Abstract
Ambient particulate matter (PM) from air pollution is associated with exacerbation of asthma. The immunological basis for the adjuvant effects of PM is still not well understood. The generation of reactive oxygen species (ROS) and the resulting oxidative stress has been identified as one of the major mechanisms. Using a new intranasal sensitization model in which ambient PM is used as an adjuvant to enhance allergic inflammation (Li et al., Environ. Health Perspect. 2009, 117, 1116-1123), a proteomics approach was applied to study the adjuvant effects of ambient PM. The enhanced in vivo adjuvant effect of ultrafine particles (UFP) correlates with a higher in vitro oxidant potential and a higher content of redox-cycling organic chemicals. Bronchoalveolar lavage fluid proteins from normal and sensitized mice were resolved by two-dimensional gel electrophoresis, and identified by mass spectrometry. Polymeric immunoglobulin receptor, complement C3, neutrophil gelatinase-associated lipocalin, chitinase-3-like protein 3, chitinase-3-like protein 4, and acidic mammalian chitinase demonstrated significantly enhanced up-regulation by UFP with a polycyclic aromatic hydrocarbon (PAH) content and a higher oxidant potential. These proteins may be the important specific elements targeted by PM in air pollution through the ability to generate ROS in the immune system, and may be involved in allergen sensitization and asthma pathogenesis.
Keywords: proteomics, asthma pathogenesis, particulate matter, ultrafine particles, bronchoalveolar lavage fluid, chitinase
1 Introduction
Vehicular traffic and combustion result in increased cardiorespiratory morbidity and mortality. Epidemiological studies have demonstrated a statistical association between air pollutants and outcome of asthma [1]. Asthma is a chronic inflammatory disease of the airways that involves antigen-presenting cells, Th2 lymphocytes, IgE-producing plasma cells, mast cells, eosinophils, mucus-secreting goblet cells, smooth muscle and endothelial cells. Ambient particulate matter (PM) has been demonstrated in both animal and human studies as an adjuvant that promotes sensitization to common environmental allergens [2-8].
A number of biological mechanisms have been proposed to explain PM-induced adverse health effects [9], including cytokine release, inflammation, endotoxin-mediated damage, free radical production, oxidative stress, stimulation of capsaicin receptors, altered autonomic nervous system activity, and the covalent modification of important cellular biomolecules. Thus, the exact mechanisms with which PM causes adverse health effects are complex and are not completely understood.
Perhaps among the best characterized variable is the effect of PM on airway inflammation and the generation of reactive oxygen species (ROS) [10]. We have previously demonstrated that excessive ROS production could lead to oxidative stress that activates a number of redox-sensitive signaling cascades [11-14]. This may include the generation of oxidative stress by organic chemicals, leading to the expression of specific cellular elements involved in the immune system and the skewing of immune response towards Th2 differentiation [15, 16].
Recently, we attempted to conduct a proteomic profiling of bronchoalveolar lavage fluid (BALF) and lung tissue from the classical OVA-sensitization murine asthma model [17]. Using two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) and liquid chromatography-tandem mass spectrometry (LC-MS/MS), we demonstrated that the protein levels of chitinase 3-like protein 3 (Ym1), chitinase 3-like protein 4 (Ym2), pulmonary surfactant-associated protein D (SP-D), and resistin-like molecule α or “found in inflammatory 1” (FIZZ1) are increased in response to OVA/alum sensitization and are subtracted by the thiol antioxidant, N-acetylcysteine (NAC), strongly suggesting that these proteins may be considered as the potential oxidative stress markers that can be used to follow the role of oxidant injury in asthma [17].
As a continuation of this work, we asked the question whether there is a specific and unique proteome profile that is associated with the adjuvant effect of pro-oxidative PM on allergic sensitization. In the present study, we used a highly sensitive murine intranasal sensitization model to determine if the adjuvant effect of an ambient UFP collection, which has been shown to have strong oxidant potential by our previous report [18], could lead to a modified proteome profile in the BALF. There are two major differences between this model and the classical OVA sensitization model previously described by us: (a) ambient pro-oxidative UFP was used as an adjuvant for OVA sensitization, instead of alum and (b) intranasal instillation was used for UFP and OVA exposure instead of intraperitoneal injection [17, 18].
We hypothesize that intranasal exposure to an extremely low dose of ambient pro-oxidative UFP, together with a low dose of allergen OVA is sufficient to change the proteome profile in the BALF and this alteration may be used to develop biomarkers for screening the adjuvant effect of pro-oxidative PM. We show that intranasal exposure to a precise amount of ambient PM was able to promote a Th2 immune response characterized by enhanced allergic airway inflammation and that the adjuvant effect of UFP was closely correlated to a significant change in the proteome profile in BALF.
2 Materials and methods
2.1 Animal treatment and sample collection
Six- to eight-week old female BALB/c mice were obtained from Charles River Laboratories (Hollister, CA, USA). Mice were housed under standard laboratory conditions and maintained on autoclaved food and acidified water. Endotoxin-free OVA and ultrafine particles (<0.15 μm), collected in downtown Los Angeles [19] were used as the allergen and adjuvant (Figure 1). All animal procedures were approved by the UCLA Animal Research Committee and were performed as described previously [18]. Mice were sacrificed by intraperitoneal injection of pentobarbital. Bronchoalveolar lavage (BAL) and differential BAL cell counts were performed as previously described [20]. The BALF supernatants were kept at -80°C. The left lung was kept in liquid nitrogen, and the right lung was stored in 4% formaldehyde (Sigma-Aldrich, St. Louis, MO USA).
Figure 1.
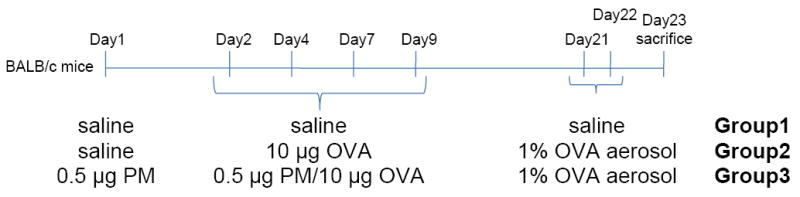
Outline of protocol for the murine intranasal sensitization model. Endotoxin-free OVA and PM (UFP) were used to sensitize the animals (6 mice/group) for demonstration of the adjuvant effect of ambient UFP. On day 1, mice from the PM exposure group received 0.5 μg PM suspension intranasally. On day 2, endotoxin-free OVA (10 μg) was used together with PM (UFP) (0.5 μg) to sensitize the PM exposure group by intranasal instillation, which was repeated on days 4, 7 and 9. The OVA and control groups only received OVA and saline. All animals in the PM and OVA groups were subsequently challenged with 1% OVA aerosol on day 21 and 22.
2.2 Sample preparation for 2D-PAGE analysis
Aliquots of BALF supernatants corresponding to 6 mice in each group were pooled and precipitated with 75% ethanol and incubated overnight at -20°C. After washing with cold 75% ethanol, the pellet was dried at room temperature and resuspended in rehydration buffer (7 M urea, 2 M thiourea, 50 mM DTT, 4% CHAPS, 5% glycerol, 10% isopropanol, 1% ampholytes). Protein concentrations were quantified using a modified Bradford protein assay procedure [21].
2.3 2D-Gel electrophoresis
All chemicals used for 2D gel electrophoresis were of electrophoresis grade. BALF protein (100 μg) of each pooled group was dissolved in 300 μL rehydration buffer and applied to 17-cm pH 3-10 immobilized IPG strips (Bio-Rad, Hercules, CA USA). IPG strips were active rehydrated for 14 hr at 50 V (22°C) in the Protean IEF Cell (Bio-Rad) and then subjected to isoelectric focusing (linear ramp to 100 V in 2 hr, linear ramp to 250 V in 2 hr, linear ramp to 4000 V in 5 hr, hold at 4000 V for 23000 vh and hold at 8000 V for 50000 vh). After isoelectric focusing, the IPG strips were equilibrated at room temperature for 20 min in SDS-equilibration buffer (0.375 mM Tris/HCl pH 8.8, 6 M urea, 20% (v/v) glycerol, 2% (w/v) SDS) supplemented with 2% DTT and for another 20 min with the same buffer supplemented with 2.5% iodoacetamide. After equilibration, the IEF strips were applied to 8-16% SDS-PAGE gels (Protean II ready gel, Jule, Inc., Milford, CT USA). Electrophoresis was carried out at 200 V for 2 hr and followed by 300 V for 3 hr. Gels were stained using Sypro Ruby (Invitrogen, Carlsbad, CA USA). For gel-image analysis, gels were scanned at high resolution with an FX Molecular Imager FX (Bio-Rad). PDQuest software version 7.2 (Bio-Rad) was used for protein spots analysis. Spots showing a greater than 1.8-fold change in intensity were selected for mass spectrometry analysis.
