Abstract
A homogeneous protein with a subunit apparent molecular mass of ~50 kDa that catalyzes the previously described mitochondrial NADH-supported ammonium-stimulated hydrogen peroxide production (Grivennikova, V.G., Gecchini, G. and Vinogradov, A.D. (2008) FEBS Letts. 583, 1287-1291) was purified from the mitochondrial matrix of bovine heart. Chromatography of partially purified protein showed that the peaks of ammonium-stimulated NADH-dependent H2O2 production and that of NADH:lipoamide oxidoreductase activity coincided. The catalytic properties and mass spectrometry of the trypsin-digested protein revealed peptides that allowed identification of the protein as the Bos taurus dihydrolipoyl dehydrogenase.
Keywords: Hydrogen peroxide, ammonium, reactive oxygen species, dihydrolipoyl dehydrogenase, mitochondria
1. Introduction
It is well recognized that mitochondria are significant producers of cellular hydrogen peroxide [1-5]. Two components of the respiratory chain, NADH: ubiquinone oxidoreductase (complex I) [6-8] and ubiquinol:cytochrome c oxidoreductase (complex III) [9-11], have been shown to generate superoxide anion (), the immediate precursor of H2O2 [12]. Bovine heart submitochondrial particles (SMP) which are essentially free of the mitochondrial matrix enzymes produce superoxide during coupled steady-state oxidation of either succinate or NAD-dependent substrates, at rates of about 0.7 or 0.3 % of the total oxygen consumed, respectively, in the overall state 4 coupled oxidase reactions [13]. About two-third of the succinate-supported generation by SMP is sensitive to the complex I inhibitor rotenone [13]. These data indicate that complex I is a major contributor to the respiratory chain-mediated superoxide production. A peculiar feature of complex I catalyzed superoxide production is that it has an optimum activity at relatively low (~50 μM) NADH concentration, whereas it is significantly decreased at higher physiologically relevant levels of NADH [13]. The superoxide generating activity of complex I is also strongly inhibited by NAD+ [13]. Thus, it seems unlikely that complex I is capable of significant superoxide production under physiologically relevant conditions where the total pool of NADH/NAD+ is in the millimolar range and a substantial amount of NAD+ is always present. We have proposed that rather than complex I some other mitochondrial enzyme(s) is poised in equilibrium with the NADH/NAD+ couple and that this enzyme(s) is the major producer of hydrogen peroxide. This proposal was confirmed by demonstration of NADH-supported ammonium-stimulated H2O2 formation catalyzed by a crude soluble fraction of matrix-located proteins [14]. Quantitation of the NADH-supported hydrogen peroxide formation by permeabilized rat heart mitochondria (in the presence of rotenone) showed that more than 70% of the total reaction was catalyzed by protein(s) other than complex I. When stimulating ammonium was present almost all (~95%) of the H2O2 was produced by those protein(s) [15].
The nature of the soluble NADH-dependent, ammonium stimulated, hydrogen peroxide producing enzyme(s) of mitochondrial matrix remained obscure. Therefore we report here the data showing that the E3 component of α-oxoacids dehydrogenases, dihydrolipoyl dehydrogenase is the protein responsible for this activity.
