Abstract
Acute anterior uveitis (AAU) is the most common form of autoimmune uveitis in the eye with few known autoantigens. Identification of autoantigens will improve our understanding of the molecular mechanisms and capability for disease diagnosis. Phage display is a powerful technology for autoantigen identification. However, because of uncontrollable reading frames, phage display with conventional cDNA libraries identifies high percentage of non-open reading frames (non-ORFs) with minimal implications for autoantigen identification. We recently developed ORF phage display technology with minimal reading frame problem. Herein we used ORF phage display to identify 18 patient-specific clones, including 16 ORFs encoding endogenous proteins as candidate autoantigens for AAU. One of the identified antigens was heterogeneous nuclear ribonucleoprotein H3 (Hnrph3) that was further characterized for AAU relevance and independently verified by Western blot. These results demonstrate that ORF phage display is a valuable approach for identification of unknown autoantigens.
Keywords: Acute anterior uveitis, heterogeneous nuclear ribonucleoprotein H3, Hnrph3, autoantigen, ORF phage display, disease biomarker
Introduction
Uveitis is a group of heterogeneous inflammatory eye diseases and an important cause of visual impairment and blindness. Uveitis and its complications are responsible for more than 10% of visual impairment in people registered legally blind under age 65 [1, 2]. Acute anterior uveitis (AAU) is the most common form of uveitis, accounting for 50–92% of total uveitis cases [1]. The human major histocompatibility complex (MHC) class I HLA-B27 is associated with an estimated 50% of AAU cases and is one of the most identifiable causes for AAU [1]. Despite extensive clinical and research investigations, however, the molecular mechanisms of AAU remain elusive. Identification of autoantigens directly for AAU patients will not only improve our understanding of the disease mechanisms, but also offer valuable biomarkers for the diagnosis and prognosis.
However, autoantigens are traditionally identified on case-by-case basis with daunting challenges. Efforts from multiple groups have identified less than a handful of autoantigens for anterior uveitis. Melanin-associated antigen (MAA) is a well-characterized antigen for the induction of experimental autoimmune anterior uveitis (EAAU) [3]. The N-terminal amino acid sequence of MAA is identical to type I collagen α-2 chain, which was also validated as an autoantigen in EAAU [2, 4]. A 15 kDa nuclear antigen with unknown identity was described for patients with juvenile chronic arthritis and uveitis [5]. β-B1-crytallin was identified as a primary ciliary body antigen for anterior uveitis [6].
Several technologies are being developed to facilitate identification of unknown autoantigens, including phage display [7, 8]. However, a major problem for phage display with conventional cDNA libraries is that high percentage of identified phage clones are non-open-reading-frame (non-ORFs) encoding short unnatural peptides, which have minimal implication in identification of genuine autoantigens [8–10]. To address the problem, we developed a new technology of ORF phage display capable of identifying real endogenous proteins with minimal reading frame problem [11]. We demonstrated ORF phage display as a new technology of functional proteomics by identifying unknown binding proteins for a protein bait, a non-protein molecule and even a whole-cell bait [11–13]. Its versatile applications, including identification of autoantigens, were discussed in a recent review [10].
In this study, we used ORF phage display to identify unknown AAU autoantigens. A total of 18 patient-specific clones were identified, including 16 ORFs encoding endogenous proteins. One of the antigens, heterogeneous nuclear ribonucleoprotein H3 (Hnrph3), was further characterized for the disease relevance and independently verified by Western blot. These results suggest that ORF phage display is a valuable technology for systematic identification of unknown autoantigens. The potential of ORF phage display in combination of other emerging technologies to accelerate identification of disease-relevant autoantigens in large scales is discussed.
Materials and methods
Subjects
Thirty-one patients diagnosed with AAU were enrolled, including 24 HLA-B27+, 3 HLA-B27, 1 HLA-B51+ and 3 with unknown HLA-B27 status. AAU was defined as the sudden onset of a primarily anterior inflammation in one eye with clinical symptoms of pain, redness and sensitivity to light. Additional 13 healthy subjects were recruited as controls without bias of HLA-B27. Blood was collected according to a protocol approved by the Institutional Review Board at the University of Miami.
