Abstract
Understanding the links between developmental patterning mechanisms and force-producing cytoskeletal mechanisms is a central goal in studies of morphogenesis. Gastrulation is the first morphogenetic event in the development of many organisms. Gastrulation involves the internalization of surface cells, often driven by the contraction of actomyosin networks that are deployed with spatial precision -- both in specific cells and in a polarized manner within each cell. These cytoskeletal mechanisms rely on different cell fate and cell polarity regulators in different organisms. C. elegans gastrulation presents an opportunity to examine the extent to which diverse mechanisms may be used by dozens of cells that are internalized at distinct times within a single organism. We identified 66 cells that are internalized in C. elegans gastrulation, many of which were not known previously to gastrulate. To gain mechanistic insights into how these cells internalize, we genetically manipulated cell fate, cell polarity and cytoskeletal regulators and determined the effects on cell internalization. We found that cells of distinct lineages depend on common actomyosin-based mechanisms to gastrulate, but different cell fate regulators, and, surprisingly, different cell polarity regulators. We conclude that diverse cell fate and cell polarity regulators control common mechanisms of morphogenesis in C. elegans. The results highlight the variety of developmental patterning mechanisms that can be associated with common cytoskeletal mechanisms in the morphogenesis of an animal embryo.
Keywords: morphogenesis, gastrulation, C. elegans
INTRODUCTION
Morphogenesis is the process by which cells of an embryo become organized in a way that can produce a functioning organism. One key goal in studying morphogenesis is to understand the links between patterning mechanisms and the cytoskeletal mechanisms that drive embryonic shape changes (Wieschaus, 1995). Such links are being identified in various developmental systems (Harris et al., 2009; Sawyer et al., 2010). One theme emerging is that diverse animals employ a handful of common cytoskeletal mechanisms, deployed with spatial precision by varied and often organism-specific patterning mechanisms. For example, contraction of an apical actomyosin network internalizes cells in diverse systems. In Drosophila gastrulation, an actomyosin network is spatially regulated by an apically-localized guanine nucleotide exchange factor for Rho (RhoGEF). Recruitment of RhoGEF to the apical sides of cells depends on the secreted protein Fog and the transmembrane protein T48 (Barrett et al., 1997; Kolsch et al., 2007; Nikolaidou and Barrett, 2004; Rogers et al., 2004). Fog and T48 homologs are not known to exist in vertebrates, where instead the F-actin binding protein Shroom3 acts as an apical determinant in epithelial morphogenesis. Shroom3 recruits a more direct myosin activator, Rho kinase, to the apical sides of cells, and also affects the apical localization of myosin and F-actin (Haigo et al., 2003; Hildebrand, 2005; Hildebrand and Soriano, 1999; Nishimura and Takeichi, 2008). C. elegans uses yet another mechanism for apico-basal polarization of myosin-dependent forces. Here, a putative GTPase activating protein for Cdc42 localizes basolaterally in response to cell contacts, where it prevents the basolateral localization of apical PAR proteins that are important for apical myosin localization (Anderson et al., 2008; Nance et al., 2003). These cases illustrate that different apico-basal patterning mechanisms can control apical constriction in different systems. Similar data exist for cell fate: a diverse set of fate regulators control the apical constriction machinery in various animal systems (see Sawyer et al. 2010 for review).
The degree of such diversity within an organism is less clear. There are cases where different cells internalize by morphologically distinct processes in a single animal system, for example between gastrulation and tracheal tube formation in Drosophila. In these cases, different patterning mechanisms are indeed known to act upstream of common cytoskeletal mechanisms (Brodu and Casanova, 2006; Leptin and Grunewald, 1990). However, it is not well understood to what extent such diversity of spatial patterning mechanisms might regulate repeated, superficially similar morphogenetic processes within a single organism. When many cells move in the same direction and in apparently similar patterns, do they use the same cytoskeletal mechanisms to do so, and are these mechanisms associated with common upstream spatial patterning mechanisms? Answering these questions will be a key step toward outlining the mechanistic links between patterning and cytoskeletal-based force production. Answers may provide insights into which kinds of mechanisms discovered in model systems will be most directly relevant to understanding morphogenesis-related birth defects in humans. This information is valuable because morphogenesis-related birth defects like neural tube closure defects are common and yet mechanistically poorly-understood human birth defects (Copp and Greene, 2010).
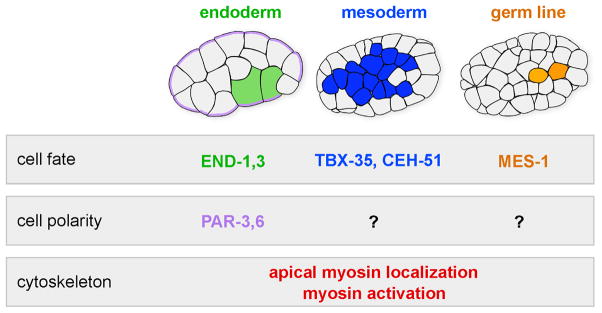
Gastrulation is the earliest morphogenetic process in many animal embryos. During gastrulation, cells that will establish the internal germ layers – the endoderm, the mesoderm and the germ line – move from the outside of the embryo to the interior, leaving just the ectoderm at the surface. C. elegans gastrulation is a valuable model for dissecting the cellular and molecular mechanisms involved in cell internalization because internalization begins early in development, at the 28 cell stage, soon after cell fates are acquired. C. elegans gastrulation starts with the internalization of two endodermal precursor cells by actomyosin-dependent apical constriction (Lee et al., 2006; Lee and Goldstein, 2003; Nance et al., 2003; Nance and Priess, 2002; Anderson et al., 2008). Cell fate, cell polarity and cytoskeletal mechanisms control the timely internalization of the endoderm (Fig. 1). Endodermal cell fate appears necessary and sufficient for the endodermal precursors to internalize on schedule (Lee et al., 2006; Maduro et al., 2005; Zhu et al., 1997). Apically-localized PAR polarity proteins are required for enrichment of a nonmuscle myosin II motor (NMY-2) on the apical surface of each endodermal precursor cell (Nance et al., 2003). These cells undergo an actomyosin-driven apical constriction: Phosphorylation of the regulatory light chains of myosin at a conserved site is thought to activate the contraction of the actomyosin network at the apical cortex in the endodermal precursor cells, resulting in the movement of neighboring cells under the endodermal precursors and hence the internalization of the endoderm (Lee et al., 2006). In addition to the endodermal precursor cells, other cells internalize at distinct times, nearly all from the ventral surface of the embryo (Nance and Priess, 2002; Sulston et al., 1983;). The relatively small number of cells involved suggests that an understanding of the regulation of gastrulation at the level of individual cells is possible.
