Abstract
The mechanisms of pyridoxal 5′-phosphate (PLP) dependent enzymes require substrates to form covalent “external aldimine” intermediates, which absorb light strongly between 410 nm and 430 nm. Aspartate aminotransferase (AAT) is a prototypical PLP dependent enzyme that catalyzes the reversible interconversion of aspartate and α-ketoglutarate with oxalacetate and glutamate. From kinetic isotope effects, it is known that deprotonation of the aspartate external aldimine Cα-H bond to give a carbanionic quinonoid intermediate is partially rate limiting in the thermal AAT reaction. We show that excitation of the 430 nm external aldimine absorption band increases the steady-state catalytic activity of AAT, which is attributed to the photoenhancement of Cα-H deprotonation based on studies with Schiff bases in solution. Blue light (250 mW) illumination gives an observed 2.3-fold rate enhancement for WT AAT activity, a 530-fold enhancement for the inactive K258A mutant, and a 58,600-fold enhancement for the PLP-Asp Schiff base in water. These different levels of enhancement correlate with the intrinsic reactivities of the Cα-H bond in the different environments, with the less reactive Schiff bases exhibiting greater enhancement. Time-resolved spectroscopy, ranging from femtoseconds to minutes, was used to investigate the nature of the photoactivation of Cα-H bond cleavage in PLP-amino acid Schiff bases both in water and bound to AAT. Unlike the thermal pathway, the photoactivation pathway involves a triplet state with a Cα-H pKa that is estimated to be between 11 and 19 units lower than the ground state for the PLP-Val Schiff base in water.
I. Introduction
Although light-activated enzymes such as DNA photolyase1 and protochlorophyllide oxidoreductase (POR)2 exist, most enzymes do not require the absorption of light for catalytic activity. However, many enzymes do require cofactors (e.g. hemes, flavins, metal centers, etc.) that absorb light in the ultraviolet (UV) and visible regions of the spectrum. In some cases, light can initiate biological activity. For example, CO dissociation can be initiated in hemoglobin and myoglobin by blue light absorption,3-5 as can Co-C bond cleavage in adenosyl cobalamin (vitamin B12).6 This raises the question of whether light excitation can generally affect the catalytic activity of chromophoric enzymes by accelerating cofactor-dependent rate-limiting steps. If so, what molecular mechanisms can couple light absorption to enzymatic activity, can light enhancement be a useful tool to explore enzymatic mechanisms, and what broader biological role might this play for organisms (e.g., plants and bacteria) that grow under solar radiation?
Pyridoxal 5′-phosphate (PLP; vitamin B6) is a chromophoric cofactor required for catalytic activity by a wide variety of enzymes.7,8 It typically exhibits absorption bands at ~430, ~360, and ~330 nm with extinction coefficients of ~5000 M−1 cm−1. While PLP enzymes are thermally activated in vivo, it has been reported that some PLP enzymes can be activated by UV light.8-11 Previous studies9 with aspartate aminotransferase (AAT) have suggested that the carbanionic quinonoid intermediate, which is on the thermal reaction pathway of most PLP enzymes, is photogenerated by UV laser excitation. 5-Hydroxytryptophan decarboxylase, which produces serotonin, has also been reported to have increased steady-state catalytic activity when exposed to light.10 To date, no detailed mechanism has been proffered for these interesting observations.
AAT catalyzes the reversible reaction of L-aspartate with α-ketoglutarate to give oxaloacetate and L-glutamate via two independent half-reactions (eq. 1).7,8,12
| (1a) |
| (1b) |
AAT is central to nitrogen metabolism in all living systems, and is a component of the malate/aspartate shuttle that ushers reducing equivalents into mitochondria of eukaryotes.8 There is a large, informative body of literature on AAT, making it a useful prototype for fundamental studies on this class of enzymes, including light activation.7-9,13,14
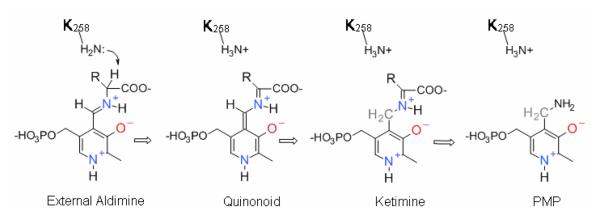
The reaction mechanism of AAT begins with PLP bound as a Schiff base with K258 forming the “internal aldimine”. Amino acid substrates (i.e., L-aspartate or L-glutamate) bind and form the covalent “external aldimine” (Scheme 1) by displacing K258 from the internal aldimine. It is generally thought that the L-Asp external aldimine is deprotonated at Cα to form the carbanionic quinonoid intermediate, which is then protonated at C4' of the coenzyme to give the oxalacetate ketimine intermediate. The ketimine is then hydrolyzed to give the pyridoxamine 5′-phosphate (PMP) enzyme form and free oxalacetate. The second half-reaction converts E-PMP back to E-PLP using α-ketoglutarate as the second substrate, generating L-glutamate as the second product, through the reverse of this sequence of steps.15,16
Scheme 1.
Mechanism of the transamination half-reaction from the external aldimine intermediate onward.
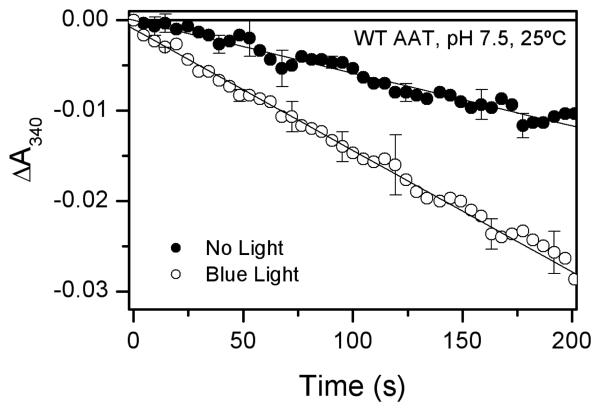
It is demonstrated here that blue light (440 nm) increases the overall catalytic activity of wild-type AAT 2.3 ± 0.3 fold (Figure 2). In the K258A mutant, where the lysine general base catalyst is absent causing a dramatic reduction in activity under thermal conditions, catalysis is increased 530 ± 15 fold at the same excitation power (250 mW), and the rate constant for deprotonation of the L-Asp Schiff base in water is increased 58,600 ± 200 fold. The kinetics of the light-induced processes are characterized with time-resolved transient absorption spectroscopic techniques that span a time scale from femtoseconds to minutes. A detailed mechanism is proposed, involving formation of a reactive triplet state in which weakening of the Cα-H bond facilitates rapid deprotonation. In the enzyme, the light-induced deprotonation produces a higher steady-state concentration of the quinonoid intermediate than in the thermal reaction, resulting in an increase in the overall reaction rate.