Phage selection by competitive subtraction
IgG was purified from patient and control sera using protein G agarose columns, as previously described [7]. A new scheme of phage selection by competitive subtraction was developed as the following. Patient IgG was coated on ELISA plates (10 μg/ml, 100 μl/well) and blocked with 1% polyvinyl alcohol, as described [11]. An ORF phage display library [11] (1 × 1011 pfu in 0.5 ml) was pre-incubated with pooled control IgGs (0.2 mg/ml) in the presence of 0.2% Tweeen-20 for 30 min at 4°C, followed by incubation with the immobilized patient IgG for 1 h at 4°C. The wells were washed with phosphate-buffered saline (PBS) containing 0.2% Tween-20, 6 times with 5 minutes each. The bound phages were eluted by 3C protease cleavage (~1 unit/100 l) (Novagen, Madison, WI) at 4°C for 16 h, as described [11]. An aliquot of eluted phages was quantified by plaque assay, and the remaining eluted phages were amplified in BLT5615 bacteria and used as input for the next round of selection. To improve the subtraction efficiency, the control IgG was increased to 0.4 and 0.8 mg/ml for the second and third rounds of selection with the enriched phage lysates reduced to 1 × 1010 and 1 × 109 pfu, respectively. After three rounds of phage selection, post-panning ORF selection with immobilized streptavidin was performed to enrich ORF clones, as described [10, 11].
Colorimetric screening for patient-specific clones
The colorimetric screening was carried out as previously described [11]. Briefly, individual phage clones were randomly picked from plates of enriched phages and amplified in BLT7FLAG bacteria. The phage lysates (50 l/well) were incubated with immobilized patient IgG or control IgG in the presence of Tween-20 (0.2%) for 1 h. After washing, bound phages were quantified with anti-FLAG M2 monoclonal antibody (mAb) (Sigma, St. Louis, MO), followed by horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG and colorimetric assay as described [11]. The clones with increased binding specificity to patient IgG versus control IgG were subject to further validation by phage binding assay.
Phage binding assay
The phage binding assay was performed as the colorimetric screening with the following modifications. The clonal phages were amplified in BLT5615 bacteria. The phage lysates (50 l/well) were incubated with immobilized patient IgG or control IgG as described in the colorimetric screening. After washing, the bound phages were eluted by 3C protease cleavage and quantified by plaque assay [11]. Only the clones with more than ~10-fold increase in binding specificity to patient IgG versus control IgG were identified by DNA sequencing. Identified clones were further analyzed for their binding specificity to all other patient IgGs and control IgGs by phage binding assay.
Western blot
Maltose binding protein-Hnrph3 fusion protein (MBP-Hnrph3) was prepared as follows. The cDNA insert of cloned Hnrph3-phage was amplified by PCR using primers of 5′-TCGGATCCGACAGGATGCGAGATGGAAG-3′ and 5′-AACTCGAGTTAAGAACCTCC CATTCCAGAGC-3′ (underline sequences for BamHI and XhoI sites). After digestion with BamHI and XhoI, the PCR product was inserted into pMAL-c4E plasmid (New England Biolab, Piscataway, NJ) at BamHI and SalI sites. The resulting plasmid was verified by sequencing. MBP-Hnrph3 and control MBP were expressed in BL21(DE3) cells, as described [11], and purified with amylose columns (New England Biolab) according to the manufacturer’s protocol. The purified recombinant proteins were verified by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Commassie blue staining. MBP-Hnrph3 and MBP were used to analyze the immunoreactivity of patient or control IgG by Western blot with 12% SDS-PAGE, as described previously [7].
Data analysis
The sensitivities and specificities of the binding data of Hnrph3-phage for assessing the disease relevance was summarized with ROC (receiver operating characteristic) curve analysis as previously described [14, 15]. Additionally, the averages for patients and controls were compared with the two-sample t-test (the unpooled variance version was used due to variance heterogeneity).
