Abstract
Circadian rhythms exist in most living organisms. The general molecular mechanisms that are used to generate 24-hour rhythms are conserved among organisms, although the details vary. These core clocks consist of multiple regulatory feedback loops, and must be coordinated and orchestrated appropriately for the fine-tuning of the 24-hour period. Many levels of regulation are important for the proper functioning of the circadian clock, including transcriptional, post-transcriptional and post-translational mechanisms. In recent years, new information about post-transcriptional regulation in the circadian system has been discovered. Such regulation has been shown to alter the phase and amplitude of rhythmic mRNA and protein expression in many organisms. Therefore, this Commentary will provide an overview of current knowledge of post-transcriptional regulation of the clock genes and clock-controlled genes in dinoflagellates, plants, fungi and animals. This article will also highlight how circadian gene expression is modulated by post-transcriptional mechanisms and how this is crucial for robust circadian rhythmicity.
Keywords: Circadian, mRNA decay, Translation, Rhythmic, mRNA stability, Splicing
Introduction
The rotation of the earth creates a daily fluctuation of environmental cues. Organisms have evolved internal timing systems, called circadian clocks (see Box 1 for a glossary of terms), to coordinate their daily activities to anticipate and prepare for these environmental changes. Circadian rhythms are endogenous and persist under constant conditions, such as constant darkness, with a period (Box 1) of ~24 hours. The clock can also be reset by external cues, such as light and temperature, to maintain synchrony with the environment. Circadian clocks govern many biological processes, ranging from molecular and biochemical pathways to physiological and behavioral rhythms. Conceptually, there are three main components to a functional circadian clock (Fig. 1A). In order for an organism to properly synchronize its internal clock to its external environment, it must perceive a signal from the environment (input) and adjust the timing of the clock accordingly. These signals feed into the molecular clock (oscillator), which consists of several clock genes that form transcriptional-translational feedback loops. This oscillator generates self-sustained rhythms of ~24 hours. Finally, the timing information is transmitted to different pathways downstream of the molecular clock, which control a variety of different molecular and physiological processes that exhibit circadian rhythmicity (output).
Box 1. Glossary of terms used in circadian biology.
Amplitude. The variation between the peak and trough of the oscillation.
Circadian clock. Internal timing mechanism that coordinates physiological events with the external environment.
Free running. Self-sustained rhythms not influenced by external cues.
Pacemaker. An oscillator that coordinates the timing of other oscillators.
Period. The time for one complete cycle.
Phase. The time of a point in the rhythm (usually the peak or trough) with reference to some external point, such as local time or the onset of activity.
Suprachiasmatic nucleus (SCN). Area within the hypothalamus that receives light input from the optic nerve and is responsible for synchronizing the clocks in peripheral tissues.
Temperature compensation. Retaining relatively constant periodicity over a broad physiological range of temperatures.
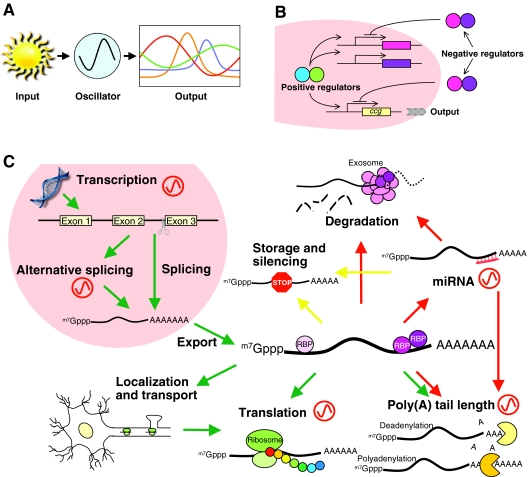
Fig. 1.
Circadian clocks and their contribution to post-transcriptional events. (A) The circadian system is generally considered to comprise three major components: input, oscillator and output. Light is the most powerful input signal, but other cues, such as temperature and nutrient availability, are also used. Examples of output include hormone secretion, body temperature, metabolic activity, sleep and wake cycle (mammals), eclosion, locomotor activity, olfactory responses (Drosophila), conidiation, carbon and nitrogen metabolism (Neurospora), flowering, germination, leaf movement and photosynthesis (Arabidopsis). (B) The generic structure of the molecular clock. Heterodimeric positive regulators (blue and green) activate the expression of negative regulators (purple and pink) by binding cis-regulatory elements in their promoters. Upon translation, these negative regulators inhibit their own transcription by blocking the activity of the positive regulators to complete the cycle. This cycle takes about 24 hours and thus generates circadian oscillation. Positive regulators also activate transcription of ccgs to drive rhythmic gene expression and generate circadian output. (C) The involvement of circadian rhythms in post-transcriptional regulation. Red oscillators indicate the steps that are known to be regulated by circadian clocks. Green, yellow and red arrows indicate pathways leading towards translation, translational silencing and mRNA degradation pathways, respectively. m7Gppp, 7-methylguanosine cap.
Genetic and molecular approaches have identified cellular regulatory mechanisms that are necessary for accurate clock function in many organisms. One common property of an oscillation is to escape from equilibrium; forming a negative feedback loop is one way to achieve this temporary departure from equilibrium. Although the details vary among organisms, circadian clocks generally utilize interlocking transcriptional-translational feedback loops as a mechanism to generate rhythms (Fig. 1B). The similarity among all of these systems strongly suggests that this feedback loop reflects a common lineage of circadian oscillators (Wijnen and Young, 2006). The molecular clock drives the rhythmic expression of core clock genes, together with a broad array of other circadian-controlled genes (ccgs) that are involved in various output pathways. Transcriptome analyses revealed that the expression of ~5–10% of genes exhibits circadian rhythmicity at steady-state mRNA levels in any given organism or tissue (Duffield, 2003). For transcripts to be rhythmic, they are required to have relatively short half-lives. A theoretical model of how mRNA stability impacts their amplitude (Box 1) shows that the more stable the transcript, the lower the amplitude of its cycling (Wuarin et al., 1992). Therefore, the regulation of mRNA stability, or its decay rate, plays an important role in the cycling of transcripts. In addition to mRNA decay, other post-transcriptional mechanisms also play an important role in rhythmic expression. For example, in some cases, protein synthesis rhythms can be uncoupled from mRNA rhythms, suggesting that translation can also be a key regulatory node. Proteomic analysis in mouse liver revealed that up to 20% of soluble proteins show rhythmic expression, although almost half of these cycling proteins did not exhibit corresponding rhythmic mRNA expression (Reddy et al., 2006). In this Commentary, we will review the evidence for post-transcriptional regulation in dinoflagellates, plants, fungi and animal circadian clocks to elucidate why and how circadian clocks have incorporated post-transcriptional regulation into the clock-generating machinery.