2. Materials and methods
2.1. Protein purification
Bovine heart mitochondria were prepared as described [16], separated for “heavy” and “light” fractions and stored at -20°C. Pilot experiments showed that the specific activities of NADH-supported, ammonium-stimulated H2O2 production were essentially the same for “heavy” and “light” mitochondria and the latter fraction was used for protein purification. All operations were carried out at 0°C. Mitochondria were thawed, diluted to a protein content of 20 mg/ml, EDTA (1 mM final concentration) and NH4OH was added, to adjust the pH to 8.6, and the mixture was subjected to sonication (Soniprep 150-MSE, maximal power output) under argon flow 6 times for 30 sec with 1 min intervals. The suspension was centrifuged for 60 min at 200 000 g. The clear supernatant was collected and used as crude matrix protein fraction. Solid ammonium sulphate (0.17 g per ml, 30% saturation) was added and the mixture was centrifuged for 20 min at 10 000 g. The dialyzed supernatant solution was heated in a water bath (20 min, 60° C) and after separation of precipitated protein the supernatant (~170 ml) was concentrated to a final volume of 50 ml (Centriprep 30). The solution was applied to a 5 ml High Trap Q-Sepharose column (AKTA FPLC system, GE Healthcare) equilibrated with 15 mM Tris/acetate buffer containing 0.1 mM EDTA (pH 7.6). The column was washed and the protein was then eluted by a linear gradient of 0 – 0.4 M NaCl. The active fractions (NADH-supported H2O2 production, 1 ml each) were collected. Ammonium sulphate (7.3 g) was added to the 25 ml pooled fractions and the sample was applied to 5 ml High Trap Phenyl HP column equilibrated with 30 mM Tris/acetate, 0.1 mM EDTA and 1.8 M ammonium sulphate. The column was washed, the proteins were eluted by a linear descending gradient of ammonium sulphate (40 – 20%) in the same buffer. The peak fractions (1 ml each) showing activity were eluted by 20 – 0% linear descending gradient of ammonium sulphate. They were pooled in a total volume of 8 ml and dialyzed two times for 1 h against 1-liter of 15 mM Tris/Cl-, 0.1 mM EDTA solution, pH 7.6. The protein thus obtained was stored in liquid nitrogen as small aliquot samples.
2.2. SDS-PAGE and protein determination
Electrophoresis was carried out using 12.5 % SDS-polyacrylamide gels according to standard procedure [17]. Sigma Dalton Mark VII-L Standard Mixture, 14,000 – 66,000 for SDS-PAGE was used for molecular mass calibration. Gels were stained with Coomassie Brilliant Blue (R250). Protein content was determined by the biuret reagent with bovine serum albumin as the standard. All catalytic activities are expressed on a protein concentration basis. At some steps of purification when diluted protein samples were assayed the biuret method could not be applied due to its low sensitivity. To overcome this difficulty the following procedure was used. At the early steps of purification the protein content was determined by both biuret method and A280 absorption. An arbitrary coefficient for the protein content determined as A280 absorption based on the values determined by the biuret method was calculated (0.63 for 1 mg of protein per ml) and used for the assays in the samples with low protein content.
2.3. The catalytic assays
All activities were assayed at 30°C in the standard reaction mixture composed of 5 mM KH2PO4, 10 mM KCl, 0.25 M sucrose, 0.1 mM EDTA (pH 7.5) and appropriate substrates (concentration are indicated in the legends to the Figures and Tables). Hydrogen peroxide formation was monitored at 572 nm (resorufin formation, ε572 = 54 mM-1cm-1 [18] in the standard mixture supplemented with Amplex Red (10 μM), horseradish peroxidase (2 U/ml) and bovine erythrocyte superoxide dismutase (SOD) (6 U/ml). Superoxide production was measured as SOD-sensitive cytochrome c (10 μM) reduction at 550 nm (ε550 = 20 mM-1cm-1). Transhydrogenase activity was measured with acetyl-NAD+ as the second substrate by following the absorption change at 375 nm (ε375 = 5 mM-1cm-1). Lipoamide reductase activity was determined as NADH oxidation by 1 mM lipoamide. All the activities were initiated by the enzyme additions. The background reaction seen in the presence of all components before the addition of protein was negligible (not more than 1-2% of the catalytic rates).
2.5. Mass-spectrometry
MS experiments were performed at the Institute of Physico-Chemical Medicine, Federal Medical-Biological Academy, Moscow, Russian Federation on an Ultraflex tandem time-of-flight instrument (Bruker Daltonics, Germany). The precision of monoisotopic masses after calibration by trypsin autolysis peaks was 0.005%. The destained dehydrated gel samples were digested by modified trypsin (Promega) in 50 mM NH4HCO3 for 14 h at 37° C and hydrolyzed material was extracted by 0.5% trifluoroacetic acid and 10% (v/v) acetonitrile. The extracts were used for matrix (2,5-dihydroxibenzoic acid)-assisted laser desorption/ionization (MALDI) spectra. Fingerprint protein identification search was done in the NCBI database using the Mascot program (www.matrixscience.com).