Results
Autoantigen identification
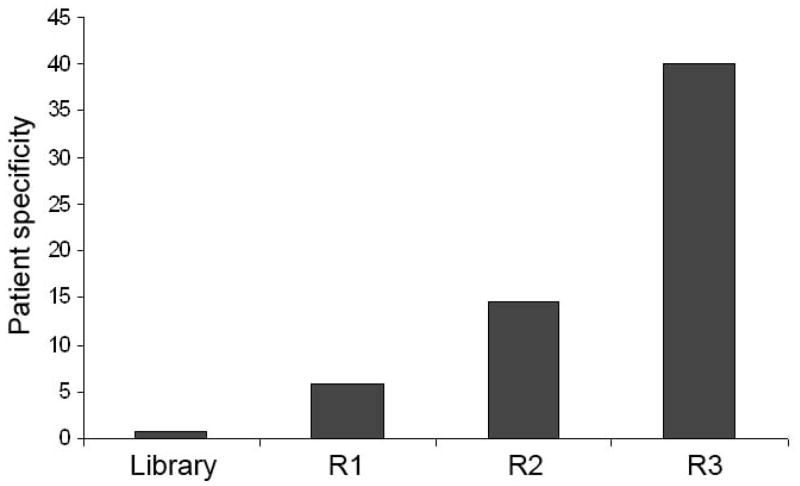
One of the most daunting challenges for autoantigen identification is that only a tiny fraction of total IgG is disease relevant. Thus, it is critical to minimize the interference of vast majority of disease-irrelevant common antibodies in patient blood for efficient identification of patient-specific autoantigens. This study designed a new scheme of competitive subtraction to improve the patient specificity of enriched phage clones. In the new scheme, patient IgG was immobilized on ELISA plates, while the excessive amount of control IgG was in the binding solution to compete for the phage clones binding to common IgG. The advantage of competitive subtraction is that the amount of control IgG and enriched phage lysates can be conveniently adjusted to improve the subtraction efficiency. The ratio of control IgG versus immobilized IgG was at least 80:1 at Round 3 selection. The increased ratio of control IgG versus enriched phage lysate further enhanced the subtraction efficiency. After 3 rounds of selection, the enriched phages showed ~40-fold increase in binding specificity to patient IgG versus control IgG (Fig. 1).
Fig. 1.
Patient specificity of enriched phages. The ORF phage display cDNA library was enriched by phage selection with competitive subtraction for three rounds. Enriched phages at each round were reanalyzed for their binding activity to patient IgG versus control IgG. The patient specificity was expressed as the ratio of total bound phages to patient IgG versus control IgG. Unselected ORF library was used as a negative control.
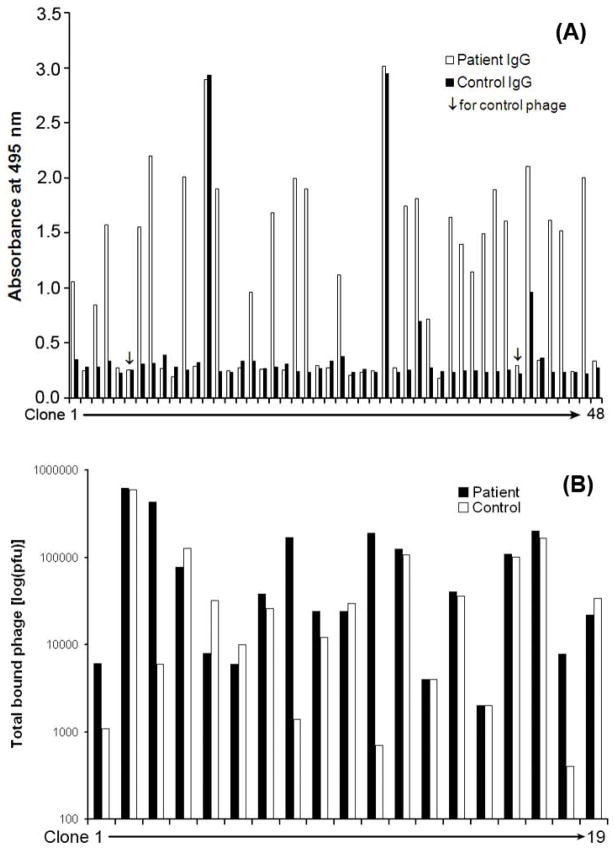
The screening of individual phage clones with binding activity to patient IgG, but not the control IgG, was carried out by a recently developed colorimetric assay [11]. Compared with plaque assay for T7 phage, the colorimetric assay substantially increased the screening efficiency. The results showed that 23 out of 46 clones (~50%) enriched by competitive subtraction were patient-specific (Fig. 2A). Only two clones (clone 13 and 29) had binding activity to both patient IgG and control IgG. Additionally, two other clones (clone 32 and 42) showed partial binding activity to control IgG. We included our earlier data for patient specificity by conventional subtractive phage display with absorption [7], which only identified 4 out of 19 clones (~21%) with patient specificity (Fig. 2B). The results suggest that competitive subtraction is more efficient than absorptive subtraction in term of enriching patient-specific clones. All positive clones with 3-fold increase in binding activity to patient IgG versus control IgG were reverified for their patient specificity by more sensitive and reliable plaque assay. A total of 18 patient-specific clones were isolated with at least 10-fold increase in the binding specificity. DNA sequencing identified 16 ORFs encoding real endogenous proteins (Table 1).