Figure 2.
AAT catalytic activity measured in the dark (closed circles) and under 250 mW of 440-nm irradiation (open circles). Conditions: 20 nM AAT, 10 × KM Asp and 5 × KM α-ketoglutarate, pH 7.5, 25 °C. Error bars represent one standard deviation calculated from three trials. For clarity, only every other data point is shown.
Finally, analysis of the dependence of AAT catalytic activity on excitation power provides insight into the thermal reaction pathway. Although the quinonoid intermediate is considered central to most PLP catalyzed mechanisms, its existence in AAT has been debated. The present work provides support for the generally accepted stepwise mechanism, in which the quinonoid is an obligatory intermediate, rather than an alternative concerted mechanism in which Cα-H deprotonation and C4' protonation occur simultaneously.16,17
II. Materials and Methods
All reagents were obtained from Fischer Scientific in the highest quality available and used without further purification unless otherwise noted. HEPES (0.1 M, adjusted with NaOH) buffer was used in the appropriate pH range. PLP-Schiff base samples were prepared as previously described.18 Based on the known equilibrium constants and pKa values for the PLP-Asp Schiff base in solution,8,19 it is difficult to form a homogeneous ionization-state population near neutral pH. In contrast, PLP-Val has a more favorable equilibrium constant with fewer protonation states, allowing for the generation of samples of homogenous populations for study. Therefore, in several experiments PLP-Val was used rather than PLP-Asp. The samples were purged with N2 (30 min per mL of sample) prior to study. KI (1 M) was added as a triplet quencher to some samples and KCl was used at the same concentration to control for ionic strength effects. HPLC was performed with an Agilent 1100 instrument using a Supelco Supelcosil C-18 column (4.6 × 250 mm, 5 μm). Product separation employed an isocratic elution with 2% CH3CN/0.1 M potassium phosphate buffer pH 7.5 and a flow rate of 0.5 mL/min with absorbance monitored at 330 nm.
WT and K258A AAT were expressed and purified as previously described.20,21 The steady-state transamination of l-Asp and α-ketoglutarate catalyzed by WT AAT was monitored by coupling the production of oxaloacetate (OAA) to malate dehydrogenase (MDH), which converts OAA to malate with concurrent oxidation of NADH.21 Samples were maintained at 25 °C via a temperature-controlled water jacket. Steady-state rate measurements were made with an Ocean Optics S2000 spectrometer. The enzyme assay was conducted at pH 7.5, 100 mM HEPES with 100 mM KCl, 150 μM NADH, 20 nM AAT, 10 × KM Asp and 5 × KM α-ketoglutarate, and 10 units/mL MDH. As a control, the light dependence of MDH activity was measured using 25 mM OAA in MHP buffer pH 7.5 with 100 mM KCl; no light-induced change in activity was observed.22 The K258A mutant AAT transamination half-reaction was monitored by the spectral changes in the coenzyme: the K258A-PLP external aldimine with L-Asp (λmax = 430 nm) generates K258A-PMP (λmax = 335 nm).21 The reaction was conducted under saturating conditions, with L-Asp in 100-fold molar excess over K258A-PLP in 100 mM HEPES buffer pH 8.0 and 100 mM KCl. The half-reaction maximal rate constant, kmax, was obtained by fitting a single exponential to the 430 nm absorbance decay.
The light dependence of the activity of glutathione reductase, which uses the blue-absorbing chromophore FAD for activity, was also measured using the same experimental setup. The enzyme-bound FAD, in both the oxidized and reduced forms, has strong absorption in the 440 nm region similar to PLP. The reduction of oxidized glutathione by NADPH was monitored at 340 nm. Reactions were performed at 25 °C in 100 mM potassium phosphate buffer pH 7.6 with 3 mM oxidized glutathione, 150 μM NADPH and 0.3 nM glutathione reductase.
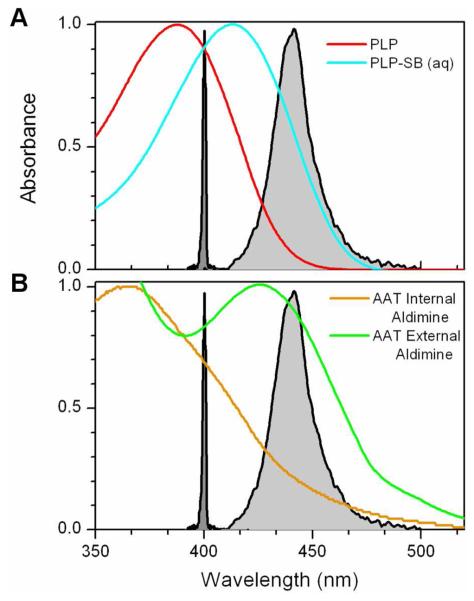
All experiments used a variable-intensity 440 nm LED array (Roithner Labs, Austria L435-66-60-590) as the actinic light source, since it overlaps well with the external aldimine absorption (Figure 1).16 The K258A reactions were also measured with a Varian Cary 50 Bio with an orthogonal xenon light source filtered to FWHM of 25 nm centered at 450 nm. The degree of light enhancement measured with this setup was identical to that from the LED array light source but showed less noise.
Figure 1.
Comparison of absorption spectra for: A) PLP (red), PLP-amino acid Schiff base in pH 9.0 water (blue); B) Internal aldimine of AAT (orange), external aldimine of AAT with α-methylaspartate (green). Spectra for excitation sources are shown for comparison: 440 nm LED (light grey), 400 nm laser (dark grey).
Dispersed femtosecond transient absorption measurements were conducted as previously described.18,23 Samples were excited with 50 fs, 400 nm pulses produced by frequency doubling the output of an amplified 800 nm Ti:Sapphire laser system. Pump-induced changes in the transmission of a white-light continuum (320-650 nm) were monitored with 50 fs resolution from 50 fs to ~8 ns; measurements on this time scale are referred to in this text as “ultrafast transient absorption.” The Ti:Sapphire laser setup was modified23 to probe dynamics up to 300 μs with 30 ns resolution, and the resulting measurements referred to as “μs transient absorption.” In both transient absorption measurements, the polarization of the probe light was set at the magic angle (54.7°) with respect to the pump polarization. The pump intensity was limited to prevent multi-photon processes.24-26 Multidimensional global fitting27,28 was used to analyze the time- and wavelength-dependent experimental data.