2.4 Protein identification by liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Protein spots were excised from the Sypro Ruby-stained gel gels by the ProteomeWorks spot cutter (Bio-Rad). The in-gel trypsin digestion and peptide extraction was accomplished manually using standard protocols [17]. The resulting tryptic peptides were identified by using an LC-quadrupole time-of-flight (QTOF) system (Dionex/LC Packings nano-LC and Applied Biosystems/Sciex QSTAR Pulsar XL mass spectrometer). Mascot search program (Matrix Science, London, UK) was used to search the MS/MS spectra against the Swiss-Prot murine protein sequence database. For searching the sequence database, the following variable modifications were set: carbamidomethylation of cysteines, oxidization of methionines, conversion of N-terminal glutamate and aspartate to pyro-Glu, and cyclization of N-terminal cysteine. One missed tryptic cleavage was tolerated and the peptide and MS/MS mass tolerance was set as ±0.3 Da. Positive protein identification was based on standard Mascot criteria for statistical analysis of the MS/MS data. A -10Log (P) score, where P is the probability that the observed match is a random event, of 72 was regarded as significant.
2.5 Western blot analysis
Pooled BAL fluid from each group was precipitated with 75% ethanol overnight at -20°C. After washing with cold 75% ethanol, protein was dissolved in 7 M urea containing protease inhibitors and subjected to protein quantification. Lungs from different mice were sonicated in 7 M urea, containing protease inhibitors in ice bath for 10 sec × 3 times and were subsequently centrifuged at 13,000g for 15 min at 4°C. After protein quantification, supernatants in the same group were pooled and stored at −80°C. BAL fluid protein (20 μg) or lung protein (50 μg) from pooled sample was electrophoresed on 10-20% SDS polyacrylamide gels before transfer to PVDF membranes. Horseradish peroxidase-conjugated secondary antibodies (GE Healthcare/Amersham Biosciences, Uppsala, Sweden) were used followed by ECL reaction to develop the blots according to the manufacturer’s instructions. Primary antibodies were used to detect the expression of the following proteins: Ym1/Ym2 (Dr. Shioko Kimura, National Cancer Institute, Bethesda, MD, USA), γ-actin (Sigma-Aldrich), SP-D (Chemicon International, Temecula, CA, USA), FIZZ1 (Alpha Diagnostic International, Inc. San Antonio, TX, USA), Complement C3 (Cedarlane Laboratories Lt., Burlington, Ontario, Canada), PIGR (R&D System, Inc., Minneapolis, MN, USA), NGAL (Abcam Inc., Cambridge, MA, USA), and AMCase (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). Band intensities from film were analyzed by IMAGEQUANT 5.2 software (Molecular Dynamics, Sunnyvale, CA, USA).
2.6 Real-time PCR
Total RNA was isolated from lung tissue using the RNeasy kit (Qiagen, Valencia, CA USA). Reverse transcription was carried out with the iScript cDNA Synthesis kit (BioRad) for RT-PCR. One μg RNA from each sample was used for the 20 μL cDNA reaction. For real-time PCR, a Bio-Rad iQ™ SYBR® Green Supermix and a myiQ™ machine were used. The PCR primer sets used were:
FIZZ1: 5’-AGGAACTTCTTGCCAATCCA-3’ and 5’-CAGTAGCAGTCATCCCAGCA-3’
PIGR: 5’-AGCCCCATATTTGGTCCCCAGGAGG-3’ and 5’-TCCAGGCTGACATCGAAGGACAGGC-3’
Complement C3: 5’-CTGCTGTTGGCCAGCTCCCCATTAG-3’ and 5’-CCCCGAAGTTTGCCACCACTGTCAC-3’
NGAL: 5’-TGGCCCTGAGTGTCATGTGTCYGGG-3’ and 5’-TGGTCCCTGACCAGGATGGAGGTGA-3’
Ym1/Ym2: 5’-CATGAGCAAGACTTGCGTGACTATGAAGCA-3’ and 5’-TGTGGAAGTGAGTAGCAGCCTTGGAATGTC-3’, 5’-GTAGCAGCCTTGGAATGTGGTTCAAAGTG-3’
SPD: 5’-CCAACAGAGAATGGCCTGCCTGGTC-3’
AMCase: 5’-TGCGTGAAGCTTTTGAGCAGGAGGC-3’ and 5’-CAGCTGGGGCTCCATTGTTCTTCCA-3’
Samples and reference (actin) were run in triplicate and the standard curves were duplicated on the 96-well PCR plate. PCR data was analyzed using the Pfaffl method [22, 23].
2.7 Histological staining
Lung tissue sections were immunohistochemically stained with antibodies against FIZZ1 and Ym1/2. Unstained hydrated paraffin sections were incubated with a blocking solution (Vectastain ABC-AP kit, Vector Laboratories, Burlingame, CA USA) following the manufacturer’s instruction. The sections were then incubated with the antibodies followed by treatment with a secondary antibody. Some tissue sections were incubated without the secondary antibody to check for nonspecific binding. Immunoreactive proteins were microscopically detected after treatment with Vectastain ABC-AP complex followed by Vector Red AP substrate solution.
2.8 Statistical analysis
Data were expressed as the mean ± SME and analyzed by Student’s t-test. All differences of p < 0.05 were considered significant and showed as *. A p < 0.01 is represented as **.
3 Results
3.1 Adjuvant effect of ambient UFP
The ambient UFP (collected in Los Angeles) was used in the OVA intranasal instillation model. The mice received saline (Group 1), OVA (10 μg; Group 2) or OVA (10 μg) plus UFP (0.5 μg) (Group 3) for allergic sensitization (Figure 1). BAL analysis showed that UFP is effective in enhancing OVA sensitization. Compared to saline and OVA alone, UFP plus OVA induced a statistically significant increase in the BAL eosinophils (Figure 2A), oxidant-sensitive cells considered relevant in allergic inflammation. Consistent with allergic inflammation, the levels of OVA-specific IgG1 and IgE in the serum were also significantly increased (Figures 2B and 2C). These immunoglobulin classes reflect Th2 immunity. Taken together, these data suggest that ambient UFP can acts as an adjuvant to promote Th2 polarization and to enhance allergic sensitization.
Figure 2.
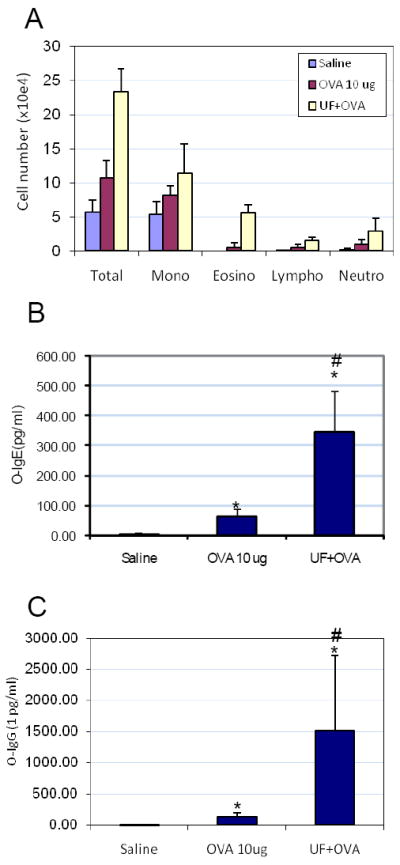
The adjuvant effect of UFP on OVA-induced allergic sensitization. BAL collections and differential BAL cell counts were performed as previously described [19]. Serum OVA-specific IgG1 and IgE were measured by ELISA. (A) Ambient UFP increased OVA-induced allergic inflammation in the lung. (B) Enhanced OVA-IgE production by 0.5 μg UFP instillation. (C) Enhanced OVA-IgG1 production by 0.5 μg UFP instillation. *p < 0.05 compared to control; #p < 0.05 compared to OVA alone.