Fig. 1. Cell fate, cell polarity, and cytoskeletal mechanisms regulating internalization of the endoderm in C. elegans.
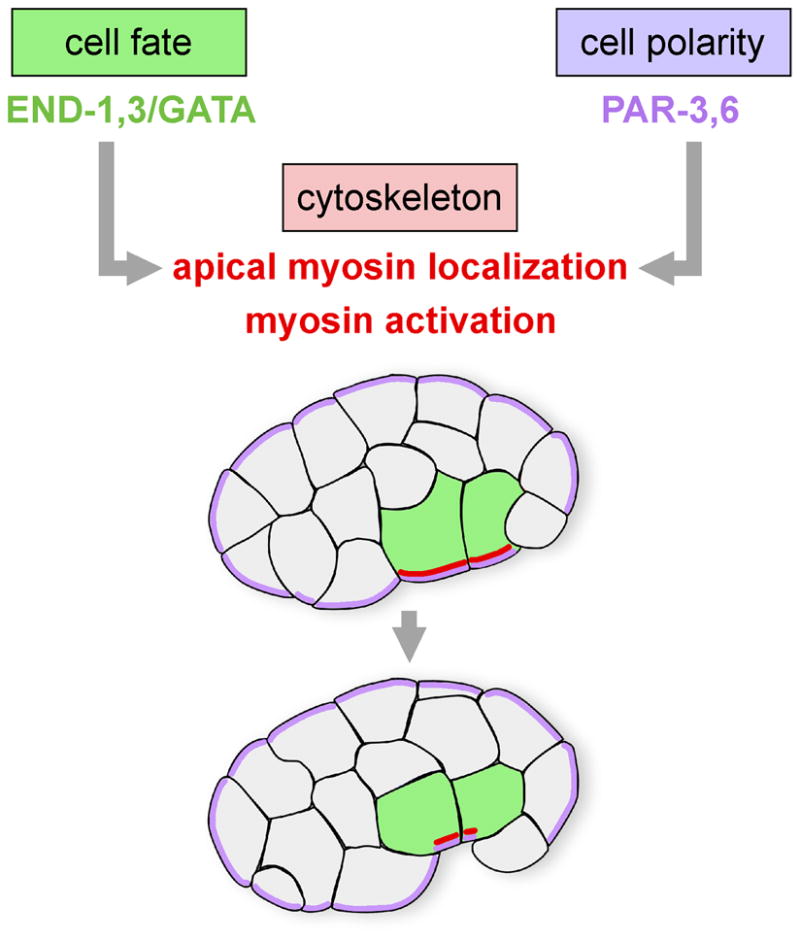
GATA-like transcription factors END-1,-3 and the cell polarity proteins PAR-3,-6 regulate internalization of two endodermal precursors (green) at the 28-cell stage by actomyosin-dependent apical constriction. PAR-3 and PAR-6 (purple) are required for apical myosin localization (red), and the endoderm-specifying GATA factors are predicted to do the same, because myosin is enriched apically only in these cells at this stage.
Here, we have examined cell fate and cell polarity regulators as well as cytoskeletal mechanisms that are used by multiple cells during C. elegans gastrulation. Before we began to study mechanism, some descriptive work was necessary. Six founder cells are traditionally recognized in C. elegans: AB, MS, E, C, D, and P4 (Sulston et al., 1983). Time intervals have been reported during which progeny of each of these founder cells become internalized (Nance and Priess, 2002; Sulston et al., 1983;), but not all of the gastrulating cells had been identified. We show that sixty-six cells gastrulate in C. elegans – the two endodermal precursors and then sixty-four additional cells that contribute primarily to the nervous system, the mesoderm and the germ line. We addressed the mechanisms by which these cells gastrulate using gene disruption, live imaging, and protein localization experiments to identify key cell fate regulators, cell polarity mechanisms, and cytoskeletal mechanisms. Our results demonstrate that actomyosin-based cytoskeletal mechanisms function to internalize diverse cells, but that cell internalization is under the control of different cell fate and cell polarity mechanisms in different cells. The results lead to a mechanistic outline of C. elegans gastrulation in which multiple patterning mechanisms are associated with common cytoskeletal mechanisms that internalize diverse cells at distinct times.
MATERIALS AND METHODS
Strains and Worm Maintenance
Nematodes were cultured as described (Brenner, 1974). Wild-type N2 (Bristol) and strains in Supplementary Table 1 were used. Strains were maintained at 20°C except nmy-2 (ne3409), which was maintained at 15°C and filmed at 25°C, and mes-1 (bn7), which was maintained at 15°C and shifted to 25°C 1–2 days before each experiment.
DIC and Confocal Time-Lapse Microscopy
Embryos were mounted and DIC images were acquired as by Roh-Johnson and Goldstein (2009). Time-lapse images were acquired at 1 μm optical sections every 1 minute. Internalization was scored when a cell was beneath the surface of the embryo, fully covered by other cells, before division. Spinning disk confocal images were acquired and processed as described previously (Roh-Johnson and Goldstein, 2009). Lateral surfaces of NMY-2::GFP or PH::mCherry; NMY-2::GFP embryos were filmed by DIC until cells of interest were born. For NMY-2::GFP embryos, images were acquired every 30 seconds under both DIC and fluorescence in several planes once cells of interest that would internalize were born. One plane was chosen for analysis. To analyze PH::mCherry; NMY-2::GFP dynamics, one to three planes 0.5 μm apart were acquired every 5 seconds in each channel. One plane was chosen for analysis. The embryonic cell lineage was drawn by a custom-written program (available on request) using timing data from Wormbase release 181, from wormbase.org. Analysis including kymographs was done in Metamorph software (Molecular Devices).
Cell Fate Transformation
tbx-35;ceh-51 embryos were filmed and then assessed for lethality and mCherry expression associated with a rescuing plasmid several hours later. Only films of embryos that both died and lacked mCherry fluorescence were analyzed for MS descendant internalization defects. For mes-1, only embryos where P4 and D divided within 1 minute of each other were included in analysis.
Polarity Regulators
par-3(ZF1) and par-6(ZF1) embryos were filmed by DIC microscopy,. Timing of MS and D divisions and internalization timing in the MS and D lineages were recorded. p values from unpaired Student’s t-tests were obtained by comparing the timing of internalization of sister cell pairs between wild-type, par-3(ZF1), and par-6(ZF1) embryos, using a Bonferroni correction for multiple comparisons. Because 24 comparisons were made, values were considered significantly different if p<0.0021 (0.05/24). Among all MS and C descendants, the internalization timing of one pair of cells in a par-6(ZF1) mutant was marginally statistically different than wild-type (MSpppa/MSpppp, p=0.002).