2.6. Reagents
Amplex Red was from AnaSpec, Inc. (U.S.A.), Tris and sucrose were from MP Biomedical Inc. (France). Other fine chemicals were from Sigma-Aldrich (U.S.A.).
3. Results
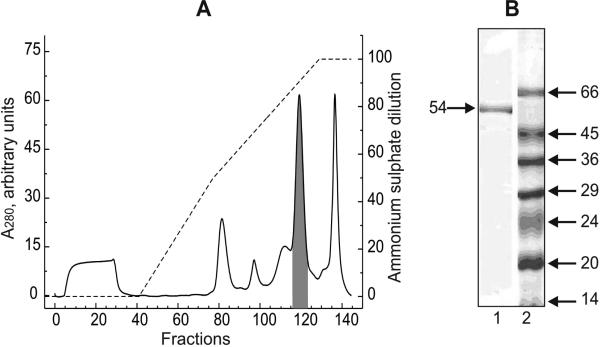
Table 1 provides a summary of the results of the purification of the protein responsible for the mitochondrial NADH-supported hydrogen peroxide production. About one-third of the total ammonium-stimulated activity seen in the crude soluble supernatant was recovered in the fractions eluted from Phenyl-Sepharose as a single peak (Fig. 1A). The pooled fractions had a specific activity of 1.8 μmol·min-1·mg-1 (in the presence of ammonium chloride) corresponding to a 240-fold enrichment of the protein. The active protein that eluted from Phenyl-Sepharose column appeared as a single band in SDS-electrophoresis corresponding to an apparent molecular mass of about 50 kDa (Fig. 1B). This is similar to the molecular size of the E3 component (dihydrolipoyl dehydrogenase) of α-oxoacid dehydrogenases and glycine cleavage complex. It was found that the activity for the NADH-supported ammonium-stimulated hydrogen peroxide generation and NADH: lipoamide oxidoreductase coincided (Fig. 1A) in the protein eluting from the column. The relative oxidoreductase activities of the purified protein with different electron acceptors are depicted in Table 2. Neither transhydrogenase, nor lipoamide reductase activities were affected by ammonium chloride whereas more than 10-fold stimulation of hydrogen peroxide production was evident. Only about two-fold stimulation of superoxide production by ammonium was observed. The stimulatory effect of ammonium was specific: guanidine chloride (10 mM), methyl-, dimethyl-, trimethyl-amines (30 mM each), lysine, arginine and spermine (10 mM each) were tested and none of these compounds affected hydrogen peroxide producing activity. When 1 mM NADPH was used as the substrate a formation of small amount (~1 μM) of resorufin in the Amplex Red assay was observed (not shown). This was evidently due to contamination of NADH in the NADPH samples.
Table 1.
Purification of the mitochondrial protein catalyzing NADH-supported, ammonium-stimulated H2O2 production
| Fraction | Protein (mg) | Total activitya (nmol·min-1) | Specific activity (nmol·min-1·mg-1) |
Purification | Yield (%) | |
|---|---|---|---|---|---|---|
| -NH4Cl | +NH4Cla | |||||
| Mitochondriab | 2,540 | 19,400 | 1.9 | 7.7 | 1 | 100 |
| Supernatant (200,000 g) | 448 | 6,000 | 3.7 | 13.4 | 1.7 | 31 |
| After heat treatment | 54 | 3,830 | 9.2 | 70.9 | 9.2 | 20 |
| Q-Sepharose pool | 5 | 2,600 | 55.0 | 532 | 69 | 13 |
| Phenyl-Sepharose pool | 1 | 1,850 | 232 | 1,850 | 240 | 9.5 |
Measured with 50 μM NADH in the presence of 30 mM NH4Cl.