Post-transcriptional regulation
The control of gene expression is a complex process. Even after mRNA is transcribed from DNA, mRNAs can undergo many processing and regulatory steps that influence their expression (Keene, 2010). This type of regulation, at the post-transcriptional level, is beneficial, because it allows cells to alter protein levels rapidly without requiring transcript synthesis or processing. mRNA processing begins immediately following transcription with splicing and, in some cases, results in multiple isoforms of the transcript. Upon translocation into the cytoplasm, some mRNAs directly undergo deadenylation and degradation, whereas others can be stored in cytoplasmic compartments, such as processing bodies or stress granules, until they are degraded or re-polyadenylated. Other transcripts interact with the translational machinery and are translated into protein immediately after localization to the cytoplasm. Other mRNAs interact with localization machinery to be transported to a specific compartment within the cell for localized translation (Fig. 1C). All of these steps must be coordinated to ensure the appropriate expression profiles of mRNAs and proteins. In general, these events are mediated by cis-elements commonly found in 5′ or 3′ untranslated regions (UTRs) of mRNAs, which serve as specific binding sites for trans-acting factors, such as RNA-binding proteins (RBPs) or microRNAs (miRNAs). The fate of the mRNA is therefore largely determined by the composition of these trans-acting factors and the timing of the interaction.
Post-transcriptional regulation in dinoflagellates
The very first evidence regarding post-transcriptional regulation of the circadian clock came from the study of Lingulodinium (formerly known as Gonyaulax). In this primitive eukaryotic unicellular organism, several different physiological processes are regulated by circadian rhythms, such as photosynthesis, cell motility, cell division and luminescence flashing (Hastings, 2007). Lingulodinium has a highly developed capacity for bioluminescence and is one of the main species contributing to nocturnal luminescence of the sea. This nocturnal bioluminescence is produced from specialized bioluminescent organelles, the scintillons, which contain three components that are necessary for the chemical reaction: the substrate luciferin, a luciferin-binding protein (LBP) and the enzyme luciferase (LCF) (Morse et al., 1989). The clock controls the synthesis of these proteins, and both LBP and LCF are rhythmically expressed and peak at night, causing night-time flashing. However, the expression of lbp mRNA is not rhythmic, demonstrating that LBP and LCF expression is post-transcriptionally controlled (Johnson et al., 1984; Morse et al., 1989). Further research showed that an RNA-binding protein, clock controlled translational regulator (CCTR), interacts with a UG-repeat sequence in the lbp 3′-UTR and represses lbp translation during the day (Mittag et al., 1994). A similar protein, CHLAMY1, has been identified in Chlamydomonas. It also interacts with an UG repeat in a circadian-dependent manner (Mittag, 1996), indicating that this phenomenon is conserved among at least some algae.
Post-transcriptional regulation in plants
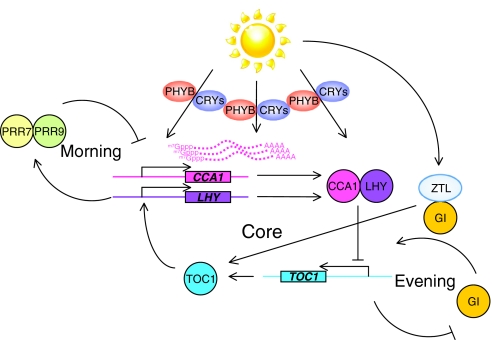
One of the first circadian phenomena recorded was in plants, in the 18th century, when leaf movement was found to vary between night and day, even in constant conditions. Since then, many more circadian phenomena have been described and Arabidopsis has emerged as an important model system for the discovery of plant clock mechanisms. The Arabidopsis clock consists of at least three interlocking feedback loops, the so-called ‘morning’, ‘evening’ and ‘core’ loops (Pruneda-Paz and Kay, 2010) (Fig. 2). The core loop is driven by two negative regulators, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), and one positive regulator, TIMING OF CAB 1 (TOC1). There are several photoreceptors in plants and, among them, CRYPTOCHROME 1 and 2 as well as PHYTOCHROME B are regulated by the circadian clock to provide light information to the central oscillator (Chen et al., 2004). Light induces the expression of CCA1 and LHY (as well as a number of ccgs), and this induction can entrain the clock (Martinez-Garcia et al., 2000; Wang and Tobin, 1998). Light also regulates post-transcriptional events. For example, the stability of CCA1 mRNA is different between the dark and light phases, indicating that light (or lack of light) controls the transcript stability (Yakir et al., 2007). Furthermore, light upregulates the level of LHY protein under conditions in which it does not alter the level of LHY transcript (Kim et al., 2003). Lastly, blocking the ZEITLUPE (ZTL)-mediated light input pathway alters the amplitude of rhythmic TOC1 protein without changing its mRNA amplitude. ZTL is another blue-light photoreceptor and forms a protein complex with GIGANTEA (GI) to regulate TOC1 protein expression (Kim et al., 2007b). These results indicate that light not only plays a pivotal role in activating transcription, but also regulates the stability of the transcript and its rhythmic expression pattern.
Fig. 2.
The effect of light on post-transcriptional regulation of clock genes in Arabidopsis. Light is perceived by photoreceptors (PHYB, red light; CRY1 and CRY2; blue light) and this input signal is transmitted to the circadian core loop to entrain the clock. ZTL is also a blue-light photoreceptor, affecting an evening loop, and regulates TOC1 expression together with GIGANTEA (GI). Light also provokes several post-transcriptional events, such as CCA1 mRNA degradation, translational activation of LHY and the enhancement of the amplitude of TOC1 rhythmic expression. PRR7 and PRR9 are PSEUDO RESPONSE REGULATOR 7 and PSEUDO RESPONSE REGULATOR 9.
Recent microarray studies revealed that clock-controlled genes in Arabidopsis have short half-lives and their rapid mRNA decay is mediated by a downstream (dst) element in the 3′-UTR (Gutierrez et al., 2002; Perez-Amador et al., 2001). Moreover, a mutant strain (dst1) was recently isolated, in which the dst-mediated decay pathway is disrupted. This mutant shows abnormal daily cotyledon movement (Lidder et al., 2005), further indicating the importance of the regulation of transcript stability to the circadian clock. The circadian clock also controls splicing in Arabidopsis (see section below on temperature-induced alternative splicing).
Post-transcriptional regulation in Neurospora
Neurospora crassa has been an important fungal model system for understanding the molecular control of circadian rhythms, combining powerful genetics with easily measured circadian outputs such as the daily conidiation rhythm – the asexual reproduction of spores. The molecular clock in Neurospora is composed of two positive regulators, White Collar-1 (WC-1) and White Collar-2 (WC-2), which together form the WC protein complex (WCC) and activate the transcription of frq, a core component of the clock that encodes a negative regulator, as well as a number of ccgs. Upon translation, FRQ forms a complex with FRQ-interacting RNA helicase (FRH) and inhibits WCC activity by promoting the phosphorylation of WCC. This leads to inhibition of its own mRNA synthesis, thereby closing the feedback loop (Brunner and Kaldi, 2008). Among these four core clock genes, only frq exhibits rhythmic expression at both mRNA and protein levels (Brunner and Kaldi, 2008), and the rhythmic expression of FRQ contributes to the rhythmic expression of WC-1 protein (Lee et al., 2000). The peak of cycling FRQ levels is 4–6 hours after its mRNA peak (Garceau et al., 1997). This led to the hypothesis that FRQ itself might also play a role in the degradation of its own mRNA, particularly when the mRNA level is declining and the protein level is increasing. In fact, the FRQ–FRH protein complex (FFC) destabilizes the frq mRNA by shortening the length of its poly(A) tail and also recruits the exosome. It is not known whether FFC formation or its mRNA-destabilizing activity is phase dependent (Guo et al., 2009). The exosome has several functions, including RNA degradation (Belostotsky, 2009), and it is often referred to as an analog of the proteasome, which is part of the protein degradation machinery. The idea that the exosome is essential for appropriate regulation of the Neurospora circadian clock is supported by the following data. Firstly, the rhythmic expression of rrp44, one of the core catalytic subunits of the exosome, is controlled by WCC. Secondly, knocking down rrp44 abolishes the conidation rhythm and lengthens the period of rhythmic expression of frq and FRQ. Finally, RRP44 interacts with FFC (Guo et al., 2009). None of the identified core clock proteins in other organisms are known to directly interact with the exosome, but this has not been tested in other systems and it would be interesting to see whether this mechanism is conserved.