III. Results and Discussion
Photochemistry of Free PLP in Solution
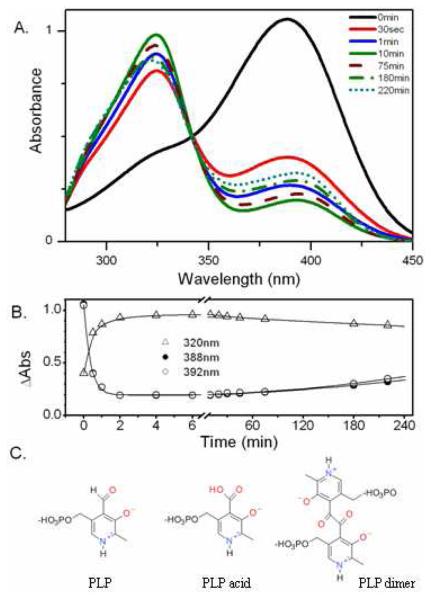
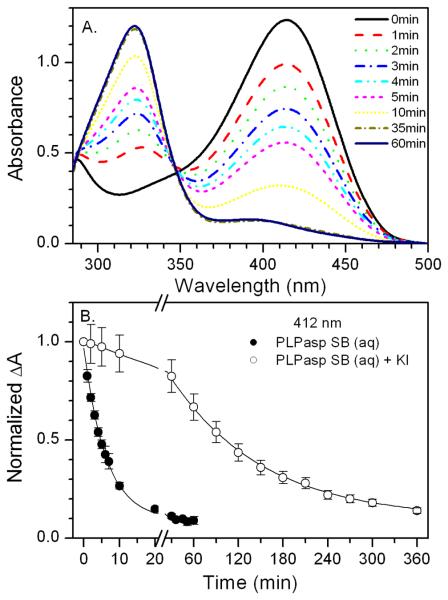
The photochemical reactions of free PLP in water under continuous irradiation were monitored by time-resolved changes in the UV-vis absorbance spectra. At the onset of irradiation (50 mW, 440 nm), the 388 nm band of free PLP decreased (k > 0.03 s−1) with a concurrent increase in absorption at 320 nm (Figure 3A). Within two minutes, essentially all PLP was converted to the oxidized form (pyridoxic acid) identified in previous studies.29-31 Continued irradiation promoted slow formation of a new product (k ≈ 8 × 10−5 s−1, Figure 3A and 3B), which is ascribed to the PLP dimer with known absorption at 392 nm (ε = 5300 M−1cm−1, pH > 9; ε = 700 M−1cm−1, neutral pH).31 These results agree well with experiments in which PLP photoproducts isolated after exposure to UV light revealed that the aldehyde group first oxidizes to the acid before dimerizing (Figure 3C).30,31
Figure 3.
UV-visible absorption spectra of PLP in solution at pH 7.0, 25 °C. A) Under exposure to 50 mW of 440-nm irradiation, solid arrows indicate progression of free PLP to pyridoxic acid; dotted arrows indicate progression from pyridoxic acid to PLP dimer. B) Kinetics showing the slower rise in absorbance at 392 nm (open circles) compared to 388 nm (closed circles) and the decay of 320 nm (triangle). C) Structures of PLP and its dominant photoproducts.
A number of quenching experiments have suggested that free PLP forms a triplet state when excited by UV and blue light.8,18,29,30,32 To determine if the observed PLP photochemistry involves a triplet state, the dynamics were measured in the presence of the triplet quencher KI.32-34 The decay of the 388 nm PLP absorption peak is slower in the presence of KI (k = 0.0051 ± 0.0008 s−1) than in its absence (data not shown). Solutions with KCl at the same concentration exhibit nearly identical kinetics to reactions without added salt, showing that the effect is specific to iodide. These results indicate that the free PLP triplet state is a precursor to pyridoxic acid and the PLP dimer.
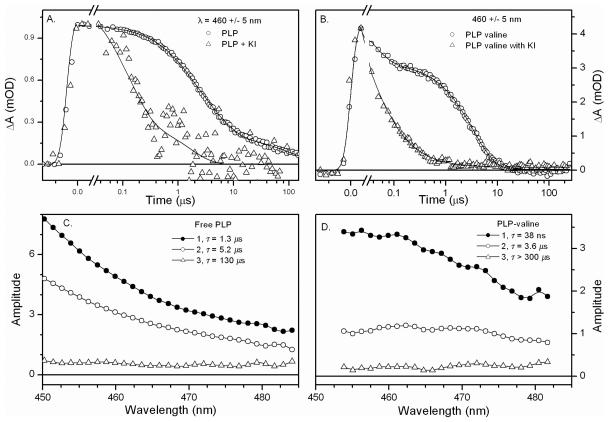
Transient absorption experiments allow characterization of triplet dynamics for free PLP following 400-nm excitation (Figure 4 A & C). The kinetics monitored at 470 nm for free PLP at pH 7.0 are shown in Figure 4A. The data were analyzed using a three exponential, sequential model resulting in Evolution Associated Difference Spectra (EADS)35 (Figure 4C) with the following rate constants: 7.6 ± 1.2 × 105 s−1, 1.9 ± 0.1 × 105 s−1, and 7.7 ± 0.6 × 103 s−1. KI increases the rate constant for the initial process by an order of magnitude (Figure 4A, open triangles) while the two slower rates were unaffected. KCl has no effect at the same concentration. Based on its sensitivity to KI, the first rate constant is assigned to a triplet state, while the latter two phases are assigned to formation and decay of a radical species.8,29,30,32
Figure 4.
Free PLP and PLP-Valine μs-transient absorption signals. A) Kinetics with (triangle) and without (circle) KI at 460 nm for free PLP. B) Kinetics with (triangle) and without (circle) KI at 460 nm for PLP-Valine Schiff base. C) Evolution associated difference spectra (EADS) for free PLP. D) EADS for PLP-Valine.
In previous flash photolysis experiments, it was concluded that the aldehyde form of PLP exhibits an n,π* triplet state that undergoes hydrogen abstraction to form a reactive ketyl radical.36 The triplet state absorbs at 440 nm and exhibits a decay rate constant of 106 s−1, comparable to the rate constant observed here.32,36 The ketyl radical has been reported36 to absorb also at 440 nm with a decay rate constant of 2.4 × 104 s−1, although Ledbetter32 reported a rate constant of 8.3 × 103 s−1 for radical decay after 337 nm excitation of PLP in water at pH 7. The discrepancy in rates could arise from the variation in the PLP forms excited in solution at the two excitation wavelengths. Both the aldehyde and the hydrate forms of PLP in H2O absorb at 337 nm,37 the excitation wavelength used in Ref 36, while only the aldehyde form of PLP absorbs 400 nm.37 Both forms of PLP are expected to undergo triplet and radical formation after 337-nm excitation, though possibly with different kinetics.