Fig. 1.
A, Chromatography of Q-Sepharose pooled fractions on Phenyl-Sepharose. The details of the chromatography are described in Materials and Methods section. Solid line, protein elution profile; broken line, ammonium sulfate descending gradient. Shadowed peak corresponds to the fractions showing NADH-supported hydrogen peroxide generation and NADH: lipoamide oxidoreductase activities. Seventy one and eighty one per cent of the total protein applied on the column NADH-supported H2O2 production and lipoamide reduction activities, respectively, were recovered in the fractions 117-127 (shadowed peak). A sample of the peak fraction was diluted 1.5-fold by denaturating solution and 5 μl of the mixture were applied on the gel. B, Lane 1, SDS-PAGE of the protein showing hydrogen peroxide generating and lipoamide reductase activities. Lane 2, molecular mass markers. The apparent molecular mass (kDa) is shown on the right.
Table 2.
Reactivity of purified protein with different electron acceptors (pH 7.5, 30° C)
| Reaction | Specific activity (μmol of NADH consumed·min-1·mg-1) |
|
|---|---|---|
| – NH4Cl | + NH4Cla | |
| NADH → O2 (H2O2 production)b | 0.17 | 1.8 |
| NADH → O2 (superoxide production)c | 0.08 | 0.19 |
| NADH → Acetyl-NAD+ (transhydrogenase)d | 7.4 | 5.6 |
| NADH → Lipoamidee | 44.0 | 49.0 |
30 mM NH4Cl.
Amplex Red assay with 200 μM NADH.
SOD -sensitive reduction of 10 μM cytochrome c by 200 μM NADH. SOD-insensitive rate of cytochrome c reduction was not more than 15% of the observed overall rate.
100 μM NADH and 100 μM Acetyl-NAD+ as the substrates.
100 μM NADH and 1 mM lipoamide as the substrates.
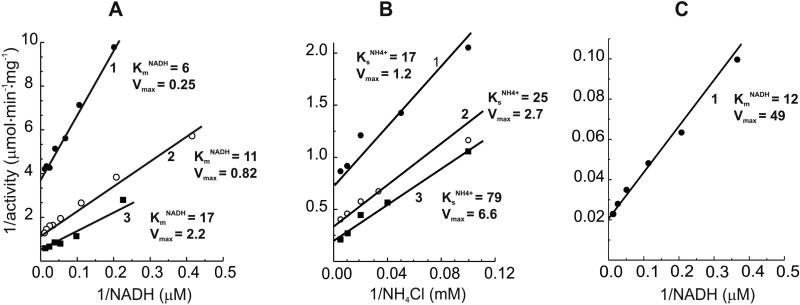
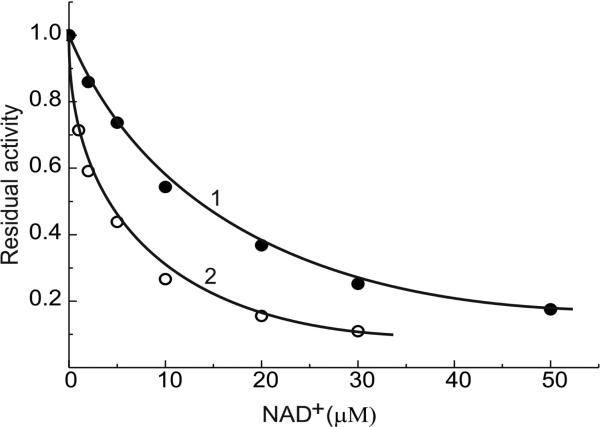
The kinetics of NADH-supported hydrogen peroxide formation were also evaluated. The activity was dependent on NADH and ammonium chloride concentrations (within 5 – 100 μM and 10 – 200 mM ranges, respectively) in a simple mutually dependent hyperbolic modes (Fig. 2 A and B). The same pattern was seen in ammonium-independent lipoamide reduction (Fig. 2C) with an apparent KmNADH equal to 12 μM. This value is close to that determined for the NADH-supported H2O2 production in the absence of ammonium. The hydrogen peroxide generating activity was strongly decreased by NAD+; half maximal inhibition at 4 and 12 μM NAD+ was observed when 20 and 100 μM NADH was used as the substrate (Fig. 3).