Fig. 2.
Screening of patient-specific clones. (A) Patient specificity by competitive subtraction. Individual phage clones were randomly isolated from phages selected by competitive subtraction, amplified in BLT7FLAG bacteria, bound to patient IgG and control IgG, and quantified by the colorimetric assay. Clone 6 and 41 were control phage with no foreign cDNA insert. (B) Patient specificity by absorptive subtraction. The data for the screening of individual phage clones enriched by phage selection with conventional absorptive subtraction [7] was included. The bound phages in (B) were quantified by plaque assay.
Table 1.
Identified candidate AAU autoantigens by ORF phage display
| Protein name | Accession number | Matched aa residues |
|---|---|---|
| Heterogeneous nuclear ribonucleoprotein H3(Hnrph3) | NM_001079824 | 167D-280S |
| Nucleolin (Ncl) | NM_010880 | 15K-108G |
| Chromodomain helicase DNA binding protein 2 (Chd2) | XM_001478970 | 1347K-1411R |
| Syndecan 3 (Sdc3) | NM_011520 | 41R-189P |
| Myosin, heavy polypeptide 4, skeletal muscle (Myh4) | NM_010855 | 1134A-1225E |
| Topoisomerase (DNA) I (Top1) | NM_009408 | 64K-191D |
| Piccolo (presynaptic cytomatrix protein) (Pclo) | NM_011995 | 573T-723G |
| ATPase, Na+/K+ transporting, beta 1 polypeptide (Atp1b1) | NM_009721 | 1M-23E |
| THO complex 2 (Thoc2) | NM_001033422 | 1205G-1257N |
| Multiple EGF-like-domains 8 (Megf8) | XM_001478942 | 2697R->2813L |
| Ubiquitin associated protein 2-like (Ubap21) | NM_028475 | 22Q-90N |
| Neurofilament, heavy polypeptide (Nefh) | NM_010904 | 85E-121T |
| Apoptotic chromatin condensation inducer 1 (Acin1) | NM_023190 | 856A-964E |
| Chromatin modifying protein 4B (Chmp4b) | NM_029362 | 72K-207K |
| Ankyrin repeat domain 12 (Ankrd12) | NM_001025572 | 760E-893D |
Disease relevance
Among the identified autoantigens was the nuclear protein of Hnrph3. The cDNA insert of the cloned phage encoded 113 amino acids of Hnrph3 with 100% homology between human and mouse. The immunoreactivity profiles of patient and control IgGs to Hnrph3 were analyzed by phage binding assay with the clonal Hnrph3-phage. The results showed that 11 out of 31 patient IgGs had high binding activity to Hnrph3. On the other hand, most control IgGs had relatively low binding activity to Hnrph3-phage (Fig. 3A). There was a significant difference between the mean logarithms of the test values with 3.3 (0.7) for patients and 2.9 (0.3) for controls (p=0.007, unpooled variance t-test). Sensitivities and specificities are summarized in an ROC curve (Fig. 3B). If we accept a specificity of ~70%, the sensitivity is ~65%. In other words, if we are willing to incur ~30% false positives in the controls, we will detect about 65% of AAU cases with Hnrph3 immunoreactivity as a biomarker.