Photochemistry of PLP-Amino Acid Schiff Bases in Solution
The photochemistry of PLP-amino acid Schiff bases was also investigated. The ketoenamine tautomer of PLP-amino acid Schiff bases, the dominant form in water,8,37 absorbs maximally at 410 nm. Transamination of the PLP-Schiff bases is observed as a decrease in absorbance at 410 nm, with a concurrent increase at 325 nm attributed to the formation of PMP (see λmax in Figure 5A). The rate of this reaction with PLP-Asp increases 8,400 fold under 50 mW blue light exposure (Figure 5A) and 58,600-fold at 250 mW. The power dependence of the observed rate constants is shown in Supporting Information (Figure S1), as well as data for PLP-Val and PLP-AIB Schiff bases (Figures S2 and S3).
Figure 5.
A) UV-visible absorption spectra of PLP-Asp in solution at pH 9.0, 25 °C under exposure to 50 mW of 440 nm excitation. B) Kinetics at 412 nm with (open circles) and without (closed circles) KI. Error bars indicate one standard deviation calculated from three trials.
PLP-amino acid Schiff base samples irradiated with 50 mW light reach the endpoint for PMP formation in ~30 min. HPLC analysis of the PLP-Asp and PLP-Val light-induced reactions confirmed that PMP is produced (Supporting Information, Figure S4). For the PLP-Asp Schiff base only, further irradiation leads to a photoproduct that absorbs at 392 nm. Since the equilibrium mixture of the PLP-Asp Schiff base has ~37% of the total PLP present as free PLP, this 392 nm absorbing photoproduct is assigned to the PLP dimer that is observed in the reactions of free PLP in the absence of amino acid (Figure 3C).19 Samples with more favorable Schiff base equilibrium constants (e.g., PLP-Val and PLP-AIB) contain much less free PLP, and do not exhibit this photoproduct (Figures S2 and S3).
KI significantly slows the light dependent reaction under otherwise identical conditions (Figure 5B), indicating that a triplet state is involved in the light-induced deprotonation reaction in solution. The changes in rate constants are summarized for three PLP-Schiff base samples in Table 1. Similar results were found for all Schiff bases tested. Like PLP-Asp and PLP-Val, the reaction rate for the PLP-AIB Schiff base in solution is increased by blue light exposure and is sensitive to KI (Figure S3). PLP-AIB Schiff base undergoes decarboxylation but cannot undergo deprotonation. Therefore, these results suggest that the increased reactivity of the triplet state is due to a general weakening of bonds to Cα and not to factors specific to deprotonation. These results are congruent with previous studies.38,39
Table 1.
Rate constants for conversion of PLP-Schiff base samples in water to PMP in the dark (k), in 50 mW of 440-nm light (λk) and in the presence of heavy atoms.
| Sample | k, s−1 | λk, s−1 | λk + KI, s−1 |
|---|---|---|---|
| PLP-Asp | 6.4 ± 0.1 × 10−7 | 5.4 ± 0.8 × 10−3 | 1.3 ± 0.1 × 10−4 |
| PLP-Val | 5.7 ± 0.1 × 10 −7 | 1.0 ± 0.2 × 10 −3 | 2.9 ± 0.3 × 10 −4 |
| PLP-AIB | 3.9 ± 0.2 × 10 −7 | 3.7 ± 0.7 × 10 −4 | -- |
The μs-transient absorption following 400 nm excitation of the PLP-Val Schiff base at pH 7.5 exhibited kinetics with multi-exponential decay similar to free PLP (Figure 4B & D). Global fitting to a sequential model with formation of a persistent photoproduct that did not decay within the 300 μs duration of the experiment (Figure 4D) identifies two rate constants: 3.2 ± 0.4 × 107 s−1 and 2.9 ± 0.1 × 105 s−1. When KI was added, the second rate constant was significantly reduced (Figure 4B) and these data can be fitted to a single exponential with a rate constant of 2.7 ± 0.2 × 106 s−1 and an offset of 0.07 ± 0.02 (Figure S6). The sensitivity of the reaction rate to KI supports involvement of a triplet in reactions of PLP Schiff bases. The EADS extracted for PLP-Val are compared in Figure 4D. Within error, there is no significant evolution of the transient spectrum (Figure S5), suggesting that transient spectrum corresponds to a single species. In comparison with the spectra obtained from the free PLP μs kinetic data (Figure 4C), PLP-Val has a greater absorbance at 475 nm, a wavelength commonly reported as the absorbance maximum for the quinonoid intermediate in solution.40,41 This species is therefore assigned as the ground state quinonoid, though the limited range of the experimental data does not rule out the possibility of a non-equilibrium precursor to quinonoid. The observed rate constant of 2.9 ± 0.1 × 105 s−1 includes both the photochemical reaction that ultimately yields quinonoid and intersystem crossing back to the singlet ground state of the external aldimine. Based on additional light-dependent studies (unpublished data), we estimate a value of at most 0.64 for the triplet to quinonoid yield, corresponding to a rate of k ~1.9 × 105 s−1. This rate sets an upper bound on the rate of deprotonation.
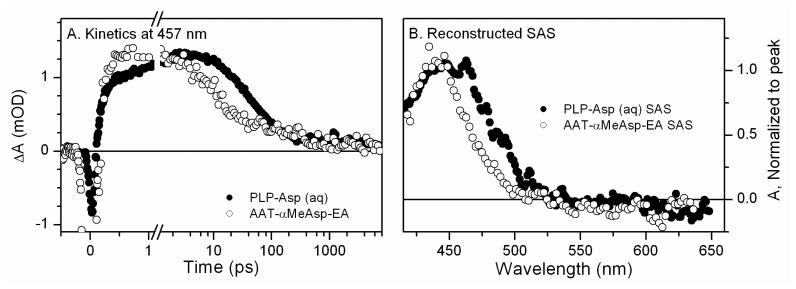
The primary photodynamics of triplet state formation for the Schiff bases was examined in ultrafast (fs-ns) experiments. The kinetics of the PLP-Asp Schiff base at 457 nm (Figure 6A) are similar to those obtained in previous ultrafast studies of PLP-Val in solution.18 An initial photoproduct with an absorption maximum at 450 nm is formed within 1 ns (Figure 6B). In the previous work, this species was incorrectly assigned to the quinonoid intermediate.18 The KI quenching results reported above instead support triplet assignment for both samples. The pure spectrum for this species was constructed by decomposing the 6 ns transient absorption spectrum into ground state bleach and a solvated electron spectrum (Figure S7) generated from multi-photon ionization.26 The triplet is formed within 500 ps with PLP-Asp and 1 ns with PLP-Val.18
Figure 6.
A) Ultrafast kinetics at 457 nm for the AAT external aldimine with α-methylaspartate and the PLP-Asp Schiff base in solution show intersystem crossing on a sub-nanosecond time scale. B) Reconstructed species associated spectra (SAS) were generated by subtracting the ground state and solvated electron spectra from the 6-ns transient absorbance spectrum.