Another unique phenomenon in Neurospora is the existence of a long antisense RNA of frq (about 5 and 5.5 kb) (Kramer et al., 2003). This antisense frq RNA shows rhythmic expression and its expression pattern is anti-phase to that of the sense frq RNA. The function of this antisense RNA is not clear, but mutant strains lacking the expression of this antisense RNA have a delayed frq mRNA rhythm and an altered response to light pulses (Kramer et al., 2003). It is also not known whether the antisense frq, or a possible RNA duplex of sense and antisense frq, is subject to FFC-mediated RNA degradation by the exosome.
Post-transcriptional regulation in animals
Genetic analysis of rhythms in Drosophila began in the early 1970s. Period (Per) was the first gene to be identified and then cloned a decade later (Bargiello and Young, 1984; Konopka and Benzer, 1971; Reddy et al., 1984). It took another decade for the mammalian homologue of Per to be identified and cloned (Sun et al., 1997; Tei et al., 1997). The Drosophila and mouse clock mechanisms are, generally speaking, conserved, although some of the genes are different and the mammalian system is more complex, containing multiple versions of some of the core clock genes. Detailed descriptions of the molecular clock structure in flies and mammals can be found elsewhere (Hastings et al., 2008; Wijnen and Young, 2006), but in brief, a heterodimeric transcription factor complex – called CLOCK and CYCLE (CYC) in flies and CLOCK and BMAL1 in mammals – activates the expression of genes that encode components of a repressive complex – PER and TIMELESS (TIM) in flies, and PER and CRYPTOCHROME (CRY) in mammals. The PER–TIM or PER–CRY complexes repress the activity of CLK–CYC (or of CLOCK–BMAL1), which closes the loop. In mammals, REV-ERBα, a member of the REV-ERB family of nuclear receptors, binds to the promoter region of Bmal1 and also regulates CLOCK–BMAL1 activity by inhibiting Bmal1 transcription. This forms a second feedback loop (Drosophila also has a similar interlocking loop, composed of different genes, but will not be discussed here).
The first examination of the post-transcriptional regulation of clock genes in animals was a detailed comparison of the active transcription and the steady-state expression profiles of Per. This analysis revealed that the half-life of Per mRNA changes throughout the circadian cycle, indicating that the stability of Per mRNA is under circadian control (So and Rosbash, 1997). It was subsequently shown that cycling mRNAs in mammals, such as Per2 and Cry1, also change their mRNA stability around the clock, and are more stable during the rising phase of mRNA cycling and less stable during the declining phase (So and Rosbash, 1997; Woo et al., 2010; Woo et al., 2009). As a similar regulation has been described for CCA1 in Arabidopsis (Yakir et al., 2007), this mechanism is conserved at least in animals and plants. The first evidence in mammals for post-transcriptional regulation came from a study showing that Per1 mRNA decays more rapidly following light induction than that of the control luciferase gene, which was driven by the Per1 promoter in the central clock in the brain (Wilsbacher et al., 2002). A later study demonstrated that an element in the 3′-UTR of Per1 promotes its instability, thereby contributing to this destabilization (Kojima et al., 2003). The 3′-UTRs of other clock genes, such as Drosophila Per, rat Aanat, and mouse Per2, Per3 and Cry1, also contribute to regulating their rhythmic expression, thereby leading to an accurate circadian period (Chen et al., 1998; Kim et al., 2005; Kim et al., 2007a; Kwak et al., 2006; Woo et al., 2010; Woo et al., 2009).
Several trans-factors (mostly RBPs) are known to regulate the clock or clock-controlled genes. LARK (or RBM4) is an RNA-binding protein that contains three RNA-binding domains (Kojima et al., 2007; Lai et al., 2003; Newby and Jackson, 1993). In Drosophila, Lark affects locomotor activity and eclosion rhythms (Huang et al., 2009; Newby and Jackson, 1996; Schroeder et al., 2003), and activates the translation of Eip74EF, an ecdysone-induced protein. The molecular function of Eip74EF has not been clarified, but an Eip74EF mutant fly exhibits an early eclosion phenotype (Huang et al., 2007). This is reminiscent of lark mutant flies, which also have an abnormally early eclosion (Newby and Jackson, 1993), suggesting that the post-transcriptional control of Eip74EF by LARK is essential for eclosion rhythms. In mammals, LARK1 (or RBM4a) activates the translation of PER1 by interacting with the Per1 3′-UTR without affecting its mRNA level (Kojima et al., 2007). The phase of LARK expression is similar to that of PER1 expression in the mouse suprachiasmatic nuclei (SCN), a circadian pacemaker (Box 1) in mammals, and peaks about 4–6 hours after its mRNA peak. The parallel timing of PER1 and LARK expression suggests that their interaction is important, although the function of mouse LARK in vivo is yet to be determined. There has also been a report that LARK activates internal ribosome entry site (IRES)-mediated translation, whereas it suppresses cap-dependent translation (Lin et al., 2007), possibly through the interaction with Argonaute proteins (Hock et al., 2007; Lin et al., 2007). Together, this further indicates the role of LARK in translational control. Interestingly, the expression of LARK itself is also under post-transcriptional control. LARK protein levels are rhythmic, whereas its mRNA levels remain unchanged around the clock in both mammals and flies (Kojima et al., 2007; McNeil et al., 1998).