Laser Delay
Cells were irradiated as by Lee et al. (2006) with minor changes. The nucleus of P4 was irradiated approximately 55 minutes after it was born with 3 nanosecond pulses at 20 Hz at a sublethal laser intensity for 1 minute. Experiments were included in analysis only if the P4 cell failed to divide before internalizing.
Immunostaining
Anti-Phospho-Ser19-MLC (1/250, Abcam) immunostaining was done as by Lee et al. (2006) with minor changes. Embryos were allowed to develop to the desired stage based on timing from the 4-cell stage. CEH-51::GFP was detected with mouse GFP antibodies (1/100; Molecular Probes). PAR-3 immunostaining was done using a freeze-crack method as described (Tenlen et al., 2008) with minor changes. Incubations with anti-PAR-3 (1/25; Developmental Studies Hybridoma Bank) were at 4°C overnight, and incubations with rabbit anti-GFP (1/1000; Abcam) were at 37°C for 1 hour. Embryos were imaged with a Zeiss LSM510 confocal microscope and LSM software, and images were processed with Metamorph. MS descendants were counted after anti-GFP staining of nuclei in CEH-51::GFP embryos. Anti-PAR-3 fluorescence intensity was measured in linescans using Metamorph software. Basolateral and apical membranes were identified by the localization borders of CEH-51::GFP levels, calculated as averages of three-pixel-wide linescans in each embryo. Apical and basolateral GFP pixel intensity fall-offs were used to align anti-PAR-3 intensity measurements between embryos.
RESULTS
Identifying each of the cells that gastrulate
We began by tracing cell lineages and identifying each cell that internalized during gastrulation from thirty-three multiplane DIC recordings of wild-type embryos and four spinning disk confocal recordings of embryos expressing a plasma membrane marker, PH::mCherry (Kachur et al., 2008). Specific lineages were followed in each embryo, taking advantage of embryo orientations to follow unambiguously specific lineages in each embryo. We mapped the identities of gastrulating cells onto the C. elegans embryonic cell lineage (Fig. 2A). We define gastrulating cells here as precursor cells that internalize before embryonic cell divisions are complete, distinguishing gastrulation from the internalization of postmitotic cells, which occurs later in embryogenesis, for example during ventral epidermal enclosure (Chisholm and Hardin, 2005; Williams-Masson et al., 1997). The identities of many of the gastrulating cells had been reported before (Nance and Priess, 2002; Sulston 1983), but a full lineage could not be drawn from published data because not all gastrulating cells had been identified. Also, ambiguity had remained as to which cells internalized at which times in the MS, AB, and C lineages.
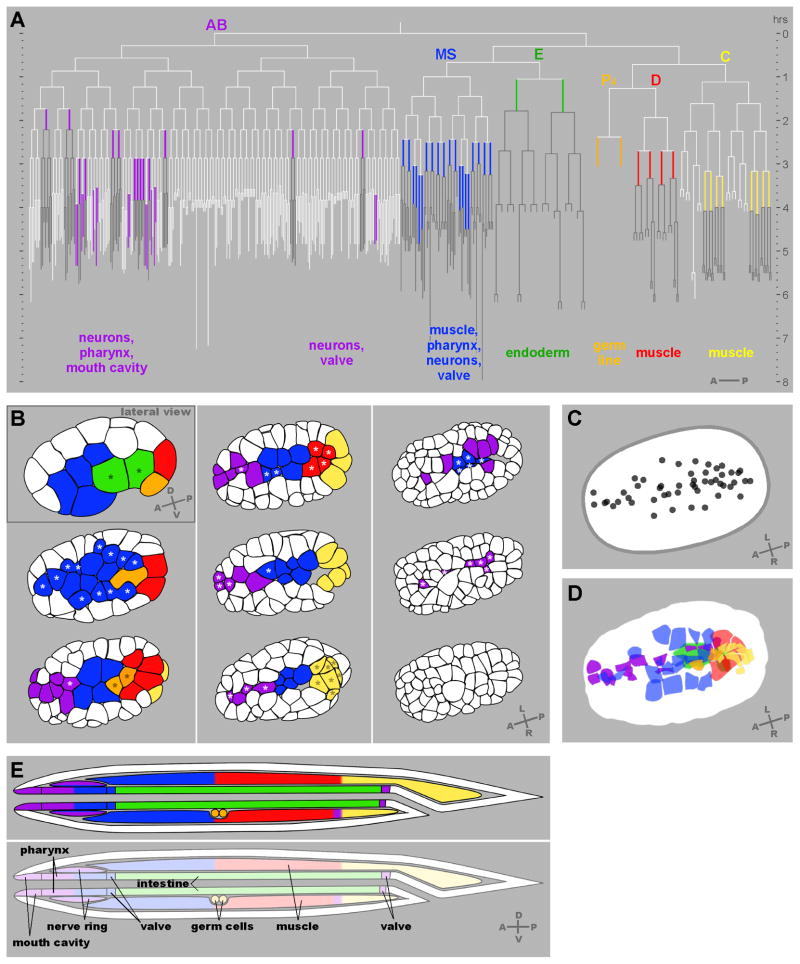
Fig. 2. Gastrulation in C. elegans.
(A) The C. elegans embryonic cell lineage with the 66 gastrulating cells each represented in colors here and in (B,D,E) corresponding to the six traditionally-recognized cell lineages. Each vertical line represents a cell, with its length indicating its cell cycle period. Horizontal lines are cell divisions. The anterior daughter at each division is to the left, posterior to the right, as indicated on the lower right. Descendants of gastrulating cells, which generally remain internalized, are drawn in dark gray. Internal structures that each set of cells produces are indicated below. (B) The positions of the gastrulating cells. Renderings of cell outlines were generated from a spinning disk confocal film of a membrane-marked embryo viewed at a plane through the middle of the top layer of cells, adjusting to match the middle of the top layer of cells as cell divisions resulted in smaller and smaller cells. All views are ventral views except the first stage shown, a lateral view. Here and in the accompanying Movies 1 and 2 in the supplementary material, E, MS, P4, D and all of their descendants are colored from the time they are born. In the AB and C lineages, only some descendants gastrulate. For these lineages, we have colored the AB cells (in purple) only during the cell cycle at which each cell internalization occurs, and we have colored the C lineage (in yellow) at the birth of C, with yellow later marking only those C lineage cells that internalize. Nine embryonic stages are shown, with asterisks marking cells that are about to internalize (that internalize before the next stage shown). (C) The center of each cell’s apical domain is marked by a dot three minutes before its internalization was complete, from a ventral view, showing that most cells internalize from a position near the embryo’s ventral midline. 50 of the 66 cells (all but 16 of the AB lineage cells) could be seen internalizing from this view. (D) As in (C), except the entire apical domain is drawn. Endoderm was drawn here based on a separate movie. One cell of the 50 indicated in (C) is not represented in (D). This cell internalized by division toward the interior in this specific embryo. (E) Diagram of a hatched, larval worm (L1 stage, after Sulston et al., 1983) indicates contributions to internal tissues from each lineage.