Photochemistry in Enzymes
AAT reactions irradiated with 250 mW of 440 nm light exhibit a 2.3-fold increase in steady-state catalytic activity compared to reactions in the dark (Figure 2). The coupling enzyme, MDH, was not affected by light exposure (Supporting Information, Figure S8). Light enhancement of the K258A mutant of AAT, in which the lysine responsible for Cα-H deprotonation is replaced with alanine,42 was also studied (Figure S9). The half-reaction kmax for K258A in the dark is 7.0 ± 0.2 × 10−6 s−1 for L-Asp at pH 8.0, 25 °C.21 Excitation with 250 mW of 450 nm light increases the rate constant 530-fold. The rate of light enhancement in both WT and K258A AAT is power dependent (Figure S10), as discussed below.
Although photoexcitation of PLP Schiff bases both in solution and on AAT generates PMP, the same photochemical mechanisms are not necessarily in effect for both. Unfortunately, heavy atom studies are inconclusive with enzymes since the reactive species are removed from bulk solvent. However, ultrafast triplet state formation dynamics of PLP-Asp in solution can be directly compared to that obtained with AAT. The ultrafast transient absorption signals of the internal and external aldimines in AAT at pH 8.0 after 400 nm both resulted in long-lived photoproduct formation within 55 ps and 110 ps, respectively (Figure 6A, open circles), that have an absorption maximum at 450 nm (Figure 6B, open circles). Due to the strong resemblance of the long-lived intermediate in the transient spectrum of AAT and PLP-Asp in solution (Figure S11), the population at 6 ns is assigned to a triplet state.18 This assignment suggests that the protein does not significantly alter the intersystem crossing mechanism that leads to the reactive triplet species, and that Schiff bases in both water and AAT follow similar photochemical mechanisms.
An alternative explanation of the light enhancement of the enzymatic reactions observed here is that absorption of a photon simply leads to generation of a vibronically hot chromophore43 that heats the local environment through vibrational relaxation, thereby increasing the rate of the thermal mechanism. This alternative hypothesis was tested by measuring the light dependence of the activity of the FAD dependent enzymes glutathione reductase and diaphorase (Supporting Information). Both enzymes absorb in the 410 – 500 nm region with extinction coefficients similar to PLP enzymes.44 Ultrafast transient absorption investigations on free flavins have shown generation of a triplet state within 1-2 ns, and in numerous light-dependent flavoenzymes, formation of a triplet is necessary to enzymatic activity.45 However, the activities of these two enzymes were not increased by blue light (Figure S12 for glutathione reductase; data not shown for diaphorase). These results suggest that local heating is not the mechanism for the increase in AAT activity.
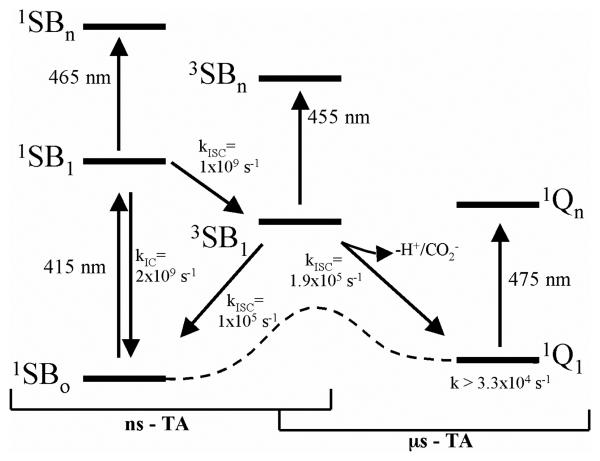
Incorporating the results of both the steady-state and transient absorption measurements, we arrive at a scheme for the primary photochemistry underlying the light dependent formation of PMP from Schiff bases in solution and in AAT (Figure 7). In this scheme, blue light excites the external aldimine from the ground state to an excited singlet state (λmax = 465 nm) that decays within 1 ns (k = 1 × 109 s−1) to an excited triplet state (λmax = 455 nm). The triplet state decays in 3.5 μs (k = 2.9 × 105 s−1) during which Cα-H cleavage generates the carbanionic quinonoid intermediate (λmax = 475 nm) in its ground state. The decay of the absorbing species persisting longer than 300 μs (k = 3.3 × 104 s−1) is assumed to form the ketimine intermediate or revert back to the external aldimine ground state.
Figure 7.
Proposed scheme for PLP Schiff base photodynamics in solution and on AAT. Rate constants refer to PLP-Val in water at pH 7.5, 25 °C. Dashed line schematically represents the thermal reaction.
The light enhancement of catalytic activity is proposed to result from an increase in the rate of deprotonation of the external aldimine to form the quinonoid intermediate due to higher acidity of the Cα-H bond in the triplet state. Therefore, the ΔpKa between the ground and triplet state for the aldimine Cα-H bond is a quantity of fundamental interest to calculate. Pearson and Dillon demonstrated a correlation between the rate constant for deprotonation of carbon acids and their pKa values.46 One can use this correlation to estimate the ΔpK for the aldimine Cα-H bond in the aqueous Schiff bases from the difference in the rate constant for deprotonation. Although the rate constant for quinonoid formation was not obtained in the present experiments, upper and lower bounds on this rate constant are set by those for triplet decay and PMP formation, respectively. The triplet decay rate constant obtained from μs-transient absorption provides an upper bound on the rate constant for deprotonation of 2.9 × 105 s−1 for PLP-Val. The rate constant for deprotonation cannot be slower than that of the light-dependent transamination reaction, which involves both deprotonation and quinonoid reprotonation. Triplet formation is fast (i.e., at equilibrium) compared to product formation under continuous illumination, therefore a pre-equilibrium treatment can be applied. The program COPASI47 was used to simulate, at a fixed excitation power, the steady-state concentration of the triplet state. By dividing the observed rate constant for the light-dependent transamination of PLP-Val (1 × 10−3 s−1; Table 1) by the fraction of the population in the triplet state (0.0008 at 50 mW excitation power), the maximal rate constant for PMP formation and the lower bound on the rate of deprotonation is estimated to be 1.25 s−1. Comparing these estimates for the rate constant for triplet deprotonation to the thermal deprotonation of the PLP-Val ground state (5.7 × 10−7 s−1; Table 1), a decrease of between 11 and 19 units in the pKa of Cα-H in the triplet state of the PLP-Val Schiff base in water is estimated. Previous reports have estimated the Cα-H pKa in the ground state of the PLP-ethylamine Schiff base in water to be ~ 17;8,48-50 a ΔpKa in the range 11-19, would then result in a pKa* between 6 and −2. These estimates of ΔpKa for the aqueous Schiff bases are strikingly large, but are consistent with the experimental pKa* value of less than −2.5 reported for free PLP in solution.51
One can propose that the observed trend in optical enhancement (water > K258A > WT) is due to the greater reactivity of the ground state Schiff base in the active site environment compared to water. Unfortunately, the correlation of Pearson and Dillon is for deprotonation of carbon acids by water, and is therefore inappropriate for AAT, where deprotonation is performed by the K258 amino group in the active site. Although the K258A mutant presumably employs water as the base for Cα-H deprotonation, the steric barrier to water access presented by the protein contributes significantly to the kinetics and precludes use of the correlation (which only includes reactions with insignificant steric effects) in calculating ΔpKa.52 The active site certainly facilitates the thermal reaction by decreasing the energy between the external aldimine and quinonoid intermediates (i.e. by lowering the pKa of the Cα-H bond) compared to reaction in water, and the activation effect of light is mitigated by this intrinsic protein activation.
