest that a canonical aerobic metabolism can be buttressed by an expansive capacity for anaerobic ATP production, too (Fritz-Laylin et al. 2010; Hug et al. 2010). In addition, a robust and surprisingly complex network for anaerobic metabolism has been characterised in the green alga Chlamydomonas reinhardtii (Atteia et al. 2006; Dubini et al. 2009; Mus et al. 2007; Posewitz et al. 2009). Collectively, these observations provide a novel biochemical perspective on the molecular ecology of ubiquitous protists. One can also argue that the blend of aerobic and anaerobic metabolism seen in unrelated taxa speaks directly to several theories posited for eukaryogenesis (discussed in Embley and Martin 2006; Lopez-Garcia and Moreira 1999; Martin et al. 2001, 2003). A genome-revised view of core metabolism in protists is the subject of this review. We start with a short summary of the canonical (textbook) organisation of central energy metabolism in eukaryotes.
Outline of a ‘Conventional’ Eukaryotic Energy Metabolism
In a simple model of energy metabolism in heterotrophic eukaryotes (Fig. 2), a cytosolic Embden-Meyerhof (glycolytic) pathway is used to catabolise glucose to pyruvate, yielding a net gain of 2 ATP from substrate-level phosphorylation and 2 NADH per molecule of metabolised glucose. In order to regenerate the NAD+ necessary to sustain metabolic flux through glycolysis, cytosolic NADH can be oxidized to regenerate NAD+ through a variety of routes involving either cytosolic fermentation of pyruvate (to produce excreted end-products, such as lactate and ethanol) or the shuttling of electrons from NADH into the mitochondrial respiratory chain (with the subsequent carriage of these electrons to the terminal electron acceptor O2; examples of classic, well-known electron shuttling systems include the malate-aspartate and glycerol-3-phosphate shuttles).
Figure 2.
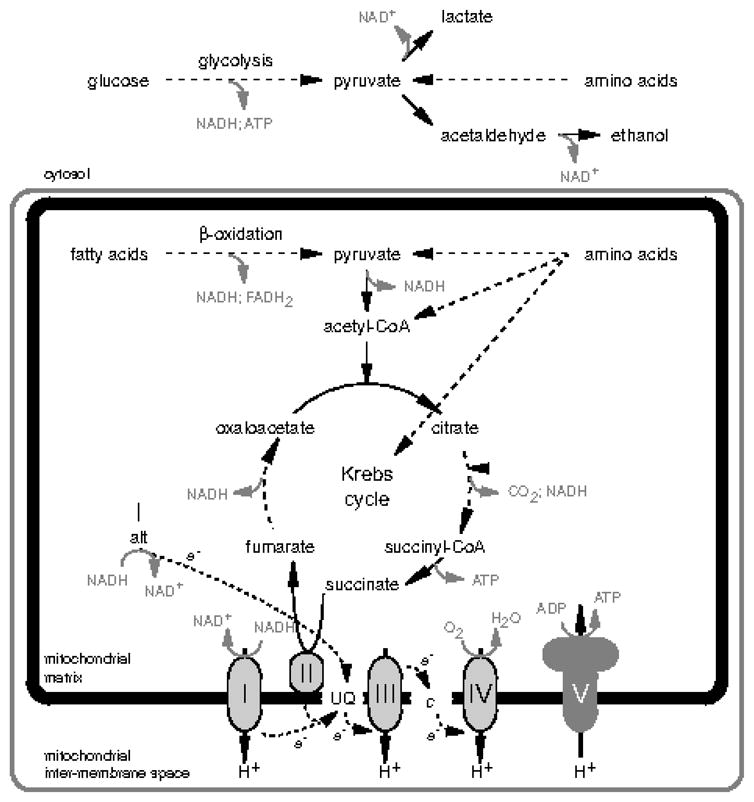
A simple ‘textbook’ model for central energy metabolism in eukaryotes. Dotted lines indicate intermediary metabolites are linked by multiple enzyme-catalysed reactions. Abbreviations: I, II, III, IV, V, mitochondrial complexes I, II, III, IV, and V, respectively; Ialt, alternative mitochondrial NADH:ubiquinone oxidoreductase; c, mitochondrial cytochrome c; UQ, ubiquinone.
Continuing the simple, yet classic textbook model of eukaryotic metabolism, the mitochondrial matrix is where pyruvate is first decarboxylated to produce acetyl-CoA (typically by the multi-subunit enzyme complex pyruvate dehydrogenase). Acetyl-CoA remains in the mitochondrial matrix, and is completely oxidized to CO2 via the citric acid (or Krebs) cycle. Mitochondrial metabolism of fatty acids, as well as many amino acids, also produces acetyl-CoA or other Krebs cycle intermediates. During the mitochondrial oxidation of these carbon sources and the oxidation of pyruvate to acetyl-CoA, electrons are transferred to either NAD+ or FAD, and thence to ubiquinone, a mobile electron carrier of the mitochondrial respiratory chain. The energy released during the transfer of electrons from NADH to ubiquinone by mitochondrial complex I, from ubquinol to cytochrome c by complex III, and from reduced cytochrome c to O2 by complex IV is coupled to the transfer of protons across the inner mitochondrial membrane to produce an electrochemical gradient. This protonmotive force is used to drive (i) the reversal of another proton pump, ATP synthase, thereby generating ATP from ADP and Pi – i.e. oxidative phosphorylation – (ii) mitochondrial matrix protein import, (iii) metabolite (e.g. Pi) import, and (iv) heat production through the activity of uncoupling proteins.
Deviations from the Classic Core Eukaryotic Metabolism
Traditional biochemistry and more recent molecular studies have uncovered numerous examples of protists, fungi, and animals that deviate from the classic ‘textbook’ paradigm for eukaryotic metabolism. Some deviations are due to dramatic re-compartmentalisation of core metabolism. For example, some glycolytic enzymes have been re-localised to the peroxisomes in kinetoplastids (e.g. the sleeping sickness parasite Trypanosoma brucei) and to the mitochondrial matrix in diatoms (Ginger et al. 2010; Kroth et al. 2008; Liaud et al. 2000). However, other deviations are due to differences in composition of the core metabolic network, and are of greater relevance for this review. Many of these differences are found in microbial eukaryotes. For instance, the capacity for cytochrome-dependent respiration was independently lost from the ancestors of some fungi (e.g. van der Giezen et al. 1997) and various evolutionarily diverse protists, including anaerobic ciliates (Boxma et al. 2005) and the microaerophilic parasite Trichomonas vaginalis (Carlton et al. 2007; Dyall et al. 2004). In the mitochondria of these species, organellar substrate-level phosphorylation is coupled to H2-formation, a process that does not occur in the ‘classic’ mitochondria described in textbooks. Degenerate H2-forming mitochondria are classified as ‘hydrogenosomes’. Alternatively, there are organisms that switch the capacity for mitochondrial ATP synthesis up or down in response to environmental cues. The baker’s yeast, Saccharomyces cerevisiae, is a good example, as is the sleeping sickness parasite, Trypanosoma brucei. In Trypanosoma brucei, ATP production does not occur in the mitochondrion during the pathogenic stage of the life cycle, although oxidative phosphorylation is essential for life cycle forms found in the tsetse fly vector which transmits the parasite between mammals (Hellemond et al. 2005).
Among unicellular organisms, there are also organisms, such as the microsporidian parasite Encephalitozoon cuniculi and diarrhoeal pathogens Entamoeba histolytica and Giardia lamblia, that have entirely lost the capacity for organellar ATP production, leaving cytosolic substrate-level phosphorylation as the only mechanism for ATP synthesis (Katinka et al. 2001; Loftus et al. 2005; Morrison et al. 2007). Here, the simplification of the energy metabolism has also been accompanied by the loss, or downscaling, of many biosynthetic pathways. At least one microsporidian parasite appears to have taken things even further and dispensed with a glycolytic pathway, as well as mitochondrial options for energy production; this parasite is therefore wholly reliant upon the import of ATP from its host cell for survival (Keeling et al. 2010). Studies using Encephalitozoon cuniculi indicated that import of ATP from the host cytosol by microsporidians occurs using plasma membrane-localised nucleotide transporters of the kind found in intracellular bacterial parasites (Tsaousis et al. 2008).
(a) ATP Production via Fermentation in Anaerobic and Microaerophilic Protists
In the anaerobic metabolism of Trichomonas vaginalis, Entamoeba histolytica, and Giardia lamblia carbohydrate metabolism uses enzymes that were traditionally thought to be absent from ‘aerobic’ eukaryotes. These enzymes (e.g. pyruvate:ferredoxin oxidoreductase (PFO); FeFe-hydrogenase; acetyl-CoA synthetase [ADP-forming family]), as well as the absence of pyruvate dehydrogenase, were characterised using biochemical and molecular methods during the 1970s, 80s and 90s (Ellis et al. 1993; Horner et al. 1999, 2000; Reeves et al. 1977; Rodriguez et al. 1996; Sanchez et al. 2000; Williams et al. 1987). Their function in the metabolism of anaerobic protists is summarised in Figure 3 (and discussed in more detail below).
Figure 3.
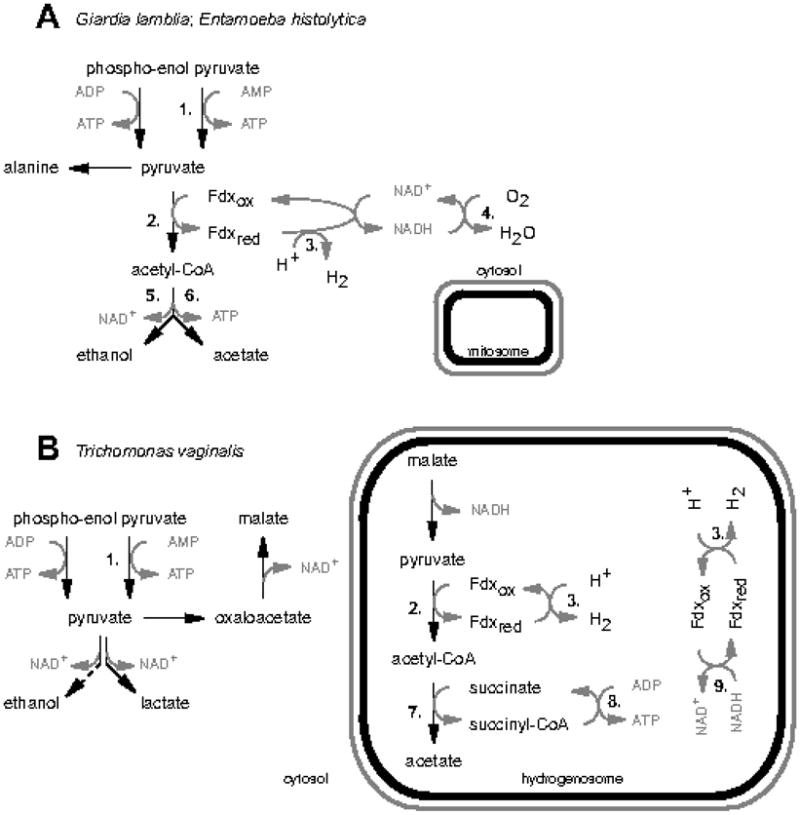
Pyruvate metabolism in anaerobic protists. A, Giardia lamblia and Entamoeba histolytica. B, Trichomonas vaginalis. The compartmentalisation of energy metabolism downstream of the formation of the glycolytic intermediate phospho-enolpyruvate is summarised. Enzymes considered specific to anaerobic energy metabolism (discussed further in the main text) are numbered as follows: 1, pyruvate phosphate dikinase (PPDK); 2, PFO; 3, FeFe-hydrogenase; 4, NADH oxidase; 5, alcohol dehydrogenase E; 6, acetyl-CoA synthetase [ADP-forming]; 7, acetate:succinate CoA transferase; 8, succinyl-CoA synthetase; 9, hydrogenosomal NADH dehydrogenase (derived from 51kDa and 24 kDa sub-units of mitochondrial complex I). Note, PPDK has widely been considered to be an enzyme associated with the transition to anaerobic metabolism, and displacement of pyruvate kinase, but is in fact found in a wide variety of eukaryotes, including plants, and can operate in forward and reverse directions (see Slamovits and Keeling 2006 for further discussion).
In anaerobic energy metabolism, just as in aerobic metabolism, ATP must be produced and NADH and FADH2 oxidized in order to sustain metabolic flux through trunk pathways, such as glycolysis. Although it is common to find prokaryotes that couple ATP production to facultative respiration of electron acceptors other than O2 (e.g. NO3−; SO42−; Richardson 2000), this is rarely seen in eukaryotes (see below). Instead, most microaerophilic and anaerobic protists rely on fermentation and substrate-level phosphorylation to satisfy cellular energy demands; glucose or other carbohydrates are widely considered to provide the major carbon source for energy production, with some additional contributions from the catabolism of one or more amino acids. In some of the identified examples of amino acid metabolism (e.g. the arginine dihydrolase pathway or the use of tryptophan indole-lyase; Edwards et al. 1992; Loftus et al. 2005), pyruvate, acetyl-CoA, and/or ATP are produced without the additional reduction of NAD+.
In instances where the major carbon source glucose is not in limiting supply, the small yield of ATP produced from glycolysis can be sufficient to support cellular energy demands. In these instances the end-product of glycolysis, pyruvate, can be fermented (in order to re-generate NAD+ from reduced NADH) without any requirement to also couple pyruvate metabolism to continued ATP generation via the production of acetate (next paragraph). Indeed, there are some parasites where under aerobic conditions glycolysis is used as the sole or predominant pathway for ATP production, and pyruvate is either fermented to lactate (e.g. the asexual blood stages of malarial parasites; Sherman 1979) or is itself excreted from cells (e.g. the African trypanosome Trypanosoma brucei, which uses a dihydroxyacetone-glycerol-3-phosphate shuttle to support NADH oxidation; Brohn and Clarkson 1978; Helfert et al. 2001; Hellemond et al. 2005). This emphasis of glycolysis as an energy source in some protists is reminiscent of the situation often seen in proliferating mammalian cells, including tumour cells, where aerobic glycolysis and the production of lactate is used to sustain cell growth (‘the Warburg effect’); here, the preference for aerobic glycolysis over oxidative phosphorylation to sustain ATP production may even be a pre-requisite for sustaining flux through the biosynthetic pathways necessary to sustain rapid cell proliferation (Vander Heiden et al. 2009).
In obligate microaerophilic or anaerobic protists, the metabolism of the end-product of glycolysis, pyruvate, is coupled to ATP production and NADH oxidation using enzymes that are not often discussed in textbook descriptions of eukaryotic energy metabolism. One characteristic deviation is the replacement of pyruvate dehydrogenase by PFO, an enzyme that decarboxylates pyruvate to acetyl-CoA with the concomitant transfer of electrons to the small redox protein ferredoxin, rather than NAD+ (Charon et al. 1999). The reason for this displacement is not immediately apparent, although it may be relevant that pyruvate dehydrogenase activity is reliant upon the use of the eight-carbon fatty acid lipoic acid as a co-enzyme, and organisms such as Giardia, Entamoeba, and Trichomonas have apparently lost the ability to synthesise fatty acids.
In trichomonads, PFO is found in hydrogenosomes (Williams et al. 1987; Fig. 3B), but in Giardia and Entamoeba this enzyme, like the rest of energy metabolism, PFO is considered to be cytosolic (Fig. 3A). In Giardia and Entamoeba, the fermentation of acetyl-CoA to ethanol is catalysed by a bi-functional alcohol dehydrogenase E (adhE), thereby regenerating the two molecules of NAD+ that are oxidised (per glucose molecule consumed) during glycolysis (Dan and Wang 2000; Espinosa et al. 2001; Sanchez 1998). This potentially spares one molecule of acetyl-CoA for ATP production through the reaction catalysed by acetyl-CoA synthetase [ADP-forming]. Further acetyl-CoA can also be spared for ATP production by using NADH oxidases for NAD+ regeneration (Brown et al. 1996; Nixon et al. 2002; Weinbach et al. 1977; Fig. 3A).
The acetyl-CoA synthetase [ADP-forming] homologues (also known as acetate thiokinase) found in Giardia, Entamoeba, and several other protists (Fig. 4) are unrelated to the ubiquitous acetyl-CoA synthetase [AMP-forming], an enzyme that catalyses the reverse reaction of acetyl-CoA formation under physiological conditions. It is, however, homologous to the TCA cycle enzyme succinyl-CoA synthetase (Sanchez et al. 2000). In Entamoeba histolytica, the acetate kinase pathway was also considered to be used for acetate production (Loftus et al. 2005). In this two step pathway, ATP and acetate are produced from acetyl-CoA. This is a common pathway in prokaryotes, but has only been found in a very small number of eukaryotes. Entamoeba histolytica contains a putative acetate kinase pathway, but there is no evidence for the presence of the enzyme that converts acetyl-CoA to acetyl-phosphate, phosphate acyltransferase (PTA), in the completed genome sequence (Tielens et al. 2010). The apparent absence of PTA suggests that the Entamoeba acetate kinase homologue has an alternative physiological function.
Figure 4.