Fig. 3.
Hnrph3 is an AAU-relevant autoantigen. (A) Hnrph3-phage binding to patient IgGs versus control IgGs. Bound phages were quantified by plaque assay. (B) ROC curve analysis summarizing sensitivities and specificities of the binding results in (A) to identify patients versus controls with area under the curve of 0.624 (SE=0.079). A significant difference was detected between the mean logarithms of the test values (p=0.007, unpooled variance t-test). With an assumed specificity of ~70%, the expected sensitivity of Hnrph3 as an AAU biomarker is ~65%. The diagonal segments are produced by ties.
The patient specificity of Hnrph3 was further validated by Western blot. Hrnph3 was expressed as a MBP fusion protein and purified using an amylose column. Purified protein MBP-Hnrph3 and control MBP were verified by SDS-PAGE (Fig. 4A). Western blot analysis revealed that patient IgG recognized only MBP-Hnrph3, but not MBP (Fig. 4B). The control IgG had no binding activity to both proteins.
Fig. 4.
Independent validation of Hnrph3 as an autoantigen. (A) Purified MBP-Hnrph3 and MBP control (~5 g/lane) were analyzed by SDS-PAGE. (B) Immunoreactivity of patient IgG. MBP-Hnrph3 and MPB were analyzed in duplicates by Western blot using patient or control IgG.
Discussion
Autoantigens are critical for in-depth understanding of molecular mechanisms of autoimmune diseases. Several antigens, including endotoxin, MAA and myelin basic protein (MBP), have been used to induce AAU in animals [1, 4, 16]. These animal models are valuable to provide molecular insights into immunological mechanisms for the pathogenesis. However, the drawback is that an antigen may be uveitogenic in one particular mouse strain, but not in others [17]. Thus, it may not always be appropriate to extrapolate the autoantigens directly from animal models to patients. Although transgenic mouse models expressing human MHC class I, such as HLA-B27, may bridge the gap [18], identification of antigens directly in patients is critical for our understanding of the disease pathogenesis. It is worth noting that not all identified autoantigens in patients are disease-triggering autoantigens. Nonetheless, autoantigens passively associated with AAU is still valuable as biomarkers for disease diagnosis and prognosis. Despite the importance, few human autoantigens for AAU have been identified.
Hnrph3 as an autoantigen is yet to be further characterized, including more reliable analysis for specificity and sensitivity with a larger number of patients and controls and quantitative analysis of antibody activity change over the course of AAU. Regardless of the role of Hnrph3 in the disease pathogenesis, the autoantigen could be used as a biomarker for AAU and should be investigated for the relevance to other HLA-B27-associated spondyloarthropathies.
Serological disease biomarkers in general can be classified into two major categories: immunoglobulins and non-immunoglobulin molecules. The latters are various proteins and non-protein molecules in the plasma of patients, but not healthy subjects, and can be identified by different technologies, including mass spectrometry (MS) [19]. The challenge of this method is that huge amount of disease-irrelevant plasma proteins, such as albumin and transferrin, will interfere with the detection of tiny amount of disease-relevant biomarkers, which are usually 106–1010-fold less than albumin [20]. Alternatively, antigens or autoantigens specifically recognized by patient immunoglobulins may be identified by technologies [21], such as SEREX (serological identification of antigens by recombinant expression cloning) or phage display [7, 22]. Identified antigens can be used as biomarkers to profile the reactivity of patient immunoglobulins for disease diagnosis [8, 23]. Phage display with multi-round selection and amplification is capable of enriching and identifying less abundant antigens. Consequently, it should be more efficient and sensitive to identify autoantigens than SEREX.
One of the major challenges for phage display to identify unknown autoantigens is the reading frame problem. Because of unpredictable reading frames for cellular proteins, it is difficult to fuse cDNA libraries of cellular proteins to phage capsid in correct reading frames [10]. Only an estimated 5% of cDNA library clones constructed by random priming method may be in the correct reading frames due to three protein reading frames, two orientations, 5′- and 3′-untranslated regions. As a result, previous studies showed that only ~6–10% of identified phage clones encoded real endogenous proteins [8, 9].