3.2 Two-dimensional gel electrophoresis and mass spectrometry reveal significant proteome profile changes
Proteome profiles of BALF displayed by two-dimensional PAGE were measured in duplicate, and yielded >400 individual protein spots for each gel (Figure 3). Protein spots of the control (Group 1; average number of spots, 449), OVA (Group 2; average number of spots, 656) and OVA plus UFP (Group 3; average number of spots, 406) were matched, and their intensities were compared.
Figure 3.
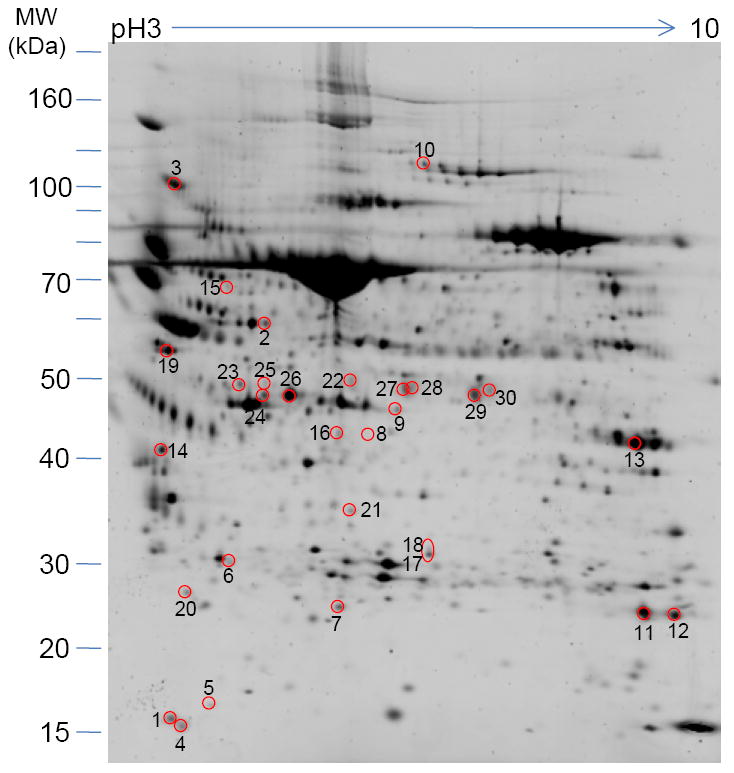
Two-dimensional gel electrophoresis profile of murine BAL fluid. Aliquots of BALF supernatants corresponding to 6 mice in each group were pooled and precipitated with 75% ethanol. BALF protein (100 μg) was isoelectric focused on 17-cm IPG (pH 3-10) strips followed by 8-10% SDS-PAGE. After Sypro-Ruby staining, intensity of the spots was determined by PDQuest software (Bio-Rad). Proteins with enhanced or decreased expression levels in the presence of OVA plus UF compare to OVA alone were selected and identified by LC-MS/MS. Those proteins are numbered and their identities disclosed in Table 1.
BALF proteins from mice exposed to OVA (Group 2) showed significant changes in intensity compared to those of the control group. Moreover, there were markedly greater changes in the spot patterns found for the UFP/OVA group (Group 3) compared to those found for the OVA-alone group. Differential protein expression analysis showed a total of 27 gel spots with a significant increase in intensity and 3 gel spots with a significant decrease in intensity for Group 3 compared to the control group (Group 1). These 30 protein spots are marked in Figure 3. After washing, trypsin digestion, and extraction, the proteins from the individual spots were identified by LC-MS/MS and sequence database searching. From these 30 protein gel spots, 16 unique proteins were identified (Table 1). Many of the protein spots represent the same family of proteins or gene products with multiple potential modifications. Among the unique proteins, 7 proteins exhibited a statistically significant increase in intensity (approximately >4 fold) for Group 3 compared with the control group animals. These proteins include polymeric immunoglobulin receptor (PIGR), chitinase 3-like protein 4 (Ym2), chitinase 3-like protein 3 (Ym1), neutrophil gelatinase-associate lipocalin (NGAL), complement C3, acidic mammalian chitinase (AMCase), and pulmonary surfactant-associated protein D.
Table 1.
Proteins with significant expression level changes in bronchoalveolar lavage fluid from mice challenged with OVA and OVA plus ultra fine particles
| Protein assigned | Spot No. | Accession Number | Mr (kDa) | Cal. pI | Sequence Coverage (%) | Mascot Score | Peptides Matched | Fold changed (OVA vs. PBS) | Fold changed (OVA/UFP vs. PBS) |
|---|---|---|---|---|---|---|---|---|---|
| Haptoglobin | 1 | HPT_MOUSE | 38,7 | 5.9 | 10 | 108 | 4 | +2.49 | +3.82 |
| 2 | 22 | 349 | 10 | -1.78 | -3.31 | ||||
| Polymeric-immunoglobulin receptor (Poly-Ig receptor) (PIGR) | 3 | PIGR_MOUSE | 84,9 | 5.3 | 19 | 511 | 15 | +5.77 | +6.77 |
| Thioredoxin (Trx) (ATL-derived factor) (ADF) | 4 | THIO_MOUSE | 11,7 | 4.8 | 20 | 64 | 2 | -1.55 | -2.81 |
| Coactosin-like protein | 5 | COTL1_MOUSE | 15.9 | 5.3 | 21 | 131 | 5 | +1.88 | +3.2 |
| Pulmonary surfactant-associated protein A (SP-A) | 6 | SFTPA_MOUSE | 26,1 | 5.3 | 14 | 173 | 2 | +1.94 | +2.52 |
| Ferritin light chain 1 | 7 | FRIL1_MOUSE | 20,8 | 5.7 | 22 | 238 | 4 | -1.64 | -1.95 |
| Chitinase 3-like protein 4 (Ym2) | 8 | CH3L4_MOUSE | 44,9 | 5.8 | 15 | 155 | 6 | +1.25 | +46.29 |
| Transcobalamin-2 | 9 | TCO2_MOUSE | 47,6 | 5.9 | 12 | 202 | 6 | +1.23 | +1.68 |
| Plasminogen | 10 | PLMN_MOUSE | 90,7 | 6.2 | 1 | 290 | 11 | -1.88 | -1.42 |
| Neutrophil gelatinase-associated lipocalin (NGAL) | 11 | NGAL_MOUSE | 22,9 | 9.0 | 13 | 132 | 9 | +5.24 | +5.50 |
| 12 | 13 | 76 | 6 | +5.69 | +5.25 | ||||
| Chitinase-3-like protein 1 (Cartilage glycoprotein 39) (GP-39) | 13 | CH3L1_MOUSE | 43 | 8.7 | 27 | 348 | 24 | +1.44 | +1.89 |
| Complement C3 (HSE-MSF) | 14 | CO3_MOUSE | 186,4 | 6.4 | 5 | 203 | 13 | +3.79 | +4.78 |
| 15 | 11 | 224 | 19 | +2.70 | +4.00 | ||||
| 16 | 4 | 173 | 7 | +37.61 | +83.33 | ||||
| 17 | 5 | 402 | 10 | +6.35 | +13.95 | ||||
| 18 | 5 | 146 | 6 | +5.33 | +6.95 | ||||
| Acidic mammalian chitinase | 19 | CHIA_MOUSE | 52 | 4.9 | 18 | 250 | 15 | +1.39 | +4.96 |
| 20 | 1 | 49 | 1 | +1.72 | +4.8 | ||||
| Serum albumin | 21 | ALBU_MOUSE | 68,6 | 5.8 | 9 | 124 | 5 | +8.97 | +17.04 |
| 22 | 15 | 224 | 11 | +1.49 | +4.14 | ||||
| Chitinase 3-like protein 3 (Ym1) | 23 | CH3L3_MOUSE | 44,4 | 5.4 | 25 | 274 | 13 | +1.49 | +4.38 |
| 24 | 27 | 282 | 14 | +5.89 | +20.15 | ||||
| 25 | 22 | 159 | 9 | +6.32 | +26.72 | ||||
| 26 | 30 | 368 | 22 | +2.34 | +5.27 | ||||
| Pulmonary surfactant-associated protein D (PSP-D) | 27 | SFTPD_MOUSE | 37,7 | 7.6 | 30 | 234 | 10 | +3.48 | +7.35 |
| 28 | 13 | 99 | 4 | +5.04 | +13.36 | ||||
| 29 | 18 | 250 | 9 | +2.60 | +4.27 | ||||
| 30 | 18 | 243 | 9 | +5.99 | +21.94 | ||||
3.3 RT-PCR, immunoblotting analysis, and histology confirm adjuvant effect of UFP
To examine the fidelity of these increased proteins and to confirm the 2D-PAGE measurements and analysis, Western blotting and real-time PCR were performed for those proteins that exhibited significant changes in intensity on the 2D-gels. To confirm equal loading of the protein on western blots, anti-β-actin antibody was used. Primers for β-actin were used for normalization in real-time RT-PCR experiments.