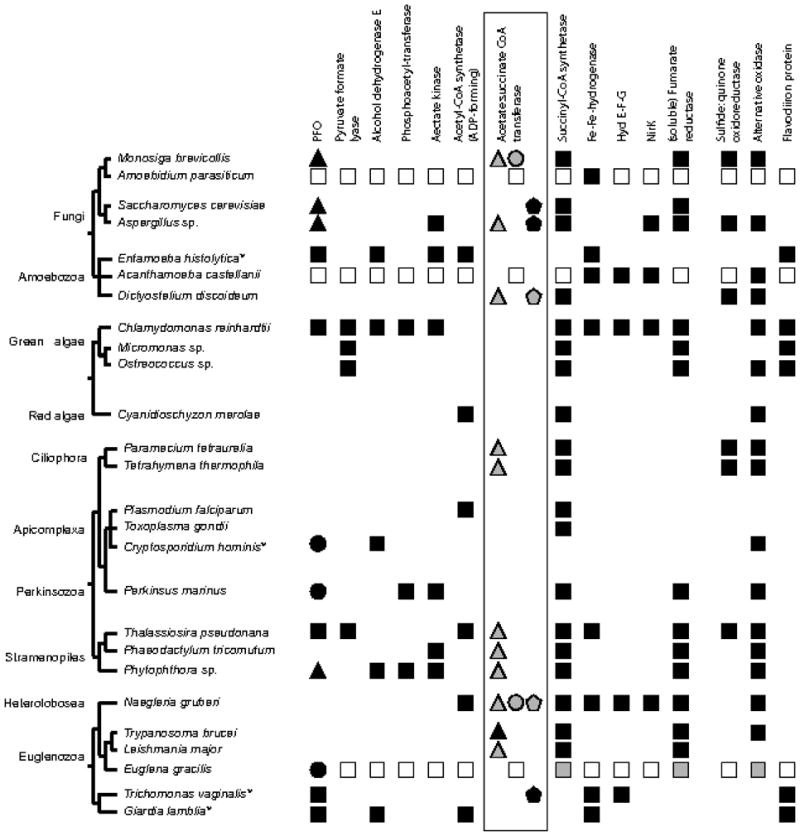
Phylogenetic distribution of enzymes associated with anaerobic energy metabolism in unicellular eukaryotes. Published draft or complete genome sequences were analysed using BLAST for the presence or absence of homologues related to each of the enzymes listed. Except for two exceptions (PFO and acetate:succinate CoA transferase), a black square indicates a homologue in a genome that detects the query as the top hit in a reciprocal BLAST analysis; open square, no genome sequence available; grey square, biochemical evidence for the presence of the stated enzyme; no symbol indicates that no homologue was detected in a complete or draft genome sequence. For PFO homologues, a black square indicates a homologue with the canonical eukaryotic/eubacterial PFO domain architecture (Rotte et al. 2001); black circle, homologue where the PFO domain architecture is C-terminally fused to a NADPH-cytochrome P450 reductase module providing either known or putative (Perkinsus marinus) PNO activity (as described in the main text); black triangle, homologue exhibits similarity to eukaryotic/eubacterial PFO domain architecture and contains a c-terminal ‘cysI’ domain. CysI is the domain characterising one of the sub-units of multi-meric assimilatory sulfite reductase; a requirement for the PFO-related CysI-containing homologue from Saccharomyces cerevisiae (Met5) in sulphur assimilation has recently been reported (Cordente et al. 2009). Phototrophs, including all of the algae surveyed here, possess CysI-containing polypeptides that lack the associated PFO-related domain architecture. The ‘mix’n’match’ combinations of domains in PFO-related proteins has been noted previously, and possibly reflects the co-option of the PFO domain for use in numerous redox reactions (Hug et al. 2010; Rotte et al. 2001). For acetate:succinate CoA transferase homologues, a black triangle indicates cloning and biochemical verification of a ‘sub-family 1A’ homologue (related to mammalian SCOT transferase; Tielens et al. 2010); grey triangle, homologue of a ‘sub-family 1A’ enzyme detected in a genome; grey circle, homologue of a ‘sub-family 1B’ acetate:succinate CoA transferase detected in a genome; black pentagon, cloning and biochemical verification of a ‘sub-family 1C’ acetate:succinate CoA transferase; grey pentagon, ‘sub-family 1C’ homologue detected in a genome. Note: Amoebidium parasiticum, Acanthamoeba castellanii, and Euglena gracilis have no genome sequence available, but are shown here because biochemical and/or molecular data indicate the presence of stated enzymes, and thus, point towards intriguing signatures of anaerobic metabolism. In contrast to the widespread distribution of enzymes required for anaerobic ATP production, the only heterotrophs that contain flavo-diiron protein as defence against oxygen toxicity are obligate anaerobes/microaerophiles. The presence of flavo-diiron proteins in some oxygenic phototrophs, and their putative role in photosystem II maintenance was noted previously (Zhang et al. 2009).
Trichomonas vaginalis also couples the metabolism of acetyl-CoA to acetate with the concomitant production of ATP via substrate-level phosphorylation, but it does so using a different system from Entamoeba and Giardia. Here, acetate:succinate CoA-transferase catalyses the production of succinyl-CoA and acetate from acetyl-CoA and succinate (van Grinsven et al. 2008). ATP is then formed in the subsequent recycling of succinyl-CoA back to succinate by succinyl-CoA synthetase, which is better known for its role as a TCA cycle enzyme in aerobes. This acetyl-CoA:succinate shuttle is widely conserved within anaerobic mitochondria (including hydrogenosomes), and curiously it occurs in the aerobic mitochondria of trypanosomatid parasites, too (Tielens et al. 2010; van Hellemond et al. 1998). However, although the shuttle is well conserved, there are three sub-families of acetate:succinate CoA-transferase. There is little similarity in amino acid sequence between the members of these different sub-families, and each sub-family has a different phylogenetic distribution in eukaryotes. The biochemistry of the CoA-transferase family and, more generally, acetate formation within parasitic protists and helminths was recently the subject of a comprehensive review (Tielens et al. 2010); the reader is directed towards this article for further, more specific discussion of ‘acetate formation and energy metabolism’.
Acetate, glycerol, succinate, lactate, and ethanol have been identified as the organic end-products of carbohydrate metabolism in Trichomonas species (Paget and Lloyd 1990; Steinbüchel and Müller 1986; ter Kuile 1996), with lactate and ethanol obvious end-products from fermentation. Cytosolic NADH is also oxidised by the conversion of phosphoenol-pyruvate to malate, and a hydrogenosomal NADH-dehydrogenase (Dyall et al. 2004; Hrdy et al. 2004) provides an alternative for oxidation of mitochondrial NADH (Fig. 3B). This enzyme probably evolved from the 51 kDa and 24 kDa sub-units of mitochondrial complex I (Hrdy et al. 2004). [2Fe-2S] Ferredoxin, the same protein that is reduced in the PFO-catalysed reaction, is a likely physiological electron acceptor used by hydrogenosomal NADH-dehydrogenase during NADH oxidation (Do et al. 2009). Reduced ferredoxin then serves as the electron donor for H2 production catalysed by FeFe-hydrogenase, the defining enzyme present in all eukaryotic hydrogenosomes studied to date (Hjort et al. 2010; Shiflett and Johnson 2010).
H2 formation and a putative FeFe-hydrogenase have also been detected in Giardia trophozoites, albeit with H2 production occurring at a ~10-fold lower rate than observed in Trichomonas vaginalis (Lloyd et al. 2002). At least one bona fide FeFe-hydrogenase is encoded in the Entamoeba histolytica genome (Nixon et al. 2003). In both of these parasites FeFe-hydrogenase is thought to be cytosolic. Given the pervasive minimalism of many aspects of giardial cell biology and metabolism (Morrison et al. 2007), it is likely that FeFe-hydrogenase plays a significant physiological role during Giardia’s colonisation and passage through the digestive tract of a mammalian host, while in Entamoeba the FeFe-hydrogenase appears to be one component within an extended portfolio of enzymes (Loftus et al. 2005) that leaves this parasite well-suited to the fermentation of a wide-range of carbon sources. However, there is little insight into how the relative use of different metabolic enzymes in Giardia or Entamoeba changes as a consequence of environmental cues (although it is known that in both organisms the end-products of glucose metabolism change as a function of O2 tension; Paget et al. 1990; Pineda et al. 2010). It is possible that under the observed culture conditions FeFe-hydrogenase activity is limited by the stability or rate of post-translational insertion of the Fe-S clusters that are necessary for catalytic function, or that competition from other redox reactions (Nixon et al. 2002; Fig. 3A) limits the availability of reduced ferredoxin, the co-enzyme necessary for FeFe-hydrogenase-catalysed H2 formation.
(b) Anaerobic Respiration and Fumarate Reduction in Unicellular Eukaryotes
As an alternative to fermentation, anaerobic respiration can be used to sustain ATP production. Indeed, anaerobic respiration is commonplace among prokaryotes, but hitherto much rarer amongst eukaryotes where published descriptions are limited to: (i) complete denitrification in foraminifers and an aquatic sister taxon Gromiida (Pina-Ochoa et al. 2010; Risgaard-Petersen et al. 2006); (ii) nitrate respiration by the ciliate Loxodes in an anaerobic lake (Finlay et al. 1983) and by some fungi during hypoxia (Kobayashi et al. 1996; Takaya et al. 2003); (iii) the use of fumarate as an electron sink in some marine invertebrates and helminths (Tielens et al. 2002). Sulphur reduction by anoxic Fusarium oxysporum has also been reported (Abe et al. 2007), but it seems more likely that the resultant production of H2S is coupled to ATP synthesis via substrate level phosphorylation and acetate production, akin to the NH3 fermentation seen in Fusarium and some other fungi (see below), rather than F1Fo-ATP synthase activity (Takasaki et al. 2004; Zhou et al. 2002). Since fumarate provides an electron sink of endogenous origin, the reduction of fumarate to succinate could be viewed as a type of fermentation, but in helminths and marine organisms reduction is coupled to proton-pumping across the inner-mitochondrial membrane and ATP synthesis in a process called ‘malate dismutation’ (Tielens et al. 2002). Crucially, however, irrespective of whether NO3−, NO2−, or fumarate is used as an electron sink, these descriptions of anaerobic respiration in eukaryotes pertain to organisms that can switch between the use of oxidative phosphorylation and O2-independent energy metabolism, rather than obligate anaerobes or microaerophiles.
Biochemical and molecular insight into NO3−/NO2− respiration by eukaryotes is still rather scant. The gene(s) encoding eukaryotic NO3− reductase(s) are yet to be identified, but the available evidence outlined below points towards a mitochondrial location for the respiration of NO3− and/or NO2−.
Scanning and transmission electron microscopy analyses indicate that the number and distribution of bacterial ecto- and endo-symbionts are not sufficient to produce the observed rates of NO3− respiration in ciliates and denitrification in foraminifers (Finlay et al. 1983; Risgaard-Petersen et al. 2006). Further, the number of mitochondria in NO3− -respiring Loxodes is ~70% higher and their organelles are more cristate than under oxic conditions, consistent with the lower energy output classically associated with anaerobic respiration versus the maximal ATP output available from using O2 as the terminal electron acceptor (Finlay et al. 1983). Biochemical sub-fractionation reveals that anaerobic respiratory activity is present within purified fungal mitochondria and is also coupled to ATP production (Kobayashi et al. 1996). Moreover, biochemical analysis of NO2− reduction in mitochondria isolated from Fusarium oxysporum indicates reduced mitochondrial cytochrome c is a likely physiological electron donor for the reduction of NO2− to NO (Takaya et al. 2003). The identity of eukaryotic NO3− reductase(s) is not known, but a recombinant NO2− reductase with similar biochemical properties to the enzyme purified from denitrifying cells was recently cloned from Fusarium (Kim et al. 2009). Significantly, this fungal gene is homologous to NirK, one of the NO2− reductase sub-families that are widely distributed in bacteria. Moreover, the analysis of protist genome sequences and EST databases reveals NirK homologues are also found in various, evolutionarily distant protists that utilise oxidative phosphorylation as a more obvious ATP-production strategy (Kim et al. 2009; Fig. 4 discussed in the next sub-section). Complete denitrification occurs in benthic foraminifers, but in fungi the end-point for NO3−/NO2− respiration is reduction of NO to N2O, which is catalysed in species examined thus far by a class of mitochondrial cytochrome P450 that is widely distributed among the fungi (Takaya et al. 1999). Nothing is known about the molecular identity of the enzymes responsible for the respiration of NO3− to NO2− in Loxodes or denitrification in rhizarians.
Amongst fungi, the best biochemically characterised of the NO3− -respiring eukaryotes, NO3− respiration occurs under micro-oxic conditions. Yet, under more anoxic conditions another type of NO3− -dependent metabolism is observed in Fusarium and Aspergillus nidulans, namely NH3 fermentation (Takasaki et al. 2004; Zhou et al. 2002). Here, cytosolic NAD(P)H-dependent assimilatory NO3−- and NO2−-reductases (niaD and niiA in Aspergillus nidulans) oxidise NADH, and ATP synthesis is supported by substrate-level phosphorylation and acetate production. The amount of ATP produced using NH3 fermentation is sufficient to support an increase in Fusarium biomass, albeit at a slow rate. Analytical measurements of metabolism have revealed anoxic NH3 fermentation is widely conserved amongst other soil-dwelling fungi (Zhou et al. 2002), but it is not clear whether most of these species can use NH3-fermentation as a viable strategy for long-term increases in biomass or if it more usefully provides a neat short-term adaptation to anoxia. Fusarium oxysporum is also able to grow using sulphur as an exogenous electron acceptor, too, in a pathway that is likely to share the same enzymes for ATP production as NH3 fermentation (Abe et al. 2007; Zhou et al. 2002).
Much of the molecular insight into the use of fumarate as a terminal electron acceptor in eukaryotes has come from studies of animal mitochondria that make use of fumarate reduction in the absence of O2. The reduction of fumarate to succinate is not simply a reversal of the more familiar TCA cycle reaction catalysed by succinate dehydrogenase (or mitochondrial complex II; Fig. 2) in which succinate is oxidised to fumarate, although the evolutionary histories of fumarate reductase and complex II are entwined (van Hellemond et al. 2003). In fact, membrane-bound fumarate reductase and succinate dehydrogenase co-exist, and are used in the flexible metabolism of many prokaryotes. Succinate dehydrogenase activity and often the activity of fumarate reductase are each dependent upon an electron transport chain, although under standard conditions different quinones act as the electron donor for fumarate reduction or the electron acceptor in succinate oxidation, respectively (van Hellemond and Tielens 1994). Reduction uses a quinone electron donor of lower standard electron potential than the quinone used as the electron acceptor for succinate oxidation. In prokaryotes, menaquinone is typically used as the electron donor for fumarate reductase, and ubiquinone is the electron acceptor used for succinate oxidation, as with the mitochondrial respiratory chain. The membrane-bound fumarate reductase and succinate dehydrogenase catalysing these redox reactions have a common evolutionary ancestry. However, in parasitic helminths that use a membrane-bound fumarate reductase for anaerobic ATP production, the biochemical properties and phylogenetic affinity of the eukaryotic reductase are more similar to mitochondrial complex II than they are to prokaryotic fumarate reductases (van Hellemond et al. 2003). This strongly suggests that these mitochondrial quinone-dependent fumarate reductases evolved from mitochondrial complex II during metazoan evolution. Structurally similar fumarate reductases are predicted to exist in coastal marine invertebrates that experience tidal variations in O2 availability. In these marine organisms and helminths, rhodoquinone, rather than menaquinone, is the quinone of low redox potential that donates electrons for fumarate reduction (van Hellemond et al. 1995). Rhodoquinone is also found in anaerobically-functioning Euglena mitochondria where it is linked to redox reactions involving fumarate reduction and lactate oxidation (Castro-Guerrero et al. 2005; Hoffmeister et al. 2004). It is also found in the hydrogenosomes of the ciliate Nyctotherus ovalis (Boxma et al. 2005).
As with other taxa discussed above, the mitochondrion of the excavate alga Euglena gracilis switches between aerobic and anaerobic modes of metabolism depending upon environmental cues. Euglenids belong to the phylum Euglenozoa, where they are one of the three major sub-groups currently recognised – the others being the kinetoplastids, which include the medically relevant trypanosomatids, and the diplonemids, which are a group of little-studied aquatic flagellates (von der Heyden et al. 2004). As with many other protist groups phagotrophy represents an important nutritional mode that characterises both past and current evolution within the group (Leander 2004), but some euglenids, such as Euglena gracilis, are secondarily photosynthetic due to engulfment of and endosymbiosis with a chlorophyte alga in a common ancestor (Leander 2004; Turmel et al. 2009). The flexibility of the Euglena mitochondrion in response to dynamic environmental change therefore mirrors the metabolism of other phototrophs that respond rapidly to the onset of hypoxia within natural (Steunou et al. 2006) or laboratory-controlled environments (e.g. Mus et al. 2007; see next sub-section). In response to anaerobic conditions, Euglena gracilis stops metabolising pyruvate to acetyl-CoA using the ‘classic’ pyruvate dehydrogenase complex, and instead switches to using an unusual variant of PFO, called pyruvate:NADP oxidoreductase (PNO), that transfers the electrons from pyruvate oxidation to NADP, rather than ferredoxin (Inui et al. 1987; Nakazawa et al. 2000; Rotte et al. 2001). Under dark anaerobic conditions, acetyl-CoA (and potentially the reducing power generated from the PNO-catalysed reaction) is used in the anaerobic production of wax esters (Inui et al. 1982; Tucci et al. 2010). An unusual trans-2-enoyl-CoA reductase is deployed for O2-independent synthesis of fatty acids under anaerobic conditions (Hoffmeister et al. 2005). This synthesis is ATP-sparing since acetyl-CoA is used directly for acyl-chain extension, rather than first requiring ATP-dependent activation to malonyl-CoA. Anaerobic fumarate reduction is linked to the provision of propionyl-CoA for odd-chain fatty acid production in Euglena’s anaerobic biosynthesis wax esters (Tucci et al. 2010). In the laboratory, some Euglena strains grow well using the wax ester pathway for ATP production, but it is unclear whether this is strictly a fermentation pathway using only substrate-level phosphorylation to generate ATP, or if rhodoquinone-dependent reduction of fumarate is also coupled to ATP production by ATP synthase. If mitochondrial complex I is one of the enzymes that reduces rhodoquinone in Euglena mitochondria, then, as with the example of malate dismutation in helminth mitochondria (Tielens et al. 2002), the establishment of a proton motive force across the inner mitochondrial membrane is conceivably coupled to ATP synthesis by mitochondrial complex V. It is unlikely that mechanisms of protein and metabolite import differ substantially between aerobic and anaerobic Euglena mitochondria. Thus, under anaerobic conditions maintenance of a mitochondrial inner membrane potential is presumably still required to sustain mitochondrial protein import and the import of metabolites such as Pi. If electron transfer to rhodoquinone does not contribute to setting membrane potential, then presumably the hydrolysis of ATP by complex V is used to set this electrochemical gradient2. There are precedents which support this possibility: the use of ATP synthase to set membrane potential across the cytoplasmic membrane is not uncommon in bacteria, and in bloodstream form Trypanosoma brucei, which does not express the proton-pumping respiratory chain complexes, ATP hydrolysis by complex V is used to set mitochondrial membrane potential (Schnaufer et al. 2005).