Power Dependence of kcat and Implications for the Thermal Mechanism
The light enhancement of AAT catalyzed transamination exhibits a clear nonlinear dependence on excitation power (Figure 8). While the rate constant for the microscopic step that is affected by light (i.e., L-Asp external aldimine Cα-H deprotonation) is expected to have a linear dependence on excitation power, two factors contribute to the observed nonlinear dependence of kcat. First, the balance of rate-limiting steps changes at high excitation power, where Cα-H deprotonation is no longer rate-limiting. Second, the electronic transition in the external aldimine optically saturates (discussed below).The first phenomenon is a result of the multistep nature of the enzymatic reaction mechanism. The thermal reaction is only partially rate-limited by Cα-H deprotonation, as judged from kinetic isotope effects16,53 and this partial rate limitation disappears as the rate for the deprotonation step increases with excitation power, with subsequent, light-independent steps becoming rate-limiting. Equation 2 (see Supporting Information for derivation) models the light-dependent increase in kcat, relative to the thermal reaction, for a simplified model of the AAT reaction.
| (2) |
Here, k3 is the net rate constant for conversion of enzyme bound aspartate (which includes Michaelis complex and external aldimine) to the quinonoid, λk3 is the net rate constant for the light-dependent pathway for quinonoid formation, and k4 is the net rate constant for all steps subsequent to quinonoid formation (i.e., reprotonation at C4', ketimine hydrolysis, and the second half-reaction with α-ketoglutarate). The model used here subsumes the equilibrium constant for Michaelis complex-external aldimine interconversion into k3 along with external aldimine deprotonation. This equilibrium constant is known to be near unity.15
Figure 8.
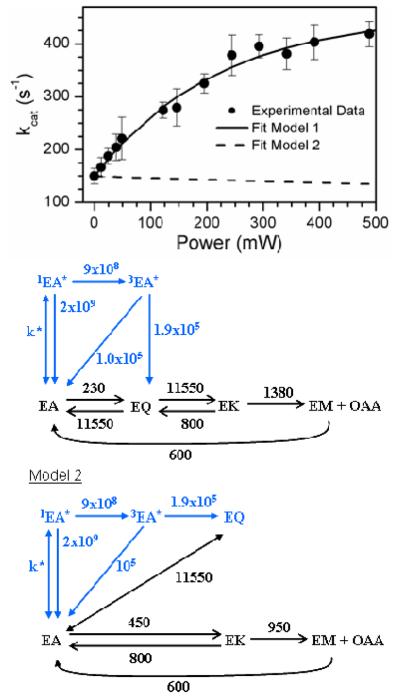
Power dependence of the AAT catalytic activity. Simulations with two kinetic models are shown. Model 1 has the quinonoid on the productive pathway, while model 2 has it off the productive pathway. Error bars indicate one standard deviation calculated from three trials. Conditions: 20 nM AAT, 10 × KM Asp and 5 × KM α-ketoglutarate pH 7.5, 25 °C.
Values for k3 and k4 for the simplified model of the thermal reaction were determined by fitting Equation 2 to the experimental values of kcat as a function of excitation power (Figure 8). To do this, values of λk3 were first calculated for each experimental excitation power, using rate constants obtained directly from the transient absorption experiments reported above, with where B12 is the Einstein B coefficient, I is the irradiance per unit frequency interval in W m−2 Hz−1 and c is the speed of light (see Supporting Information), and assuming a 0.64 yield for quinonoid from triplet. The λk3 values were then used as the independent variable against which the ratio λkcat /kcat was fitted. The fit resulted in values of k3 = 230 ± 45 s−1, and k4 = 425 ± 73 s−1. The value of kcat for the overall thermal reaction calculated from the fitted values of k3 and k4 is 149 ± 23 s−1, in good agreement with the present and literature values.15,16 These results demonstrate the power of selectively perturbing a single step by a known amount in a multistep process such as the AAT reaction with aspartate. Here, it allows one, from purely steady-state rate measurements, to define the value of the thermal rate constant for the perturbed step as well as for subsequent steps.
The rate constants for both K258A and Schiff bases in water saturate at high excitation power due to saturation of the external aldimine optical transition. When the rate of excitation equals the rate of de-excitation due to stimulated emission of the excited state (i.e., ), the steady-state concentration of excited singlet and the triplet states becomes independent of excitation power. Because the singlet excited state is longer lived for K258A (k ~ 1 × 108 s−1; data not shown) compared to Schiff bases in solution or on AAT (k ~ 9 × 108 s−1), the optical transition saturates at a higher excitation power (Figure S13). COPASI simulations of the AAT reaction indicate that optical saturation has a significant effect on kcat only at much higher excitation powers than reported here.
The existence of the quinonoid intermediate on the productive thermal pathway in AAT has been contested in the literature.16,53 The quinonoid is observed at low concentration in equilibrium mixtures of AAT with aspartate or glutamate and their respective α-keto acids.16,40,54 The alternative substrate erythro-β-hydroxyaspartate gives a higher fraction of AAT in the quinonoid state.40,55 The quinonoid has been established as a kinetically competent intermediate in the reaction of AAT with the alternative substrate L-cysteine sulfinate,56 and in numerous other PLP enzymes including Citrobacter freundii tyrosine phenol-lyase57 and dialkylglycine decarboxylase.58 An alternative AAT mechanism was suggested by Goldberg and Kirsch in which the 1,3-prototropic shift from external aldimine to ketimine is concerted, with the quinonoid intermediate off the productive pathway.16 They speculated that the 1,3-prototropic shift proceeds through a nonlinear transition state with simultaneous Cα-H deprotonation and C4' protonation catalyzed by K258.
COPASI47 was used to simulate steady-state kinetic data from which kcat values for AAT catalyzed reactions at various light intensities were calculated. Simulations include the microscopic rate constants in model 1 of Figure 8, which has the quinonoid intermediate on the thermal pathway. Rate constants in black were obtained from the literature15,16 or obtained from the analysis above. The values of 11,550 s−1 and 800 s−1 were estimated from the percentage population present on AAT saturated with both aspartate and oxalacetate (20% external aldimine and 1% quinonoid intermediate).16 The simulated data clearly reproduces the nonlinear power dependence of the observed value of kcat. In contrast, model 2, with the quinonoid off the thermal pathway,16 predicts an inhibition of catalytic activity since photoexcitation would decrease the population on the productive pathway. Thus, the photoenhancement results presented here strongly support the quinonoid being on the productive thermal pathway, as well as the photochemical pathway. Corroborating this, it is also found that 440 nm irradiation increases the population of the quinonoid intermediate in stopped-flow experiments with AAT (unpublished results).