PIGR
Secretary immunoglobulin (SIgA, and to a lesser extent, SIgM) is the first line of specific immunological defense against environmental antigens. Epithelial transcytosis of pIgA is mediated by pIgR, a key component of the mucosal immune system. Regulation of PIGR expression in mucous membranes involve complex interactions among host, microbial, and environmental-derived factors such as cytokine, hormones, dietary and microbial factors. Both western blot analysis (Figure 4B, left panel) and real time PCR (Figure 4C) of lung tissue from OVA challenged mice exhibited significantly enhanced expression of PIGR compared to the control group; this up-regulation is even more significant in the UFP plus OVA group. These results indicate that, in an allergic inflammation condition, expression of PIGR is up-regulated at the transcriptional and translational level and the UFP exerted its effect on the transcription and translation of PIGR. However, the level of secretary component (SC) of PIGR in the OVA plus UFP group did not exhibit a more significantly increase compared with OVA alone as showed by 2D gel electrophoresis (Figure 4A) and western blot analysis (Figure 4B, right panel) of BALF. Transcription and translation of the PIGR gene is clearly an important determinant of the regulation, but post-translational mechanisms are also involved. The secretary component is a proteolytic fragment of the integral membrane protein PIGR. Compared to the membrane form of PIGR from the lungs, the SC of PIGR in lungs and BALF are produced by multiple post-translational steps such as glycosylation, binding of ligands to PIGR, cleavage of PIGR to SC and subcellular localization. From our results, UFP may not have influenced such post-translational steps.
Figure 4.
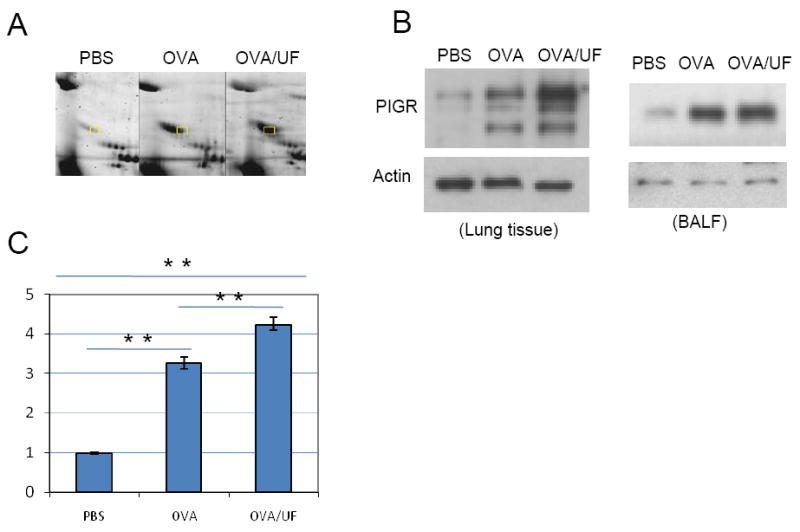
Differential expression of polymeric immunoglobulin receptor (PIGR). (A) Two-dimensional gel electrophoresis profile of PIGR. BAL fluid protein (100 μg) was separated on 17-cm IPG (pH 3-10) strips, followed by 8–16% SDS-PAGE. Proteins were stained with Sypro-Ruby. LC–MS/MS identified PIGR were labeled. (B) Western blot analysis of PIGR level in the lung (left panel) and in the BALF (right panel). Lung tissue protein (50 μg) and BALF protein (20 μg) from three different groups were electrophoresed on 8-16% SDS polyacrylamide gels before transfer to nitrocellulose membranes. Goat anti-mouse PIGR polyclonal antibody was used at 0.1 μg/mL. (C) Real-time RT-PCR analysis. Samples were run in triplicate and the standard curves were duplicated on a single 96-well PCR plate. PCR data was analyzed using the Pfaffl method. **p<0.01.
AMCase
Acidic mammalian chitinase is known to be expressed during Th2-mediated inflammation. AMCase is a member of the glycosyl hydrolase 18 family and has structural similarity with other family members, including the lectins Ym1 and Ym2, cartilage glycoprotein-39 and chitotriosidase. Compared to the control group, 2D-PAGE, western blotting analysis and real-time PCR all showed a significantly increased expression of AMCase in lung tissue and BALF from OVA and UFP/OVA challenged animals (Figure 5). In addition, AMCase levels are significantly higher in UFP/OVA-exposed mice than in OVA-alone mice.
Figure 5.
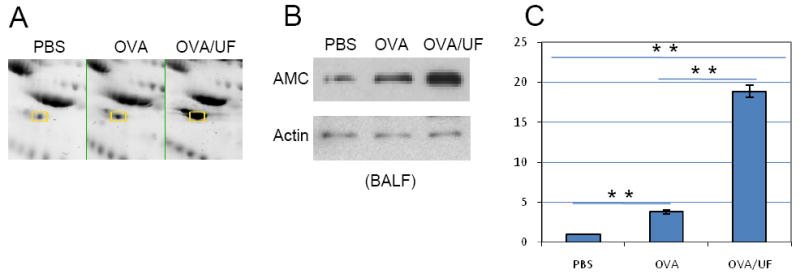
Differential expression of acidic mammalian chitinase (AMCase). (A) Two-dimensional gel electrophoresis profile of AMCase. AMCase spots are labeled. (B) Western blot analysis of AMCase level in the BALF. Polyclonal goat anti-mouse AMC antibody was used at 1:1000. (C) Real-time RT-PCR. **p<0.01.
Complement C3
Complement C3 plays a central role in the activation of the complement system. C3 is an acute-phase reactant; an increase in C3 synthesis is induced during acute inflammation [24]. After translation, it is processed and cleaved into 10 chains. Using monoclonal anti-C3 antibodies, we detected 5 C3 protein bands on the western blot for mouse BALF (Figure 6B). 2D-gel profiles, real-time PCR and western blot analysis clearly showed that C3 expression level in lung and BALF is significantly increased in the OVA and OVA plus UFP group compared to the control group. Furthermore, the expression level increased more significantly in OVA plus UFP than in OVA alone (Figure 6).
Figure 6.
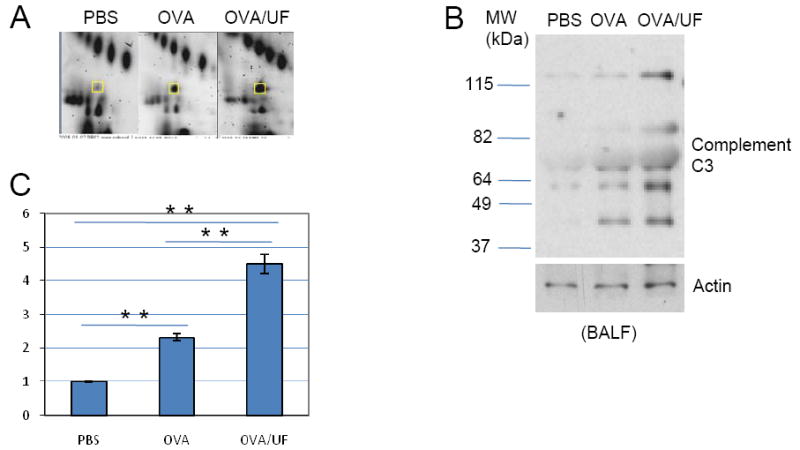
Complement C3 profiling. (A) Two-dimensional gel electrophoresis profile of Complement C3. (B) Western blot analysis of Complement C3 level in the BALF. Anti-mouse complement component C3 monoclonal antibody was used at 1:1000. (C) Real-time RT-PCR. *p<0.05; **p<0.01.