Rhodoquinone and the production of succinate as an end-product of glucose catabolism have been both been detected in Nyctotherus ovalis. This anaerobic ciliate is unusual as it possesses hydrogenosomes which retain an organellar genome – a remnant from when the organelle had a more typical mitochondrial function (Boxma et al. 2005). Partial genome sequencing of this organellar genome, EST analysis of nuclear gene expression, and the ablation of organellar membrane potential (by treatment of Nyctotherus with rotenone, piercidin, fenzaquin, or 1-methyl-4-phenylpyridinium) suggest mitochondrial complex I catalyses the physiological reduction of rhodoquinone, and that rhodoquinol is then used as the electron donor for fumarate reduction. Moreover these analyses also strongly suggest that the ciliate couples NADH:quinone oxidoreductase activity to proton pumping across the hydrogenosomal inner membrane, although the lack of evidence for complex V activity argues against the possibility that the generation of proton motive force is coupled to ATP synthesis. It is likely that Nyctotherus requires a membrane potential across the inner hydrogenosomal membrane for protein and metabolite import into the organelle. On a basis of organellar DNA sequencing and nuclear EST profiling (specifically, the identification of mitochondrial complex I and complex II homologues, but not complex III, complex IV or complex V sub-unit homologues; Perez-Brocal and Clark 2008; Stechmann et al. 2008; Wawrzyniak et al. 2008), as well as a demonstration of hydrogenosomal putative FeFe-hydrogenase, one would predict that the mitochondria-related organelles in the anaerobic stramenopile parasite Blastocystis also function in exactly the same way as the Nyctotherus hydrogenosomes with regard to fumarate reduction. With regard to our understanding of how mitochondrial function devolves during adaptation to an obligate anaerobic life style, this represents a most interesting example of convergent evolution.
A final mitochondrial respiratory chain component we will mention here as a candidate component in anaerobic ATP metabolism in eukaryotes is sulphide:quinone oxidoreductase (SQO) (Theissen et al. 2003). SQO transfers electrons from sulphide to ubiquinone thereby providing a lithotrophic source of electrons to support mitochondrial respiration and thence oxidative phosphorylation. SQO has been described as a mitochondrial protein in a handful of eukaryotes (e.g. annelid lugworms, chickens, the fission yeast Schizosaccharomyces pombe). Although there is no indication that the electron transfer from sulphide to ubiquinone is itself coupled directly to proton-pumping across the inner mitochondrial membrane, mitochondrial sulphide oxidation can be coupled to oxidative phosphorylation through the subsequent transfer of electrons from ubquinol to a terminal electron acceptor via the intermediacy of cytochrome c (Doeller et al. 2001). However, because sulphide exerts a concentration-dependent effect upon mitochondrial complex IV activity, SQO’s primary function in many organisms is thought to be in sulphide detoxification. In any event, Martin and co-workers have argued that the presence of this enzyme in extant eukaryotes is reflective of ancestral metabolic flexibility in the early eukaryotes that were evolving when the Earth’s oceans were largely sulphidic, rather than oxic (Theissen et al. 2003). SQO is therefore an enzyme in eukaryotes that is germane to the discussion in subsequent sections of this review.
(c) A Footprint of Anaerobic Metabolism in the Nuclear Genomes of Protists with ‘Aerobic’ Mitochondria
From the examples discussed above, several alternative modes of eukaryotic energy metabolism emerge: (i) the classic ‘textbook’ model of aerobic metabolism, (ii) fermentation in anaerobic or microaerophilic protists that is generally linked to the production of ethanol and acetate, plus in some instances H2 production, too; (iii) a facultative switch to anaerobic mitochondrial respiration in some species of ciliates, fungi, and foraminifers, as well as the excavate alga Euglena gracilis; and (iv) mitochondrial fumarate reduction that can either be coupled to ATP synthesis or is coincident with H2 production. Significantly, the unicellular eukaryotes that exhibit an ability to respire nitrate as well as O2 are neither obscure nor found in restricted, extreme environments. In fact, they are ubiquitous aquatic or soil-dwelling organisms. Unfortunately, none of these organisms, except for Aspergillus, have been subject to genome sequencing, but intriguingly the genome sequencing and expression profiling of another three ubiquitous mud-dwellers – the chlorophyte alga Chlamydomonas reinhardtii, the amoeboflagellate Naegleria gruberi and the evolutionarily distant amoeba Acanthamoeba castellanii – have identified a more expansive capacity for anaerobic metabolism than seen previously in eukaryotes capable of oxidative phosphorylation. This capacity combines homologues of enzymes characterised initially in anaerobic protists and nitrate-respiring Fusarium.
The genome sequence of Naegleria gruberi (Fritz-Laylin et al. 2010) and published ESTs from Acanthamoeba castellanii (Hug et al. 2010) each provide a strong prediction of an ability to toggle between aerobic and anaerobic modes of metabolism. The physiological interplay between aerobic and anaerobic modes of ATP production has been studied more extensively in the chlorophyte alga Chlamydomonas reinhardtii. Distributed worldwide, this ubiquitous alga is found in a diversity of aquatic, soil and forest environments (Harris 2009). Known more for growing as a photoautotroph, Chlamydomonas nonetheless responds to dark anaerobic conditions by fermenting the plastidic starch that accumulates in the light (Kreuzberg 1984; Mus et al. 2007; Posewitz et al. 2004). In laboratory-grown algae, the end-products of fermentation are formaldehyde, acetate, ethanol, and H2, plus a little malate (Gfeller and Gibbs 1984; Kreuzberg 1984; Mus et al. 2007). This metabolism is compartmentalised. Pyruvate is predicted to be decarboxylated in the algal cytosol by pyruvate decarboxylase, and the resultant acetaldehyde fermented to ethanol with concomitant NADH oxidation. In the mitochondrion and the chloroplast, pyruvate is predicted to be anaerobically oxidised by the O2-sensitive enzyme pyruvate:formate lyase (Atteia et al. 2006; Kreuzberg et al. 1987; Mus et al. 2007), thereby accounting for the accumulation of formate in the culture medium of anaerobically-maintained cells. In the chloroplast, pyruvate is additionally oxidised by O2-sensitive PFO (Atteia et al. 2006; Mus et al. 2007); the activity of this enzyme in anaerobic cells is evident in the electron transfer from reduced ferredoxin to produce H2. Chloroplast- and mitochondrial-derived acetyl-CoA is then predicted to be used for acetate and concomitant ATP production using the acetate kinase pathway (Atteia et al. 2006; Mus et al. 2007) although, to date, activity of the acetate kinase pathway has only been reported in the mitochondrion (Kreuzberg et al. 1987).
The ability of Chlamydomonas to toggle between aerobic and anaerobic modes of metabolism should come as little surprise. Not only are the moist, organic-rich environments where Chlamydomonas is readily found subject to fluctuating O2 tension (when high rates of microbial respiration exceed the rate of oxidative photosynthesis or O2 solubility), but the secretion of formaldehyde, acetate and ethanol production have been described for decades, and the production of H2 by green algae was first documented almost seventy years ago (Gaffron and Rubin 1942). Moreover, FeFe-hydrogenase, formaldehyde production and growth within anoxic environments are evident in related algae, such as Scenedesmus (Florin et al. 2001; Kreuzberg 1984; Vincent 1980; Winkler et al. 2002), suggesting that a facultative anaerobic metabolism may be widespread among the Chlorophyceae. However, molecular descriptions of the enzymes responsible for anaerobic metabolism in Chlamydomonas only became available during the last decade, and the full repertoire of anaerobic metabolism available to Chlamydomonas reinhardtii was evident only from the complete genome sequence (Merchant et al. 2007), and subsequent post-genomic expression profiling.
Transcript profiling of cells exposed to anaerobic conditions (Mus et al. 2007) revealed increased mRNA expression of amylases (for starch breakdown), pyruvate:formate lyase, PFO, FeFe-hydrogenase, phosphoacetyl transferase and acetate kinase, and one plastidic ferredoxin isoform (FDX5 – although this is reported not to be a physiological electron donor for the FeFe-hydrogenase; Jacobs et al. 2009). These results support the predictions made from the annotation of the draft Chlamydomonas genome sequence, and suggest Chlamydomonas reinhardtii deploys its ‘anaerobic’ fermentation capacity in a manner that is largely similar to obligate microaerophilic/anaerobic protists (Mus et al. 2007). However, a study of the effect of gene inactivation upon changes in anaerobic metabolism suggests this model of metabolic flexibility may be too simplistic (Dubini et al. 2009).
The study of algal metabolism is of fundamental interest, particularly at an interface of microbial biochemistry and ecology, but there is also a potential commercial interest in studying algal metabolism, too. This is due to interest in algal-derived H2, lipids, and alcohols as commercially exploitable sources of biofuels, including biohydrogen (reviewed in Beer et al. 2009; Greenwell et al. 2010). One could therefore anticipate that a dark-acclimatised, anaerobically-grown mutant that does not produce H2, due to the loss of one the maturation enzymes that participate in the maturation of the dual Fe moiety within the catalytic H-centre of FeFe-hydrogenase (Shepard et al. 2010), would increase flux through one of the other fermentation pathways evident in wild type algae. In contrast to expectation, however, the Chlamydomonas ΔHYDEF mutant responds to anaerobic conditions by activating the expression of alternative metabolic enzymes, which allow the alga to use fumarate as an additional electron sink, resulting in the excretion of succinate into the culture medium (Dubini et al. 2009).
The compartmentalisation of fumarate reduction in Chlamydomonas is not known, and attempts to predict localisation are frustrated by numerous examples of dual targeting of enzymes to multiple organelles (Karniely and Pines 2005; Martin 2010), including both mitochondria and plastids (Carrie et al. 2009; Pino et al. 2007), as well as the complex interplay and metabolite shuttling that occurs between chloroplasts and mitochondria in the metabolism of plants and algae (e.g. Noguchi and Yoshida 2008). It is clear, however, that the putative fumarate reductases activated in response to dark O2 deprivation by a Chlamydomonas reinhardtii ΔHYDEF mutant are more closely related to the soluble fumarate reductases characterised in the γ-proteobacterium Shewanella and in trypanosomatid protists (Besteiro et al. 2002; Coustou et al. 2005; Leys et al. 1999; Taylor et al. 1999), rather than the complex II-related enzymes found in parasitic helminths (van Hellemond et al. 2003) and predicted in organisms such as Nyctotherus (Boxma et al. 2005). In eukaryotes possessing soluble fumarate reductase, NADH is considered to act directly as the electron donor for fumarate reduction, and this has been shown for the T. brucei enzyme (Besteiro et al. 2002).
There is clearly much to learn about how a ubiquitous alga such as Chlamydomonas switches between aerobic and unexpectedly diverse anaerobic modes of metabolism. The complexity of the anaerobic metabolic network in Chlamydomonas reinhardtii is further underscored by photochemical activation of FeFe-hydrogenase activity and sustained H2 production by sealed laboratory cultures in response to sulphur-deprivation (reviewed in Posewitz et al. 2009).
The recently published nuclear genome of the heterolobosean amoeboflagellate Naegleria gruberi (Fritz-Laylin et al. 2010) identifies a heterotrophic protist in which classical oxidative phosphorylation is predicted to be complemented by an expansive capacity for anaerobic ATP production, as judged by the pathways identified in other eukaryotes and discussed in the previous two sub-sections. As with Chlamydomonas, the geographical distribution of Naegleria is broad; Naegleria gruberi is commonly found within both oxic and micro-oxic environments in freshwater and wet soils across the globe (de Jonckheere 2002; Fulton 1993). It grows and divides as an amoeba that hunts, captures and phagocytoses its bacterial food, and it also can form synchronously and rapidly (within 90 min), a de novo microtubule cytoskeleton that includes two basal bodies and flagella with the canonical 9-triplet centriole and ‘9+2’ microtubule axoneme configurations (Dingle and Fulton 1966). The flagellate form is only a transitory response to environmental stress – in the laboratory such stress includes temperature or osmotic changes, or removal of nutrients (Fulton 1977) – and provides a mechanism for the organism to locomote quickly in search of more favourable local conditions. As an alternative response to environmental stress (also apparently an unrefined form of starvation) Naegleria can also form walled, resting cysts in the environment.
Prior to genome sequencing, little was known about the central metabolism of Naegleria, although a minimal medium had been described (Fulton et al. 1984) and the sterol composition and biochemistry of growing amoebae had been subject to some analysis (Raederstorff and Rohmer 1987). The results from these studies indicated the presence of lipid biosynthetic pathways and pointed towards the absence of a purine biosynthetic pathway and an inability to synthesise de novo sufficient quantities of several amino acids (Fulton et al. 1984; Raederstorff and Rohmer 1987). The sequence of the mitochondrial genome (deposited in Genbank with accession number NC_002573) indicates the presence and function of a typical mitochondrial respiratory chain. The only hint of anaerobic biochemistry was the characterization of PPi-dependent, rather than ATP-dependent, phosphofructokinase (Wessberg et al. 1995). On the one hand, PPi-dependency of phosphofructokinase is a common trait in anaerobic or microaerophilic microbes that minimizes the amount of ATP that needs to be invested at the start of the glycolytic pathway. However, it is also a trait seen in some parasites that can utilize mitochondrial oxidative phosphorylation (Denton et al. 1996). The nuclear genome sequence of Naegleria gruberi confirmed the presence of a full suite of pathways that can be deployed for O2-dependent metabolism of carbon sources through the Krebs cycle and electron transfer through the mitochondrial respiratory chain, but a big surprise was the presence of a bona fide-appearing FeFe-hydrogenase (i.e. an enzyme that looks capable of H2-production), together with homologues for the three enzymes, HydE, HydF, and HydG, required for post-translational maturation of the H-centre that is part of the FeFe-hydrogenase catalytic site. Significantly, there are good predictions for mitochondrial targeting signals on these four proteins, suggesting that Naegleria is a rare example of a heterotroph with a mitochondrial metabolism that is capable of both oxidative phosphorylation and H2-formation under appropriate conditions.
The characterization of mitochondrial genomes from the ciliate Nyctotherus ovalis and the stramenopile Blastocystis provide unequivocal evidence that mitochondria can and do undergo reductive evolution to hydrogenosomes in the presence of an organelle-targeted FeFe-hydrogenase, but there is no biochemical or EST evidence to suggest that these organisms are capable of cytochrome c-dependent respiration and oxidative phosphorylation (Boxma et al. 2005; Stechmann et al. 2008) using O2, NO3− or NO2− as terminal electron acceptors. The only other protists of which we are aware that are likely to be able to show the same sort of mitochondrial flexibility as Naegleria gruberi are (i) Acanthamoeba castellanii, which catalyses both cytochrome c-dependent and alternative oxidase-dependent reduction of O2 (Edwards and Lloyd 1978; Jarmuszkiewicz et al. 1998) and possesses genes encoding FeFe-hydrogenases and at least two of its three associated maturases (HydE and HydG) (Hug et al. 2010) and (ii) Amoebidium parasiticum, an aquatic protist closely related to the metazoan lineage and often found on or within the hind-gut of marine invertebrates (Benny and O’Donnell 2000). Amoebidium parasiticum also possesses a FeFe-hydrogenase gene (Hug et al. 2010), and from its partially characterized mitochondrial genome Amoebidium parasiticum can also be confidently predicted to assemble a typical cytochrome-dependent respiratory chain (Burger et al. 2003). We therefore await with interest the results from future biochemical and/or genome analyses of these protists. Intriguingly, Naegleria gruberi, Acanthamoeba castellanii, and Amoebidium parasiticum all belong to different eukaryotic supergroups, suggesting that although we still await formal experimental confirmation of an aerobic mitochondrion that can, under suitable environmental conditions, generate H2, it could turn out that this unexpected trait is relatively common in microbial eukaryotes. Indeed, perhaps it should not be surprising given that (a) the reductive evolution of mitochondria into hydrogenosomes has occurred independently on multiple occasions in evolutionarily distant organisms (Hackstein et al. 2006) and (b) many prokaryotic organisms are able to toggle aerobic metabolism with anaerobic fermentation and respiration that often use O2-labile enzymes (e.g. Richardson 2000). That we have had to wait so long for the likely glimpse of a mitochondrion capable of oxidative phosphorylation and H2 production is probably best seen as testimony of limited microbial sampling.