Fig.2.
Double reciprocal plots for the substrate (NADH, A) and activator (NH4+, B) dependencies of the enzyme hydrogen peroxide generating activity. A, lines 1, 2 and 3, ammonium concentration in the standard assay mixture were 0, 5, and 30 mM, respectively. The kinetic parameters (KmNADH and Vmax,) corresponding to each lines are expressed in μM and μmol·min-1·mg-1, respectively. B, lines 1, 2, and 3, NADH concentrations in the assay mixture were: 10, 50, and 100 μM, respectively. The kinetic parameters (KsNH4+ and Vmax) are expressed in mM and μmol·min-1·mg-1, respectively. C, lipoamide reductase activity. KmNADH and Vmax are expressed as in A.
Fig. 3.
Inhibition of NADH-supported hydrogen peroxide formation by NAD+. Ammonium concentration in the standard assay mixture was 30 mM. Arbitrary unit activity corresponds to 1.9 μmol·min-1·mg-1 (line 1) and 1.3 μmol·min-1·mg-1 (line 2) at 100 and 20 μM NADH, respectively.
Taken together the results presented above strongly suggested that the protein responsible for the mitochondrial ammonium-stimulated hydrogen peroxide production is identical to dihydrolipoyl dehydrogenase. This was confirmed by mass spectrometry analysis of the peptides obtained after tryptic digestion of the single protein band shown in Fig. 1B. Table 3 depicts the peptides identified by the mass-spectral analysis as they correspond to the amino acid sequence of bovine dihydrolipoyl dehydrogenase. Further confirmation of the identification was obtained by using cloned and purified human dihydrolipoyl dehydrogenase which showed essentially the same catalytic activities as those depicted in Table 2 (data not shown).
Table 3.
Correspondence between peptide fragments of bovine dihydrolipoamide dehydrogenase and the mol. masses of the peptides after tryptic digestion of the mitochondrial protein catalyzing NADH-supported, ammonium-stimulated H2O2 production as determined by MALDI analysisa
| Amino acid residue numbersb | Mr of matched peptidesc | Peptide assigned | |
|---|---|---|---|
| Observed | Calculated | ||
| 60–66 | 734.3933 | 733.4123 | K.AAQLGFK.T |
| 90–104 | 1721.9225 | 1720.8797 | K.ALLNNSHFYHLAHGK.D |
| 90–109 | 2298.1630 | 2297.1454 | K.ALLNNSHFYHLAHGKDFASR.G |
| 110–117 | 920.4668 | 919.4433 | R.GIEMSEVR.L |
| 110–117 | 936.4639 | 935.4382 | R.GIEMSEVR.L Oxidation (M) |
| 110–122 | 1533.8313 | 1532.7868 | R.GIEMSEVRLNLEK.M Oxidation (M) |
| 133–143 | 1127.6952 | 1126.6499 | K.ALTGGIAHLFK.Q |
| 316–334 | 2201.1873 | 2200.1488 | R.RPFTQNLGLEELGIELDTR.G |
| 347–365 | 1995.1301 | 1994.0659 | K.IPNIYAIGDVVAGPMLAHK.A Oxidation (M) |
| 405–417 | 1581.8074 | 1580.7569 | K.SEEQLKEEGIEYK.V |
| 418–428 | 1193.6822 | 1192.6353 | K.VGKFPFAANSR.A |
| 421–428 | 909.4682 | 908.4504 | K.FPFAANSR.A |
| 483–495 | 1467.7734 | 1466.7089 | R.VCHAHPTLSEAFR.E |
| 483–495 | 1538.7980 | 1537.7460 | R.VCHAHPTLSEAFR.E Propionamide (C) |
| 496–505 | 1007.5454 | 1006.5083 | R.EANLAASFGK.S |
Protein score of 142 for bovine dihydrolipoamide dehydrogenase (accession in NCBI database gil76615127) with cut-off value of 82 for p<0.05 confidence. Sequence coverage was 26%. No search for specific posttranslation modifications was done.