NGAL
Neutrophil gelatinase–associated lipocalin is expressed and secreted by immune cells. NGAL is well known for its function as a shuttle for iron and siderophores. Elevated tissue NGAL expression has been documented under diverse infectious and inflammatory conditions such as inflammatory bowel disease, appendicitis, diverticulitis, and urinary tract infections [25]. However, there is no previous report about its increased expression under asthma conditions. Compared to the control group, the OVA and OVA plus UFP groups showed increased expression level of NGAL (Figure 7). Moreover, the OVA plus UFP group showed a more significant increase compared to Group 2 (OVA alone).
Figure 7.
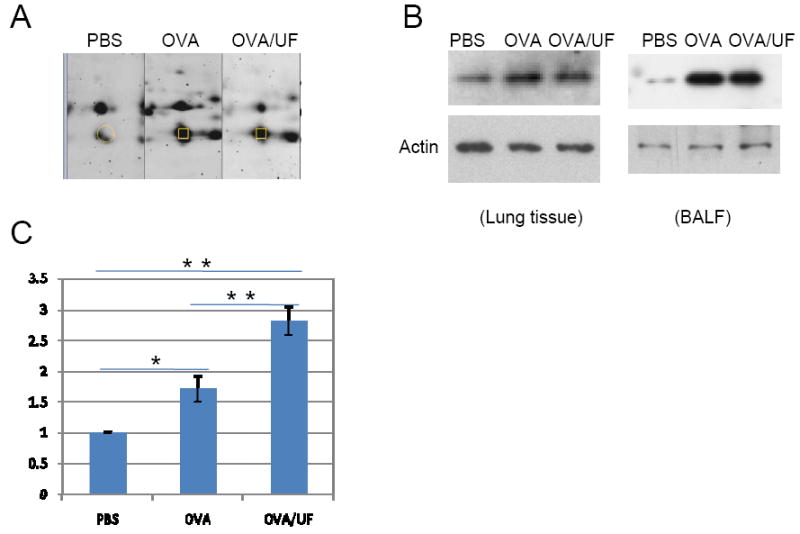
Polymeric Neutrophil gelatinase-associated lipocalin (NGAL) profiling. (A) Two-dimensional gel electrophoresis profile of NGAL. (B) Western blot analysis of NGAL level in the lung (left panel) and in the BALF (right panel). Polyclonal rabbit antibody to NGAL was used at 1 μg/μl. (C) Real-time RT-PCR. *p<0.05; **p<0.01.
Chitinase-like Proteins
Ym1/Ym2 is involved in allergic inflammation through the IL-4/STAT 6 signal transduction pathway [26]. Figure 8 shows the differential expression of chitinase-3-like protein 3 (Ym1) and chitinase-3-like protein 4 (Ym2). Both 2D-gel analysis and western blotting demonstrated a significantly increased level of Ym1/Ym2 in OVA-challenged mice. Introduction of UFP highly enhanced expression of Ym1/Ym2 compared to OVA alone. As showed by Figure 8, Panel C and D, Moreover, the mRNA level of Ym1 and Ym2 significantly increased 35 and 584 fold, respectively (Figure 8C and 8D). The level of Ym2 was higher than that of Ym1.
Figure 8.
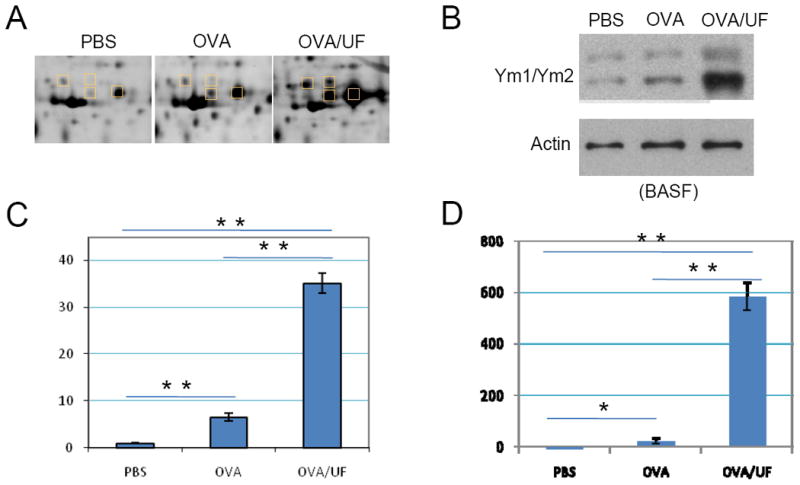
Ym1 and Ym2 proteome profiling. (A) Two-dimensional gel electrophoresis profile of Ym1/Ym2. (B) Western blot analysis of Ym1/Ym2 level in the BALF. The Ym1/Ym2 antibody was used at 1:10000. (C) Real-time RT-PCR of Ym1, **p<0.01. (D) Real-time RT-PCR of Ym2, *p<0.05; **p<0.01.
Pulmonary Surfactant-associated Protein D
SPD is known to play an important role in the innate immunity of mucosal surfaces, especially in lungs, protecting against inhaled microorganisms and allergen challenges. It is important in regulating allergic responses [27]. In our allergic sensitization model, expression of SP-D appears to be stimulated by OVA. However, OVA plus UFP treatment did not show a more significant stimulation at both translational and transcriptional level compared to OVA alone (Figure 9).
Figure 9.
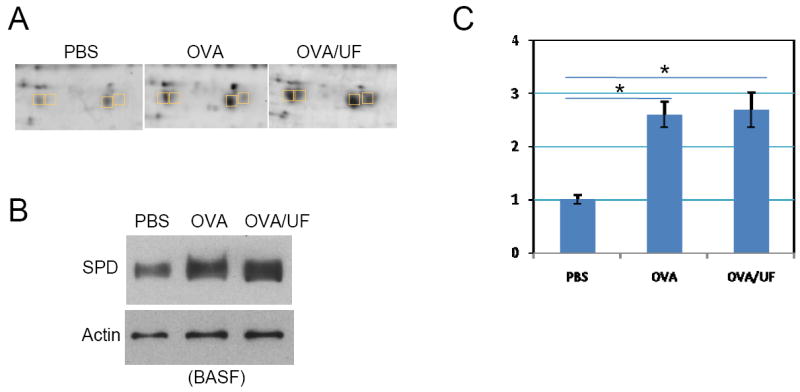
Differential expression of pulmonary surfactant-associated protein D. (A) Two-dimensional gel electrophoresis profile of SP-D. (B) Western blot analysis of SP-D level in the BALF. Rabbit polyclonal anti-mouse SP-D antibody was used at 1:1000. (C) Real-time RT-PCR. *p<0.05.
Ym1/2 and FIZZ1 have been reported by us previously to be potential marker proteins for OVA-induced allergic airway disease [17]. In our new intranasal sensitization model, the extent of the allergic sensitization was determined by lung histology similarly. In UFP-exposed mice, there was a markedly greater mucous cell metaplasia in the surface epithelium lining the conducting airways compared to those in the control or OVA-alone groups (Figure 10, AB/PAS staining). The mucosa changes were accompanied by immunohistological evidence of over expression of Ym1/2 and FIZZ1 (Figure 10, Ym1/2, FIZZ staining).
Figure 10.
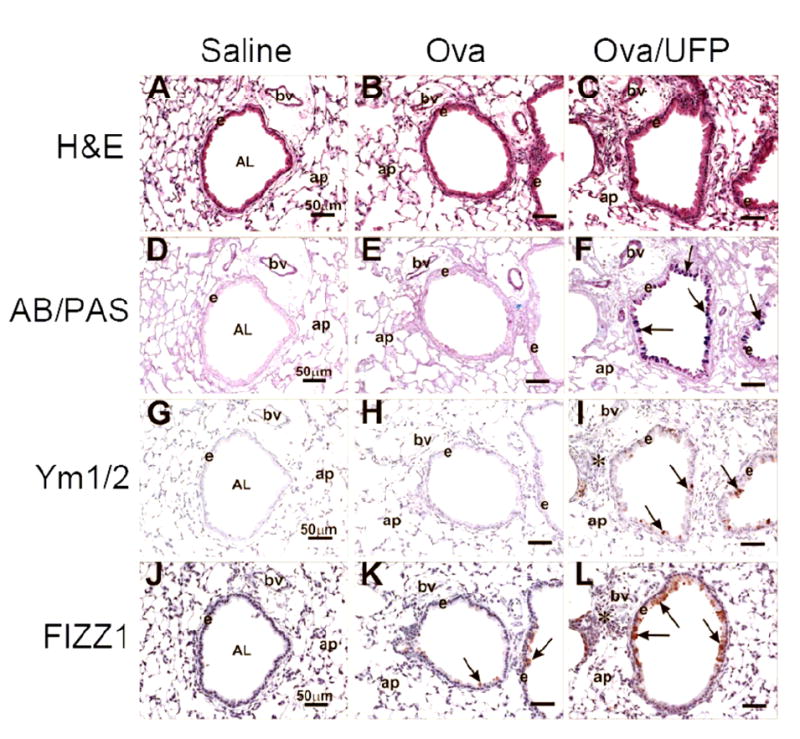
Histological examination of lung tissue. Tissues from pre-terminal bronchioles were analyzed for morphologic features, mucosubstances in mucous cells and the presence of some proteins associated with allergic lung inflammation. (A-C) H&E staining. (D-E) AB/PAS staining. Arrows depict AB/PAS-stained mucosubstances in airway epithelium. (G-I) Immunohistological staining using anti Ym1/Ym2 antibodies. Arrows depict stained Ym1/Ym2. (J-L) Immunohistological staining using anti FIZZ1 antibodies. Arrows depict FIZZ in the epithelium. Abbreviations: AB/PAS, Alcian blue-Periodic acid schiff double stain; AL, airway lumen; ap, alveolar parenchyma; bv, blood vessel; e, airway surface epithelium; H&E, hematoxylin and eosin stain.