Intriguingly, the mitochondrial flexibility of Acanthamoeba and Naegleria mitochondria can be predicted to extend beyond oxidative phosphorylation and H2 production since both organisms possess homologues of the NirK enzyme used in fungal NO3−/NO2− respiration. In the case of Naegleria gruberi, a NirK homologue is encoded in the nuclear genome; in the case of Acanthamoeba castellanii an EST has been deposited in Genbank (accession number DQ384327.1; cited by Kim et al. (2009) and first described in Watkins and Gray (2006)). This raises the possibility that NO3− or NO2− respiration may occur in these soil-dwelling amoebae. In Naegleria gruberi, the presence of the NirK homologue tentatively correlates with the occurrence of four putative membrane-bound adenylate cyclases associated with NIT domains in the genome (Fritz-Laylin et al. 2010). NIT domains are linked to NO3− and NO2− sensing in some bacteria (Shu et al. 2003). With a NO3−/formate class of transporter also predicted to be a lateral gene transfer candidate in Naegleria, a tentative picture for NO3− or NO2− usage by this protist under anoxic or hypoxic conditions can start to be reconciled. Two genes encoding proteins with putative fumarate reductase domains are also evident in the Naegleria genome. Although no prediction of their sub-cellular localization is offered, the identification of rhodoquinone in anaerobically functioning mitochondria of another excavate protist (Euglena gracilis; Hoffmeister et al. 2004), albeit a species from another phylum (Euglenozoa), and in the hydrogenosomes of the anaerobic ciliate Nyctotherus ovalis (Boxma et al. 2005) suggest that an analysis of quinone composition in Naegleria is merited. In this way it will be possible to determine if fumarate reduction is linked to either quinone-dependency (and therefore potentially coupled to generation of a membrane potential across the inner mitochondrial membrane potential) or directly to NADH oxidation (as in trypanosomatids).
To ask if the anaerobic metabolic predictions in Acanthamoeba and Naegleria and the complex anaerobic metabolic network characterized in Chlamydomonas represent the tip of an unrealized iceberg, we surveyed published nuclear genome sequences and EST surveys for the presence or absence of enzymes we have discussed already in the context of ‘anaerobic energy metabolism’. The chosen sequences covered, as best as possible, the breadth of known protist diversity. The results from the survey are shown in Figure 4. On the basis of this survey several predictions can be offered.
In protists other than Chlamydomonas reinhardtii, Naegleria gruberi, and Acanthamoeba castellanii, genome- or EST-based evidence for an ability to combine both aerobic (oxidative phosphorylation) with an extensive anaerobic metabolism (i.e. anaerobic respiration and/or H2 production) is weak. However, the anaerobic metabolic network in Chlamydomonas reinhardtii is potentially further extended by the identification of a gene with NirK homology. Of the other green or red algae for which genome sequences were published at the time of writing, there was no obvious capacity for anaerobic function in oceanic picoplankton of the genera Ostreococcus and Micromonas, other than the prediction of putative fumarate reductase and pyruvate:formate lyase. In the extremophile red alga Cyanidioschyzon merolae, which lives in acidic hot sulphur springs, the only hint of an enzyme classically linked with anaerobic energy metabolism was a homologue of acetyl-CoA synthetase [ADP-forming]. The physiological function of these putative enzymes in algae has not been tested.
Despite only a faint hint of significant anaerobic potential in Micromonas, Ostreococcus, and the pennate diatom Phaeodactylum tricomutum, a plastid-localised FeFe-hydrogenase homologue is predicted in the centric diatom Thalassiosira pseudonana. The presence of this homologue has been noted previously (Hug et al. 2010; Stechmann et al. 2008), but its function is unclear. The absence of HydE, HydF, and HydG homologues from the Thalassiosira genome sequence could suggest that the protein has no catalytic function in H2 function. However, the FeFe-hydrogenase maturases are also absent from Giardia, which does produce H2 (Lloyd et al. 2002), and Entamoeba histolytica possesses a FeFe-hydrogenase that was expressed as an active recombinant enzyme in the cytoplasm of Escherichia coli in the absence of HydE, HydF, and HydG (Nixon et al. 2003). FeFe-hydrogenases are thought to be irreversibly inactivated by their reaction with O2 (Cracknell et al. 2009). One speculative possibility therefore, is that the diatom FeFe-hydrogenase does catalyse H2 production, but that the absence of the usual maturases is linked to the evolution of an O2-tolerant enzyme adapted to function in aerobic oceanic environments.
Some protists – the ciliates Paramecium tetraurelia and Tetrahymena thermophila and the social amoeba Dictyostelium discoideum – appear to conform to a simple ‘textbook’ model of aerobic heterotrophic metabolism. Dictyostelium discoideum seems to have a very limited capacity to stray beyond the aerobic leaf litter during bacterial predation. There are, however, suggestions for anaerobic ATP production in some heterotrophic protists, including the pathogens Perkinsus marinus and Phytophthora where both ‘soluble’ fumarate reductase and acetate kinase are predicted. Some Phytophthora species disseminate and transfer between plants through the flagellar motility of zoospores in soil; a potential link between the likely composition of the central metabolic network and fluctuating local O2 tensions in a soil environment is therefore once again evident. Expression profiling of the zoospore and other life cycle stages will spread further light on the possible role of anaerobic metabolism in Phytophthora and disease dissemination and pathogen virulence.
The sequence-based predictions (Fritz-Laylin et al. 2010; Hug et al. 2010) and experimental validation (Dubini et al. 2009; Mus et al. 2007) of metabolic flexibility in Naegleria, Acanthamoeba, and Chlamydomonas extend the earlier biochemical demonstrations of facultative anaerobic respiration in some fungi, ciliates, and foraminifers (Finlay et al. 1983; Kobayashi et al. 1996; Pina-Ochoa et al. 2010; Risgaard-Petersen et al. 2006; Takaya et al. 2003). However, it is important to remember that genome sequences and EST profiles in isolation only facilitate a prediction of function. Collectively, we strongly believe that the signature of anaerobic metabolism seen in the Naegleria genome and hinted at from the initial EST profiling of Acanthamoeba is extensive enough to be unequivocal with regard to conclusions drawn, particularly given that this signature not only mirrors the metabolic biochemistry of chlorophyte algae found in similar ecological niches, but is also absent from another soil-dwelling protist (Dictyostelium discoideum) that hunts bacteria within a somewhat different trophic niche on forest floors. Further support for the prediction of an extensive anaerobic metabolism in Naegleria comes from unpublished observations that Naegleria gruberi will grow in microaerophilic conditions under N2 when fed on bacteria, but not axenically in a defined medium (C. Fulton, unpublished observations).
Despite confidence in predictions made above, the scientific literature is littered with examples of enzymes that display either alternative or broader substrate specificities than those ascribed from bioinformatic annotation. Moreover, in a genome-sequencing context, many annotations are automated only on a basis of top BLAST hit identities, meaning propagation of a false annotation, a bioinformatic version of ‘Chinese whispers’, is all too easy. Thus, in the absence of additional experimental data, caution or alternative explanations might be invoked for some of the patterns of gene presence/gene absence highlighted in Figure 4. For example, the presence of both an acetate kinase homologue and a phosphoacetyl-transferase in both Perkinsus and Phytophthora is a strong predictor that the acetate kinase pathway operates in these protists. In contrast, the presence of only an acetate kinase homologue in the pennate diatom Phaeodactylum tricomutum suggests that this homologue has an alternative function. This situation is analogous to Entamoeba histolytica, which also possesses an acetate kinase homologue, but no obvious PAT (Tielens et al. 2010). Similarly, the substrate specificity of different acetyl-CoA synthetase homologues, including the biochemically characterized family members from Entamoeba histolytica and Giardia lamblia, often extends beyond the acceptance of acetyl-CoA as a substrate for the forward reaction (Sanchez et al. 2000). Thus, the function of the acetyl-CoA synthetase homologue found in Plasmodium species, but not found to date in any other apicomplexans (Fig. 4 and M. Ginger, unpublished observations), is intriguing, not least because pyruvate dehydrogenase, which provides the only obvious route to significant acetyl-CoA formation in Plasmodium, is found only in the parasite’s non-photosynthetic plastid relic (Fleige et al. 2007; Foth et al. 2005). This pyruvate dehydrogenase is thought only to contribute to the provision of a two-carbon building block for biosynthetic pathways in the apicoplast (Pei et al. 2010). Since the central energy metabolism of different apicomplexan parasites has clearly been shaped by patterns of differential gene loss, presumably as a consequence of adapting to different parasitic niches in mammalian host or vector (e.g. Ginger 2006; Seeber et al. 2008), one can anticipate that the acetyl-CoA synthetase homologue conserved in Plasmodium will turn out to have a physiologically important function. Perhaps the clearest example of where the bioinformatic survey (Fig. 4) gives equivocal results is the distribution of acetate:succinate CoA transferase homologues in protists. Three classes of acetate:succinate CoA transferase exist, but the enzyme characterized from trypanosomatids (Riviere et al. 2004) is very clearly orthologous (i.e. a homologue separated by speciation) to the SCOT family of mammalian acyl:CoA transferases. These animal enzymes are linked not to acetate formation, but to the physiologically important formation of a 3-oxoacyl-CoA and succinate from succinyl-CoA and a 3-oxoacid (Tielens et al. 2010, and references cited therein). Trypanosomatids have little or no capacity for survival under anaerobic conditions (van Hellemond et al. 1997; van Weelden et al. 2003); it is therefore unclear whether the coupling of acetate formation to ATP production is an ancient, derived, or even re-acquired trait in the kinetoplastid lineage. In contrast, Naegleria gruberi contains homologues of all three classes of acetate:succinate CoA transferase and a homologue of the acetyl-CoA synthetase [ADP-forming] enzyme. Thus, one can be confident that the predicted capacity for anaerobic respiration and H2 formation is also buttressed by substrate level phosphorylation of ADP and concomitant production of acetate as a metabolic end-product even though the identity of the candidate enzyme(s) responsible for acetate production is not easily predicted. This capacity will require experimental verification, as will the function of the diverse assortment of other candidate CoA transferases found in other organisms.
Central Metabolism in Microbial Eukaryotes – Retention of an Ancient Legacy or Sculpted by Lateral Transfer?
It is important to stress, as others have done (e.g. Martin et al. 2001; Mentel and Martin 2008; Richardson 2000), that the flexibility in energy metabolism seen in eukaryotes pales when compared to the metabolic flexibility seen in individual bacteria3. In eukaryotes examined thus far, only a handful of additional enzymes are required to facilitate the anaerobic fermentation or respiration of glucose and other carbon sources. The origin(s) of the genes encoding some of these enzymes, notably PFO and FeFe-hydrogenase, have been the subject of debate over a number of years (see e.g. Embley et al. 1998; Horner et al. 2002, 1999; Hug et al. 2010; Martin and Müller 1998; Rotte et al. 2001 for early and Hug et al. [2010] for more recent discussions of the topic), but unfortunately it is a debate that remains hindered by PFO and FeFe-hydrogenase phylogenies that lack resolution, consequences, at least in part, of limited taxon sampling and a saturation of variable sites (Embley 2006; Hug et al. 2010). The issue of whether PFO is likely to be of monophyletic origin in eukaryotes remains unresolved (Hug et al. 2010), but despite problems in attaining robust resolution for FeFe-hydrogenase phylogenies, several independent analyses lend support to a conclusion that FeFe-hydrogenases have entered the eukaryotic lineage on multiple occasions through lateral gene transfer (LGT) from bacterial sources (Boxma et al. 2007; Fritz-Laylin et al. 2010; Horner et al. 2000; Hug et al. 2010).
Certainly, there is little doubt that LGT on a small scale4 of one or several genes is emerging as an important factor in shaping not just the central and secondary metabolism of many protists, but also an enhanced capacity for environmental perception and signalling in some microbial eukaryotes (Anantharaman et al. 2007; Andersson 2009; Bowler et al. 2008; Keeling 2009; Keeling and Palmer 2008), and even operational processes such as gene expression and replication, too (Andersson et al. 2005). Also relevant is the concept of ‘gene-sharing’ whereby genes with patchy distributions in eukaryotes and prokaryotes generally, also appear to be recurrently found in unrelated microbes that occupy similar ecological niches. This suggests independent fluxes of common genetic material from a given environment into unrelated organisms: the distribution of enzymes adapted for the digestion of complex carbohydrates among the microbial fauna of the gut rumen (Ricard et al. 2006) and the presence in Naegleria gruberi and Dictyostelium discoideum, but not in most other eukaryotic lineages, of twenty five examples of laterally-transferred bacterial gene families (Andersson 2009) provide recent examples of the gene-sharing phenomenon involving protists. However, just as LGT facilitates innovation of novel metabolic traits, it can also facilitate gene displacement. Mosaic origins of some glycolytic enzymes in anaerobic protists provide good examples of displacement (Liapounova et al. 2006; Stechmann et al. 2006). Differences in Vmax, substrate affinities, etc, which conceivably effect changes in metabolic flux along a pathway such as glycolysis provide the obvious selective advantages that could result in the retention of a gene encoding a laterally transferred enzyme within a recipient genome, followed by an eventual displacement and loss of the ancestral endogenous gene that encoded the original isoform.
In contrast to invoking LGT as an explanation for the distribution of PFO and FeFe-hydrogenase genes in diverse eukaryotes, there is an opposing view, which contends that the presence of PFO, FeFe-hydrogenase, and other enzymes of anaerobic metabolism in unrelated protists reflects the differential retention of genes that were present at an early stage of eukaryotic evolution. Thus, PFO, FeFe-hydrogenase, and Nirk homologues have been suggested as enzymes that were encoded in the α-proteobacterial endosymbiont that gave rise to the proto-mitochondrion (Kim et al. 2009; Martin et al. 2001, 2003; Mentel and Martin 2008; van der Giezen 2009). Indeed, the facultative anaerobic metabolism of a hypothetical H2-producing α-proteobacterium possessing PFO and FeFe-hydrogenase was even posited as a driving force in the process of eukaryogenesis (the ‘Hydrogen Hypothesis’ for eukaryotic origins; Martin and Müller 1998). If the integration of PFO, FeFe-hydrogenase, Nirk, and potentially other enzymes into the anaerobic metabolic networks of extant eukaryotes, is a legacy of ancestral metabolic versatility in the earliest eukaryotes, then (i) the patchy distributions of homologues in extant taxa, (ii) the common ancestry of ‘classic’ aerobic mitochondria, hydrogenosomes, and mitosomes, and (iii) the apparent frequency with which mitochondrial degeneracy gives way to hydrogenosomes and mitosomes are all explained by complex patterns of gene loss, as either aerobic or anaerobic modes of metabolism become dispensable during the process of niche adaptation. Failure to recover monophyly of eukaryotic FeFe-hydrogenases, in this instance, could reflect either gene displacement or a secondary re-acquisition of an ability to generate H2. There is little doubt that the loss of aerobic metabolism has occurred on numerous occasions during eukaryotic evolution; the unresolved issue is whether loss of anaerobic metabolism is an equally widespread phenomenon. If, by way of comparison, one looks within eukaryotic groups for the presence or absence of the iconic elements of eukaryotic cell biology, then there are numerous observations that can only be explained by surprisingly complex patterns of gene loss, rather than secondary gain via LGT. For example, Fungi and Amoebozoa are often mis-conceived as classically aflagellate eukaryotic groups, but present views of fungal and amoebozoal phylogeny (Fiore-Donno et al. 2010; James et al. 2006; Pawlowski and Burki 2009) indicate that loss of the ability to build a flagellum has occurred independently on several occasions within each of these groups. Similarly, in the trypanosomatids, the American trypanosome Trypanosoma cruzi retains components for a more elaborate actin-based cytoskeleton that do the African trypanosome or Leishmania (Berriman et al. 2005). Since the Trypanosoma are now widely accepted to be monophyletic to the exclusion of other trypanosomatid genera (Simpson et al. 2006), streamlining of the actin-based cytoskeleton has occurred at least twice during trypanosomatid evolution.
Despite the appeal of the hydrogen hypothesis as an elegant biochemical model to explain eukaryogenesis, the likelihood that PFO and FeFe-hydrogenase were present in the α-proteobacterial ancestor of mitochondria has legitimately been challenged using the argument that homologues of both enzymes are very seldom encoded in the genomes of extant α-proteobacteria (Hug et al. 2010; Nixon et al. 2003). Notwithstanding the observation that gene content within prokaryotic genomes is most certainly not static over evolutionary time, and that the gene inventory of the α-proteobacterial endosymbiont that gave rise to mitochondria may therefore be significantly different from gene inventories within contemporary α-proteobacteria sampled to date (Dagan and Martin 2009; Esser et al. 2007), the arguments raised by Samuelson and Roger with their respective co-workers is a strong one, and is also an argument that sits well alongside current phylogenetic support for multiple origins of FeFe-hydrogenase (Boxma et al. 2007; Fritz-Laylin et al. 2010; Horner et al. 2000; Hug et al. 2010), and perhaps PFO (Hug et al. 2010), in eukaryotes. However, it is not easy to dismiss the case for an early origin of mitochondrial FeFe-hydrogenase, not least because an FeFe-hydrogenase-related protein, NARF, is a universally conserved component (Allen et al. 2008b) of the essential cytosolic Fe-S cluster assembly pathway (CIA) found in all eukaryotes examined to date, including protists discussed in this review (Lill 2009). The CIA pathway appears to be a eukaryotic-specific invention. FeFe-hydrogenases are at least as poorly represented in Archaeal genomes as they are in α-proteobacterial genomes, a relevant observation given the similarities between replication, transcription, and translation machineries in Archaea and eukaryotes, and the possibility that eukaryotes evolved as a sister group to the Crenarchaeota (Foster et al. 2009). The origin of the ancestral NARF protein is therefore an enigma, but its strict conservation in eukaryotes is a strong indicator that FeFe-hydrogenase-dependent biochemistry featured at a very early point in eukaryotic evolution – a point also discussed in Horner et al. (2002) when the wide phylogenetic distribution of NARF homologues was first becoming apparent, but its function was unknown. The possibility that a FeFe-hydrogenase acquired mitochondrial targeting signals during early mitochondrial evolution (i.e. before the divergence of the last common ancestor of extant eukaryotes) and became integrated into the redox metabolism of the organelle (only to be lost again on one or more occasions) should therefore be considered. It is also relevant to note that the origin of the plastidic FeFe-hydrogenase in chlorophytes is also a mystery: there is no indication (yet) that hydrogenases are ubiquitous in algal chloroplasts. Even though bi-directional NiFe-hydrogenases are common in cyanobacteria, it is curious that no NiFe-hydrogenase homologue has yet been described in an alga, and there is no indication of FeFe-hydrogenases in extant cyanobacteria (Horner et al. 2002; Ludwig et al. 2006; Vignais et al. 2001; MLG unpublished observations from searches of genome sequences publicly available in Summer 2010).
