Abstract
In this issue of Molecular Cell, Winkler and coworkers (2007) describe in atomic detail the structural changes that allow a specific RNA aptamer (the M box) to regulate transcription in response to changing levels of magnesium.
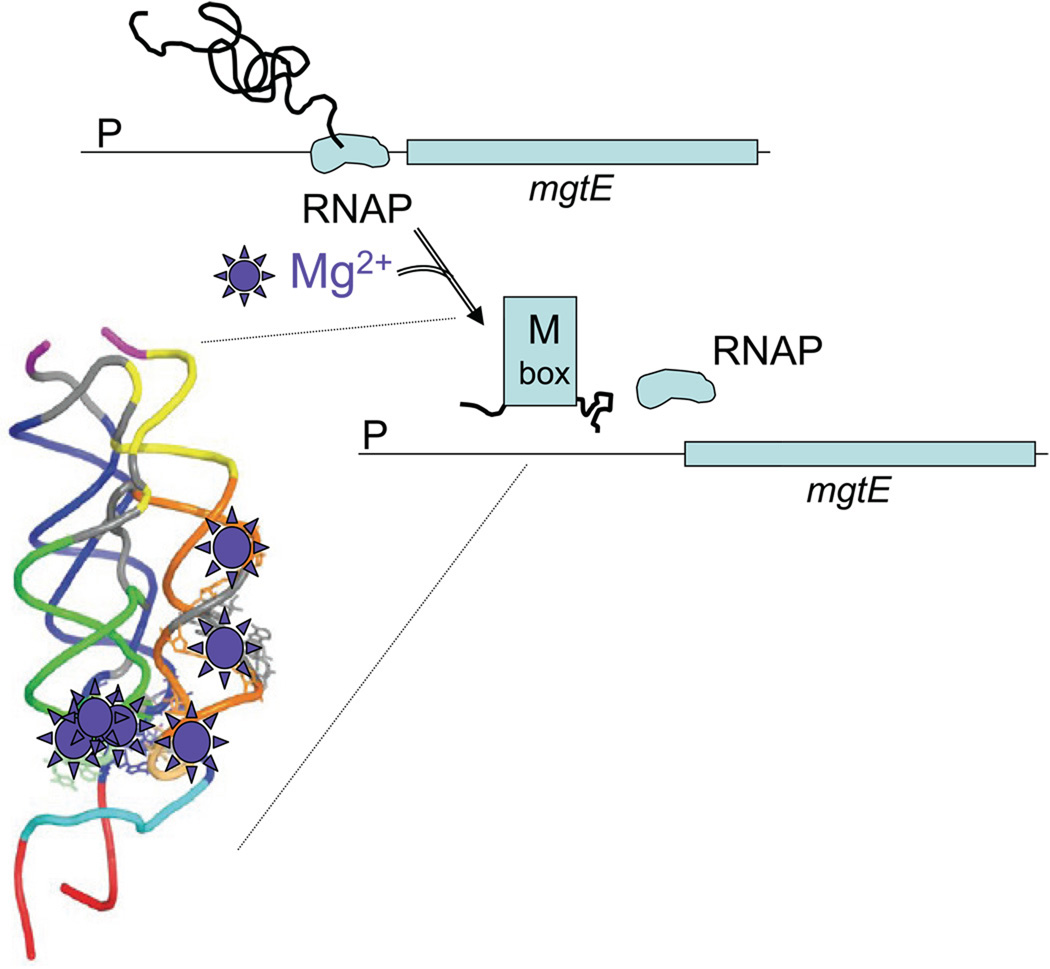
In a landmark study, Winkler and coworkers (Dann III et al. 2007) describe the structure of an intracellular Mg(II)-sensing riboswitch. When intracellular Mg(II) levels are sufficient, the “M box” aptamer mediates transcription termination in the leader region of the mgtE gene encoding a Mg(II) uptake channel (Fig. 1). This study provides the first detailed insights into the role of RNA elements in metalloregulation. Although metalloregulation has traditionally been associated with proteins, the chemical constraints associated with development of selective metal-responsive elements are universal (Finney and O'Halloran 2003; Tottey et al. 2005; Helmann et al. 2007).