Previous work had identified in the AB lineage only pharyngeal and buccal cavity precursors that gastrulate (Nance and Priess, 2002; Sulston et al., 1983). We found twelve cells of the AB lineage gastrulating that had not been recognized to do so previously, including eight precursors of much of the AB-derived portion of the nervous system (Table 1). To our knowledge, none of the AB-derived nervous system precursors had been previously reported to internalize during gastrulation. The remaining four cells function as precursors to other internal structures: ABalpaapp (which produces four anterior buccal cavity cells, including two hypodermal cells just inside the mouth), ABprpapppp (the two intestino-rectal valve cells), ABaraapap (four pharyngeal cells), and ABaraappp (four pharyngeal cells). Our findings above reveal that much of the AB-derived nervous system -- eight precursor cells that give rise to 60 neurons -- becomes internalized during gastrulation, instead of internalizing postmitotically.
Table 1.
Eight gastrulating neuronal precursors give rise to 60 neurons.
| gastrulating neuronals precursor cells | neurons |
||||
|---|---|---|---|---|---|
| ring ganglion cells | ventral ganglion cells | retrovesicular ganglion cells | ventral cord cells | lateral CANR neuron | |
| ABalaap | 8 | ||||
| ABalapp | 11 | 1 | |||
| ABalpppa | 6 | ||||
| ABalpppp | 7 | ||||
| ABarappp | 7 | 1 | |||
| ABprpaap | 7 | 1 | |||
| ABplppap | 5 | 4 | |||
| ABprpapppa | 1 | 1 | |||
| totals | 46 | 3 | 6 | 4 | 1 |
Although C. elegans gastrulation involves many cells moving in at distinct times, we noticed nearly complete invariance with respect to which cells move in at which division round. Cells of the MS lineage internalize after 4 or 5 divisions of the MS founder cell (Nance and Priess, 2002). We found that the specific MS descendants that internalized after the fourth MS lineage cell division were the same in all embryos, and the specific descendants that remained on the surface until after the fifth MS lineage cell divisions were the same (n=8 embryos where all of these cells were followed) (Fig. 2A). These cells that remained on the surface until after this fifth MS cell division internalized in 8/9 wild-type embryos, and there was one embryo where 6/8 remaining MS cells did not internalize within 90 minutes of their last division, a rare case of variation in the lineage in wild-type embryos. The internalization of the germ line precursors Z2 and Z3 (n=8) and the four D lineage cells (n=7) was invariant (Fig. 2A). In the C lineage, we noticed one case with variability in the Capp lineage: In two out of three embryos in which the C lineage was traced, Cappa and Cappp internalized, and in the other embryo, their mother cell, Capp, internalized instead. All other C lineage cells that internalized (Fig. 2A) did so at the same division round in each case.
We discovered that nearly all of the gastrulating cells, including the newly-identified AB lineage cells, comprise a continuous stripe along the ventral side of the embryo running from the anterior pole to the posterior pole (Fig. 2B,C,D and Supplemental Movie 1). The position of cells in each lineage during internalization correlates well with antero-posterior positions along the body later, in the hatching worm (Fig. 2B,D,E), suggesting that gastrulation plays a large part in positioning cells, and that little large-scale movement of cells along the antero-posterior axis occurs later. Consistent with this, we noticed that the germ line precursors Z2 and Z3 internalized at the same time as the two somatic gonad precursors from the MS lineage, the grandparents of Z1 and Z4, with the four cells close to each other (Fig. S1). Together with the work of Sulston et al. (1983) and Nance and Priess (2002), this descriptive work serves as a platform for dissecting mechanisms of cell internalization.
Different lineages use distinct fate regulators to control internalization
We hypothesized that cell fate regulators might be required for timely internalization of diverse cell types. This issue was previously addressed by examining two embryos from a mutant in which AB lineage cells were transformed into MS lineage cells (Nance and Priess, 2002). In these embryos, some cells internalized that were not known to do so in wild-type. However, the interpretation of this result is complicated by our finding that more AB lineage cells internalize than were known at the time of the earlier experiments. Therefore, we revisited the effect of cell fate on cell internalization timing. We used mutations in key cell fate regulators to alter cell fates, examining the consequences of these mutations on the timing of cell internalization. We focused on two lineages with well-studied fate specification regulators: the MS lineage, which produces mostly mesodermal cells, and the P4 lineage, which produces the germ line.
Two transcription factors are known to play important roles in the initial establishment of MS lineage fate: the T-box protein TBX-35, and the NK-2 homeodomain protein CEH-51 (Broitman-Maduro et al., 2009). tbx-35;ceh-51 double mutant embryos show an incomplete MS to C lineage transformation: they generally fail to make MS-derived tissue, and the MS lineage often develops C lineage markers (Broitman-Maduro, 2009). We analyzed gastrulation timing in each cell of the MS lineage in five tbx-35;ceh-51 embryos (Fig. 3A–J, Supplementary Fig. S2). In some embryos, at least one cell internalized one cycle early, after only three rounds of MS divisions (Supplementary Fig. S2). As presented above, this rarely occurred in the wild-type C lineage and never in the MS lineage. Some cells that would normally internalize after five rounds of MS lineage divisions in wild-type embryos instead internalized one division round earlier in the double mutants, after four rounds of division (Supplementary Fig. S2). This is similar to what occurs in nearly all cases in the C lineage (Fig. 2). We conclude that the MS cell fate specification regulators TBX-35 and CEH-51 are important for the normally invariant temporal pattern of MS cell internalization.
Fig. 3. The MS cell fate regulators TBX-35 and CEH-51 are required for normal gastrulation timing in the MS lineage.