In bovine dihydrolipoamide dehydrogenase.
For peptides after tryptic digestion of the band shown in Fig. 1 B. Seven peptides not matched to dihydrolipoamide dehydrogenase sequence were present in small amount thus suggesting slight contamination of the band by other protein(s).
4. Discussion
We have identified the mitochondrial matrix located ammonium-stimulated H2O2 producing protein as dihydrolipoyl dehydrogenase. Although these studies were focused on the problem of identification, not on the development of preparative procedure, the data of Table 1 merit brief discussion. About 4-fold stimulation of the specific activity by ammonium was seen when all matrix-located proteins, except for complex I, participated in the overall NADH-supported H2O2 production. The stimulatory effect was significantly increased (up to 8-10-fold) when purified protein was assayed. This suggests that in addition to dihydrolipoyl dehydrogenase other not yet identified matrix located protein(s) contribute to the mitochondrial NADH- and/or NADPH-supported hydrogen peroxide production. No detergents or other strong solubilizing agents were used to obtain the crude soluble matrix fraction where only about 30% of the original total activity was recovered. This suggests that some amount of dihydrolipoyl dehydrogenase and, perhaps other hydrogen peroxide producing protein(s) in addition to the fraction captured in unbroken mitochondria are bound to the inside-out vesicles. We were unable to see any -oxoglutarate or pyruvate dehydrogenase activities in the 200 000 g supernatant (data not shown). This observation may reasonably be interpreted that the protein purified as shown in Table 1 is free, soluble dihydrolipoyl dehydrogenase (hydrogen peroxide producing NADH oxidase) not associated with supramolecular complexes of α-oxoglutarate, branched chain oxoacids and pyruvate dehydrogenases or glycine-cleavage system. If this interpretation is correct, the ammonium-dependent hydrogen peroxide production is another intriguing “moonlight function” for dihydrolipoyl dehydrogenase [21-24].
We have shown previously that mitochondrial hydrogen peroxide formation is strongly activated by ammonium [14, 15]. As reported here this effect appears specific: none of the tested compounds that contain positively charged nitrogen atoms stimulate the reaction catalyzed by the purified enzyme (Table 2). A gradual increase of ammonium concentration was accompanied by an increase of both Vmax and apparent Km for NADH (Fig. 2A). A plausible, although speculative explanation of the phenomenon is that binding of ammonium near the isoalloxazine moiety of FAD decreases the midpoint redox potential of the flavin. This would be expected to shift the equilibrium of intramolecular transformation: Eox·NADH ↔ Ered·NAD+, the step which contributes to apparent affinity of the enzyme to NADH (see Ref. [25] for more discussion of this point). On the other hand, at saturating NADH a decrease of flavin redox potential is expected to increase its reactivity to oxygen. This explanation is also applicable to the data shown in Fig. 2B that shows a decrease of apparent affinity to ammonium upon saturation of the enzyme by NADH.
Perhaps the most interesting problem concerning the strong ammonium-dependence of hydrogen peroxide production is whether the phenomenon is relevant to mitochondrial physiology. The apparent stimulatory effect of ammonium as reported here is seen at very high concentration (30 mM) which exceeds that suggested as the physiologically relevant level. It should be noted, however, that the actual steady-state concentrations of ammonium in the mitochondrial matrix during oxidation of ammonium producing substrates is not known. The ammonium concentration may rise if a kinetic limitation of the inner membrane for permeability for ammonium exists when mitochondria oxidize glutamine or glutamate. Such a limitation for “free permeable” hydrogen peroxide has recently been demonstrated [15]. It would be of obvious interest to compare hydrogen peroxide production by intact mitochondria oxidizing “classical” Krebs cycle intermediates (succinate, pyruvate plus malate) with that observed during ammonium producing substrate oxidation. Studies on the respiratory and hydrogen peroxide producing activities in the presence of glutamine, the most abundant free amino-acid would seem of particular interest [26, 27].