Another group of RBPs that play significant roles in the circadian system is the heterogeneous nuclear ribonucleoproteins (hnRNPs). More than 30 proteins have been categorized as hnRNPs; each individual hnRNP has more than one function in the post-transcriptional events described in Fig. 1C (Chaudhury et al., 2010). Among these, three hnRNPs seem to be particularly relevant to the circadian clock system: hnRNP I, hnRNP D and hnRNP Q. hnRNP I, also called polypyrimidine-tract-binding protein (PTB), has four RNA-recognition motifs (RRMs) and binds to sequences that contain 15 to 25 pyrimidines, with a preference for pyrimidine tracts that have a UCUU sequence element (Singh et al., 1995). Interestingly, hnRNP I shuttles between the cytoplasm and nucleus (Sawicka et al., 2008), and the protein level of hnRNP I in the cytoplasm is rhythmic, even though its overall level is not (Kim et al., 2010). hnRNP I interacts with the 3′-UTR of Per2 mRNA and promotes its degradation (Woo et al., 2009) (Fig. 3A). hnRNP I also interacts with an IRES of Rev-erbα mRNA and activates its translation (Kim et al., 2010). The fluctuation of cytoplasmic hnRNP I is almost anti-phase to Per2 mRNA expression (Fig. 3A) and to REV-ERBα mRNA and protein expression (Kim et al., 2010; Woo et al., 2009). Therefore, the activity of hnRNP I might only have a minor effect on REV-ERBα translation, but this is a good example of hnRNP proteins having multiple functions. A similar observation has been reported for the interaction between hnRNP D and Cry1 (Woo et al., 2010) (Fig. 3A). hnRNP D, also known as ARE-binding factor 1 (AUF1), recognizes a U-rich sequence of the Cry1 3′-UTR and destabilizes its mRNA. The rhythmic expression of cytoplasmic hnRNP D is anti-phase to Cry1 mRNA expression (Woo et al., 2010), supporting the idea that Cry1 mRNA stability is negatively controlled by hnRNP D (Fig. 3A). Although these data are intriguing, the roles of hnRNP proteins are complex and a complete understanding of their functions in the clock system will require additional mechanistic insights. For example, AUF1 has been implicated in both stabilization and destabilization of target mRNAs, depending on the circumstances, and, moreover, it exists in four alternatively spliced isoforms, which have different activities (Raineri et al., 2004). It will be interesting to learn whether one or more of these isoforms is circadian and, if so, whether this varies between tissues, as well as which of these isoforms contribute to the control of clock gene expression.
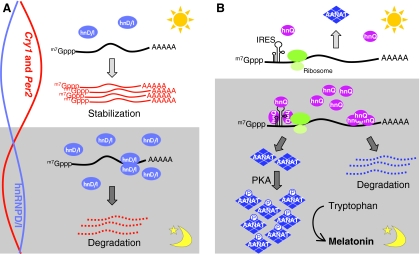
Fig. 3.
Post-transcriptional control of mammalian clock genes. (A) The regulation of Cry1 or Per2 mRNA stability by hnRNP D or I (hnD/I). During the night-time, the expression of hnRNP proteins in the cytoplasm is relatively high and more Cry1 and Per2 mRNAs are subject to degradation, leading to lower levels of mRNA (relative RNA levels depicted by curves on the left). By contrast, the level of hnRNPs is relatively low during the day; therefore, both Cry1 and Per2 mRNA can escape from the degradation caused by hnRNPs. Thus, these mRNAs become more stable, leading to higher mRNA levels. (B) Post-transcriptional and post-translational regulation of AANAT protein expression. During the night-time, hnRNP Q (hnQ) expression increases and this promotes the translation of AANAT through the interaction with IRESs. AANAT protein is phosphorylated by a PKA-mediated pathway and this prevents it from being degraded by the proteasome. Both of these post-transcriptional and post-translational regulatory steps contribute to the extreme amplitude of AANAT expression during the night. At the same time, hnRNP Q also binds to the 3′-UTR of Aanat mRNA and promotes degradation of Aanat mRNA. During the day, Aanat mRNA expression and the level of cytoplasmic hnRNP Q are low, and little or no AANAT protein is made. Melatonin is synthesized from tryptophan by four enzymatic steps, of which AANAT catalyzes the third to convert serotonin to N-acetylserotonin. The effects of hnRNP L and hnRNP R are not depicted here for simplicity. m7Gppp, 7-methylguanosine cap.
hnRNPs also function in the melatonin synthesis pathway, a key rhythmic output of the circadian clock. Melatonin is produced by the pineal gland and its levels exhibit circadian rhythmicity in amphibians, birds, reptiles and mammals, with a peak during the dark phase (Iuvone et al., 2005). The nocturnal increase in circulating melatonin is mirrored by the expression of arylalkylamine N-acetyltransferase (AANAT). AANAT is the rate-limiting enzyme in the melatonin biosynthetic pathway and its expression is rhythmic, with a more than 100-fold increase in amplitude during the night (Roseboom et al., 1996). However, the expression pattern of Aanat mRNA is species specific; it is rhythmic in rodents and peaks a few hours prior to the AANAT protein, but remains constant around the clock in primates and ungulates. Regardless of this difference, one common principle is that mRNA expression is out of phase compared with its protein levels, indicating that Aanat is under post-transcriptional control. Phosphorylation of AANAT mediated by protein kinase A (PKA) and cyclic AMP (cAMP)-dependent pathways stabilizes the protein. It is thought that this is one mechanism that regulates its nocturnal peak protein levels (Ganguly et al., 2002) (Fig. 3B). Recent new evidence has now shown that AANAT translation is also activated by hnRNP Q, which interacts with an IRES in the Aanat 5′-UTR (Kim et al., 2007a). The expression of hnRNP Q is rhythmic and its expression profile coincides with the phase of AANAT expression (Kim et al., 2005). Additionally, hnRNP Q, together with hnRNP L and hnRNP R, interacts with the 3′-UTR of Aanat mRNA and promotes the degradation of the Aanat transcript (Kim et al., 2005) (Fig. 3B). Although the expression of these hnRNP proteins is high when Aanat mRNA expression is relatively low, mathematical modeling predicts that hnRNP-mediated Aanat degradation affects the phase and amplitude of Aanat mRNA fluctuation (Kim et al., 2005). Taken together, these post-transcriptional regulatory mechanisms, in conjunction with the resulting post-translational modifications, shape the extreme AANAT expression pattern.
The circadian clock also exerts control over other post-transcriptional events in animals, such as splicing (Belanger et al., 2006), polyadenylation (Cagampang et al., 1994; Robinson et al., 1988), deadenylation (Baggs and Green, 2003; Garbarino-Pico et al., 2007) and translation (Fujimoto et al., 2006; Kim et al., 2002; Nishii et al., 2006; Yamamoto et al., 2005), although the mechanistic details of many of these events have not been fully elucidated.