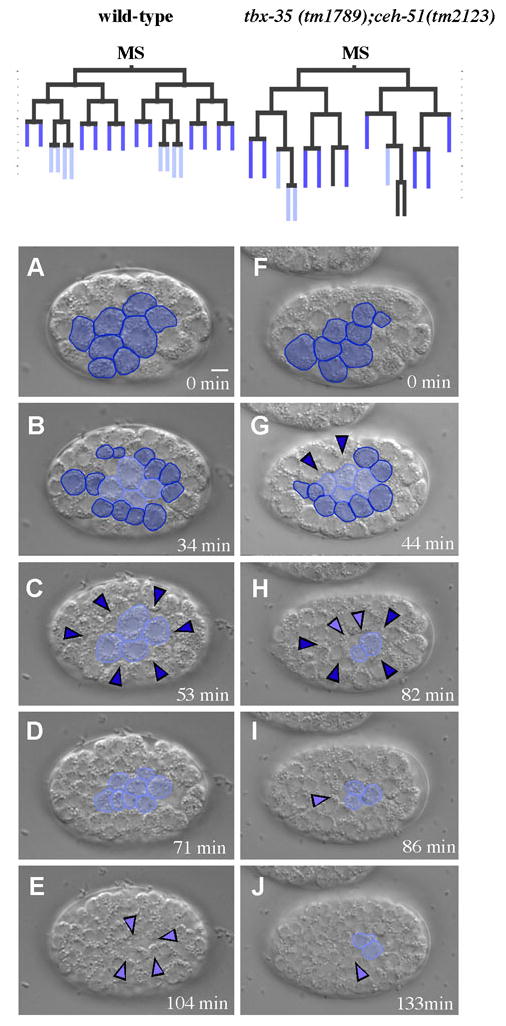
Images from a movie of a wild-type embryo (A–E) and a tbx-35(tm1789);ceh-51(tm2123) embryo (F–J). Dark blue represents the MS descendants that normally internalize after the fourth MS cell division, and the light blue represents the MS descendants that normally internalize after the fifth MS cell division. Colored arrowheads mark positions where cells internalized. (A) MS8 stage. (B) MS16 stage (C) 12 MS descendants have internalized (arrowheads). 4 MS descendants remain on the surface. (D) After the 4 MS descendants remaining at the ventral surface have divided. (E) These MS descendants have internalized. (F–J) tbx-35(tm1789);ceh-51(tm2123) MS descendants at similar stages, showing both premature and late cell internalizations compared to wild-type. Lineages at the top of the figure are drawn from these two embryos. Times are minutes after beginning of MS8 stage. Scale bar: 5 μm.
Specification of the germ line depends on MES-1, a receptor tyrosine kinase-like protein (Berkowitz and Strome, 2000; Capowski et al., 1991; Strome et al., 1995). mes-1 loss of function mutant worms produce embryos where germ line ribonucleoprotein particles termed P granules do not become partitioned properly to the germ line blastomeres, and the primordial germ cell P4 adopts the fate of its sister cell, D, a muscle precursor (Strome, 1995). The cell division that establishes P4 and D is normally asymmetric, and the larger D cell normally divides before the smaller P4 cell (Sulston, 1983). In mes-1 mutant embryos, cell size and division order are equalized to various degrees (Strome, 1995). We examined recordings of twelve embryos from mes-1 hermaphrodites. In five of these embryos, P4 and D cells divided synchronously, suggesting an effective fate transformation of P4. These five embryos were used for further analysis. In wild-type embryos, P4 divides once, and its two daughter cells internalize, and D divides twice before the resulting four cells internalize (Fig. 4A–D). In 3/5 mes-1 mutant embryos examined, P4 divided twice before its four descendants internalized, as the D lineage normally does (Fig. 4E–H, Supplementary Fig. S3). In the other two cases, some of the four descendants internalized at this same stage, and others did not (Supplementary Fig. S3). We conclude that MES-1, which is important for P4 cell fate, is also important for internalization of cells at the appropriate division round. To assess the specificity of this transformation, we determined if the loss of MES-1 affected internalization of other lineages. In four mes-1 embryos, 44 MS descendants could be followed unambiguously during their internalization, and each one internalized during the same cell cycle as it does in wild-type embryos (data not shown). These results, together with previous results discussed above (Lee et al., 2006; Nance and Priess, 2002), demonstrate that factors responsible for cell fate specification in specific lineages also affect internalization timing in these lineages.
Fig. 4. MES-1 is required for normal gastrulation timing in the germ lineage.
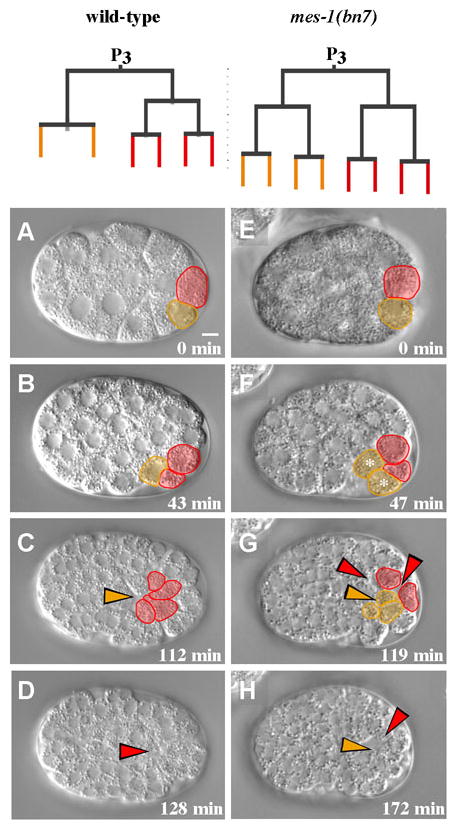
Images from a movie of a wild-type embryo (A–D), and a mes-1 (bn7) embryo (E–H). Germ line and D lineage cells are pseudo-colored. Colored arrowheads mark positions where cells internalized. (A) The P4 cell (orange) is smaller than the D cell (red) in wild-type embryos. (B) The D cell divides on the surface of the embryo before P4 divides. (C) The germ line precursor cells have internalized (arrowhead). (D) The D lineage cells have internalized (arrowhead). (E–H) The germ line precursor cell and D lineage cells were approximately the same size, divided on similar schedules, and their descendants internalized after two divisions. Times indicated are minutes after P4 birth. Scale bar: 5 μm.
Apical PAR proteins that function in endoderm internalization do not play a major role in timely internalization of two mesodermal lineages
In endodermal precursors, the apical PAR proteins PAR-3 and PAR-6 are required for apical localization of myosin II heavy chain and for cell internalization at the 28 cell stage (Nance et al., 2003). These authors also showed minor defects in internalization timing in two MS descendants that were followed in the absence of PAR-3. However, our results above reveal more variation in internalization timing among wild-type embryos than was recognized previously. Additionally, PAR-3 has been reported to disappear below detectable levels by around the 50 cell stage, earlier than MS lineage internalization (Etemad-Moghadam et al., 1995). It remains possible that PAR-3 might localize apically in MS lineage cells only briefly, for example specifically as cells internalize, or that it functions early to establish polarity that will be used later during MS lineage internalization. Therefore, we used protein localization and disruption of function experiments to assess whether apical PAR proteins are required for timely internalization of mesodermal precursors.