The origins of PFO and FeFe-hydrogenase will probably be resolved by further genome sampling of carefully selected free-living protists from each of the major eukaryotic groups, it is interesting that another enzyme that is poorly represented in α-proteobacteria but which is no longer considered an unusual occurrence in protist and animal mitochondria is the alternative oxidase (Fig. 4). This enzyme transfers electrons directly from ubiquinol to O2 without contributing to the generation of inner-mitochondrial membrane potential. Only a decade ago, the presence of alternative oxidase in heterotrophic eukaryotes was somewhat of a rarity (Atteia et al. 2004), but today alternative oxidase provides a classic example of a ubiquitous mitochondrial enzyme that just so happens to be absent from the mammals and yeast upon which textbook views of intermediary metabolism in eukaryotes are often based (Atteia et al. 2004; Williams et al. 2010). In contrast, NO3− respiration is a common feature of α-proteobacterial metabolism, including in species such as Paracoccus denitrificans that have long been known to develop a mitochondrial-type electron transport chain and possess other characteristics also seen in mitochondria (John and Whatley 1975). Without more detailed analysis of the NirK and ‘soluble’ fumarate reductase homologues in eukaryotes, it is too early to speculate on whether these enzymes really are the legacy of metabolic flexibility and anaerobic capacity in the proto-mitochondrion. However, the distribution of homologues in microbial eukaryotes examined thus far by either genome sequencing or EST profiling is intriguing.
Closing Perspectives
A combination of comparative genomics and experimental analysis is facilitating the provision of metabolic maps for an increasing diversity of microbial eukaryotes. Initially, maps that predict holistic metabolic potential were mostly available for different fungi and parasitic protists. The human benefit from the identification and subsequent development of novel, much-needed drug targets against parasites that cause serious diseases in some of the world’s poorest countries (e.g. the causal agents of malaria, African sleeping sickness, leishmaniasis, amoebic dysentery) was heralded as one of the significant likely outcomes from whole genome sequencing efforts (e.g. Ash and Jasny 2005; Berriman et al. 2005; Wirth 2002). Striking examples of where EST sequencing and genome annotation have helped fast-track our understanding of protist metabolism are the functional characterisations of relic mitochondria in Giardia, Entamoeba and microsporidian parasites, and a non-photosynthetic plastid relic in apicomplexan parasites. The presence of these organelles was originally inferred from the molecular characterisation of mitochondrial-type chaperones (degenerate mitochondria; Clark and Roger, 1995; Horner et al. 1998 and references cited therein) and sequencing of a cryptic organellar genome (the apicomplexan plastid or ‘apicoplast’; Wilson et al. 1996; 1994; Wilson and Williamson, 1997 for further discussion). Using comparisons with conventional mitochondria and chloroplasts to aid bioinformatic predictions of organellar function and targeting pathways, pivotal cellular roles played by these organelles were quickly identified. The chronology and current understanding arising from these discoveries have been the subject of many excellent reviews (e.g. Hackstein et al. 2006; Hjort et al. 2010; Lim and McFadden 2010; Ralph et al. 2004).
Several years on from the initial releases of genome data, reverse genetics and metabolomic approaches are readily applied to some parasites (notably the trypanosomatids and various apicomplexans) to test predictions arising from genome annotation and to probe the functional importance of numerous individual metabolic pathways in a life cycle-specific context – i.e. relevance of specific pathways to pathogenicity in the host (e.g. Crawford et al. 2006; Gibellini et al. 2009; Gunther et al. 2009; Mazumdar et al. 2006; Pei et al. 2010; Stokes et al. 2008; Vaughan et al. 2009; van Weelden et al. 2003; Vega-Rodriguez et al. 2009; Yu et al. 2008). In a recent study, the use of [13C]-labelled glucose and glutamine tracers identified a bifurcated Krebs cycle in Plasmodium falciparum, whereby acetyl-CoA is not the substrate for oxidation within a cyclic pathway, but is the product liberated from the reductive metabolism of the 2-oxoglutarate intermediate from glutamine metabolism (Olszewski et al. 2010). Not only does this work identify a completely unexpected and novel rewiring of a core metabolic pathway, but it illustrates how (i) the puzzle posed by the annotation of an ‘incomplete’ pathway in a genome sequence – the presence of all the canonical Krebs cycle enzymes except NAD+-dependent isocitrate dehydrogenase – and (ii) the conflict of a expression profile that at first glance challenges an accepted dogma – expression of Krebs cycle enzymes, but the reliance on aerobic glycolysis in blood stage parasites – can be resolved by the application of analytical approaches.
Increasingly, metabolic maps are becoming available for a variety of unrelated free-living protists, including organisms where there is already some biochemical data that reveals how ecological constraints, such as nutrient limitation, influence metabolic organisation and more critically, metabolic fluxes (e.g. Allen et al. 2008a; Moseley et al. 2002; Peers and Price 2006; Strzepek and Harrison 2004; Terauchi et al. 2010). Like the example of the bifurcated Plasmodium Krebs cycle characterised using metabolomics (Olszewski et al. 2010), these example citations illustrate the type of dynamic parameters that cannot be provided from the static metabolic map predicted from a genome sequence. As with parasites, metabolic maps from free-living protists also point towards unexpected examples of pathway diversity and metabolic function, even in species such as Chlamydomonas reinhardtii that have traditionally provided important models for experimental studies in metabolism. In this review, we discussed the composition and the likely ecological and evolutionary relevance of an unexpected blend of anaerobic and aerobic metabolism that is evident from the recent genome sequencing and transcript profiling of several free-living protists. We have glimpsed this metabolism despite only a sampling a very narrow fraction of protist diversity. Further sampling of carefully selected protists, notably organisms such as Palpitomonas or the Apusomonadida (which each combine characteristics seen in multiple eukaryotic lineages and likely branch deep within currently accepted eukaryotic supergroups; Cavalier-Smith and Chao 2010; Yabuki et al. 2010) or the jakobids (a group which includes both micro-aerophiles, such as Andalucia incarcerata [Lara et al. 2006; Simpson and Patterson 2001], as well as aerobic protists with the most eubacterial-like mitochondrial genomes characterised to date [Lang et al. 1997]), should help provide insight into how much of this anaerobic metabolism is reflective of the ancestral metabolic capabilities present in the first eukaryotes.
There are also other important examples of unexpected and sometimes enigmatic metabolic diversity that have been uncovered through genome sequencing. For example, the discovery that trypanosomatid parasites synthesize their fatty acids using a microsomal elongase-based pathway was, in part, informed by the absence of the classic type I fatty acid synthase from Trypanosoma and Leishmania genome sequences, and the realisation that type II fatty synthesis in trypanosomes is, as in other heterotrophs, a mitochondrial pathway used primarily for the biosynthesis of a restricted set of fatty acids, including lipoic acid (a co-enzyme for pyruvate dehydrogenase). This work on trypanosomatid fatty acid biosynthesis provided a new paradigm for the core biosynthetic pathway of fatty acid biosynthesis (Lee et al. 2006, 2007), but carefully scrutiny of the genome sequences and nutritional requirements of Dictyostelium and Naegleria suggests this so-called ‘type III’ pathway of fatty acid synthesis (Cronan 2006) has a wider phylogenetic distribution (Fritz-Laylin et al. 2010). Furthermore, genome sequencing identified diatoms as the only known phototrophs to possess an intact urea cycle although the function of the cycle is unclear (Armbrust et al. 2004; Parker et al. 2008); it presumably does not function in nitrogen excretion given the competition for bioavailable nitrogen in the ocean (Elser et al. 2007). We look forward to the continued experimental interrogation of the metabolic predictions provided by the metabolic annotations of protist genomes, and probably the sequence-led discovery of further metabolic novelties, too.
Acknowledgments
MLG is a Royal Society University Research Fellow and gratefully acknowledges support from The Royal Society and The Leverhulme Trust for his work on protist metabolism. LKFL was supported by a Tien Scholars Fellowship in Environmental Sciences and Biodiversity. WZC and LKFL acknowledge the support of grant NIH AI054693. SCD acknowledges grant NIH/NIAID 1R01AI77571-01A1.
Footnotes
Or even five clades, if the Amoebozoa (a clade composed exclusively of protists) and Opisthokonta (Metazoa, Fungi, some protists) are confirmed to be monophyletic with respect to other eukaryotes.
In microbial eukaryotes without any capacity for mitochondrial electron transport – i.e. lacking quinones – and lacking the mitochondrial ATP synthase, a streamlining of conserved mitochondrial protein import machinery is consistently observed (Dolezal et al. 2010; Regoes et al. 2005; Waller et al. 2009). It is not clear if such streamlined import machineries require mitochondrial membrane potential or indeed how electrochemical gradients across inner-mitosomal or inner-hydrogenosomal membranes are set.
Where flexibility can be defined either by the ability of a single species (e.g. Rhodospirillum rubrum; Rhodobacter capsulatus) to differentially regulate the use of aerobic respiration, anaerobic respiration, fermentation and photosynthesis depending upon environmental conditions (discussed in Mentel and Martin 2008) or extreme respiratory flexibility, such as that observed in species from the γ-proteobacterial genus Shewanella; many Shewanella species can respire more than 20 organic and inorganic electron acceptors in the absence of O2 (Hau and Gralnick 2007).
I.e. distinct from the large-scale endosymbiotic gene transfer of genes from the genomes of mitochondrion and primary and secondary plastids to the nucleus.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe T, Hoshino T, Nakamura A, Takaya N. Anaerobic elemental sulfur reduction by fungus Fusarium oxysporum. Biosci Biotechnol Biochem. 2007;71:2402–2407. doi: 10.1271/bbb.70083. [DOI] [PubMed] [Google Scholar]
- Allen AE, Laroche J, Maheswari U, Lommer M, Schauer N, Lopez PJ, Finazzi G, Fernie AR, Bowler C. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc Natl Acad Sci USA. 2008a;105:10438–10443. doi: 10.1073/pnas.0711370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JW, Ferguson SJ, Ginger ML. Distinctive biochemistry in the trypanosome mitochondrial intermembrane space suggests a model for stepwise evolution of the MIA pathway for import of cysteine-rich proteins. FEBS Lett. 2008b;582:2817–2825. doi: 10.1016/j.febslet.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Amaral Zettler LA, Gomez F, Zettler E, Keenan BG, Amils R, Sogin ML. Microbiology: eukaryotic diversity in Spain’s River of Fire. Nature. 2002;417:137. doi: 10.1038/417137a. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Iyer LM, Aravind L. Comparative genomics of protists: new insights into the evolution of eukaryotic signal transduction and gene regulation. Annu Rev Microbiol. 2007;61:453–475. doi: 10.1146/annurev.micro.61.080706.093309. [DOI] [PubMed] [Google Scholar]
- Andersson JO. Gene transfer and diversification of microbial eukaryotes. Annu Rev Microbiol. 2009;63:177–193. doi: 10.1146/annurev.micro.091208.073203. [DOI] [PubMed] [Google Scholar]
- Andersson JO, Sarchfield SW, Roger AJ. Gene transfers from Nanoarchaeota to an ancestor of diplomonads and parabasalids. Mol Biol Evol. 2005;22:85–90. doi: 10.1093/molbev/msh254. [DOI] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kroger N, Lau WW, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Ash C, Jasny BR. Trypanosomatid genomes. Introduction. Science. 2005;309:399. doi: 10.1126/science.309.5733.399. [DOI] [PubMed] [Google Scholar]
- Atteia A, van Lis R, Gelius-Dietrich G, Adrait A, Garin J, Joyard J, Rolland N, Martin W. Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J Biol Chem. 2006;281:9909–9918. doi: 10.1074/jbc.M507862200. [DOI] [PubMed] [Google Scholar]
- Atteia A, van Lis R, van Hellemond JJ, Tielens AG, Martin W, Henze K. Identification of prokaryotic homologues indicates an endosymbiotic origin for the alternative oxidases of mitochondria (AOX) and chloroplasts (PTOX) Gene. 2004;330:143–148. doi: 10.1016/j.gene.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Beer LL, Boyd ES, Peters JW, Posewitz MC. Engineering algae for biohydrogen and biofuel production. Curr Opin Biotechnol. 2009;20:264–271. doi: 10.1016/j.copbio.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Benny GL, O’Donnell K. Amoebidium parasiticum is a protozoan, not a trichomycete. Mycologia. 2000;92:1133–1137. [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, Macleod A, Mooney PJ, Moule S, Martin DM, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream MA, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CM, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE, El-Sayed NM. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Besteiro S, Biran M, Biteau N, Coustou V, Baltz T, Canioni P, Bringaud F. Succinate secreted by Trypanosoma brucei is produced by a novel and unique glycosomal enzyme, NADH-dependent fumarate reductase. J Biol Chem. 2002;277:38001–38012. doi: 10.1074/jbc.M201759200. [DOI] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret JP, Chiovitti A, Choi CJ, Coesel S, De Martino A, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kroger N, Kroth PG, La Roche J, Lindquist E, Lommer M, Martin-Jezequel V, Lopez PJ, Lucas S, Mangogna M, McGinnis K, Medlin LK, Montsant A, Oudot-Le Secq MP, Napoli C, Obornik M, Parker MS, Petit JL, Porcel BM, Poulsen N, Robison M, Rychlewski L, Rynearson TA, Schmutz J, Shapiro H, Siaut M, Stanley M, Sussman MR, Taylor AR, Vardi A, von Dassow P, Vyverman W, Willis A, Wyrwicz LS, Rokhsar DS, Weissenbach J, Armbrust EV, Green BR, Van de Peer Y, Grigoriev IV. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Boxma B, de Graaf RM, van der Staay GW, van Alen TA, Ricard G, Gabaldon T, van Hoek AH, Moon-van der Staay SY, Koopman WJ, van Hellemond JJ, Tielens AG, Friedrich T, Veenhuis M, Huynen MA, Hackstein JH. An anaerobic mitochondrion that produces hydrogen. Nature. 2005;434:74–79. doi: 10.1038/nature03343. [DOI] [PubMed] [Google Scholar]
- Boxma B, Ricard G, van Hoek AH, Severing E, Moon-van der Staay SY, van der Staay GW, van Alen TA, de Graaf RM, Cremers G, Kwantes M, McEwan NR, Newbold CJ, Jouany JP, Michalowski T, Pristas P, Huynen MA, Hackstein JH. The [FeFe] hydrogenase of Nyctotherus ovalis has a chimeric origin. BMC Evol Biol. 2007;7:230. doi: 10.1186/1471-2148-7-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohn FH, Clarkson AB., Jr Quantitative effects of salycylhydroxamic acid and glycerol on Trypanosoma brucei glycolysis in vitro and in vivo. Acta Trop. 1978;35:23–33. [PubMed] [Google Scholar]
- Brown DM, Upcroft JA, Upcroft P. A H2O-producing NADH oxidase from the protozoan parasite Giardia duodenalis. Eur J Biochem. 1996;241:155–161. doi: 10.1111/j.1432-1033.1996.0155t.x. [DOI] [PubMed] [Google Scholar]