Photoenhancement of the activity of other PLP dependent enzymes (dialkylgycine decarboxylase and alanine racemase) occurs with magnitudes similar to that with AAT (unpublished results). Thus, the acceleration of PLP enzymes by light may well be a general phenomenon. If this is the case, then the light dependence of the activity of PLP dependent enzymes may play a heretofore unrecognized role in the physiology of organisms that live under solar irradiation (e.g. plants, fungi, algae, bacteria). PLP enzymes are ubiquitous in the physiology of all organisms, being central to amino acid metabolism. In E. coli, PLP enzymes constitute ~2% of the proteins encoded in the genome.7 Under the solar standard AM 1.5, approximately 30% of solar spectrum is resonant with the external aldimine absorption band in AAT. If the full resonant flux were absorbed, the results reported here suggest that there would be a generalized 50% increase in the activity of PLP enzymes. Even a fraction of this calculated increase in the activity of such a large number of enzymes could have very significant biological effects. One can speculate the involvement of this phenomenon in the coordination of day vs. night metabolism, blue light sensing, and other regulatory phenomena.
V. Conclusions
Photoexcitation of free PLP, PLP Schiff bases, and PLP bound to AAT leads to the formation of a reactive triplet state. The triplet state lowers the pKa of the Cα-H bond by 11-19 units for Schiff bases in water. The increased acidity of this bond in the triplet state increases the rate of nonenzymatic transamination by 58,600-fold, the rate of transamination catalyzed by the K258A mutant of AAT by 530-fold, and the rate of the AAT reaction by 2.3-fold under 250 mW of 440 nm irradiation. The reaction mechanism of AAT is complex, and the rate of AAT is only partially limited by Cα-H deprotonation,15,16 resulting in a nonlinear dependence of the rate enhancement on light intensity. Selective perturbation of the deprotonation step by light is a powerful tool that allows this individual rate constant in the complex mechanism to be defined from steady-state kinetics only, and demonstrates for AAT that the quinonoid intermediate is on the thermal reaction pathway. Photoinitiation of PLP reactions is possible on the 1-100 μs time scale, with pulsed excitation generating a synchronized population. If other enzymes are found to show increased catalytic activity due to increased reactivity of singlet excited states, then ultrafast photoinitiation of enzymatic reactions that are not intrinsically light dependent could lead to new and interesting insights into enzyme catalysis, in a similar vein to ultrafast studies of ligand binding to heme proteins.4,59
Supplementary Material
ACKNOWLEDGMENTS
This work was supported with a Career Development Award (CDA0016/2007-C) from the Human Frontiers Science Organization (to DSL) and a grant (GM54779) from the National Institutes of Health (to MDT). We thank Dr. Nathan C. Rockwell, Rob Griswold, Justin Foust and Kristin Ziebart for laboratory assistance and constructive discussions.
Footnotes
SUPPORTING INFORMATION AVAILABLE: Additional data are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Sancar A. Chem. Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 2.Heyes DJ, Hunter CN. Trends. Biochem. Sci. 2005;30:642–649. doi: 10.1016/j.tibs.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Cao WX, Ye X, Sjodin T, Christian JF, Demidov AA, Berezhna S, Wang W, Barrick D, Sage JT, Champion PM. Biochemistry. 2004;43:11109–11117. doi: 10.1021/bi049077g. [DOI] [PubMed] [Google Scholar]
- 4.Sage JT, Morikis D, Champion PM. Biochemistry. 1991;30:1227–1237. doi: 10.1021/bi00219a010. [DOI] [PubMed] [Google Scholar]
- 5.Tian WD, Sage JT, Champion PM. J. Mol. Biol. 1993;233:155–166. doi: 10.1006/jmbi.1993.1491. [DOI] [PubMed] [Google Scholar]
- 6.Walker LA, Jarrett JT, Anderson NA, Pullen SH, Matthews RG, Sension RJ. J. Am. Chem. Soc. 1998;120:3597–3603. [Google Scholar]
- 7.Toney MD. Arch. Biochem. Biophys. 2005;433:279–287. doi: 10.1016/j.abb.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 8.Kallen RG, Korpela T, Martell AE, Matsushima Y, Metzler CM, Metzler DE, Morozov YV, Ralston IM, Savin FA, Torchinsky YM, Ueno H. In: Transaminases. Christian P, Metzler DE, editors. John Wiley and Sons; New York: 1985. [Google Scholar]
- 9.Cornish TJ, Ledbetter JW. IEEE J. Quantum Electron. 1984;20:1375–1379. [Google Scholar]
- 10.Fraikin GY, Strakhovskaya MG, Ivanova EV, Rubin AB. Photochem. Photobiol. 1989;49:475–477. doi: 10.1111/j.1751-1097.1989.tb09197.x. [DOI] [PubMed] [Google Scholar]
- 11.Stepuro II, Konovalova NV, Solodunov AA, Tyshchenko AS. Mol. Biol. 1993;27:483–487. [PubMed] [Google Scholar]
- 12.Jenkins WT, Yphantis DA, Sizer IW. J. Bio. Chem. 1959;234:51–57. [PubMed] [Google Scholar]
- 13.Toney MD, Kirsch JF. Protein Sci. 1992;1:107–119. doi: 10.1002/pro.5560010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunathan HC. Proc. Natl. Acad. Sci. U. S. A. 1966;55:712. doi: 10.1073/pnas.55.4.712. &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuramitsu S, Hiromi K, Hayashi H, Morino Y, Kagamiyama H. Biochemistry. 1990;29:5469–5476. doi: 10.1021/bi00475a010. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg JM, Kirsch JF. Biochemistry. 1996;35:5280–5291. doi: 10.1021/bi952138d. [DOI] [PubMed] [Google Scholar]