We examined endogenous PAR-3 distribution by immunostaining, using CEH-51::GFP to identify MS descendants at the MS16 stage (when 16 MS descendants are present). We compared apical and basal localization of PAR-3 at the stage of MS cell internalization, and, for comparison at the MS4 stage, when PAR-3 is known to be apically enriched (Etemad-Moghadam et al., 1995). PAR-3 appeared apically enriched as expected at the MS4 stage, but not in MS descendants at the MS16 stage (Fig. 5A–B). Quantification of fluorescence intensities (Fig. 5C) revealed no apical peak of PAR-3 in MS16 cells. Apical PAR-3 levels were statistically indistinguishable from basal PAR-3 levels in MS16 cells (p>0.5) and significantly lower than the apical level at the MS4 stage (p<0.005). We conclude that there is no detectable apical enrichment of PAR-3 in the MS lineage cells at the time that these cells internalize.
Fig. 5. PAR proteins do not play a major role in the timely internalization of MS and D lineage cells.
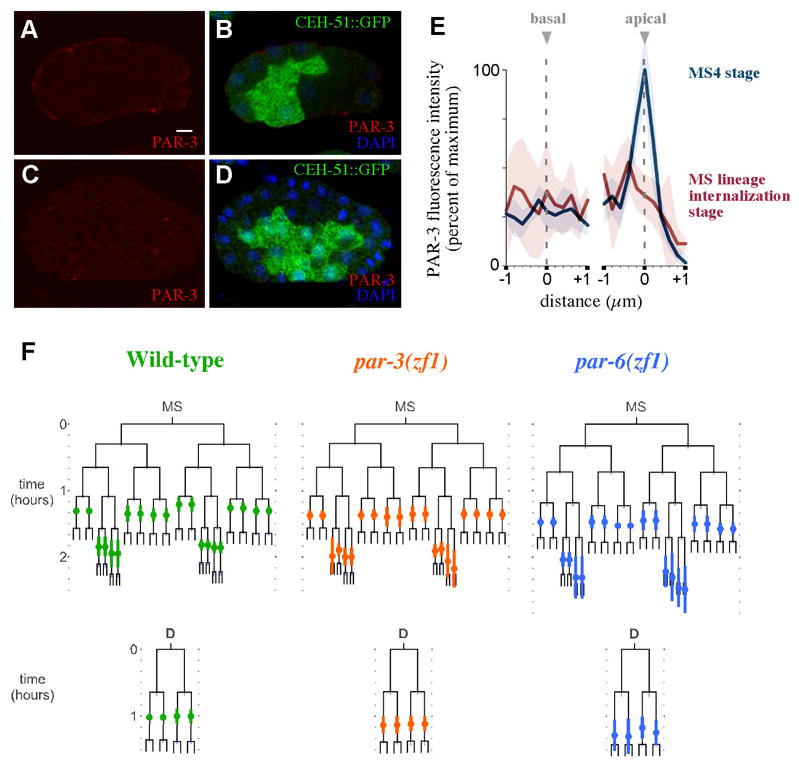
(A) CEH-51::GFP embryo at the MS4 cell stage immunostained for PAR-3 (left) and merged with GFP and DAPI images (right). (B) Same at MS16 stage. (C) Anti-PAR-3 fluorescence intensity quantified in MS4 stage (n=6, blue) and MS16 stage (n=5, red). Fluorescence intensities were calculated within 1 μm on either side of the apical and basal membranes. Shading indicates 95% confidence intervals. (D) MS and D lineages for wild-type (n=7), par-3(ZF1) (n=9), par-6(ZF1) (n=10) embryos. The timing of cell internalization for cells where internalization was observed is indicated by colored circles on the lineage, along with standard deviation bars. Scale bar: 5 μm.
Although PAR-3 is not apically enriched at this time, it is possible that apical PAR proteins establish polarity in MS lineage cells at an earlier stage, and that this is required later for cell internalization. To test roles for apical PAR proteins, DIC recordings were generated from par(ZF1) strains in which PAR-3 or PAR-6 became degraded specifically in somatic cells (Nance et al., 2003) (Fig. 5D). We examined complete MS and D lineages. We found that internalization timing in both lineages was almost indistinguishable from that in wild-type embryos. The six pairs of MS descendants that internalize after the fourth MS cell division in wild-type embryos internalized during the same cell cycle in all par-3(ZF1) and par-6(ZF1) embryos. The difference in timing of these MS cells between wild-type and par-3(ZF1) and par-6(ZF1) embryos was small and marginally statistically significant in only one pair of these cells (see Materials and Methods). Four pairs of MS descendants that normally internalize after the fifth MS cell division frequently showed variation that mirrors natural variation in wild-type embryos. In 5/9 par-3(ZF1) embryos and 5/10 par-6(ZF1) embryos, the four pairs of MS lineage cells that normally internalize after the fifth MS cell division did so in these embryos as well. Consistent with slight variation of internalization of these cells in 1/9 wild-type embryos (discussed above), we found in the remaining cases that most of these eight MS lineage cells in par-3(ZF1) and par-6(ZF1) did not internalize during the duration of our films either during this cell cycle or after another division. In two of these embryos, some of these eight MS lineage cells did internalize after one more round of division. In all par-3(ZF1) and 6/7 par-6(ZF1) embryos, the cells in the D lineage internalized as 4 cells, with timing that was statistically indistinguishable from D lineage internalization in wild-type embryos (see Materials and Methods, Fig. 5D). We conclude that PAR-3 and PAR-6 do not play a major role in the timely internalization of MS and D descendants, or that if they do, they must do so redundantly. This is in contrast to what has been established for the E lineage (Nance et al., 2003), suggesting that regulation of cell internalization by polarity proteins works differently in different lineages.