- Burger G, Forget L, Zhu Y, Gray MW, Lang BF. Unique mitochondrial genome architecture in unicellular relatives of animals. Proc Natl Acad Sci USA. 2003;100:892–897. doi: 10.1073/pnas.0336115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Pawlowski J. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol Lett. 2008;4:366–369. doi: 10.1098/rsbl.2008.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, Sicheritz-Ponten T, Noel CJ, Dacks JB, Foster PG, Simillion C, Van de Peer Y, Miranda-Saavedra D, Barton GJ, Westrop GD, Muller S, Dessi D, Fiori PL, Ren Q, Paulsen I, Zhang H, Bastida-Corcuera FD, Simoes-Barbosa A, Brown MT, Hayes RD, Mukherjee M, Okumura CY, Schneider R, Smith AJ, Vanacova S, Villalvazo M, Haas BJ, Pertea M, Feldblyum TV, Utterback TR, Shu CL, Osoegawa K, de Jong PJ, Hrdy I, Horvathova L, Zubacova Z, Dolezal P, Malik SB, Logsdon JM, Jr, Henze K, Gupta A, Wang CC, Dunne RL, Upcroft JA, Upcroft P, White O, Salzberg SL, Tang P, Chiu CH, Lee YS, Embley TM, Coombs GH, Mottram JC, Tachezy J, Fraser-Liggett CM, Johnson PJ. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Giraud E, Whelan J. Protein transport in organelles: Dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 2009;276:1187–1195. doi: 10.1111/j.1742-4658.2009.06876.x. [DOI] [PubMed] [Google Scholar]
- Castro-Guerrero NA, Jasso-Chavez R, Moreno-Sanchez R. Physiological role of rhodoquinone in Euglena gracilis mitochondria. Biochim Biophys Acta. 2005;1710:113–121. doi: 10.1016/j.bbabio.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6:342–345. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE. Phylogeny and evolution of Apusomonadida (Protozoa: Apusozoa): new genera and species. Protist. 2010;161:549–576. doi: 10.1016/j.protis.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Charon MH, Volbeda A, Chabriere E, Pieulle L, Fontecilla-Camps JC. Structure and electron transfer mechanism of pyruvate:ferredoxin oxidoreductase. Curr Opin Struct Biol. 1999;9:663–669. doi: 10.1016/s0959-440x(99)00027-5. [DOI] [PubMed] [Google Scholar]
- Clark CG, Roger AJ. Direct evidence for secondary loss of mitochondria in Entamoeba histolytica. Proc Natl Acad Sci USA. 1995;92:6518–6521. doi: 10.1073/pnas.92.14.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordente AG, Heinrich A, Pretorius IS, Swiegers JH. Isolation of sulfite reductase variants of a commercial wine yeast with significantly reduced hydrogen sulfide production. FEMS Yeast Res. 2009;9:446–459. doi: 10.1111/j.1567-1364.2009.00489.x. [DOI] [PubMed] [Google Scholar]
- Coustou V, Besteiro S, Riviere L, Biran M, Biteau N, Franconi JM, Boshart M, Baltz T, Bringaud F. A mitochondrial NADH-dependent fumarate reductase involved in the production of succinate excreted by procyclic Trypanosoma brucei. J Biol Chem. 2005;280:16559–16570. doi: 10.1074/jbc.M500343200. [DOI] [PubMed] [Google Scholar]
- Cracknell JA, Wait AF, Lenz O, Friedrich B, Armstrong FA. A kinetic and thermodynamic understanding of O2 tolerance in [NiFe]-hydrogenases. Proc Natl Acad Sci USA. 2009;106:20681–20686. doi: 10.1073/pnas.0905959106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford MJ, Thomsen-Zieger N, Ray M, Schachtner J, Roos DS, Seeber F. Toxoplasma gondii scavenges host-derived lipoic acid despite its de novo synthesis in the apicoplast. EMBO J. 2006;25:3214–3222. doi: 10.1038/sj.emboj.7601189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE. Avant garde fatty acid synthesis by trypanosomes. Cell. 2006;126:641–643. doi: 10.1016/j.cell.2006.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W. Getting a better picture of microbial evolution en route to a network of genomes. Philos Trans Roy Soc Lond B Biol Sci. 2009;364:2187–2196. doi: 10.1098/rstb.2009.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan M, Wang CC. Role of alcohol dehydrogenase E (ADHE) in the energy metabolism of Giardia lamblia. Mol Biochem Parasitol. 2000;109:25–36. doi: 10.1016/s0166-6851(00)00233-4. [DOI] [PubMed] [Google Scholar]
- Dawson SC, Pace NR. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc Natl Acad Sci USA. 2002;99:8324–8329. doi: 10.1073/pnas.062169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf RM, Duarte I, van Alen TA, Kuiper JW, Schotanus K, Rosenberg J, Huynen MA, Hackstein JH. The hydrogenosomes of Psalteriomonas lanterna. BMC Evol Biol. 2009;9:287. doi: 10.1186/1471-2148-9-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonckheere JF. A century of research on the amoeboflagellate genus Naegleria. Acta Protozool. 2002;41:309–342. [Google Scholar]
- Denton H, Roberts CW, Alexander J, Thong KW, Coombs GH. Enzymes of energy metabolism in the bradyzoites and tachyzoites of Toxoplasma gondii. FEMS Microbiol Lett. 1996;137:103–108. doi: 10.1111/j.1574-6968.1996.tb08090.x. [DOI] [PubMed] [Google Scholar]
- Dingle AD, Fulton C. Development of the flagellar apparatus of Naegleria. J Cell Biol. 1966;31:43–54. doi: 10.1083/jcb.31.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do PM, Angerhofer A, Hrdy I, Bardonova L, Ingram LO, Shanmugam KT. Engineering Escherichia coli for fermentative dihydrogen production: potential role of NADH-ferredoxin oxidoreductase from the hydrogenosome of anaerobic protozoa. Appl Biochem Biotechnol. 2009;153:21–33. doi: 10.1007/s12010-008-8508-5. [DOI] [PubMed] [Google Scholar]
- Doeller JE, Grieshaber MK, Kraus DW. Chemolithoheterotrophy in a metazoan tissue: thiosulfate production matches ATP demand in ciliated mussel gills. J Exp Biol. 2001;204:3755–3764. doi: 10.1242/jeb.204.21.3755. [DOI] [PubMed] [Google Scholar]
- Dolezal P, Dagley MJ, Kono M, Wolynec P, Likic VA, Foo JH, Sedinova M, Tachezy J, Bachmann A, Bruchhaus I, Lithgow T. The essentials of protein import in the degenerate mitochondrion of Entamoeba histolytica. PLoS Pathog. 2010;6:e1000812. doi: 10.1371/journal.ppat.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubini A, Mus F, Seibert M, Grossman AR, Posewitz MC. Flexibility in anaerobic metabolism as revealed in a mutant of Chlamydomonas reinhardtii lacking hydrogenase activity. J Biol Chem. 2009;284:7201–7213. doi: 10.1074/jbc.M803917200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall SD, Yan W, Delgadillo-Correa MG, Lunceford A, Loo JA, Clarke CF, Johnson PJ. Non-mitochondrial complex I proteins in a hydrogenosomal oxidoreductase complex. Nature. 2004;431:1103–1107. doi: 10.1038/nature02990. [DOI] [PubMed] [Google Scholar]
- Edwards MR, Schofield PJ, O’Sullivan WJ, Costello M. Arginine metabolism during culture of Giardia intestinalis. Mol Biochem Parasitol. 1992;53:97–103. doi: 10.1016/0166-6851(92)90011-8. [DOI] [PubMed] [Google Scholar]
- Edwards SW, Lloyd D. Properties of mitochondria isolated from cyanide-sensitive and cyanide-stimulated cultures of Acanthamoeba castellanii. Biochem J. 1978;174:203–211. doi: 10.1042/bj1740203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Kouni MH. Potential chemotherapeutic targets in the purine metabolism of parasites. Pharmacol Ther. 2003;99:283–309. doi: 10.1016/s0163-7258(03)00071-8. [DOI] [PubMed] [Google Scholar]
- Ellis JE, Williams R, Cole D, Cammack R, Lloyd D. Electron transport components of the parasitic protozoon Giardia lamblia. FEBS Lett. 1993;325:196–200. doi: 10.1016/0014-5793(93)81072-8. [DOI] [PubMed] [Google Scholar]
- Elser JJ, Bracken ME, Cleland EE, Gruner DS, Harpole WS, Hillebrand H, Ngai JT, Seabloom EW, Shurin JB, Smith JE. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett. 2007;10:1135–1142. doi: 10.1111/j.1461-0248.2007.01113.x. [DOI] [PubMed] [Google Scholar]
- Embley TM. Multiple secondary origins of the anaerobic lifestyle in eukaryotes. Philos Trans Roy Soc Lond B Biol Sci. 2006;361:1055–1067. doi: 10.1098/rstb.2006.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Embley TM, Horner DA, Hirt RP. Anaerobic eukaryote evolution: hydrogenosomes as biochemically modified mitochondria? Trends Ecol Evol. 1998;12:437–441. doi: 10.1016/s0169-5347(97)01208-1. [DOI] [PubMed] [Google Scholar]
- Espinosa A, Yan L, Zhang Z, Foster L, Clark D, Li E, Stanley SL., Jr The bifunctional Entamoeba histolytica alcohol dehydrogenase 2 (EhADH2) protein is necessary for amebic growth and survival and requires an intact C-terminal domain for both alcohol dahydrogenase and acetaldehyde dehydrogenase activity. J Biol Chem. 2001;276:20136–20143. doi: 10.1074/jbc.M101349200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C, Martin W, Dagan T. The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biol Lett. 2007;3:180–184. doi: 10.1098/rsbl.2006.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Dacks JB. First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr Opin Cell Biol. 2009;21:4–13. doi: 10.1016/j.ceb.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Span ASW, Harman JMP. Nitrate respiration in primitive eukaryotes. Nature. 1983;303:333–336. [Google Scholar]
- Fiore-Donno AM, Nikolaev SI, Nelson M, Pawlowski J, Cavalier-Smith T, Baldauf SL. Deep phylogeny and evolution of slime moulds (Mycetozoa) Protist. 2010;161:55–70. doi: 10.1016/j.protis.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Fischer WW, Pearson A. Hypotheses for the origin and early evolution of triterpenoid cyclases. Geobiology. 2007;5:19–34. doi: 10.1111/j.1472-4669.2007.00096.x. [DOI] [PubMed] [Google Scholar]
- Fleige T, Fischer K, Ferguson DJ, Gross U, Bohne W. Carbohydrate metabolism in the Toxoplasma gondii apicoplast: localization of three glycolytic isoenzymes, the single pyruvate dehydrogenase complex, and a plastid phosphate translocator. Eukaryot Cell. 2007;6:984–996. doi: 10.1128/EC.00061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin L, Tsokoglou A, Happe T. A novel type of iron hydrogenase in the green alga Scenedesmus obliquus is linked to the photosynthetic electron transport chain. J Biol Chem. 2001;276:6125–6132. doi: 10.1074/jbc.M008470200. [DOI] [PubMed] [Google Scholar]
- Foster PG, Cox CJ, Embley TM. The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods. Philos Trans Roy Soc Lond B Biol Sci. 2009;364:2197–2207. doi: 10.1098/rstb.2009.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foth BJ, Stimmler LM, Handman E, Crabb BS, Hodder AN, McFadden GI. The malaria parasite Plasmodium falciparum has only one pyruvate dehydrogenase complex, which is located in the apicoplast. Mol Microbiol. 2005;55:39–53. doi: 10.1111/j.1365-2958.2004.04407.x. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, Shu S, Neupane R, Cipriano M, Mancuso J, Tu H, Salamov A, Lindquist E, Shapiro H, Lucas S, Grigoriev IV, Cande WZ, Fulton C, Rokhsar DS, Dawson SC. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Fulton C. Cell differentiation in Naegleria gruberi. Annu Rev Microbiol. 1977;31:597–629. doi: 10.1146/annurev.mi.31.100177.003121. [DOI] [PubMed] [Google Scholar]
- Fulton C. Naegleria: a research partner for cell and developmental biology. J Eukaryot Microbiol. 1993;40:520–532. [Google Scholar]
- Fulton C, Webster C, Wu JS. Chemically defined media for cultivation of Naegleria gruberi. Proc Natl Acad Sci USA. 1984;81:2406–2410. doi: 10.1073/pnas.81.8.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T. Peroxisome diversity and evolution. Philos Trans Roy Soc Lond B Biol Sci. 2010;365:765–773. doi: 10.1098/rstb.2009.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffron H, Rubin J. Fermentative and photochemical production of hydrogen in algae. J Gen Physiol. 1942;26:219–240. doi: 10.1085/jgp.26.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulin E, Bottin A, Dumas B. Sterol biosynthesis in oomycete pathogens. Plant Signal Behav. 2010;5:258–260. doi: 10.4161/psb.5.3.10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller RP, Gibbs M. Fermentative metabolism of Chlamydomonas reinhardtii: I. Analysis of fermentative products from starch in dark and light. Plant Physiol. 1984;75:212–218. doi: 10.1104/pp.75.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibellini F, Hunter WN, Smith TK. The ethanolamine branch of the Kennedy pathway is essential in the bloodstream form of Trypanosoma brucei. Mol Microbiol. 2009;73:826–843. doi: 10.1111/j.1365-2958.2009.06764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill EE, Diaz-Trivino S, Barbera MJ, Silberman JD, Stechmann A, Gaston D, Tamas I, Roger AJ. Novel mitochondrion-related organelles in the anaerobic amoeba Mastigamoeba balamuthi. Mol Microbiol. 2007;66:1306–1320. doi: 10.1111/j.1365-2958.2007.05979.x. [DOI] [PubMed] [Google Scholar]
- Ginger ML. Niche metabolism in parasitic protozoa. Philos Trans Roy Soc Lond B Biol Sci. 2006;361:101–118. doi: 10.1098/rstb.2005.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger ML, McFadden GI, Michels PA. Rewiring and regulation of cross-compartmentalized metabolism in protists. Philos Trans Roy Soc Lond B Biol Sci. 2010;365:831–845. doi: 10.1098/rstb.2009.0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell HC, Laurens LM, Shields RJ, Lovitt RW, Flynn KJ. Placing microalgae on the biofuels priority list: a review of the technological challenges. J R Soc Interface. 2010;7:703–726. doi: 10.1098/rsif.2009.0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther S, Matuschewski K, Muller S. Knockout studies reveal an important role of Plasmodium lipoic acid protein ligase A1 for asexual blood stage parasite survival. PLoS One. 2009;4:e5510. doi: 10.1371/journal.pone.0005510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstein JH, Tjaden J, Huynen M. Mitochondria, hydrogenosomes and mitosomes: products of evolutionary tinkering! Curr Genet. 2006;50:225–245. doi: 10.1007/s00294-006-0088-8. [DOI] [PubMed] [Google Scholar]
- Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AGB, Roger AJ. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci USA. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. Introduction to Chlamydomonas and its Laboratory Use. 2. Vol. 1. Academic Press; Oxford: 2009. The Chlamydomonas Sourcebook. [Google Scholar]
- Hau HH, Gralnick JA. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol. 2007;61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- Helfert S, Estevez AM, Bakker B, Michels P, Clayton C. Roles of triosephosphate isomerase and aerobic metabolism in Trypanosoma brucei. Biochem J. 2001;357:117–125. doi: 10.1042/0264-6021:3570117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemond JJ, Bakker BM, Tielens AG. Energy metabolism and its compartmentation in Trypanosoma brucei. Adv Microb Physiol. 2005;50:199–226. doi: 10.1016/S0065-2911(05)50005-5. [DOI] [PubMed] [Google Scholar]
- Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans Roy Soc Lond B Biol Sci. 2010;365:713–727. doi: 10.1098/rstb.2009.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister M, Piotrowski M, Nowitzki U, Martin W. Mitochondrial trans-2-enoyl-CoA reductase of wax ester fermentation from Euglena gracilis defines a new family of enzymes involved in lipid synthesis. J Biol Chem. 2005;280:4329–4338. doi: 10.1074/jbc.M411010200. [DOI] [PubMed] [Google Scholar]
- Hoffmeister M, van der Klei A, Rotte C, van Grinsven KW, van Hellemond JJ, Henze K, Tielens AG, Martin W. Euglena gracilis rhodoquinone:ubiquinone ratio and mitochondrial proteome differ under aerobic and anaerobic conditions. J Biol Chem. 2004;279:22422–22429. doi: 10.1074/jbc.M400913200. [DOI] [PubMed] [Google Scholar]
- Horner DS, Foster PG, Embley TM. Iron hydrogenases and the evolution of anaerobic eukaryotes. Mol Biol Evol. 2000;17:1695–1709. doi: 10.1093/oxfordjournals.molbev.a026268. [DOI] [PubMed] [Google Scholar]
- Horner DS, Hirt RP, Embley TM. A single eubacterial origin of eukaryotic pyruvate: ferredoxin oxidoreductase genes: implications for the evolution of anaerobic eukaryotes. Mol Biol Evol. 1999;16:1280–1291. doi: 10.1093/oxfordjournals.molbev.a026218. [DOI] [PubMed] [Google Scholar]
- Horner DS, Heil B, Happe T, Embley TM. Iron hydrogenases – ancient enzymes in modern eukaryotes. Trends Biochem Sci. 2002;27:148–153. doi: 10.1016/s0968-0004(01)02053-9. [DOI] [PubMed] [Google Scholar]
- Hrdy I, Hirt RP, Dolezal P, Bardonova L, Foster PG, Tachezy J, Embley TM. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- Hug LA, Stechmann A, Roger AJ. Phylogenetic distributions and histories of proteins involved in anaerobic pyruvate metabolism in eukaryotes. Mol Biol Evol. 2010;27:311–324. doi: 10.1093/molbev/msp237. [DOI] [PubMed] [Google Scholar]
- Inui H, Miyatake K, Nakano Y, Kitaoka S. Wax ester fermentation in Euglena gracilis. FEBS Lett. 1982;150:89–93. doi: 10.1002/1873-3468.13276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui H, Ono K, Miyatake K, Nakano Y, Kitaoka S. Purification and characterization of pyruvate:NADP+ oxidoreductase in Euglena gracilis. J Biol Chem. 1987;262:9130–9135. [PubMed] [Google Scholar]