- 17.Julin DA, Kirsch JF. Biochemistry. 1989;28:3825–3833. doi: 10.1021/bi00435a031. [DOI] [PubMed] [Google Scholar]
- 18.Hill MP, Carroll EC, Toney MD, Larsen DS. J. Phys. Chem. B. 2008;112:5867–5873. doi: 10.1021/jp7117345. [DOI] [PubMed] [Google Scholar]
- 19.Metzler CM, Cahill A, Metzler DE. J. Am. Chem. Soc. 1980;102:6075–6082. [Google Scholar]
- 20.Smith DL, Almo SC, Toney MD, Ringe D. Biochemistry. 1989;28:8161–8167. doi: 10.1021/bi00446a030. [DOI] [PubMed] [Google Scholar]
- 21.Toney MD, Kirsch JF. Biochemistry. 1993;32:1471–1479. doi: 10.1021/bi00057a010. [DOI] [PubMed] [Google Scholar]
- 22.Geck MK, Kirsch JF. Biochemistry. 1999;38:8032–8037. doi: 10.1021/bi983029c. [DOI] [PubMed] [Google Scholar]
- 23.Carroll EC, Hill MP, Madsen D, Malley KR, Larsen DS. Rev. Sci. Instrum. 2009;80:026102–3. doi: 10.1063/1.3070516. [DOI] [PubMed] [Google Scholar]
- 24.Larsen DS, van Stokkum IHM, Vengris M, van der Horst MA, de Weerd FL, Hellingwerf KJ, van Grondelle R. Biophys. J. 2004;87:1858–1872. doi: 10.1529/biophysj.104.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blandame MJ. Chem. Rev. 1970;70:59. [Google Scholar]
- 26.Larsen DS, Vengris M, van Stokkum IHM, van der Horst MA, de Weerd FL, Hellingwerf KJ, van Grondelle R. Biophys. J. 2004;86:2538–2550. doi: 10.1016/S0006-3495(04)74309-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holzwarth AR. In: Biophysical Techniques in Photosynthesis. Amesz J, Hoff AJ, editors. Kluwer; Dordrecht, The Netherlands: 1996. [Google Scholar]
- 28.van Stokkum IHM, Larsen DS, van Grondelle R. Biochimica Et Biophysica Acta-Bioenergetics. 2004;1657:82–104. doi: 10.1016/j.bbabio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Bazhulina NP, Kirpichnikov MP, Morozov YV, Savin FA, Sinyavina LB, Florentiev VL. Molecular Photochemistry. 1974;6:367–396. [Google Scholar]
- 30.Reiber H. Biochim. Biophys. Acta. 1972;279:310. doi: 10.1016/0304-4165(72)90148-1. [DOI] [PubMed] [Google Scholar]
- 31.Morrison AL, Long RF. J. Chem. Soc. 1958:211–215. [Google Scholar]
- 32.Cornish TJ, Ledbetter JW. Photochem. Photobiol. 1985;41:15–19. [Google Scholar]
- 33.McGlynn SP, Azumi T, Kinoshita M. Molecular Spectroscopy of the Triplet State. Vol. 1. Prentice Hall; Englewood Cliffs, N.J.: 1969. [Google Scholar]
- 34.Porter G, Windsor MW. P. Roy. Soc. Lond. A Mat. 1958;245:238–258. [Google Scholar]
- 35.van Stokkum IHM, Larsen DS, van Grondelle R. Biochimica Et Biophysica Acta-Bioenergetics. 2004;1658:262–262. [Google Scholar]
- 36.Ledbetter JW, Schaertel S. J. Photoch. Photobio. B. 1998;47:12–21. [Google Scholar]
- 37.Harris CM, Johnson RJ, Metzler DE. Biochim. Biophys. Acta. 1976;421:181–194. doi: 10.1016/0304-4165(76)90284-1. [DOI] [PubMed] [Google Scholar]
- 38.Kurauchi Y, Ohga K, Morita S, Nagamura T, Matsuo T. Chem. Lett. 1983:349–352. [Google Scholar]
- 39.Kurauchi Y, Ohga K, Yokoyama A, Morita S. Agric. Biol. Chem. 1980;44:2499–2500. [Google Scholar]
- 40.Metzler CM, Harris AG, Metzler DE. Biochemistry. 1988;27:4923–4933. doi: 10.1021/bi00413a050. [DOI] [PubMed] [Google Scholar]
- 41.Zabinski RF, Toney MD. J. Am. Chem. Soc. 2001;123:193–198. doi: 10.1021/ja0026354. [DOI] [PubMed] [Google Scholar]
- 42.Malcolm BA, Kirsch JF. Bioche. Bioph. Res. Co. 1985;132:915–921. doi: 10.1016/0006-291x(85)91894-7. [DOI] [PubMed] [Google Scholar]
- 43.Sibert EL, Reinhardt WP, Hynes JT. J. Chem. Phys. 1984;81:1115–1134. [Google Scholar]
- 44.Bohme CC, Arscott LD, Becker K, Schirmer RH, Williams CH. J. Bio. Chem. 2000;275:37317–37323. doi: 10.1074/jbc.M007695200. [DOI] [PubMed] [Google Scholar]
- 45.Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. J. Bio. Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 46.Pearson RG, Dillon RL. J. Am. Chem. Soc. 1953;75:2439–2443. [Google Scholar]
- 47.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. Bioinformatics. 2006;22:3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 48.Richard JP, Amyes TL, Crugeiras J, Rios A. Curr. Opin. Chem. Biol. 2009;13:475–483. doi: 10.1016/j.cbpa.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weng SH, Leussing DL. J. Am. Chem. Soc. 1983;105:4082–4090. [Google Scholar]
- 50.Crugeiras J, Rios A, Riveiros E, Richard JP. J. Am. Chem. Soc. 2009;131:15815–15824. doi: 10.1021/ja906230n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bridges JW, Davies DS, Williams RT. Biochem. J. 1966;98:451. doi: 10.1042/bj0980451. &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toney MD, Kirsch JF. Science. 1989;243:1485–1488. doi: 10.1126/science.2538921. [DOI] [PubMed] [Google Scholar]
- 53.Gloss LM, Kirsch JF. Biochemistry. 1995;34:3999–4007. doi: 10.1021/bi00012a018. [DOI] [PubMed] [Google Scholar]
- 54.Jenkins WT, Taylor RT. J. Bio. Chem. 1965;240:2907. [PubMed] [Google Scholar]
- 55.Jenkins WT. J. Bio. Chem. 1961;236:1121. [PubMed] [Google Scholar]
- 56.Furumo NC, Kirsch JF. Arch. Biochem. Biophys. 1995;319:49–54. doi: 10.1006/abbi.1995.1265. [DOI] [PubMed] [Google Scholar]
- 57.Chen HY, Phillips RS. Biochemistry. 1993;32:11591–11599. doi: 10.1021/bi00094a016. [DOI] [PubMed] [Google Scholar]
- 58.Zhou XZ, Jin XG, Medhekar R, Chen XY, Dieckmann T, Toney MD. Biochemistry. 2001;40:1367–1377. doi: 10.1021/bi001237a. [DOI] [PubMed] [Google Scholar]
- 59.Petrich JW, Lambry JC, Kuczera K, Karplus M, Poyart C, Martin JL. Biochemistry. 1991;30:3975–3987. doi: 10.1021/bi00230a025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.