Myosin localizes apically, becomes activated, and functions in cell internalization in diverse lineages
The data above suggest that cell fate and even cell polarity mechanisms that control gastrulation vary from lineage to lineage, raising the question of whether associated cytoskeletal mechanisms vary as well. To address this, we first examined whether myosin accumulates at the apical sides of internalizing cells. We filmed embryos expressing a GFP-tagged form of myosin II heavy chain, NMY-2::GFP, and analyzed places where cells of the MS lineage, the D lineage, and the germ line precursor cells were internalizing. We found that NMY-2::GFP accumulated near the apical sides of internalizing cells: Among 9 embryos analyzed, NMY-2::GFP accumulated near the surface of 43/43 MS lineage cells that we observed as they internalized, 8/8 internalizing germ line precursor cells, 12/13 internalizing D lineage cells, and 11/11 internalizing cells whose lineal origin was not traced, consistent with an embryo shown by Nance and Priess (2002) where a pair of MS lineage cells could be seen to accumulate myosin apically. We found no cases of internalizing cells that failed to accumulate apical NMY-2::GFP. No other cells visible to us at these stages accumulated myosin similarly, with the exception of places where we saw clear myosin accumulation at the cytokinetic rings of dividing cells, as expected (Fig. 6A–D).
Fig. 6. Internalizing cells accumulate myosin II heavy chain and activated myosin regulatory light chain near their apical surfaces.
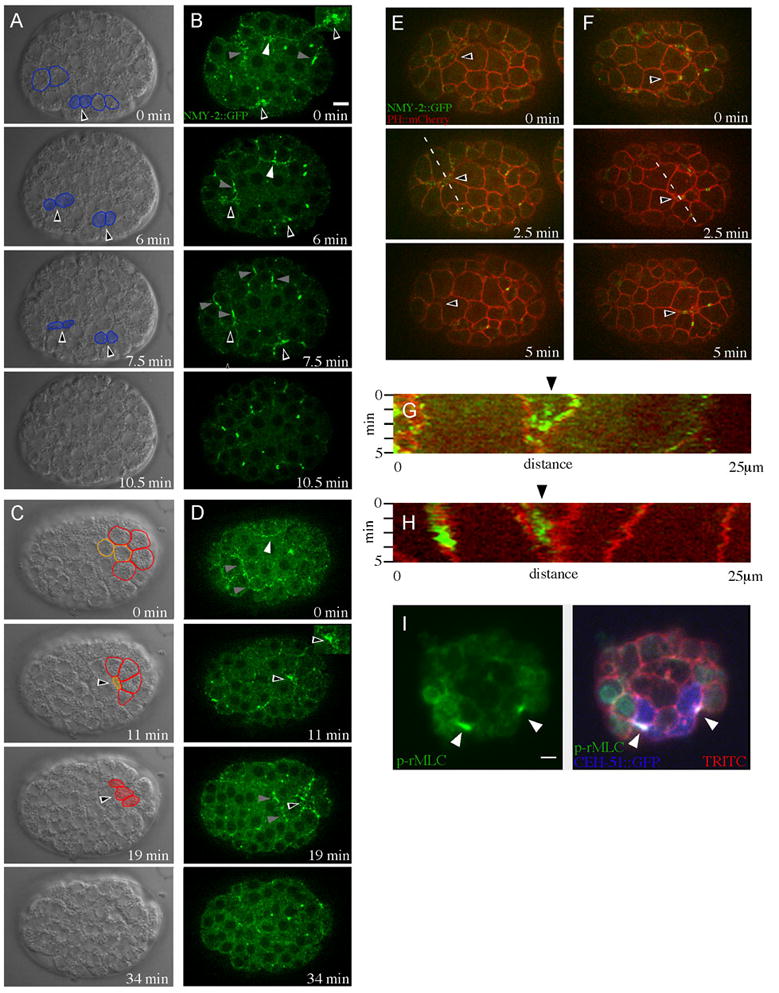
(A–D) NMY-2::GFP embryos, DIC (A,C) and GFP (B,D) views. Black arrowheads outlined in white indicate cells that are internalizing. White arrowheads mark internalizing cells that were not identified to a specific lineage in these embryos. Cells undergoing cytokinesis are marked by gray arrowheads. Cells that are internalizing are shaded in DIC images in blue (MS lineage), orange (germ cell precursors) or red (D lineage), and those descendants that will internalize in later frames are outlined in the same colors. Insets are magnified views of myosin accumulation. (A,B) Internalization of MS descendants. (C,D) Internalization of germ line precursors and D descendants. (E,F) Double-labeled PH::mCherry; NMY-2::GFP embryos. Dotted lines indicate places from which kymographs in (G) and (H) were generated. (E) Arrowheads indicate an internalizing MS descendent cell. (F) Arrowheads indicate an internalizing germ line precursor cell. (G) Kymograph analysis of the MS descendant from (E). Arrowhead indicates a narrowing apical cell surface, showing NMY-2::GFP enrichment here. (H) Kymograph analysis of the germ line precursor cell in (F). Arrowhead indicates a narrowing apical cell surface, showing NMY-2::GFP enrichment here. (I) CEH-51::GFP (blue) embryos stained with p-rMLC antibody (green) and TRITC-phalloidin (red), projection of three spinning disk confocal planes, each 0.5 μm apart. White arrowheads show apical accumulation of p-rMLC in internalizing MS descendants. View of embryo is from the anterior. Nuclear staining from p-rMLC is a background signal (Lee et al., 2006). Scale bars: 5 μm.
To determine if the myosin accumulation seen occurred within each internalizing cell or on extensions from neighboring cells, we examined embryos expressing both NMY-2::GFP and a plasma membrane marker, PH::mCherry. We imaged near the surface of live embryos at five second intervals by spinning disk confocal microscopy. Myosin accumulation occurred within the internalizing cells, rather than their neighbors, in 50/50 MS descendants in 9 embryos, 5/6 D lineage cells in 2 embryos, 8/8 germ line precursor cells in 4 embryos, and 5/5 unidentified internalizing cells in 3 embryos (Fig. 6E–H).
To determine if apical myosin became activated in internalizing cells of diverse lineages, we examined a conserved marker for myosin activation. Myosin II complexes comprise two heavy chains, two essential light chains and two regulatory light chains. The phosphorylation of the two regulatory light chains at a conserved serine (p-rMLC) is required for the formation of bipolar myosin filaments and association with actin filaments (Somlyo and Somlyo, 2003). This residue in myosin regulatory light chain is phosphorylated in endodermal precursor cells for a short period of the cell cycle, during cell internalization (Lee et al., 2006). We immunostained embryos expressing CEH-51::GFP, a marker for MS lineage fate (Maduro, 2009) at the MS16 stage, when MS lineage cell internalization begins, using cell position and morphology to identify 34 cells that were internalizing in 10 fixed embryos. 28 of these cells were enriched for apical p-RMLC (Fig. 6I), indicating that activated apical myosin was associated with cell internalization in these cells.