- Jacobs J, Pudollek S, Hemschemeier A, Happe T. A novel, anaerobically induced ferredoxin in Chlamydomonas reinhardtii. FEBS Lett. 2009;583:325–329. doi: 10.1016/j.febslet.2008.12.018. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schussler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lucking R, Budel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Jarmuszkiewicz W, Sluse-Goffart CM, Hryniewiecka L, Michejda J, Sluse FE. Electron partitioning between the two branching quinol-oxidizing pathways in Acanthamoeba castellanii mitochondria during steady-state state 3 respiration. J Biol Chem. 1998;273:10174–10180. doi: 10.1074/jbc.273.17.10174. [DOI] [PubMed] [Google Scholar]
- John P, Whatley FR. Paracoccus denitrificans and the evolutionary origin of the mitochondrion. Nature. 1975;254:495–498. doi: 10.1038/254495a0. [DOI] [PubMed] [Google Scholar]
- Kaneshiro ES. Lipids of Paramecium. J Lipid Res. 1987;28:1241–1258. [PubMed] [Google Scholar]
- Karniely S, Pines O. Single translation--dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 2005;6:420–425. doi: 10.1038/sj.embor.7400394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivares CP. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. Functional and ecological impacts of horizontal gene transfer in eukaryotes. Curr Opin Genet Dev. 2009;19:613–619. doi: 10.1016/j.gde.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Corradi N, Morrison HG, Haag KL, Ebert D, Weiss LM, Akiyoshi DE, Tzipori S. The reduced genome of the parasitic microsporidian Enterocytozoon bieneusi lacks genes for core carbon metabolism. Genome Biol Evol. 2010;2:304–309. doi: 10.1093/gbe/evq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Kim E, Simpson AG, Graham LE. Evolutionary relationships of apusomonads inferred from taxon-rich analyses of 6 nuclear encoded genes. Mol Biol Evol. 2006;23:2455–2466. doi: 10.1093/molbev/msl120. [DOI] [PubMed] [Google Scholar]
- Kim SW, Fushinobu S, Zhou S, Wakagi T, Shoun H. Eukaryotic nirK genes encoding copper-containing nitrite reductase: originating from the protomitochondrion? Appl Environ Microbiol. 2009;75:2652–2658. doi: 10.1128/AEM.02536-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Matsuo Y, Takimoto A, Suzuki S, Maruo F, Shoun H. Denitrification, a novel type of respiratory metabolism in fungal mitochondrion. J Biol Chem. 1996;271:16263–16267. doi: 10.1074/jbc.271.27.16263. [DOI] [PubMed] [Google Scholar]
- Kolisko M, Silberman JD, Cepicka I, Yubuki N, Takishita K, Yabuki A, Leander BS, Inouye I, Inagaki Y, Roger AJ, Simpson AG. A wide diversity of previously undetected free-living relatives of diplomonads isolated from marine/saline habitats. Environ Microbiol. 2010 doi: 10.1111/j.1462-2920.2010.02239.x. in press (Epub May 13) [DOI] [PubMed] [Google Scholar]
- Koonin EV. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010;11:209. doi: 10.1186/gb-2010-11-5-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzberg K. Starch fermentation via a formate producing pathway in Chlamydomonas reinhardii, Chlorogonium elongatum and Chlorella fusca. Physiol Plantarum. 1984;61:87–94. [Google Scholar]
- Kreuzberg K, Klock G, Grobheiser D. Subcellular distribution of pyruvate-degrading enzymes in Chlamydomonas reinhardtii studied by an improved protoplast fractionation procedure. Physiol Plantarum. 1987;69:481–488. [Google Scholar]
- Kroth PG, Chiovitti A, Gruber A, Martin-Jezequel V, Mock T, Parker MS, Stanley MS, Kaplan A, Caron L, Weber T, Maheswari U, Armbrust EV, Bowler C. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS One. 2008;3:e1426. doi: 10.1371/journal.pone.0001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang BF, Burger G, O’Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- Lara E, Chatzinotas A, Simpson AGB. Andalucia (n. gen.) – the deepest branch within jakobids (Jakobida; Excavata), based on morphological and molecular study of a new flagellate from soil. J Eukaryot Microbiol. 2006;53:112–120. doi: 10.1111/j.1550-7408.2005.00081.x. [DOI] [PubMed] [Google Scholar]
- Leander BS. Did trypanosomatid parasites have photosynthetic ancestors? Trends Microbiol. 2004;12:251–258. doi: 10.1016/j.tim.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Lee SH, Stephens JL, Englund PT. A fatty-acid synthesis mechanism specialized for parasitism. Nat Rev Microbiol. 2007;5:287–297. doi: 10.1038/nrmicro1617. [DOI] [PubMed] [Google Scholar]
- Lee SH, Stephens JL, Paul KS, Englund PT. Fatty acid synthesis by elongases in trypanosomes. Cell. 2006;126:691–699. doi: 10.1016/j.cell.2006.06.045. [DOI] [PubMed] [Google Scholar]
- Leys D, Tsapin AS, Nealson KH, Meyer TE, Cusanovich MA, Van Beeumen JJ. Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR-1. Nat Struct Biol. 1999;6:1113–1117. doi: 10.1038/70051. [DOI] [PubMed] [Google Scholar]
- Liapounova NA, Hampl V, Gordon PM, Sensen CW, Gedamu L, Dacks JB. Reconstructing the mosaic glycolytic pathway of the anaerobic eukaryote Monocercomonoides. Eukaryot Cell. 2006;5:2138–2146. doi: 10.1128/EC.00258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaud MF, Lichtle C, Apt K, Martin W, Cerff R. Compartment-specific isoforms of TPI and GAPDH are imported into diatom mitochondria as a fusion protein: evidence in favor of a mitochondrial origin of the eukaryotic glycolytic pathway. Mol Biol Evol. 2000;17:213–223. doi: 10.1093/oxfordjournals.molbev.a026301. [DOI] [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Lim L, McFadden GI. The evolution, metabolism and functions of the apicoplast. Philos Trans Roy Soc Lond B Biol Sci. 2010;365:749–763. doi: 10.1098/rstb.2009.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd D, Ralphs JR, Harris JC. Giardia intestinalis, a eukaryote without hydrogenosomes, produces hydrogen. Microbiology. 2002;148:727–733. doi: 10.1099/00221287-148-3-727. [DOI] [PubMed] [Google Scholar]
- Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, Nozaki T, Suh B, Pop M, Duchene M, Ackers J, Tannich E, Leippe M, Hofer M, Bruchhaus I, Willhoeft U, Bhattacharya A, Chillingworth T, Churcher C, Hance Z, Harris B, Harris D, Jagels K, Moule S, Mungall K, Ormond D, Squares R, Whitehead S, Quail MA, Rabbinowitsch E, Norbertczak H, Price C, Wang Z, Guillen N, Gilchrist C, Stroup SE, Bhattacharya S, Lohia A, Foster PG, Sicheritz-Ponten T, Weber C, Singh U, Mukherjee C, El-Sayed NM, Petri WA, Jr, Clark CG, Embley TM, Barrell B, Fraser CM, Hall N. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Moreira D. Metabolic symbiosis at the origin of eukaryotes. Trends Biochem Sci. 1999;24:88–93. doi: 10.1016/s0968-0004(98)01342-5. [DOI] [PubMed] [Google Scholar]
- Ludwig M, Schulz-Friedrich R, Appel J. Occurrence of hydrogenases in cyanobacteria and anoxygenic photosynthetic bacteria: implications for the phylogenetic origin of cyanobacterial and algal hydrogenases. J Mol Evol. 2006;63:758–768. doi: 10.1007/s00239-006-0001-6. [DOI] [PubMed] [Google Scholar]
- Martin W. Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos Trans Roy Soc Lond B Biol Sci. 2010;365:847–855. doi: 10.1098/rstb.2009.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- Martin W, Hoffmeister M, Rotte C, Henze K. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol Chem. 2001;382:1521–1539. doi: 10.1515/BC.2001.187. [DOI] [PubMed] [Google Scholar]
- Martin W, Rotte C, Hoffmeister M, Theissen U, Gelius-Dietrich G, Ahr S, Henze K. Early cell evolution, eukaryotes, anoxia, sulfide, oxygen, fungi first (?), and a tree of genomes revisited. IUBMB Life. 2003;55:193–204. doi: 10.1080/1521654031000141231. [DOI] [PubMed] [Google Scholar]
- Mazumdar J, Wilson EH, Masek K, Hunter CA, Striepen B. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc Natl Acad Sci USA. 2006;103:13192–13197. doi: 10.1073/pnas.0603391103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentel M, Martin W. Energy metabolism among eukaryotic anaerobes in light of Proterozoic ocean chemistry. Philos Trans Roy Soc Lond B Biol Sci. 2008;363:2717–2729. doi: 10.1098/rstb.2008.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, Dutcher S, Fernandez E, Fukuzawa H, Gonzalez-Ballester D, Gonzalez-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral JP, Riano-Pachon DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen CJ, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, Martinez D, Ngau WC, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K, Grigoriev IV, Rokhsar DS, Grossman AR. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi T, Kita K. Diversity in mitochondrial metabolic pathways in parasitic protists Plasmodium and Cryptosporidium. Parasitol Int. 2010;59:305–312. doi: 10.1016/j.parint.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Moreira D, Lopez-Garcia P. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 2002;10:31–38. doi: 10.1016/s0966-842x(01)02257-0. [DOI] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, Davids BJ, Dawson SC, Elmendorf HG, Hehl AB, Holder ME, Huse SM, Kim UU, Lasek-Nesselquist E, Manning G, Nigam A, Nixon JE, Palm D, Passamaneck NE, Prabhu A, Reich CI, Reiner DS, Samuelson J, Svard SG, Sogin ML. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Moseley JL, Allinger T, Herzog S, Hoerth P, Wehinger E, Merchant S, Hippler M. Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 2002;21:6709–6720. doi: 10.1093/emboj/cdf666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR. Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem. 2007;282:25475–25486. doi: 10.1074/jbc.M701415200. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Inui H, Yamaji R, Yamamoto T, Takenaka S, Ueda M, Nakano Y, Miyatake K. The origin of pyruvate: NADP+ oxidoreductase in mitochondria of Euglena gracilis. FEBS Lett. 2000;479:155–156. doi: 10.1016/s0014-5793(00)01882-2. [DOI] [PubMed] [Google Scholar]
- Nixon JE, Field J, McArthur AG, Sogin ML, Yarlett N, Loftus BJ, Samuelson J. Iron-dependent hydrogenases of Entamoeba histolytica and Giardia lamblia: activity of the recombinant entamoebic enzyme and evidence for lateral gene transfer. Biol Bull. 2003;204:1–9. doi: 10.2307/1543490. [DOI] [PubMed] [Google Scholar]
- Nixon JE, Wang A, Field J, Morrison HG, McArthur AG, Sogin ML, Loftus BJ, Samuelson J. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot Cell. 2002;1:181–190. doi: 10.1128/EC.1.2.181-190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi K, Yoshida K. Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion. 2008;8:87–99. doi: 10.1016/j.mito.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Olszewski KL, Mather MW, Morrisey JM, Garcia BA, Vaidya AB, Rabinowitz JD, Llinas M. Branched tricarboxylic acid metabolism in Plasmodium falciparum. Nature. 2010;466:774–778. doi: 10.1038/nature09301. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Paget TA, Lloyd D. Trichomonas vaginalis requires traces of oxygen and high concentrations of carbon dioxide for optimal growth. Mol Biochem Parasitol. 1990;41:65–72. doi: 10.1016/0166-6851(90)90097-6. [DOI] [PubMed] [Google Scholar]
- Paget TA, Raynor MH, Shipp DW, Lloyd D. Giardia lamblia produces alanine anaerobically but not in the presence of oxygen. Mol Biochem Parasitol. 1990;42:63–67. doi: 10.1016/0166-6851(90)90113-z. [DOI] [PubMed] [Google Scholar]
- Parker MS, Mock T, Armbrust EV. Genomic insights into marine microalgae. Annu Rev Genet. 2008;42:619–645. doi: 10.1146/annurev.genet.42.110807.091417. [DOI] [PubMed] [Google Scholar]
- Pawlowski J, Burki F. Untangling the phylogeny of amoeboid protists. J Eukaryot Microbiol. 2009;56:16–25. doi: 10.1111/j.1550-7408.2008.00379.x. [DOI] [PubMed] [Google Scholar]
- Payne SH, Loomis WF. Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryot Cell. 2006;5:272–276. doi: 10.1128/EC.5.2.272-276.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G, Price NM. Copper-containing plastocyanin used for electron transport by an oceanic diatom. Nature. 2006;441:341–344. doi: 10.1038/nature04630. [DOI] [PubMed] [Google Scholar]
- Pei Y, Tarun AS, Vaughan AM, Herman RW, Soliman JM, Erickson-Wayman A, Kappe SH. Plasmodium pyruvate dehydrogenase activity is only essential for the parasite’s progression from liver infection to blood infection. Mol Microbiol. 2010;75:957–971. doi: 10.1111/j.1365-2958.2009.07034.x. [DOI] [PubMed] [Google Scholar]
- Perez-Brocal V, Clark CG. Analysis of two genomes from the mitochondrion-like organelle of the intestinal parasite Blastocystis: complete sequences, gene content, and genome organization. Mol Biol Evol. 2008;25:2475–2482. doi: 10.1093/molbev/msn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina-Ochoa E, Hogslund S, Geslin E, Cedhagen T, Revsbech NP, Nielsen LP, Schweizer M, Jorissen F, Rysgaard S, Risgaard-Petersen N. Widespread occurrence of nitrate storage and denitrification among Foraminifera and Gromiida. Proc Natl Acad Sci USA. 2010;107:1148–1153. doi: 10.1073/pnas.0908440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda E, Encalada R, Rodriguez-Zavala JS, Olivos-Garcia A, Moreno-Sanchez R, Saavedra E. Pyruvate:ferredoxin oxidoreductase and bifunctional aldehyde-alcohol dehydrogenase are essential for energy metabolism under oxidative stress in Entamoeba histolytica. FEBS J. 2010;277:3382–3395. doi: 10.1111/j.1742-4658.2010.07743.x. [DOI] [PubMed] [Google Scholar]
- Pino P, Foth BJ, Kwok LY, Sheiner L, Schepers R, Soldati T, Soldati-Favre D. Dual targeting of antioxidant and metabolic enzymes to the mitochondrion and the apicoplast of Toxoplasma gondii. PLoS Pathog. 2007;3:e115. doi: 10.1371/journal.ppat.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posewitz MC, Dubini A, Meuser JW, Seibert M, Ghirardi ML. Hydrogenases, Hydrogen Production and Anoxia. In: Stern DB, editor. The Chlamydomonas Source Book. 2. Vol. 2. Organellar and Metabolic Processes Academic Press; Oxford: 2009. pp. 217–255. [Google Scholar]
- Posewitz MC, Smolinski SL, Kanakagiri S, Melis A, Seibert M, Ghirardi ML. Hydrogen photoproduction is attenuated by disruption of an isoamylase gene in Chlamydomonas reinhardtii. Plant Cell. 2004;16:2151–2163. doi: 10.1105/tpc.104.021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raederstorff D, Rohmer M. Sterol biosynthesis via cycloartenol and other biochemical features related to photosynthetic phyla in the amoeba Naegleria lovaniensis and Naegleria gruberi. Eur J Biochem. 1987;164:427–434. doi: 10.1111/j.1432-1033.1987.tb11075.x. [DOI] [PubMed] [Google Scholar]
- Ralph SA, van Dooren GG, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ, Tonkin CJ, Roos DS, McFadden GI. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- Reeves RE, Warren LG, Susskind B, Lo HS. An energy-conserving pyruvate-to-acetate pathway in Entamoeba histolytica. Pyruvate synthase and a new acetate thiokinase. J Biol Chem. 1977;252:726–731. [PubMed] [Google Scholar]
- Regoes A, Zourmpanou D, Leon-Avila G, van der Giezen M, Tovar J, Hehl AB. Protein import, replication, and inheritance of a vestigial mitochondrion. J Biol Chem. 2005;280:30557–30563. doi: 10.1074/jbc.M500787200. [DOI] [PubMed] [Google Scholar]
- Ricard G, McEwan NR, Dutilh BE, Jouany JP, Macheboeuf D, Mitsumori M, McIntosh FM, Michalowski T, Nagamine T, Nelson N, Newbold CJ, Nsabimana E, Takenaka A, Thomas NA, Ushida K, Hackstein JH, Huynen MA. Horizontal gene transfer from bacteria to rumen ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics. 2006;7:22. doi: 10.1186/1471-2164-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Richardson DJ. Bacterial respiration: a flexible process for a changing environment. Microbiology. 2000;146:551–571. doi: 10.1099/00221287-146-3-551. [DOI] [PubMed] [Google Scholar]
- Risgaard-Petersen N, Langezaal AM, Ingvardsen S, Schmid MC, Jetten MS, Op den Camp HJ, Derksen JW, Pina-Ochoa E, Eriksson SP, Nielsen LP, Revsbech NP, Cedhagen T, van der Zwaan GJ. Evidence for complete denitrification in a benthic foraminifer. Nature. 2006;443:93–96. doi: 10.1038/nature05070. [DOI] [PubMed] [Google Scholar]
- Riviere L, van Weelden SW, Glass P, Vegh P, Coustou V, Biran M, van Hellemond JJ, Bringaud F, Tielens AG, Boshart M. Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei. Gene identification and role in carbohydrate metabolism. J Biol Chem. 2004;279:45337–45346. doi: 10.1074/jbc.M407513200. [DOI] [PubMed] [Google Scholar]
- Rodriguez MA, Hidalgo ME, Sanchez T, Orozco E. Cloning and characterization of the Entamoeba histolytica pyruvate: ferredoxin oxidoreductase gene. Mol Biochem Parasitol. 1996;78:273–277. doi: 10.1016/s0166-6851(96)02613-8. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Simpson AGB. Evolution: revisiting the root of the eukaryote tree. Curr Biol. 2009;19:R165–R167. doi: 10.1016/j.cub.2008.12.032. [DOI] [PubMed] [Google Scholar]