4 Discussion
In our recent paper, a murine intranasal sensitization model was able to demonstrate the adjuvant effect of ambient pro-oxidative UFP on OVA sensitization and to correlate this effect to the particles’ organic chemical content and oxidant potential [18]. Both abiotic and biotic assays were performed to determine if the observed adjuvant effect of UFP is correlated with the oxidative stress potential of the UFP and thereby produces ROS. The polycyclic aromatic hydrocarbon (PAH) content of UFP is correlated with their ability to induce DTT consumption and heme oxygenase-1 (HO-1) expression, an oxidative stress marker induced by PM [10, 19]. The adjuvant effects of ambient UFP are determined by their oxidant potential and could be partially blocked by thiol antioxidant NAC administration; carbon black particles do not exert an adjuvant effect because of lack of redox-cycling compounds. All of these results suggested that organic chemical compounds detected in UFP, such as oxy-PAHs and quinones, known to be ROS generators, have the ability to promote oxidative stress effects in the lung. These in vivo adjuvant effects correlate with the ROS production by redox-cycling organic chemicals [18].
In the present study, we used the same allergic sensitization protocol as well as the same collection of ambient UFP to show that the adjuvant effect of UFP are reflected in an altered proteome profile of BAL fluid from mice sensitized with UFP [18]. Using 2D-PAGE, we demonstrated that the allergic sensitization by UFP leads to changes in intensity for 30 protein spots, representing 16 unique proteins. Among them, 7 proteins showed significant up-regulation in lung and/or in BALF. These proteins were further confirmed by immunoblotting and RT-PCR.
The ambient UFP used for this study has been well characterized, as described in our previous report [18]. These UFPs (as UF#1 in [18]) have stronger intrinsic oxidant potential and greater ability to induce oxidative stress compared to the larger PM collected at the same time. Consistent with previously published reports, intranasal instillation of these ambient UFP during OVA sensitization significantly increased BAL eosinophil count and OVA-specific Ab (OVA-IgG1 and IgE) production, both of which are the hallmark features of allergic airway inflammation and Th2 immune response (Figure 2). Eosinophils are important mediators of allergic airway inflammation including asthma [28]. Increased eosinophil count has been found to be associated with asthma disease severity. Eosinophils are a rich source of cytotoxic proteins, lipid mediators, ROS and pro-inflammatory Th2 cytokines and chemokines including IL-5, IL-4, IL-6, TNF-α, IL-8, and MIP-1α [28]. Its major basic protein (MBP) and cationic protein stimulate the release of histamine from basophils and mast cells, which is one of the major causes of airway hyperreactivity (AHR) during asthma.
Chitinases such as Ym1/Ym2 and AMCase showed a significant increase in their expression by UFP introduction. The role of AMCase in allergic airway inflammation in the BALF has been suggested by Zhu et al. [29], showing that AMCase is induced via a Th2-specific, IL-13-mediated pathway and expressed in exaggerated quantities in human asthma. AMCase may be a mediator of IL-13-induced responses in Th2-dominated disorders such as asthma. Recently, it has been demonstrated that inhibition of AMCase expression by RNA interference could significantly inhibit eosinophilic inflammation, OVA-specific IgE production, and AHR in the classical murine asthma model [30]. Ym1 and Ym2 synthesized by activated macrophages are homologous to chitinase and have chitinase activity [31, 32]. Ym1 is a highly induced IL-4 and IL-13 target gene in multiple macrophage populations. The multiple signal transducers and activators of transcription 6 (STAT6) response elements in the Ym1 promoter function in a combinatorial manner to mediate transcriptional responses to IL-4 [26]. Deletion of STAT6 [33] or the blockade of both IL-4 and IL-13 activity [34, 35] has been observed to prevent eosinophil accumulation and airway hyper-reactivity in murine models of allergy. Chitinases may have a direct protective role against allergens, but also may function to amplify inflammatory responses against chitin or other allergens [36].
Proteins involved in the immune system, PIGR and C3 complement, also demonstrated significant increase after UFP introduction. PIGR plays a major role in reacting with antigens over the huge area of mucosal surfaces that comprise the digestive, respiratory and urinogenital tracts [37, 38]. Analysis of regulatory regions within and flanking the human PIGR gene has identified several transcription factor-binding sites that mediate transcriptional responses to pro-inflammatory cytokines such as IFN-γ, TNF, IL-1, and IL-4. It has been shown that an IL-4-inducible STAT6 site is located in intron 1 of the human PIGR gene [39]. Moreover, IL-4 by itself and in cooperation with IFN-γ has been shown to up-regulate PIGR mRNA and protein levels in human intestinal and respiratory epithelial cell lines [40]. As for C3 complement, it has been shown that complement C3-deficient mice have reduced inflammation of asthmatic airways [41, 42]. The C3 gene is a large gene consisting of 41 exons, and contains hundreds of SNPs; Inoue et al. suggested that an SNP located in intron 31 of the C3 gene is associated with adult bronchial asthma [43]. Compared with wild-type mice, C3-deficient mice also exhibited diminished airway hyperresponsiveness and lung eosinophilia when challenged with an allergen [44]. They also showed decreased numbers of IL-4-producing lung cells and decreased serum-antigen-specific immunoglobulin E (IgE) levels. The complement system also plays an important role in mediating tissue injury after oxidative stress. It was reported that the lectin complement pathway mediates complement activation after tissue oxidative stress [45]. Nrf2-knockout mice are prone to develop C3 deposition in their kidneys, livers, hearts, and brains [46]. Nrf2 is a transcriptional activator essential for the coordinate transcriptional induction of antioxidant enzymes, suggesting that nrf2 is one of the genes regulating C3 expression under oxidative stress.
Taken together, all of the above proteins are reported to be involved in certain immune response signaling pathways, such as IL13, IFNγ, IL12 and IL4. Their expression clearly showed correlation with UFP induction. Further study on the regulation of these proteins’ expression will provide the immunological basis for PM adjuvant effects.
Also in our studies, NGAL was found with enhanced mRNA and protein levels in lung and BALF of OVA-challenged mice, with the OVA/UFP treatment inducing more significant increases compared OVA alone. NGAL is expressed and secreted by immune cells, hepatocytes, and rental tubular cells in various pathologic states. NGAL was first isolated as a 25 kDa glycoprotein covalently bound with matrix metalloproteinase-9 (MMP-9) in human neutrophils [47]. An absence of MMP-2, -8, and -9 activity is associated with altered lung inflammation during allergic sensitization. TIMP-1 KO mice, an inhibitor of MMPs, developed an asthma phenotype with enhanced Th2 cytokine mRNA and protein expression [48]. It was also reported that NGAL expression was induced under oxidative stress conditions and acted as a protective factor against H2O2 toxicity. Up-regulation of NGAL expression after H2O2 treatment was canceled by the addition of anti-oxidants, dimethylsulfoxide, or cysteamine [49, 50]. Although there is no previous report of a direct correlation between NGAL and asthma, our results suggests that NGAL may play a role in modulating allergic inflammation in lung via interaction with MMP-9 and TIMP-1. NGAL may potentially amend the toxicity induced by UFP and could be a useful biomarker to identify oxidative stress in the respiratory track.