Figure 1.
Role of the M-box aptamer in the transcriptional regulation of Bacillus subtilis mgtE gene. When Mg(II) levels are sub-optimal, transcription proceeds through the leader region RNA an into the mgtE structural gene. This process involves a stem-loop structure in which an anti-terminator element prevents formation of a transcription terminator. When Mg(II) levels are sufficient, the anti-terminator is instead sequestered into the stably folded structure of the M box aptamer, thereby allowing formation of a transcription terminator. The atomic structure reveals six bound Mg(II) ions in the folded M box aptamer that are implicated in this regulatory transition.
Metalloregulatory proteins are charged with monitoring intracellular metal status and coordinating appropriate genetic responses (Tottey et al. 2005; Helmann et al. 2007). Such proteins have metal-sensing sites that regulate activity: saturation of these sites signals metal sufficiency or excess. For example, both bacteria and eukaryotes contain metalloregulators that sense iron, zinc, or copper and regulate expression of the corresponding uptake systems. Conversely, when metals are in excess the same or a distinct metalloregulator activates storage or efflux functions.
Understanding how metalloregulatory proteins sense intracellular metal ion levels has been challenging. The emerging consensus is that metalloregulators have an affinity poised to detect a rise in free metal levels beyond that which is optimal for cell function. Escherichia coli sensors for copper (CueR) and zinc (Zur and ZntR) have affinities in the sub-picomolar range. Considering the volume of the cell, this implies no significant pool of thermodynamically free copper or zinc: these metals are bound to enzyme active sites or tightly chaperoned within the cytosol (Finney and O'Halloran 2003). Conversely, with manganese (MntR), the measured affinities are in the micromolar range, consistent with a significant intracellular pool of thermodynamically free manganous ion (Golynskiy et al. 2006). In general, metalloregulators have metal affinities that approximate the optimal intracellular concentration of free metal ion. This likely varies from none (in the case of copper), to micromolar (manganese), to millimolar (magnesium).
Three principles govern the ability of metalloregulators to respond selectively to their cognate metals: affinity, allostery, and access (Tottey et al. 2005). Affinity reflects a protein’s ability to provide appropriate chemistry and geometry to satisfy the cognate metal effector. One simple, if naive, assumption regarding selectivity is that metalloregulators should bind their cognate metal with an affinity greater than that of other metal ions. However, actual binding affinities do not always correlate with the observed physiological effects. Thus, allosteric discrimination and regulated access must have a key role in determining selectivity. Allosteric discrimination is exemplified by Mycobacterium tuberculosis NmtR which does not respond to zinc, even though zinc binds 6 orders of magnitude tighter than the effector cobalt. In this example, the allosteric transition requires a hexa-coordinate geometry (cobalt or nickel) whereas zinc binds tetrahedrally. Selectivity can also rely on access: the regulated exclusion from the cytosol of those metals that might compete for a sensing site. This principle is also illustrated by NmtR which responds to nickel in M. tuberculosis, but not in Synechococcus, which fails to accumulate sufficient levels of this ion (Tottey et al. 2005). Similarly, the ability of metalloregulators of the MntR/DtxR family to respond selectively to Mn(II) or Fe(II) depends, in part, on the tight regulation of the levels of the non-effector ions (Golynskiy et al. 2006; Helmann et al. 2007). Detailed analyses of these and other protein metalloregulators indicate that evolution has been driven by both the need to sense the cognate effector ion and to discriminate against the ambient levels of other metal ions at their normal homeostatic set points.
Metalloregulators are now known for zinc, copper, cobalt, nickel, iron, zinc, manganese and for toxic metals and metalloids such as cadmium, mercury, and arsenite. Notably absent from this list are sensors for the most abundant cations in the cell: magnesium and potassium. The intracellular levels of magnesium may be regulated, in part, by direct allosteric interaction with constitutively expressed, cognate uptake channels (Maguire 2006; Hattori et al. 2007). Sensing of magnesium has also been associated with an extracellular sensor: the PhoQ sensor kinase of the enterobacteria (Cromie et al. 2006). Clearly, magnesium is present at high levels in the cell and, unlike lower abundance “trace” metals, comprises a large and well hydrated cation pool. Thus, different principles might govern its homeostasis.