Acknowledgements
We are grateful to Dr. Alexander Kotlyar (Department of Biochemistry, Tel Aviv University, Israel) for supplying samples of NADH-OH. Valuable help of Dr. N.B. Gusev (Department of Biochemistry, School of Biology, Moscow State University) in chromatography are gratefully acknowledged. We thank Dr. Alexander Galkin (School of Biological Sciences Queen's University, Belfast, UK) for his advice and help in the peptide fingerprint analysis. This study was supported by the Russian Foundation for Fundamental Research grants 08-04-00594 to ADV and 09-04-00505 to VGG and NIH Research Grant #R03 TW07825 funded by the Fogarty International Center (ADV and GC) and the Department of Veterans Affairs (GC).
Abbreviations
- SOD
superoxide dismutase
- SDS-PAGE
polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chance B, Williams GB. The respiratory chain and oxidative phosphorylation. Adv. Enzymol. 1956;17:65–134. doi: 10.1002/9780470122624.ch2. [DOI] [PubMed] [Google Scholar]
- 2.Boveris A, Oshino N, Chance B. The cellular production of hydrogen peroxide. Biochem. J. 1972;128:617–630. doi: 10.1042/bj1280617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreev A.Yu., Kushnareva Yu.E., Starkov AA. Mitochondrial metabolism of reactive oxygen species. Biochemistry (Moscow) 2005;70:200–214. doi: 10.1007/s10541-005-0102-7. [DOI] [PubMed] [Google Scholar]
- 4.Adam-Vizi V. Production of reactive oxygen species in brain mitochondria: contribution by electron transport chain and non-electron transport chain sources. Antioxid. Redox Signal. 2005;7:1140–1149. doi: 10.1089/ars.2005.7.1140. [DOI] [PubMed] [Google Scholar]
- 5.Murphy MP. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinkle PC, Butow RA, Racker E, Chance B. Partial resolution of the enzymes catalyzing oxidative phosphorylation. XV. Reverse electron transfer in the flavin-cytochrome b region of the respiratory chain of beef heart submitochondrial particles. J. Biol. Chem. 1967;242:5169–5173. [PubMed] [Google Scholar]
- 7.Takeshige K, Minakami S. NADH- and NADPH-dependent formation of superoxide anions by bovine heart submitochondrial particles and NADH-ubiquinone reductase preparation. Biochem. J. 1979;180:129–135. doi: 10.1042/bj1800129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnamoorthy AIG, Hinkle P. Studies on the electron transfer pathway, topography of iron-sulfur centers, and site of coupling in NADH-Q oxidoreductase. J. Biol. Chem. 1988;263:17566–17575. [PubMed] [Google Scholar]
- 9.Ksenzenko M.Yu., Konstantinov AA, Khomutov GB, Tikhonov AN, Ruuge EK. Effect of electron transfer inhibitors on superoxide generation in the cytochrome bc1 site of the mitochondrial respiratory chain. FEBS Lett. 1983;155:19–24. doi: 10.1016/0014-5793(83)80200-2. [DOI] [PubMed] [Google Scholar]
- 10.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria. Central role of complex III. J. Biol. Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, Trumpower B. Superoxide anion generation by the cytochrome bc1 complex. Arch. Biochem. Biophys. 2003;419:198–206. doi: 10.1016/j.abb.2003.08.028. [DOI] [PubMed] [Google Scholar]
- 12.Loschen G, Azzi A, Richter C, Flohé L. Superoxide radicals as precursors of mitochondrial hydrogen peroxide. FEBS Lett. 1974;42:68–72. doi: 10.1016/0014-5793(74)80281-4. [DOI] [PubMed] [Google Scholar]