Various strategies have been developed to tackle the reading frame problem, including ORF libraries, with various successes [10]. Compared with filamentous phage display [24], the system of T7 ORF phage display recently developed by our group is advantageous for unbiased display of cellular proteins [11]. Filamentous phage requires the displayed proteins to be secreted through E. coli membrane into the periplasmic space and, therefore, is only suitable to display a subset of eukaryotic proteins [25]. In contrast, T7 phage can display eukaryotic proteins without such requirement [26, 27], and should be more appropriate for unbiased identification of autoantigens. However, a drawback is that T7 phage lacks commercial antibodies for phage quantification by ELISA-like colorimetric assay. Anti-T7 capsid mAb from Novagen is not recommended for phage quantification by the company. Fortunately, we recently developed a bacterial strain of BLT7FLAG, in which amplified phages are labeled with FLAG tag on capsid 10A protein without interfering with foreign protein display on capsid 10B [11]. Given that each phage particle carries more than 400 copies of capsid 10A, multiple copies of FLAG on single phage surface will substantially improve the sensitivity to quantify bound phages by anti-FLAG mAb for the colorimetric screening. A potential limitation for all phage display systems is the lack of appropriate posttranslational modifications, such as glycosylation. Consequently, polysaccharides as antigens will not be identified. Nonetheless, phage display is a powerful technology for identification of unknown protein antigens.
The new competitive subtraction should be more efficient to enrich patient-specific clones than conventional subtraction by absorption (Fig. 2), because the amount of soluble control IgG far exceeded the immobilized patient IgG for the former strategy. Moreover, the subtraction efficiency, patient specificity and phage clone diversity can be conveniently monitored by plaque assay at each round of selection and adjusted by increasing or decreasing the amount of control IgG or enriched phage lysate.
It is worth noting that an estimated ~10–40% of enriched clones were non-ORFs after three rounds of phage selection with patient IgG. Such high percentage of non-ORFs were not observed in our previous studies with other bait molecules or cells [11–13]. One possible explanation is that antibodies may preferentially recognize linear short peptide epitopes encoded by non-ORFs. However, post-panning ORF selection with immobilized streptavidin substantially improved the percentage of ORFs before the screening for individual patient-specific clones.
Efficient identification of autoantigens in large scales by phage display faces three major technical barriers: (a) protein reading frame problem; (b) patient specificity; and (c) disease relevance. The reading frame problem and patient specificity are solved in this study by ORF phage display and competitive subtraction, respectively. The remaining challenge is how to efficiently identify disease-relevant antigens in large scales. In addition to the conventional approach described in Fig. 3 for Hnrph3, several emerging technologies of phage display may facilitate efficient identification of disease-relevant antigens in large scales. For example, phage microarray is a powerful technology to screen a large number of enriched phage clones with disease relevance. The successful demonstration of phage microarray for cancer diagnosis [8, 23] implicates that the combination of ORF phage display and phage microarray will substantially improve our capability to efficiently identify disease-relevant autoantigens for the diagnosis of cancer and autoimmune diseases. However, an unanswered question is whether phage microarray with only ~5–15 copies of foreign proteins displayed on each T7 phage surface is sensitive enough to detect antigens with moderate antibody titer. In other words, is phage display sensitive enough for global mapping of all valuable antigens? An alternative strategy is to combine phage display with next generation DNA sequencing. Two recent studies showed that next generation sequencing can globally analyze the frequencies of all enriched phage clones to identify binding proteins or peptides [28, 29]. Thus, the combination of ORF phage display with next generation sequencing will drastically improve the efficiency of globally mapping all disease-relevant antigens by comparing the frequency profiles of all enriched clones for patient group versus control group without screening for individual phage clones.
Conclusion
In summary, this study identified Hnrph3 as an autoantigen for AAU. ORF phage display is a promising technology for systematic identification of unknown disease-relevant autoantigens to advance our understanding of autoimmune diseases and provide valuable biomarkers for disease diagnosis.
Acknowledgments
This project was supported by NIH R01EY016211, R01EY016211-05S1, P30-EY014801 and an institutional grant from Research to Prevent Blindness.