Temperature-induced alternative splicing
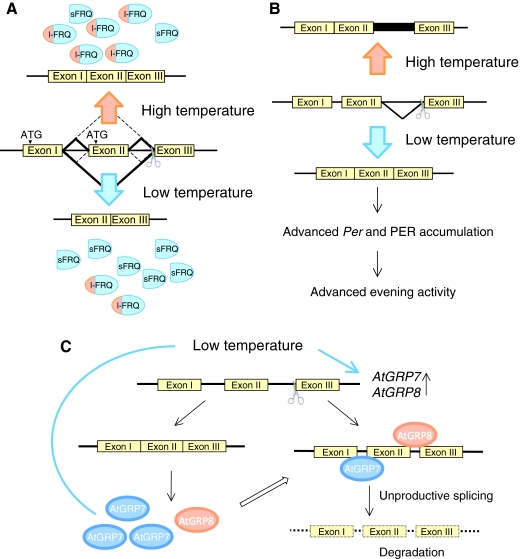
Most organisms experience significant variations in ambient temperature, but the free-running periods (Box 1) of circadian rhythms are precise and efficiently temperature compensated (Box 1) within a physiological range, even though all biochemical processes are generally temperature dependent. However, temperature shifts can often result in the resetting of circadian clocks, that is they can affect the phase of the circadian oscillation, similar to the effect of light. Comparable to light provoking several different post-transcriptional events, cold temperatures induce the alternative splicing of Per and frq in fly and fungi, respectively, and, as a result, two different protein isoforms are produced (Colot et al., 2005; Diernfellner et al., 2005; Majercak et al., 1999) (Fig. 4A,B). The ratio between one isoform and the other does not appear to be important for temperature compensation, as strains that possess only one isoform can still maintain consistent rhythms (Diernfellner et al., 2007; Majercak et al., 1999). Rather, the ratio of the two isoforms is crucial to robust free-running rhythmicity in fungi and the phase of Per and PER expression in the fly, providing a means to fine-tune the clock system in response to ambient temperature (Diernfellner et al., 2007; Diernfellner et al., 2005; Majercak et al., 1999). The molecular mechanism underlying this splicing is unknown, but one hypothesis is that trans-factors that regulate splicing are also under temperature control. In plants, the RBPs AtGRP7 and AtGRP8 are known to directly trigger splicing events. When AtGRP7 or AtGRP8 proteins reach a certain level, they bind to both of their pre-mRNAs and promote unproductive splicing coupled to degradation through the nonsense-mediated decay pathway (Schoning et al., 2008; Staiger et al., 2003), leading to the decrease in their mRNA level (Fig. 4C). AtGRP7 and AtGRP8 show robust rhythmic expression at both the mRNA and protein level, and their rhythmic expression is therefore regulated by a negative feedback loop of their own. This feedback might also contribute to the observed delay (approximately 4 hours) between mRNA and protein expression. Interestingly, mRNA expression of AtGRP7 and AtGRP8 is induced by cold temperature (Carpenter et al., 1994), although the protein level of AtGRP7, but not AtGRP8, is increased (Schoning et al., 2008). Because the splicing of AtGRP7 and AtGRP8 is regulated by their own protein level, it is likely that cold temperature also affects the splicing of these proteins, which could lead to mRNA degradation. Therefore, this cold-temperature-induced alternative splicing seems to be conserved at least in plants, Drosophila and Neurospora, and might be a common mechanism to adapt to cold temperature in other organisms as well.
Fig. 4.
Temperature-regulated alternative splicing of clock genes. (A) Temperature-sensitive splicing of frq. At low temperature, splicing at the non-canonical splice sites becomes dominant, leading to higher levels of expression of short-FRQ (sFRQ). At higher temperature, the canonical splicing site dominates, leading to higher levels of long-FRQ (l-FRQ) expression. (B) Temperature-sensitive splicing of Drosophila Per. There are two transcript forms of Per; one includes an intron sequence in the 3′-UTR and the other does not. Even though this difference does not alter the protein structure of PER, this splicing promotes the earlier accumulation of Per and PER, and advances the evening locomotor activity of fly. This might be a mechanism for adjustment to winter time, in which the temperature is lower and the photoperiod is shorter. (C) Autoregulation of AtGRP7 and AtGRP8 expression. AtGRP7 and AtGRP8 mRNAs are both induced by cold temperature. The protein level of AtGRP7, but not AtGRP8, is also increased by the cold temperature. Both AtGRP7 and AtGRP8 interact with their pre-mRNA, and promote the splicing that yields aberrant mRNA. As a consequence, these abnormal RNAs are degraded rapidly in the cell.
The role of miRNAs in the circadian clock
The discovery of miRNAs as regulators of gene expression has sparked increased interest in determining the genes that are regulated through these mechanisms and how they relate to maintaining circadian timing. miRNAs are short (19–25 nucleotides) non-coding RNAs (Shyu et al., 2008) that arise from primary transcribed miRNAs (pri-miRNAs), which are capped, adenylated and spliced just like protein-coding mRNAs. Afterwards, they are cleaved in the nucleus by the RNaseIII endonucleases Drosha and Pasha, resulting in a shorter (up to 65 nucleotides) precursor miRNA (pre-miRNA). These pre-miRNAs are then exported into the cytoplasm, where they are digested further by Dicer to form a short duplex RNA. This duplex RNA is incorporated into the functional miRNA-induced silencing complex (miRISC), which recognizes and processes target mRNAs (Carthew and Sontheimer, 2009; Chekulaeva and Filipowicz, 2009). miRNAs recognize and bind to their targets through specific sequences typically found in their 3′-UTRs. One requirement to achieve binding specificity is an almost perfect match between the 5′-proximal ‘seed’ region (position 2–8) of the miRNA and its target mRNA (Carthew and Sontheimer, 2009; Mendes et al., 2009). Once the miRNA recognizes and interacts with its target mRNA, protein synthesis is inhibited or deadenylation and degradation are triggered, but the detailed molecular mechanisms are still being debated (Carthew and Sontheimer, 2009; Chekulaeva and Filipowicz, 2009). Several algorithms have been developed that predict the interaction between miRNAs and target mRNAs, which are heavily based on the conservation of the seed region (Mendes et al., 2009). Using this information, several core clock genes, such as Period1, 2, 3 and both mammalian and Drosophila Clock, have been predicted to be miRNA targets (Kadener et al., 2009; Kiriakidou et al., 2004; Meng et al., 2006; Nagel et al., 2009; Yang et al., 2008). In addition, the effect of miR-141 on mammalian Clock was validated experimentally (Kiriakidou et al., 2004; Meng et al., 2006).
miRNAs have been found to target more than 30% of all protein-coding mRNAs; therefore, most, if not all, biological processes appear to be regulated by miRNAs to at least some degree (Chekulaeva and Filipowicz, 2009; Croce, 2009). Contributions of circadian clock function to miRNA expression or vice versa have been shown in several organisms. For example, expression of some miRNAs is found to be rhythmic in mouse retina, mouse SCN, Drosophila brain and Arabidopsis (Cheng et al., 2007; Sire et al., 2009; Xu et al., 2007; Yang et al., 2008). Other miRNAs are more directly involved in the clock system, and regulate the circadian period or response to light (Cheng et al., 2007; Jung et al., 2007; Kadener et al., 2009). For example, the expression of miR-132 is induced by a mitogen-activated protein kinase (MAPK)- and cAMP response element binding (CREB)-dependent mechanism in response to light, and facilitates the inhibition of a light-induced phase delay (Cheng et al., 2007). By contrast, the expression of miR-219 is driven by CLOCK–BMAL1 and its knockdown in vivo leads to a longer circadian period (Cheng et al., 2007). Another interesting miRNA with a role in the circadian clock is miR-122, a hepatocyte-specific miRNA. The transcription of miR-122 is rhythmic in mouse liver only at the pre-miRNA level, whereas the level of mature miRNA remains constant (Gatfield et al., 2009; Kojima et al., 2010). Knockdown of miR-122 attenuates the expression of a disproportionately high number of rhythmic genes (Gatfield et al., 2009). One of the target mRNAs of miR-122 is the circadian deadenylase Nocturnin (Kojima et al., 2010), which is a presumed key factor in circadian post-transcriptional control. Nocturnin expression is highly rhythmic and is thought to regulate the clock output pathway, rather than the core clock, most likely by deadenylating target mRNAs, resulting in changes in their stability or ability to be translated (Baggs and Green, 2003; Garbarino-Pico et al., 2007; Green and Besharse, 1996; Green et al., 2007; Wang et al., 2001).