- 13.Grivennikova VG, Vinogradov AD. Generation of superoxide by the mitochondrial Complex I. Biochim. Biophys. Acta. 2006;1757:553–561. doi: 10.1016/j.bbabio.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Grivennikova VG, Cecchini G, Vinogradov AD. Ammonium-dependent hydrogen peroxide production by mitochondria. FEBS Lett. 2008;582:2719–2724. doi: 10.1016/j.febslet.2008.06.054. [DOI] [PubMed] [Google Scholar]
- 15.Grivennikova VG, Kareyeva AV, Vinogradov AD. What are the sources of hydrogen peroxide production by heart mitochondria? Biochim Biophys. Acta. 2010;1797:939–44. doi: 10.1016/j.bbabio.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crane FL, Glenn JL, Green D. Studies on the electron transfer system. IV. The electron transfer particles. Biochem. Biophys. Acta. 1956;22:475–487. doi: 10.1016/0006-3002(56)90058-0. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal. Biochem. 1997;253:162–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- 19.Gostimskaya IS, Grivennikova VG, Zharova TV, Bakeeva LE, Vinogradov AD. In situ assay of the intramitochondrial enzymes: use of alamethicin for permeabilization of mitochondria. Anal Biochem. 2003;313:46–52. doi: 10.1016/s0003-2697(02)00534-1. [DOI] [PubMed] [Google Scholar]
- 20.Grivennikova VG, Kotlyar AB, Karliner JS, Cecchini G, Vinogradov AD. Redox-dependent change of nucleotide affinity to the active site of the mammalian complex I. Biochemistry. 2007;46:10971–10978. doi: 10.1021/bi7009822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igamberdiev AU, Bykova NV, Ens W, Hill RD. Dihydrolipoamide dehydrogenase from porcine heart catalyzes NADH-dependent scavenging of nitric oxide. FEBS Lett. 2004;568:146–150. doi: 10.1016/j.febslet.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 22.Olsson JM, Xia L, Eriksson LC, Björnstedt M. Ubiquinone is reduced by lipoamide dehydrogenase and this reaction is potently stimulated by zinc. FEBS Lett. 1999;448:190–192. doi: 10.1016/s0014-5793(99)00363-4. [DOI] [PubMed] [Google Scholar]
- 23.Chen H-JC, Chen Y-M, Chang C-M. Lipoyl dehydrogenase catalyzes reduction of nitrated DNA and protein adducts using dihydrolipoic acid or ubiquinol as the cofactor. Chem Biol. Interact. 2002;140:199–213. doi: 10.1016/s0009-2797(02)00019-4. [DOI] [PubMed] [Google Scholar]
- 24.Badaby NE, Pang Y-P, Elpeleg O, Isaya G. Cryptic proteolytic activity of dihydrolipoamide dehydrogenase. Proc. Natl. Acad. Sci. USA. 2007;104:6158–6163. doi: 10.1073/pnas.0610618104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vinogradov AD. NADH/NAD+ interaction with NADH: Ubiquinone oxidoreductase (complex I). Biochim Biophys. Acta. 2008;1777:729–734. doi: 10.1016/j.bbabio.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williamson DH, Brosnan JT. Concentrations of metabolites in animal tissues. In: Bergmeyer HU, Gawehn K, editors. Methods of Enzymatic Analysis. Verlag Chemie Weinhem, Academic Press, Inc.; NY and London: 1974. pp. 2266–2301. [Google Scholar]
- 27.Ross-Inta C, Tsai C-Y, Giulivi C. The mitochondrial pool of free amino acids reflects the composition of mitochondrial DNA-encoded proteins: indication of a post-translation quality control for protein synthesis. Biosci. Rep. 2008;28:239–249. doi: 10.1042/BSR20080090. [DOI] [PMC free article] [PubMed] [Google Scholar]