5 Concluding remarks
Our knowledge about mechanisms underlying the pathogenesis of asthma is still limited. The present study investigated alterations in global protein expressions in bronchoalveolar lavage fluid in allergic airway inflammation induced by ambient PM as an adjuvant for sensitization to OVA using a proteomics approach. Because we have shown previously that asthma (in a mouse model) involves oxidative stress and we have also shown that ambient UFP mediates its effect via ROS generation [10, 18], it is logical to hypothesize that the BAL proteome using UFP as adjuvant should reveal oxidative stress proteins. Thus we attempted to compare its protein profile to the profile measured from the standard asthma model with alum. Seven proteins were significantly altered, including surfactant protein-D, polymeric immunoglobulin receptor, complement C3, neutrophil gelatinase-associated lipocalin and a family of chitinases, including chitinase-3-like protein 3, chitinase-3-like protein 4, and acidic mammalian chitinase. Among them, Ym1, Ym2, acidic mammalian chitinase, PIGR, complement C3 and NGAL demonstrated significantly enhanced up-regulation by UFP with a polycyclic aromatic hydrocarbon (PAH) content and a higher oxidant potential. We demonstrated for the first time that the expression of NGAL is associated with allergic airway inflammation. The proteins identified in this study potentially should be useful for studying the mechanisms of the adjuvant effect of ambient PM on allergic sensitization related to air pollution. Thus, we suggest that proteomic profiling of BALF may be used as an effective approach to screen the adjuvant effect of pro-oxidative PM.
Acknowledgments
The research was supported by United States Public Health service grants (NIH), R01 ES012053 and R01 ES013432, as well as the NIAID-funded UCLA AADCRC (U19 AI070453) and the U.S. Environmental Protection Agency (EPA) Science to Achieve Results award (RD-83241301) to the Southern California Particle Center. JAL acknowledges also support from the W. M. Keck Foundation for the establishment of the UCLA Functional Proteomics Center.
Abbreviations
- PM
particulate matter
- ROS
reactive oxygen species
- UFP
ultrafine particles
- BALF
bronchoalveolar lavage fluid
- PIGR
polymeric immunoglobulin receptor (PIGR)
- NGAL
neutrophil gelatinase-associated lipocalin
- Ym1
chitinase-3-like protein 3
- Ym2
chitinase-3-like protein 4
- AMCase
acidic mammalian chitinase
- PAH
polycyclic aromatic hydrocarbon
- SP-D
pulmonary surfactant-associated protein D
- FIZZ1
found in inflammatory 1
- QTOF
quadrupole time-of-flight
References
- 1.Alexis N, Barnes C, Bernstein IL, Bernstein JA, et al. Health effects of air pollution. J Allergy Clin Immunol. 2004;114:1116–1123. doi: 10.1016/j.jaci.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 2.Diaz-Sanchez D, Tsien A, Fleming J, Saxon A. Combined diesel exhaust particulate and ragweed allergen challenge markedly enhances human in vivo nasal ragweed-specific IgE and skews cytokine production to a T helper cell 2-type pattern. J Immunol. 1997;158:2406–2413. [PubMed] [Google Scholar]
- 3.Gilliland FD, Li YF, Saxon A, Diaz-Sanchez D. Effect of glutathione-S-transferase M1 and P1 genotypes on xenobiotic enhancement of allergic responses: randomised, placebo-controlled crossover study. Lancet. 2004;363:119–125. doi: 10.1016/S0140-6736(03)15262-2. [DOI] [PubMed] [Google Scholar]
- 4.Kleinman MT, Sioutas C, Froines JR, Fanning E, et al. Inhalation of concentrated ambient particulate matter near a heavily trafficked road stimulates antigen-induced airway responses in mice. Inhal Toxicol. 2007;19(Suppl. 1):117–126. doi: 10.1080/08958370701495345. [DOI] [PubMed] [Google Scholar]
- 5.Matsumoto A, Hiramatsu K, Li Y, Azuma A, et al. Repeated exposure to low-dose diesel exhaust after allergen challenge exaggerates asthmatic responses in mice. Clin Immunol. 2006;121:227–235. doi: 10.1016/j.clim.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Steerenberg PA, van Dalen WJ, Withagen CE, Dormans JA, van Loveren H. Optimization of route of administration for coexposure to ovalbumin and particle matter to induce adjuvant activity in respiratory allergy in the mouse. Inhal Toxicol. 2003;15:1309–1325. doi: 10.1080/08958370390241786. [DOI] [PubMed] [Google Scholar]
- 7.Stevens PT, Kicic A, Sutanto EN, Knight DA, Stick SM. Dysregulated repair in asthmatic paediatric airway epithelial cells: the role of plasminogen activator inhibitor-1. Clin Exp Allergy. 2008;38:1901–1910. doi: 10.1111/j.1365-2222.2008.03093.x. [DOI] [PubMed] [Google Scholar]
- 8.Whitekus MJ, Li N, Zhang M, Wang M, et al. Thiol antioxidants inhibit the adjuvant effects of aerosolized diesel exhaust particles in a murine model for ovalbumin sensitization. J Immunol. 2002;168:2560–2567. doi: 10.4049/jimmunol.168.5.2560. [DOI] [PubMed] [Google Scholar]
- 9.Nel A. Air pollution-related illness: effects of particles. Science. 2005;308:804–806. doi: 10.1126/science.1108752. [DOI] [PubMed] [Google Scholar]
- 10.Li N, Hao M, Phalen RF, Hinds WC, Nel AE. Particulate air pollutants and asthma. A paradigm for the role of oxidative stress in PM-induced adverse health effects. Clin Immunol. 2003;109:250–265. doi: 10.1016/j.clim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Wang M, Xiao GG, Li N, Xie Y, et al. Use of a fluorescent phosphoprotein dye to characterize oxidative stress-induced signaling pathway components in macrophage and epithelial cultures exposed to diesel exhaust particle chemicals. Electrophoresis. 2005;26:2092–2108. doi: 10.1002/elps.200410428. [DOI] [PubMed] [Google Scholar]
- 12.Xiao GG, Nel AE, Loo JA. Nitrotyrosine-modified proteins and oxidative stress induced by diesel exhaust particles. Electrophoresis. 2005;26:280–292. doi: 10.1002/elps.200406145. [DOI] [PubMed] [Google Scholar]
- 13.Xiao GG, Wang M, Li N, Loo JA, Nel AE. Use of proteomics to demonstrate a hierarchical oxidative stress response to diesel exhaust particle chemicals in a macrophage cell line. J Biol Chem. 2003;278:50781–50790. doi: 10.1074/jbc.M306423200. [DOI] [PubMed] [Google Scholar]
- 14.Jung EJ, Avliyakulov NK, Boontheung P, Loo JA, Nel AE. Pro-oxidative DEP chemicals induce heat shock proteins and an unfolding protein response in a bronchial epithelial cell line as determined by DIGE analysis. Proteomics. 2007;7:3906–3918. doi: 10.1002/pmic.200700377. [DOI] [PubMed] [Google Scholar]
- 15.Chan RC-F, Wang M, Li N, Yanagawa Y, et al. Pro-oxidative diesel exhaust particle chemicals inhibit LPS-induced dendritic cell responses involved in T-helper differentiation. J Allergy Clin Immunol. 2006;118:455–465. doi: 10.1016/j.jaci.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Porter M, Karp M, Killedar S, Bauer SM, et al. Diesel-enriched particulate matter functionally activates human dendritic cells. Am J Respir Cell Mol Biol. 2007;37:706–719. doi: 10.1165/rcmb.2007-0199OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Wang M, Kang X, Boontheung P, et al. Oxidative Stress and Asthma: Proteome Analysis of Chitinase-like Proteins and FIZZ1 in Lung Tissue and Bronchoalveolar Lavage Fluid. J Proteome Res. 2009;9:1631–1638. doi: 10.1021/pr800685h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li N, Wang M, Bramble LA, Schmitz DA, et al. The adjuvant effect of ambient particulate matter is closely reflected by the particulate oxidant potential. Environ Health Perspect. 2009;117:1116–1123. doi: 10.1289/ehp.0800319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li N, Kim S, Wang M, Froines J, et al. Use of a stratified oxidative stress model to study the biological effects of ambient concentrated and diesel exhaust particulate matter. Inhal Toxicol. 2002;14:459–486. doi: 10.1080/089583701753678571. [DOI] [PubMed] [Google Scholar]