- Rotte C, Stejskal F, Zhu G, Keithly JS, Martin W. Pyruvate: NADP+ oxidoreductase from the mitochondrion of Euglena gracilis and from the apicomplexan Cryptosporidium parvum: a biochemical relic linking pyruvate metabolism in mitochondriate and amitochondriate protists. Mol Biol Evol. 2001;18:710–720. doi: 10.1093/oxfordjournals.molbev.a003853. [DOI] [PubMed] [Google Scholar]
- Sanchez LB. Aldehyde dehydrogenase (CoA-acetylating) and the mechanism of ethanol formation in the amitochondriate protist, Giardia lamblia. Arch Biochem Biophys. 1998;354:57–64. doi: 10.1006/abbi.1998.0664. [DOI] [PubMed] [Google Scholar]
- Sanchez LB, Galperin MY, Müller M. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (nucleoside diphosphate-forming) J Biol Chem. 2000;275:5794–5803. doi: 10.1074/jbc.275.8.5794. [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber F, Limenitakis J, Soldati-Favre D. Apicomplexan mitochondrial metabolism: a story of gains, losses and retentions. Trends Parasitol. 2008;24:468–478. doi: 10.1016/j.pt.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Shepard EM, McGlynn SE, Bueling AL, Grady-Smith CS, George SJ, Winslow MA, Cramer SP, Peters JW, Broderick JB. Synthesis of the 2Fe subcluster of the [FeFe]-hydrogenase H cluster on the HydF scaffold. Proc Natl Acad Sci USA. 2010;107:10448–10453. doi: 10.1073/pnas.1001937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman IW. Biochemistry of Plasmodium (malarial parasites) Microbiol Rev. 1979;43:453–495. doi: 10.1128/mr.43.4.453-495.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett AM, Johnson PJ. Mitochondrion-related organelles in eukaryotic protists. Annu Rev Microbiol. 2010 doi: 10.1146/annurev.micro.62.081307.162826. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu CJ, Ulrich LE, Zhulin IB. The NIT domain: a predicted nitrate-responsive module in bacterial sensory receptors. Trends Biochem Sci. 2003;28:121–124. doi: 10.1016/S0968-0004(03)00032-X. [DOI] [PubMed] [Google Scholar]
- Simpson AGB, Patterson DJ. On core jakobids and excavate taxa: the ultrastructure of Jakoba incarcerata. J Eukaryot Microbiol. 2001;48:480–492. doi: 10.1111/j.1550-7408.2001.tb00183.x. [DOI] [PubMed] [Google Scholar]
- Simpson AGB, Roger AJ. The real ‘kingdoms’ of eukaryotes. Curr Biol. 2004;14:R693–R696. doi: 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Simpson AGB, Stevens JR, Lukeš J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Slamovits CH, Keeling PJ. Pyruvate-phosphate dikinase of oxymonads and Parabasalia and the evolution of pyrophosphate-dependent glycolysis in anaerobic eukaryotes. Eukaryot Cell. 2006;5:148–154. doi: 10.1128/EC.5.1.148-154.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slapeta J, Moreira D, Lopez-Garcia P. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc Biol Sci. 2005;272:2073–2081. doi: 10.1098/rspb.2005.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stechmann A, Baumgartner M, Silberman JD, Roger AJ. The glycolytic pathway of Trimastix pyriformis is an evolutionary mosaic. BMC Evol Biol. 2006;6:101. doi: 10.1186/1471-2148-6-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr Biol. 2003;13:R665–R666. doi: 10.1016/s0960-9822(03)00602-x. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Hamblin K, Perez-Brocal V, Gaston D, Richmond GS, van der Giezen M, Clark CG, Roger AJ. Organelles in Blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr Biol. 2008;18:580–585. doi: 10.1016/j.cub.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbüchel A, Müller M. Glycerol, a metabolic end product of Trichomonas vaginalis and Tritrichomonas foetus. Mol Biochem Parasitol. 1986;20:45–55. doi: 10.1016/0166-6851(86)90141-6. [DOI] [PubMed] [Google Scholar]
- Steunou AS, Bhaya D, Bateson MM, Melendrez MC, Ward DM, Brecht E, Peters JW, Kuhl M, Grossman AR. In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc Natl Acad Sci USA. 2006;103:2398–2403. doi: 10.1073/pnas.0507513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Behnke A, Christen R, Amaral-Zettler L, Rodriguez-Mora MJ, Chistoserdov A, Orsi W, Edgcomb VP. Massively parallel tag sequencing reveals the complexity of anaerobic marine protistan communities. BMC Biol. 2009;7:72. doi: 10.1186/1741-7007-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MJ, Guther ML, Turnock DC, Prescott AR, Martin KL, Alphey MS, Ferguson MA. The synthesis of UDP-N-acetylglucosamine is essential for bloodstream form Trypanosoma brucei in vitro and in vivo and UDP-N-acetylglucosamine starvation reveals a hierarchy in parasite protein glycosylation. J Biol Chem. 2008;283:16147–16161. doi: 10.1074/jbc.M709581200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzepek RF, Harrison PJ. Photosynthetic architecture differs in coastal and oceanic diatoms. Nature. 2004;431:689–692. doi: 10.1038/nature02954. [DOI] [PubMed] [Google Scholar]
- Summons RE, Bradley AS, Jahnke LL, Waldbauer JR. Steroids, triterpenoids and molecular oxygen. Philos Trans Roy Soc Lond B Biol Sci. 2006;361:951–968. doi: 10.1098/rstb.2006.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki K, Shoun H, Yamaguchi M, Takeo K, Nakamura A, Hoshino T, Takaya N. Fungal ammonia fermentation, a novel metabolic mechanism that couples the dissimilatory and assimilatory pathways of both nitrate and ethanol. Role of acetyl CoA synthetase in anaerobic ATP synthesis. J Biol Chem. 2004;279:12414–12420. doi: 10.1074/jbc.M313761200. [DOI] [PubMed] [Google Scholar]
- Takaya N, Suzuki S, Kuwazaki S, Shoun H, Maruo F, Yamaguchi M, Takeo K. Cytochrome p450nor, a novel class of mitochondrial cytochrome P450 involved in nitrate respiration in the fungus Fusarium oxysporum. Arch Biochem Biophys. 1999;372:340–346. doi: 10.1006/abbi.1999.1499. [DOI] [PubMed] [Google Scholar]
- Takaya N, Kuwazaki S, Adachi Y, Suzuki S, Kikuchi T, Nakamura H, Shiro Y, Shoun H. Hybrid respiration in the denitrifying mitochondria of Fusarium oxysporum. J Biochem. 2003;133:461–465. doi: 10.1093/jb/mvg060. [DOI] [PubMed] [Google Scholar]
- Taylor P, Pealing SL, Reid GA, Chapman SK, Walkinshaw MD. Structural and mechanistic mapping of a unique fumarate reductase. Nat Struct Biol. 1999;6:1108–1112. doi: 10.1038/70045. [DOI] [PubMed] [Google Scholar]
- ter Kuile BH. Metabolic adaptation of Trichomonas vaginalis to growth rate and glucose availability. Microbiology. 1996;142:3337–3345. doi: 10.1099/13500872-142-12-3337. [DOI] [PubMed] [Google Scholar]
- Terauchi AM, Peers G, Kobayashi MC, Niyogi KK, Merchant SS. Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth Res. 2010;105:39–49. doi: 10.1007/s11120-010-9562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen U, Hoffmeister M, Grieshaber M, Martin W. Single eubacterial origin of eukaryotic sulfide:quinone oxidoreductase, a mitochondrial enzyme conserved from the early evolution of eukaryotes during anoxic and sulfidic times. Mol Biol Evol. 2003;20:1564–1574. doi: 10.1093/molbev/msg174. [DOI] [PubMed] [Google Scholar]
- Tielens AG, Rotte C, van Hellemond JJ, Martin W. Mitochondria as we don’t know them. Trends Biochem Sci. 2002;27:564–572. doi: 10.1016/s0968-0004(02)02193-x. [DOI] [PubMed] [Google Scholar]
- Tielens AG, van Grinsven KW, Henze K, van Hellemond JJ, Martin W. Acetate formation in the energy metabolism of parasitic helminths and protists. Int J Parasitol. 2010;40:387–397. doi: 10.1016/j.ijpara.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Tsaousis AD, Kunji ERS, Goldberg AV, Lucocq JM, Hirt RP, Embley TM. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature. 2008;453:553–556. doi: 10.1038/nature06903. [DOI] [PubMed] [Google Scholar]
- Tucci S, Vacula R, Krajcovic J, Proksch P, Martin W. Variability of wax ester fermentation in natural and bleached Euglena gracilis Strains in response to oxygen and the elongase inhibitor flufenacet. J Eukaryot Microbiol. 2010;57:63–69. doi: 10.1111/j.1550-7408.2009.00452.x. [DOI] [PubMed] [Google Scholar]
- Turmel M, Gagnon MC, O’Kelly CJ, Otis C, Lemieux C. The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol Biol Evol. 2009;26:631–648. doi: 10.1093/molbev/msn285. [DOI] [PubMed] [Google Scholar]
- van der Giezen M. Hydrogenosomes and mitosomes: conservation and evolution of functions. J Eukaryot Microbiol. 2009;56:221–231. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Sjollema KA, Artz RR, Alkema W, Prins RA. Hydrogenosomes in the anaerobic fungus Neocallimastix frontalis have a double membrane but lack an associated organelle genome. FEBS Lett. 1997;408:147–150. doi: 10.1016/s0014-5793(97)00409-2. [DOI] [PubMed] [Google Scholar]
- van Grinsven KW, Rosnowsky S, van Weelden SW, Putz S, van der Giezen M, Martin W, van Hellemond JJ, Tielens AG, Henze K. Acetate:succinate CoA-transferase in the hydrogenosomes of Trichomonas vaginalis: identification and characterization. J Biol Chem. 2008;283:1411–1418. doi: 10.1074/jbc.M702528200. [DOI] [PubMed] [Google Scholar]
- van Hellemond JJ, Tielens AG. Expression and functional properties of fumarate reductase. Biochem J. 1994;304:321–331. doi: 10.1042/bj3040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hellemond JJ, Opperdoes FR, Tielens AG. Trypanosomatidae produce acetate via a mitochondrial acetate:succinate CoA transferase. Proc Natl Acad Sci USA. 1998;95:3036–3041. doi: 10.1073/pnas.95.6.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hellemond JJ, van der Meer P, Tielens AG. Leishmania infantum promastigotes have a poor capacity for anaerobic functioning and depend mainly on respiration for their energy generation. Parasitology. 1997;114:351–360. [Google Scholar]
- van Hellemond JJ, van der Klei A, van Weelden SW, Tielens AG. Biochemical and evolutionary aspects of anaerobically functioning mitochondria. Philos Trans Roy Soc Lond B Biol Sci. 2003;358:205–213. doi: 10.1098/rstb.2002.1182. discussion 213–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hellemond JJ, Klockiewicz M, Gaasenbeek CP, Roos MH, Tielens AG. Rhodoquinone and complex II of the electron transport chain in anaerobically functioning eukaryotes. J Biol Chem. 1995;270:31065–31070. doi: 10.1074/jbc.270.52.31065. [DOI] [PubMed] [Google Scholar]
- van Weelden SW, Fast B, Vogt A, van der Meer P, Saas J, van Hellemond JJ, Tielens AG, Boshart M. Procyclic Trypanosoma brucei do not use Krebs cycle activity for energy generation. J Biol Chem. 2003;278:12854–12863. doi: 10.1074/jbc.M213190200. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan AM, O’Neill MT, Tarun AS, Camargo N, Phuong TM, Aly AS, Cowman AF, Kappe SH. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol. 2009;11:506–520. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Rodriguez J, Franke-Fayard B, Dinglasan RR, Janse CJ, Pastrana-Mena R, Waters AP, Coppens I, Rodriguez-Orengo JF, Srinivasan P, Jacobs-Lorena M, Serrano AE. The glutathione biosynthetic pathway of Plasmodium is essential for mosquito transmission. PLoS Pathog. 2009;5:e1000302. doi: 10.1371/journal.ppat.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais PM, Billoud B, Meyer J. Classification and phylogeny of hydrogenases. FEMS Microbiol Rev. 2001;25:455–501. doi: 10.1111/j.1574-6976.2001.tb00587.x. [DOI] [PubMed] [Google Scholar]
- Vincent WF. The physiological ecology of a Scenedesmus population in the hypolimnion of a hypertrophic pond. II. Heterotrophy. Brit Phycol J. 1980;15:35–41. [Google Scholar]
- von der Heyden S, Chao EE, Vickerman K, Cavalier-Smith T. Ribosomal RNA phylogeny of bodonid and diplonemid flagellates and the evolution of Euglenozoa. J Eukaryot Microbiol. 2004;51:402–416. doi: 10.1111/j.1550-7408.2004.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Waller RF, Jabbour C, Chan NC, Celik N, Likic VA, Mulhern TD, Lithgow T. Evidence of a reduced and modified mitochondrial protein import apparatus in microsporidian mitosomes. Eukaryot Cell. 2009;8:19–26. doi: 10.1128/EC.00313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins RF, Gray MW. The frequency of eubacterium-to-eukaryote lateral gene transfers shows significant cross-taxa variation within Amoebozoa. J Mol Evol. 2006;63:801–814. doi: 10.1007/s00239-006-0031-0. [DOI] [PubMed] [Google Scholar]
- Wawrzyniak I, Roussel M, Diogon M, Couloux A, Texier C, Tan KS, Vivares CP, Delbac F, Wincker P, El Alaoui H. Complete circular DNA in the mitochondria-like organelles of Blastocystis hominis. Int J Parasitol. 2008;38:1377–1382. doi: 10.1016/j.ijpara.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Weinbach EC, Harlow DR, Claggett CE, Diamond LS. Entamoeba histolytica: diaphorase activities. Exp Parasitol. 1977;41:186–197. doi: 10.1016/0014-4894(77)90144-8. [DOI] [PubMed] [Google Scholar]
- Wessberg KL, Skolnick S, Xu J, Marciano-Cabral F, Kemp RG. Cloning, sequencing and expression of the pyrophosphate-dependent phosphofructo-1-kinase from Naegleria fowleri. Biochem J. 1995;307:143–149. doi: 10.1042/bj3070143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol Biol Cell. 2006;17:1734–1743. doi: 10.1091/mbc.E05-11-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BA, Elliot C, Burri L, Kido Y, Kita K, Moore AL, Keeling PJ. A broad distribution of the alternative oxidase in microsporidian parasites. PLoS Pathog. 2010;6:e1000761. doi: 10.1371/journal.ppat.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Lowe PN, Leadlay PF. Purification and characterization of pyruvate: ferredoxin oxidoreductase from the anaerobic protozoon Trichomonas vaginalis. Biochem J. 1987;246:529–536. doi: 10.1042/bj2460529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M, Heil B, Heil B, Happe T. Isolation and molecular characterization of the [Fe]-hydrogenase from the unicellular green alga Chlorella fusca. Biochim Biophys Acta. 2002;1576:330–334. doi: 10.1016/s0167-4781(02)00239-7. [DOI] [PubMed] [Google Scholar]
- Wirth DF. Biological revelations. Nature. 2002;419:495–496. doi: 10.1038/419495a. [DOI] [PubMed] [Google Scholar]
- Wilson RJM, Denny PW, Preiser PR, Rangachari K, Roberts K, Roy A, Whyte A, Strath M, Moore DJ, Moore PW, Williamson DH. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- Wilson RJ, Williamson DH. Extrachromosomal DNA in the Apicomplexa. Microbiol Mol Biol Rev. 1997;61:1–16. doi: 10.1128/mmbr.61.1.1-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RJ, Williamson DH, Preiser PR. Malaria and other apicomplexans: the ‘plant’ connection. Infect Agents Dis. 1994;3:29–37. [PubMed] [Google Scholar]
- Wood SG, Gottlieb D. Evidence from mycelial studies for differences in the sterol biosynthetic pathway of Rhizoctonia solani and Phytophthora cinnamomi. Biochem J. 1978;170:343–354. doi: 10.1042/bj1700343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki A, Inagaki Y, Ishida K. Palpitomonas bilix gen. et sp. nov.: a novel deep-branching heterotroph possibly related to Archaeplastida or Hacrobia. Protist. 2010;161:523–538. doi: 10.1016/j.protis.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Yarlett N, Martinez MP, Moharrami MA, Tachezy J. The contribution of the arginine dihydrolase pathway to energy metabolism by Trichomonas vaginalis. Mol Biochem Parasitol. 1996;78:117–125. doi: 10.1016/s0166-6851(96)02616-3. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Grant J, Tekle YI, Wu M, Chaon BC, Cole JC, Logsdon JM, Jr, Patterson DJ, Bhattacharya D, Katz LA. Broadly sampled multigene trees of eukaryotes. BMC Evol Biol. 2008;8:14. doi: 10.1186/1471-2148-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Kumar TR, Nkrumah LJ, Coppi A, Retzlaff S, Li CD, Kelly BJ, Moura PA, Lakshmanan V, Freundlich JS, Valderramos JC, Vilcheze C, Siedner M, Tsai JH, Falkard B, Sidhu AB, Purcell LA, Gratraud P, Kremer L, Waters AP, Schiehser G, Jacobus DP, Janse CJ, Ager A, Jacobs WR, Jr, Sacchettini JC, Heussler V, Sinnis P, Fidock DA. The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites. Cell Host Microbe. 2008;4:567–578. doi: 10.1016/j.chom.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Allahverdiyeva Y, Eisenhut M, Aro E-M. Flavodiiron proteins in oxygenic photosynthetic organisms: photoprotection of photosystem II by Flv2 and Flv4 in Synechocystis sp. PCC 6803. Plos One. 2009;4:e5331. doi: 10.1371/journal.pone.0005331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Takaya N, Nakamura A, Yamaguchi M, Takeo K, Shoun H. Ammonia fermentation, a novel anoxic metabolism of nitrate by fungi. J Biol Chem. 2002;277:1892–1896. doi: 10.1074/jbc.M109096200. [DOI] [PubMed] [Google Scholar]