The groundbreaking study by Winkler and coworkers (Dann III et al. 2007) provides the first detailed look at an alternative metalloregulatory mechanism involving metal-dependent, allosteric regulation of RNA structure. The “M box” riboswitch functions to regulate expression of the mgtE magnesium uptake channel in Bacillus subtilis and likely other Gram positive bacteria. When Mg(II) concentrations are sufficient, the mgtE leader region adopts a compact structure that sequesters a putative anti-terminator element thereby enabling transcription termination (Fig. 1). In this study, the structural basis for Mg(II)-sensing by the M-box RNA was elucidated: the folded riboswitch contains six site-specifically associated Mg(II) ions. Magnesium-sensing riboswitches were first documented in Salmonella enterica serovar Typhimurium (Cromie et al. 2006), but this latter riboswitch is distinct in both sequence and mechanism from the M box element: it lacks an obvious terminator structure and the nature of the allosteric mechanism has yet to be elucidated.
While the detailed atomic description of the M box represents an impressive advance, it also raises numerous questions. It is not yet known which of the six Mg(II) ions visualized in the structure are responsible for sensing, and the structure under low Mg(II) conditions is not well understood. RNA, a highly charged polyelectrolyte, associates with a diffuse atmosphere of mobile, hydrated cations and a smaller number of site-specific ions (Draper 2004): either could mediate gene regulation. In this case, however, the Mg(II)-dependent folding transition occurs in the presence of 2 M monovalent cations, presumably sufficient to compete away non-specifically associated Mg(II). Thus, it is likely that one or more of the six site-specific ions is key to regulation.
As with protein regulators, affinity, allostery, and access are all important for the function of the M box switch. The affinity of the M box RNA is in the range expected for a Mg(II) sensor: an EC50 ~2.7 mM. However, the M box has little selectivity for Mg(II): both Ca(II) and Mn(II) can also increase terminator formation in vitro. This suggests a regulatory mechanism that relies in large part on access: only Mg(II) accumulates to an intracellular level sufficient to trigger regulation. It is not yet known whether riboswitches have evolved to sense other metal ions, but genome analyses indicate that related elements are found preceding presumptive Mn(II) transporters (Barrick et al. 2004). If these are in fact Mn(II)-sensing riboswitches, it will be important to define how these RNA structures employ affinity and allostery to overcome the limitations imposed by the high concentrations of Mg(II).
REFERENCES
- Barrick JE, Corbino KA, Winkler WC, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci U S A. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg2+ Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Dann CE, III, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and Mechanism of a Metal-Sensing Regulatory RNA. Molecular Cell. 2007 doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney LA, O'Halloran TV. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- Golynskiy MV, Gunderson WA, Hendrich MP, Cohen SM. Metal Binding Studies and EPR Spectroscopy of the Manganese Transport Regulator MntR. Biochemistry. 2006;45:15359–15372. doi: 10.1021/bi0607406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Tanaka Y, Fukai S, Ishitani R, Nureki O. Crystal structure of the MgtE Mg2+ transporter. Nature. 2007;448:1072–1075. doi: 10.1038/nature06093. [DOI] [PubMed] [Google Scholar]
- Helmann JD, Soonsanga S, Gabriel S. Metalloregulators: Arbiters of Metal Sufficiency. In: Nies DH, Silver S, editors. Molecular Microbiology of Heavy Metals. Berlin: Springer-Verlag; 2007. pp. 37–71. [Google Scholar]
- Maguire ME. The structure of CorA: a Mg2+-selective channel. Curr Opin Struct Biol. 2006;16:432–438. doi: 10.1016/j.sbi.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Tottey S, Harvie DR, Robinson NJ. Understanding how cells allocate metals using metal sensors and metallochaperones. Acc Chem Res. 2005;38:775–783. doi: 10.1021/ar0300118. [DOI] [PubMed] [Google Scholar]