Abbreviations
- AAU
acute anterior uveitis
- EAAU
experimental autoimmune anterior uveitis
- Hnrph3
heterogeneous nuclear ribonucleoprotein H3
- mAb
monoclonal antibody
- MBP
maltose binding protein
- ORF
open reading frame
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chang JH, McCluskey PJ, Wakefield D. Acute anterior uveitis and HLA-B27. Surv Ophthalmol. 2005;50:364–388. doi: 10.1016/j.survophthal.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Bora NS, Kaplan HJ. Intraocular diseases - anterior uveitis. Chem Immunol Allergy. 2007;92:213–220. doi: 10.1159/000099272. [DOI] [PubMed] [Google Scholar]
- 3.Bora NS, Kim MC, Kabeer NH, Simpson SC, Tandhasetti MT, Cirrito TP, Kaplan AD, Kaplan HJ. Experimental autoimmune anterior uveitis. Induction with melanin-associated antigen from the iris and ciliary body. Invest Ophthalmol Vis Sci. 1995;36:1056–1066. [PubMed] [Google Scholar]
- 4.Bora NS, Sohn JH, Kang SG, Cruz JM, Nishihori H, Suk HJ, Wang Y, Kaplan HJ, Bora PS. Type I collagen is the autoantigen in experimental autoimmune anterior uveitis. J Immunol. 2004;172:7086–7094. doi: 10.4049/jimmunol.172.11.7086. [DOI] [PubMed] [Google Scholar]
- 5.Neuteboom GH, Hertzberger-ten Cate R, de Jong J, van den Brink HG, Feltkamp TE. Antibodies to a 15 kD nuclear antigen in patients with juvenile chronic arthritis and uveitis. Invest Ophthalmol Vis Sci. 1992;33:1657–1660. [PubMed] [Google Scholar]
- 6.Stempel D, Sandusky H, Lampi K, Cilluffo M, Horwitz J, Braun J, Goodglick L, Gordon LK. BetaB1-crystallin: identification of a candidate ciliary body uveitis antigen. Invest Ophthalmol Vis Sci. 2003;44:203–209. doi: 10.1167/iovs.01-1261. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Davis JL, Li W. Identification of tribbles homolog 2 as an autoantigen in autoimmune uveitis by phage display. Mol Immunol. 2005;42:1275–1281. doi: 10.1016/j.molimm.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 8.Lin HS, Talwar HS, Tarca AL, Ionan A, Chatterjee M, Ye B, Wojciechowski J, Mohapatra S, Basson MD, Yoo GH, Peshek B, Lonardo F, Pan CJ, Folbe AJ, Draghici S, Abrams J, Tainsky MA. Autoantibody approach for serum-based detection of head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2396–2405. doi: 10.1158/1055-9965.EPI-07-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalnina Z, Silina K, Meistere I, Zayakin P, Rivosh A, Abols A, Leja M, Minenkova O, Schadendorf D, Line A. Evaluation of T7 and lambda phage display systems for survey of autoantibody profiles in cancer patients. J Immunol Methods. 2008;334:37–50. doi: 10.1016/j.jim.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Caberoy NB. New perspective for phage display as an efficient and versatile technology of functional proteomics. Appl Microbiol Biotechnol. 2010;85:909–919. doi: 10.1007/s00253-009-2277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caberoy NB, Zhou Y, Jiang X, Alvarado G, Li W. Efficient identification of tubby-binding proteins by an improved system of T7 phage display. J Mol Recognit. 2010;23:74–83. doi: 10.1002/jmr.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caberoy NB, Zhou Y, Alvarado G, Fan X, Li W. Efficient identification of phosphatidylserine-binding proteins by ORF phage display. Biochem Biophys Res Commun. 2009;386:197–201. doi: 10.1016/j.bbrc.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caberoy NB, Maiguel D, Kim Y, Li W. Identification of tubby and tubby-like protein 1 as eat-me signals by phage display. Exp Cell Res. 2010;316:245–257. doi: 10.1016/j.yexcr.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNeil BJ, Keller E, Adelstein SJ. Primer on certain elements of medical decision making. N Engl J Med. 1975;293:211–215. doi: 10.1056/NEJM197507312930501. [DOI] [PubMed] [Google Scholar]