Concluding remarks
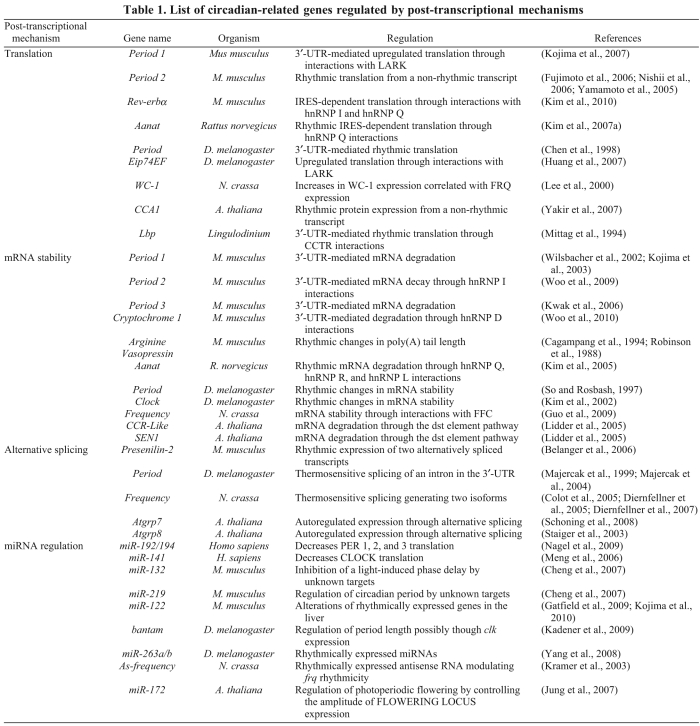
To maintain approximate 24-hour cycles at the molecular level, clocks must be tightly regulated at several steps to maintain the correct period, phase and amplitude of the rhythms of thousands of proteins that generate the wide range of rhythmic biological processes. As described above, there is now abundant evidence that post-transcriptional mechanisms play an important role in shaping these rhythms. However, much of this information is only anecdotal and additional studies need to be performed to provide a detailed knowledge of these regulatory mechanisms. A list of the publications demonstrating post-transcriptional regulation in the circadian field is presented in Table 1.
Table 1.
List of circadian-related genes regulated by post-transcriptional mechanisms
For mRNAs to be rhythmic, their half-lives must be relatively short and several reports have shown that the stability of these cycling mRNAs can change throughout the 24-hour period. However, it is not clear how broad this circadian change in mRNA stability is among cycling genes; are all cycling genes regulated in such a way or does this only occur in a subset? Furthermore, the question remains how the degradation of mRNAs is governed in a circadian-dependent manner, particularly as these rhythmic mRNAs peak at different times of the day. The recent work that demonstrates a direct role for the exosome in circadian control in Neurospora is highly exciting, because if this aspect of exosome function is conserved in other organisms, it could constitute a general mechanism for the rhythmic regulation of the stability of target mRNAs. Because hnRNPs can recruit the exosome to target mRNAs for their degradation (Hessle et al., 2009), it is tempting to speculate that rhythmically expressed RBPs might enable phase-dependent recruitment of the exosome in a target-specific manner. This possibility has not yet been widely explored, but is an area ripe for further investigation. Another interesting aspect of many cycling genes is the time lag (typically 4–6 hours) that often occurs between mRNA and protein expression, suggesting regulation at the translational level. It is not known whether this lag is caused by a universal circadian regulatory system or is the result of the functions of different RNA-specific mechanisms.
Research over the past two decades has generated a growing appreciation of the importance of gene regulatory processes at the post-transcriptional level. It has become very clear that both transcriptional and post-transcriptional processes are necessary to generate robust circadian rhythms of mRNA expression. Despite this realization, our understanding of circadian post-transcriptional mechanisms lags far behind our understanding of clock regulation at the transcriptional level. This is partially owing to the lack of well-developed methodologies to find post-transcriptionally regulated genes on a large scale. The development of such methods is likely to lead to the discovery of many more genes and mechanisms that are under post-transcriptional control.
Acknowledgments
Financial support for this work was provided by NIH grant GM076626 to C.B.G. Deposited in PMC for release after 12 months.
References
- Baggs J. E., Green C. B. (2003). Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 13, 189-198 [DOI] [PubMed] [Google Scholar]
- Bargiello T. A., Young M. W. (1984). Molecular genetics of a biological clock in Drosophila. Proc. Natl. Acad. Sci. USA 81, 2142-2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger V., Picard N., Cermakian N. (2006). The circadian regulation of Presenilin-2 gene expression. Chronobiol. Int. 23, 747-766 [DOI] [PubMed] [Google Scholar]
- Belostotsky D. (2009). Exosome complex and pervasive transcription in eukaryotic genomes. Curr. Opin. Cell Biol. 21, 352-358 [DOI] [PubMed] [Google Scholar]
- Brunner M., Kaldi K. (2008). Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol. Microbiol. 68, 255-262 [DOI] [PubMed] [Google Scholar]
- Cagampang F. R., Yang J., Nakayama Y., Fukuhara C., Inouye S. T. (1994). Circadian variation of arginine-vasopressin messenger RNA in the rat suprachiasmatic nucleus. Brain Res. Mol. Brain Res. 24, 179-184 [DOI] [PubMed] [Google Scholar]
- Carpenter C. D., Kreps J. A., Simon A. E. (1994). Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 104, 1015-1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R. W., Sontheimer E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642-655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A., Chander P., Howe P. H. (2010). Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1's multifunctional regulatory roles. RNA 16, 1449-1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M., Filipowicz W. (2009). Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 21, 452-460 [DOI] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38, 87-117 [DOI] [PubMed] [Google Scholar]
- Chen Y., Hunter-Ensor M., Schotland P., Sehgal A. (1998). Alterations of per RNA in noncoding regions affect periodicity of circadian behavioral rhythms. J. Biol. Rhythms 13, 364-379 [DOI] [PubMed] [Google Scholar]
- Cheng H. Y., Papp J. W., Varlamova O., Dziema H., Russell B., Curfman J. P., Nakazawa T., Shimizu K., Okamura H., Impey S., et al. (2007). microRNA modulation of circadian-clock period and entrainment. Neuron 54, 813-829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot H. V., Loros J. J., Dunlap J. C. (2005). Temperature-modulated alternative splicing and promoter use in the Circadian clock gene frequency. Mol. Biol. Cell 16, 5563-5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce C. M. (2009). Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704-714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diernfellner A., Colot H. V., Dintsis O., Loros J. J., Dunlap J. C., Brunner M. (2007). Long and short isoforms of Neurospora clock protein FRQ support temperature-compensated circadian rhythms. FEBS Lett. 581, 5759-5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diernfellner A. C., Schafmeier T., Merrow M. W., Brunner M. (2005). Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes Dev. 19, 1968-1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield G. E. (2003). DNA microarray analyses of circadian timing: the genomic basis of biological time. J. Neuroendocrinol. 15, 991-1002 [DOI] [PubMed] [Google Scholar]
- Fujimoto Y., Yagita K., Okamura H. (2006). Does mPER2 protein oscillate without its coding mRNA cycling? Post-transcriptional regulation by cell clock. Genes Cells 11, 525-530 [DOI] [PubMed] [Google Scholar]
- Ganguly S., Coon S. L., Klein D. C. (2002). Control of melatonin synthesis in the mammalian pineal gland: the critical role of serotonin acetylation. Cell Tissue Res. 309, 127-137 [DOI] [PubMed] [Google Scholar]
- Garbarino-Pico E., Niu S., Rollag M. D., Strayer C. A., Besharse J. C., Green C. B. (2007). Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA 13, 745-755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garceau N. Y., Liu Y., Loros J. J., Dunlap J. C. (1997). Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89, 469-476 [DOI] [PubMed] [Google Scholar]