- 20.Hao M, Comier S, Wang M, Lee JJ, Nel A. Diesel exhaust particles exert acute effects on airway inflammation and function in murine allergen provocation models. J Allergy Clin Immunol. 2003;112:905–914. doi: 10.1016/j.jaci.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Ramagli LS. Quantifying protein in 2-D PAGE solubilization buffers. Methods Mol Biol. 1999;112:99–103. doi: 10.1385/1-59259-584-7:99. [DOI] [PubMed] [Google Scholar]
- 22.Pfaffl MW. Quantification strategies in real-time PCR. IUL Biotechnol Ser. 2004;5:87–120. [Google Scholar]
- 23.Tichopad A, Dilger M, Schwarz G, Pfaffl MW. Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucl Acids Res. 2003;31:e122/121–e122/126. doi: 10.1093/nar/gng122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexander JW, Ogle CK, Stinnett JD, Macmillan BG. A sequential, prospective analysis of immunologic abnormalities and infection following severe thermal injury. Ann Surg. 1978;188:809–816. doi: 10.1097/00000658-197812000-00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu S, Venge P. Lipocalins as biochemical markers of disease. Biochim Biophys Acta. 2000;1482:298–307. doi: 10.1016/s0167-4838(00)00163-1. [DOI] [PubMed] [Google Scholar]
- 26.Welch JS, Escoubet-Lozach L, Sykes DB, Liddiard K, et al. TH2 cytokines and allergic challenge induce Ym1 expression in macrophages by a STAT6-dependent mechanism. J Biol Chem. 2002;277:42821–42829. doi: 10.1074/jbc.M205873200. [DOI] [PubMed] [Google Scholar]
- 27.Crouch E, Wright JR. Surfactant proteins A and D and pulmonary host defense. Ann Rev Physiol. 2001;63:521–554. doi: 10.1146/annurev.physiol.63.1.521. [DOI] [PubMed] [Google Scholar]
- 28.Hamid Q, Tulic M. Immunobiology of asthma. Ann Rev Physiol. 2009;71:489–507. doi: 10.1146/annurev.physiol.010908.163200. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Z, Zheng T, Homer RJ, Kim YK, et al. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 30.Yang CJ, Liu YK, Liu CL, Shen CN, et al. Inhibition of Acidic Mammalian Chitinase by RNA Interference Suppresses Ovalbumin-Sensitized Allergic Asthma. Human Gene Therapy. 2009;20:1–10. doi: 10.1089/hum.2008.092. [DOI] [PubMed] [Google Scholar]
- 31.Jin HM, Copeland NG, Gilbert DJ, Jenkins NA, et al. Genetic characterization of the murine Ym1 gene and identification of a cluster of highly homologous genes. Genomics. 1998;1:316–322. doi: 10.1006/geno.1998.5593. [DOI] [PubMed] [Google Scholar]
- 32.Sun YJ, Chang NC, Hung SI, Chang AC, et al. The crystal structure of a novel mammalian lectin, Ym1, suggests a saccharide binding site. J Biol Chem. 2001;276:17507–17514. doi: 10.1074/jbc.M010416200. [DOI] [PubMed] [Google Scholar]
- 33.Akimoto T, Numata F, Tamura M, Takata Y, et al. Abrogation of bronchial eosinophilic inflammation and airway hyperreactivity in signal transducers and activators of transcription (STAT)6-deficient mice. J Exp Med. 1998;187:1537–1542. doi: 10.1084/jem.187.9.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomkinson A, Duez C, Cieslewicz G, Pratt JC, et al. A murine IL-4 receptor antagonist that inhibits IL-4- and IL-13-induced responses prevents antigen-induced airway eosinophilia and airway hyperresponsiveness. J Immunol. 2001;166:5792–5800. doi: 10.4049/jimmunol.166.9.5792. [DOI] [PubMed] [Google Scholar]
- 35.Webb DC, McKenzie AN, Koskinen AM, Yang M, et al. Integrated signals between IL-13, IL-4, and IL-5 regulate airways hyperreactivity. J Immunol. 2000;165:108–113. doi: 10.4049/jimmunol.165.1.108. [DOI] [PubMed] [Google Scholar]
- 36.Lee CG. Chitin, chitinases and chitinase-like proteins in allergic inflammation and tissue remodeling. Yonsei Med J. 2009;50:22–30. doi: 10.3349/ymj.2009.50.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandtzaeg P, Johansen FE. Mucosal B cells: phenotypic characteristics, transcriptional regulation, and homing properties. Immunol Rev. 2005;206:32–63. doi: 10.1111/j.0105-2896.2005.00283.x. [DOI] [PubMed] [Google Scholar]
- 38.Kaetzel CS. The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunol Rev. 2005;206:83–99. doi: 10.1111/j.0105-2896.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- 39.Schjerven H, Brandtzaeg P, Johansen FE. Mechanism of IL-4-mediated up-regulation of the polymeric Ig receptor: role of STAT6 in cell type-specific delayed transcriptional response. J Immunol. 2000;165:3898–3906. doi: 10.4049/jimmunol.165.7.3898. [DOI] [PubMed] [Google Scholar]
- 40.Denning GM. IL-4 and IFN-gamma synergistically increase total polymeric IgA receptor levels in human intestinal epithelial cells. Role of protein tyrosine kinases. J Immunol. 1996;156:4807–4814. [PubMed] [Google Scholar]
- 41.Barrington R, Zhang M, Fischer M, Carroll MC. The role of complement in inflammation and adaptive immunity. Immunol Rev. 2001;180:5–15. doi: 10.1034/j.1600-065x.2001.1800101.x. [DOI] [PubMed] [Google Scholar]
- 42.Köhl J. Anaphylatoxins and infectious and non-infectious inflammatory diseases. Mol Immunol. 2001;38:175–187. doi: 10.1016/s0161-5890(01)00041-4. [DOI] [PubMed] [Google Scholar]
- 43.Inoue H, Mashimo Y, Funamizu M, Shimojo N, et al. Association study of the C3 gene with adult and childhood asthma. J Hum Genet. 2008;53:728–738. doi: 10.1007/s10038-008-0304-0. [DOI] [PubMed] [Google Scholar]
- 44.Drouin SM, Corry DB, Kildsgaard J, Wetsel RA. Cutting edge: the absence of C3 demonstrates a role for complement in Th2 effector functions in a murine model of pulmonary allergy. J Immunol. 2001;167:4141–4145. doi: 10.4049/jimmunol.167.8.4141. [DOI] [PubMed] [Google Scholar]
- 45.Collard CD, Väkevä A, Morrissey MA, Agah A, et al. Complement activation after oxidative stress: role of the lectin complement pathway. Am J Pathol. 2000;156:1549. doi: 10.1016/S0002-9440(10)65026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Stein TD, Johnson JA. Genetic dissection of systemic autoimmune disease in Nrf2-deficient mice. Physiol Genomics. 2004;18:261–272. doi: 10.1152/physiolgenomics.00209.2003. [DOI] [PubMed] [Google Scholar]
- 47.Triebel S, Bläser J, Reinke H, Tschesche H. A 25 kDa alpha 2-microglobulin-related protein is a component of the 125 kDa form of human gelatinase. FEBS Lett. 1992;314:386–388. doi: 10.1016/0014-5793(92)81511-j. [DOI] [PubMed] [Google Scholar]
- 48.Sands MF, Ohtake PJ, Mahajan SD, Takyar SS, et al. Tissue inhibitor of metalloproteinase-1 modulates allergic lung inflammation in murine asthma. Clin Immunol. 2009;130:186–198. doi: 10.1016/j.clim.2008.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roudkenar MH, Halabian R, Ghasemipour Z, Roushandeh AM, et al. Neutrophil gelatinase-associated lipocalin acts as a protective factor against H2O2 toxicity. Arch Med Res. 2008;39:560–566. doi: 10.1016/j.arcmed.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 50.Roudkenar MH, Kuwahara Y, Baba T, Roushandeh AM, et al. Oxidative stress induced lipocalin 2 gene expression: addressing its expression under the harmful conditions. J Radiat Res (Tokyo) 2007;48:39–44. doi: 10.1269/jrr.06057. [DOI] [PubMed] [Google Scholar]