- 15.Zhong L, Coe SP, Stromberg AJ, Khattar NH, Jett JR, Hirschowitz EA. Profiling tumor-associated antibodies for early detection of non-small cell lung cancer. J Thorac Oncol. 2006;1:513–519. [PubMed] [Google Scholar]
- 16.Adamus G, Amundson D, Vainiene M, Ariail K, Machnicki M, Weinberg A, Offner H. Myelin basic protein specific T-helper cells induce experimental anterior uveitis. J Neurosci Res. 1996;44:513–518. doi: 10.1002/(SICI)1097-4547(19960615)44:6<513::AID-JNR1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 17.Sun B, Sun SH, Chan CC, Wiggert B, Caspi RR. Autoimmunity to a pathogenic retinal antigen begins as a balanced cytokine response that polarizes towards type 1 in a disease-susceptible and towards type 2 in a disease-resistant genotype. Int Immunol. 1999;11:1307–1312. doi: 10.1093/intimm/11.8.1307. [DOI] [PubMed] [Google Scholar]
- 18.Hacquard-Bouder C, Ittah M, Breban M. Animal models of HLA-B27-associated diseases: new outcomes. Joint Bone Spine. 2006;73:132–138. doi: 10.1016/j.jbspin.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 20.Schiess R, Wollscheid B, Aebersold R. Targeted proteomic strategy for clinical biomarker discovery. Mol Oncol. 2009;3:33–44. doi: 10.1016/j.molonc.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan HT, Low J, Lim SG, Chung MC. Serum autoantibodies as biomarkers for early cancer detection. Febs J. 2009;276:6880–6904. doi: 10.1111/j.1742-4658.2009.07396.x. [DOI] [PubMed] [Google Scholar]
- 22.Jager D. Potential target antigens for immunotherapy identified by serological expression cloning (SEREX) Methods Mol Biol. 2007;360:319–326. doi: 10.1385/1-59745-165-7:319. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Yu J, Sreekumar A, Varambally S, Shen R, Giacherio D, Mehra R, Montie JE, Pienta KJ, Sanda MG, Kantoff PW, Rubin MA, Wei JT, Ghosh D, Chinnaiyan AM. Autoantibody signatures in prostate cancer. N Engl J Med. 2005;353:1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 24.Zacchi P, Sblattero D, Florian F, Marzari R, Bradbury AR. Selecting open reading frames from DNA. Genome Res. 2003;13:980–990. doi: 10.1101/gr.861503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paschke M. Phage display systems and their applications. Appl Microbiol Biotechnol. 2006;70:2–11. doi: 10.1007/s00253-005-0270-9. [DOI] [PubMed] [Google Scholar]
- 26.Bratkovic T. Progress in phage display: evolution of the technique and its application. Cell Mol Life Sci. 2010;67:749–767. doi: 10.1007/s00018-009-0192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krumpe LR, Atkinson AJ, Smythers GW, Kandel A, Schumacher KM, McMahon JB, Makowski L, Mori T. T7 lytic phage-displayed peptide libraries exhibit less sequence bias than M13 filamentous phage-displayed peptide libraries. Proteomics. 2006;6:4210–4222. doi: 10.1002/pmic.200500606. [DOI] [PubMed] [Google Scholar]
- 28.Di Niro R, Sulic AM, Mignone F, D’Angelo S, Bordoni R, Iacono M, Marzari R, Gaiotto T, Lavric M, Bradbury AR, Biancone L, Zevin-Sonkin D, De Bellis G, Santoro C, Sblattero D. Rapid interactome profiling by massive sequencing. Nucleic Acids Res. 2010;38:e110. doi: 10.1093/nar/gkq052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dias-Neto E, Nunes DN, Giordano RJ, Sun J, Botz GH, Yang K, Setubal JC, Pasqualini R, Arap W. Next-generation phage display: integrating and comparing available molecular tools to enable cost-effective high-throughput analysis. PLoS One. 2009;4:e8338. doi: 10.1371/journal.pone.0008338. [DOI] [PMC free article] [PubMed] [Google Scholar]