- Gatfield D., Le Martelot G., Vejnar C. E., Gerlach D., Schaad O., Fleury-Olela F., Ruskeepaa A. L., Oresic M., Esau C. C., Zdobnov E. M., et al. (2009). Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 23, 1313-1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. B., Besharse J. C. (1996). Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proc. Natl. Acad. Sci. USA 93, 14884-14888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. B., Douris N., Kojima S., Strayer C. A., Fogerty J., Lourim D., Keller S. R., Besharse J. C. (2007). Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc. Natl. Acad. Sci. USA 104, 9888-9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Cheng P., Yuan H., Liu Y. (2009). The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell 138, 1236-1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R. A., Ewing R. M., Cherry J. M., Green P. J. (2002). Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA 99, 11513-11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings J. W. (2007). The Gonyaulax clock at 50, translational control of circadian expression. Cold Spring Harb. Symp. Quant. Biol. 72, 141-144 [DOI] [PubMed] [Google Scholar]
- Hastings M. H., Maywood E. S., O'Neill J. S. (2008). Cellular circadian pacemaking and the role of cytosolic rhythms. Curr. Biol. 18, R805-R815 [DOI] [PubMed] [Google Scholar]
- Hessle V., Bjork P., Sokolowski M., Gonzalez de Valdivia E., Silverstein R., Artemenko K., Tyagi A., Maddalo G., Ilag L., Helbig R., et al. (2009). The exosome associates cotranscriptionally with the nascent pre-mRNP through interactions with heterogeneous nuclear ribonucleoproteins. Mol. Biol. Cell 20, 3459-3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J., Weinmann L., Ender C., Rudel S., Kremmer E., Raabe M., Urlaub H., Meister G. (2007). Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 8, 1052-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Genova G., Roberts M., Jackson F. R. (2007). The LARK RNA-binding protein selectively regulates the circadian eclosion rhythm by controlling E74 protein expression. PLoS ONE 2, e1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Howlett E., Stern M., Jackson F. R. (2009). Altered LARK expression perturbs development and physiology of the Drosophila PDF clock neurons. Mol. Cell Neurosci. 41, 196-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuvone P. M., Tosini G., Pozdeyev N., Haque R., Klein D. C., Chaurasia S. S. (2005). Circadian clocks, clock networks, arylalkylamine N-acetyltransferase, and melatonin in the retina. Prog. Retin. Eye Res. 24, 433-456 [DOI] [PubMed] [Google Scholar]
- Johnson C. H., Roeber J. F., Hastings J. W. (1984). Circadian changes in enzyme concentration account for rhythm of enzyme activity in gonyaulax. Science 223, 1428-1430 [DOI] [PubMed] [Google Scholar]
- Jung J. H., Seo Y. H., Seo P. J., Reyes J. L., Yun J., Chua N. H., Park C. M. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736-2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S., Menet J. S., Sugino K., Horwich M. D., Weissbein U., Nawathean P., Vagin V. V., Zamore P. D., Nelson S. B., Rosbash M. (2009). A role for microRNAs in the Drosophila circadian clock. Genes Dev. 23, 2179-2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J. D. (2010). Minireview: global regulation and dynamics of ribonucleic acid. Endocrinology 151, 1391-1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y., Woo K. C., Lee K. H., Kim T. D., Kim K. T. (2010). hnRNP Q and PTB modulate the circadian oscillation of mouse Rev-erb {alpha} via IRES-mediated translation. Nucleic Acids Res. 38, 7068-7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. Y., Bae K., Ng F. S., Glossop N. R., Hardin P. E., Edery I. (2002). Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron 34, 69-81 [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Song H. R., Taylor B. L., Carre I. A. (2003). Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J. 22, 935-944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. D., Kim J. S., Kim J. H., Myung J., Chae H. D., Woo K. C., Jang S. K., Koh D. S., Kim K. T. (2005). Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol. Cell. Biol. 25, 3232-3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. D., Woo K. C., Cho S., Ha D. C., Jang S. K., Kim K. T. (2007a). Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 21, 797-810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. Y., Fujiwara S., Suh S. S., Kim J., Kim Y., Han L., David K., Putterill J., Nam H. G., Somers D. E. (2007b). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356-360 [DOI] [PubMed] [Google Scholar]
- Kiriakidou M., Nelson P. T., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A. (2004). A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 18, 1165-1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Hirose M., Tokunaga K., Sakaki Y., Tei H. (2003). Structural and functional analysis of 3′ untranslated region of mouse Period1 mRNA. Biochem. Biophys. Res. Commun. 301, 1-7 [DOI] [PubMed] [Google Scholar]
- Kojima S., Matsumoto K., Hirose M., Shimada M., Nagano M., Shigeyoshi Y., Hoshino S., Ui-Tei K., Saigo K., Green C. B., et al. (2007). LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl. Acad. Sci. USA 104, 1859-1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Gatfield D., Esau C. C., Green C. B. (2010). MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS ONE 5, e11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka R. J., Benzer S. (1971). Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 68, 2112-2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer C., Loros J. J., Dunlap J. C., Crosthwaite S. K. (2003). Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421, 948-952 [DOI] [PubMed] [Google Scholar]
- Kwak E., Kim T. D., Kim K. T. (2006). Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J. Biol. Chem. 281, 19100-19106 [DOI] [PubMed] [Google Scholar]
- Lai M. C., Kuo H. W., Chang W. C., Tarn W. Y. (2003). A novel splicing regulator shares a nuclear import pathway with SR proteins. EMBO J. 22, 1359-1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Loros J. J., Dunlap J. C. (2000). Interconnected feedback loops in the Neurospora circadian system. Science 289, 107-110 [DOI] [PubMed] [Google Scholar]
- Lidder P., Gutierrez R. A., Salome P. A., McClung C. R., Green P. J. (2005). Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol. 138, 2374-2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. C., Hsu M., Tarn W. Y. (2007). Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proc. Natl. Acad. Sci. USA 104, 2235-2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J., Chen W. F., Edery I. (2004). Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol. Cell. Biol. 24, 3359-3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J., Sidote D., Hardin P. E., Edery I. (1999). How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219-230 [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia J. F., Huq E., Quail P. H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859-863 [DOI] [PubMed] [Google Scholar]
- McNeil G. P., Zhang X., Genova G., Jackson F. R. (1998). A molecular rhythm mediating circadian clock output in Drosophila. Neuron 20, 297-303 [DOI] [PubMed] [Google Scholar]
- Mendes N. D., Freitas A. T., Sagot M. F. (2009). Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res. 37, 2419-2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Henson R., Lang M., Wehbe H., Maheshwari S., Mendell J. T., Jiang J., Schmittgen T. D., Patel T. (2006). Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 130, 2113-2129 [DOI] [PubMed] [Google Scholar]
- Mittag M. (1996). Conserved circadian elements in phylogenetically diverse algae. Proc. Natl. Acad. Sci. USA 93, 14401-14404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag M., Lee D. H., Hastings J. W. (1994). Circadian expression of the luciferin-binding protein correlates with the binding of a protein to the 3′ untranslated region of its mRNA. Proc. Natl. Acad. Sci. USA 91, 5257-5261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D., Milos P. M., Roux E., Hastings J. W. (1989). Circadian regulation of bioluminescence in Gonyaulax involves translational control. Proc. Natl. Acad. Sci. USA 86, 172-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R., Clijsters L., Agami R. (2009). The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 276, 5447-5455 [DOI] [PubMed] [Google Scholar]
- Newby L. M., Jackson F. R. (1993). A new biological rhythm mutant of Drosophila melanogaster that identifies a gene with an essential embryonic function. Genetics 135, 1077-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby L. M., Jackson F. R. (1996). Regulation of a specific circadian clock output pathway by lark, a putative RNA-binding protein with repressor activity. J. Neurobiol. 31, 117-128 [DOI] [PubMed] [Google Scholar]
- Nishii K., Yamanaka I., Yasuda M., Kiyohara Y. B., Kitayama Y., Kondo T., Yagita K. (2006). Rhythmic post-transcriptional regulation of the circadian clock protein mPER2 in mammalian cells: a real-time analysis. Neurosci. Lett. 401, 44-48 [DOI] [PubMed] [Google Scholar]
- Perez-Amador M. A., Lidder P., Johnson M. A., Landgraf J., Wisman E., Green P. J. (2001). New molecular phenotypes in the dst mutants of Arabidopsis revealed by DNA microarray analysis. Plant Cell 13, 2703-2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J. L., Kay S. A. (2010). An expanding universe of circadian networks in higher plants. Trends Plant Sci. 15, 259-265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raineri I., Wegmueller D., Gross B., Certa U., Moroni C. (2004). Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 32, 1279-1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. B., Karp N. A., Maywood E. S., Sage E. A., Deery M., O'Neill J. S., Wong G. K., Chesham J., Odell M., Lilley K. S., et al. (2006). Circadian orchestration of the hepatic proteome. Curr. Biol. 16, 1107-1115 [DOI] [PubMed] [Google Scholar]
- Reddy P., Zehring W. A., Wheeler D. A., Pirrotta V., Hadfield C., Hall J. C., Rosbash M. (1984). Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 38, 701-710 [DOI] [PubMed] [Google Scholar]
- Robinson B. G., Frim D. M., Schwartz W. J., Majzoub J. A. (1988). Vasopressin mRNA in the suprachiasmatic nuclei: daily regulation of polyadenylate tail length. Science 241, 342-344 [DOI] [PubMed] [Google Scholar]
- Roseboom P. H., Coon S. L., Baler R., McCune S. K., Weller J. L., Klein D. C. (1996). Melatonin synthesis: analysis of the more than 150-fold nocturnal increase in serotonin N-acetyltransferase messenger ribonucleic acid in the rat pineal gland. Endocrinology 137, 3033-3045 [DOI] [PubMed] [Google Scholar]
- Sawicka K., Bushell M., Spriggs K. A., Willis A. E. (2008). Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem. Soc. Trans. 36, 641-647 [DOI] [PubMed] [Google Scholar]
- Schoning J. C., Streitner C., Meyer I. M., Gao Y., Staiger D. (2008). Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 36, 6977-6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A. J., Genova G. K., Roberts M. A., Kleyner Y., Suh J., Jackson F. R. (2003). Cell-specific expression of the lark RNA-binding protein in Drosophila results in morphological and circadian behavioral phenotypes. J. Neurogenet. 17, 139-169 [PubMed] [Google Scholar]
- Shyu A. B., Wilkinson M. F., van Hoof A. (2008). Messenger RNA regulation: to translate or to degrade. EMBO J. 27, 471-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Valcarcel J., Green M. R. (1995). Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268, 1173-1176 [DOI] [PubMed] [Google Scholar]
- Sire C., Moreno A. B., Garcia-Chapa M., Lopez-Moya J. J., San Segundo B. (2009). Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett. 583, 1039-1044 [DOI] [PubMed] [Google Scholar]
- So W. V., Rosbash M. (1997). Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 16, 7146-7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Zecca L., Wieczorek Kirk D. A., Apel K., Eckstein L. (2003). The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 33, 361-371 [DOI] [PubMed] [Google Scholar]
- Sun Z. S., Albrecht U., Zhuchenko O., Bailey J., Eichele G., Lee C. C. (1997). RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell 90, 1003-1011 [DOI] [PubMed] [Google Scholar]
- Tei H., Okamura H., Shigeyoshi Y., Fukuhara C., Ozawa R., Hirose M., Sakaki Y. (1997). Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 389, 512-516 [DOI] [PubMed] [Google Scholar]
- Wang Y., Osterbur D. L., Megaw P. L., Tosini G., Fukuhara C., Green C. B., Besharse J. C. (2001). Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev. Biol. 1, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Y., Tobin E. M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207-1217 [DOI] [PubMed] [Google Scholar]
- Wijnen H., Young M. W. (2006). Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 40, 409-448 [DOI] [PubMed] [Google Scholar]
- Wilsbacher L. D., Yamazaki S., Herzog E. D., Song E. J., Radcliffe L. A., Abe M., Block G., Spitznagel E., Menaker M., Takahashi J. S. (2002). Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice in vivo. Proc. Natl. Acad. Sci. USA 99, 489-494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K. C., Kim T. D., Lee K. H., Kim D. Y., Kim W., Lee K. Y., Kim K. T. (2009). Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 37, 26-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K. C., Ha D. C., Lee K. H., Kim D. Y., Kim T. D., Kim K. T. (2010). Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol. Cell. Biol. 30, 197-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J., Falvey E., Lavery D., Talbot D., Schmidt E., Ossipow V., Fonjallaz P., Schibler U. (1992). The role of the transcriptional activator protein DBP in circadian liver gene expression. J. Cell Sci. Suppl. 16, 123-127 [DOI] [PubMed] [Google Scholar]
- Xu S., Witmer P. D., Lumayag S., Kovacs B., Valle D. (2007). MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 282, 25053-25066 [DOI] [PubMed] [Google Scholar]
- Yakir E., Hilman D., Hassidim M., Green R. M. (2007). CIRCADIAN CLOCK ASSOCIATED1 transcript stability and the entrainment of the circadian clock in Arabidopsis. Plant Physiol. 145, 925-932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Yagita K., Okamura H. (2005). Role of cyclic mPer2 expression in the mammalian cellular clock. Mol. Cell. Biol. 25, 1912-1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Lee J. E., Padgett R. W., Edery I. (2008). Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics 9, 83 [DOI] [PMC free article] [PubMed] [Google Scholar]