To determine if myosin function is required for internalization of diverse lineages, we used a temperature-sensitive, strong loss of function nmy-2 allele (Liu et al., 2010). We shifted embryos to the restrictive temperature at a time during which many cells should be internalizing, after E lineage cell internalization should be complete, in 5 mutant embryos and 5 wild-type control embryos placed side-by-side in pairs on the same slides. In the wild-type embryos, 16 ± 3.8 (mean ± 95% CI) cells internalized within 80 minutes of the temperature shift, whereas only 3 ± 0.9 cells in the nmy-2 mutant embryos internalized in this period (Fig. 7, p<0.001, Student’s t-test). We found that cytokinesis failed in most cell divisions in the mutants after the temperature shift, although 3 cells in 5 embryos were observed to undergo cytokinesis, suggesting that a small amount of myosin function remains at the restrictive temperature in embryos of these stages, perhaps from another myosin heavy chain protein, NMY-1. We conclude that most cells examined at this stage depend on the myosin heavy chain protein NMY-2 for internalization.
Fig. 7. NMY-2 is required for internalization of many cells.
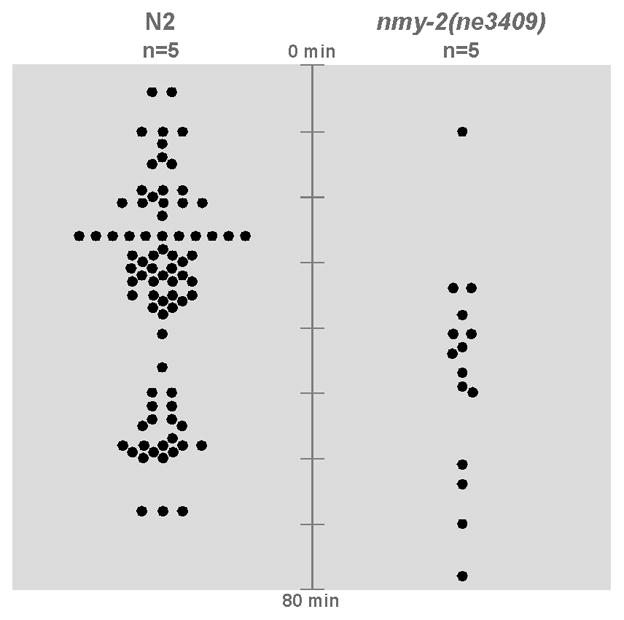
Five wild-type embryos and five nmy-2(ne3409) embryos were upshifted to 25°C 4.5 hours after the 4-cell stage and filmed side-by-side. Dots represent every cell seen to internalize after the temperature shift.
Our results reveal that internalizing cells of diverse lineages accumulate myosin apically, within each internalizing cell rather than in extensions from neighbors, that myosin becomes activated there, and that the myosin heavy chain protein NMY-2 is required in diverse cells for internalization.
Does the frequency of internalization as sister cell pairs reflect an especially efficient mode of cell internalization?
In our experiments tracking myosin dynamics above (Fig. 6A–I), and in our initial films (Fig. 2), we observed that the vast majority of gastrulating cells internalize as sister cell pairs (i.e., with pairs of sister cells internalizing synchronously) and often only one pair at a time, consistent with observations of Nance and Priess (2002). There is evidence that cell internalization in this system is temporarily prevented as cells divide (Lee et al., 2006), and this may contribute to this synchrony. However, we considered it possible that there could also be an as yet undiscovered property of the internalization mechanism that makes internalization of cell pairs more efficient than internalization of larger numbers of cells, or of single cells. Junkersdorf and Schierenberg (1992) have shown previously that a single E cell can internalize after they prevented its division by laser irradiation, and we have found that some cells in the AB lineage internalize alone during normal development (Fig. 8D). However, these results leave open the possibility that internalization of pairs of cells may be a more efficient mode of cell internalization, occurring more quickly than internalization of individual cells.
Fig 8. Pairs of cells are not required for efficient internalization.

A wild-type C. elegans embryo (A,D), a wild-type C. briggsae embryo (B), and a wild-type C. elegans embryo in which the P4 cell division has been delayed with a laser (C). Orange represents germ line precursor cells, shades of purple represent AB lineage cells. (A) Internalization of two germ line precursor cells during C. elegans gastrulation, beginning with birth of P4 cell at time 0. (B) Internalization of two germ line precursor cells during C. briggsae gastrulation, beginning with birth of P4 cell at time 0. (C) Internalization of one germ line cell after laser delay of P4 cell division. (D) The ABplppap cell internalizes as a single cell, indicated by the shaded purple cell. Its sister cell, indicated by the dark purple outline, does not internalize at this time. Other surrounding AB cells that do not internalize at this time are outlined in lighter purple. (E) Internalization timing of Z2 and Z3 in control embryos (n=7) and P4 in laser delayed embryos (n=7). Scale bar: 5 μm.
To investigate this possibility, we began by seeking to exploit a finding of Zhao et al. (2008), who showed that the P4 cell in a transgenic strain of C. briggsae, a relative of C. elegans, divided an average of 23 minutes later than in C. elegans. We reasoned that this might result in P4 internalizing alone, before it divides, in this species. However, we found no significant difference in the P4 cell cycle lengths between two C. briggsae strains and C. elegans (76±5 minutes in C. elegans wild-type N2 embryos, n=7; 77±9 minutes in C. briggsae wild-type AF16 embryos, n=7; 71±4 minutes in C. briggsae mCherry histone RW20025 embryos, n=4; p>0.05 for each C. briggsae strain compared to C. elegans, Student’s t-test). P4 divided and then internalized as a pair of cells in all cases in both species (Fig. 8A,B). We conclude that the difference reported by Zhao et al. (2008) is characteristic of their transgenic strain or imaging techniques and not of C. briggsae more generally. We therefore turned to a different method to test our hypothesis.
To investigate this issue in C. elegans, we used a sublethal dose of laser irradiation targeted to specific cells to delay cell division, and then followed cell internalization to determine if a single, laser-delayed cell could internalize efficiently. Internalization timing was recorded from 7 untreated embryos and 7 laser-irradiated embryos. Laser irradiation delayed cell division, and P4 internalized as a single cell. Internalization of P4 occurred no later than its descendants in untreated embryos (Fig. 8C,E). We conclude that although C. elegans gastrulation involves internalization of sister cell pairs in many lineages, pairs of cells are not required for efficient internalization.
DISCUSSION
C. elegans gastrulation presents an unusual opportunity to examine mechanisms of cell internalization used by cells of diverse lineages and fates. We found that proteins required for cell fate specification are also required for timely internalization of cells in specific lineages. We were surprised to learn that two proteins that define apicobasal polarity in internalizing endodermal precursors do not play a major role in timely internalization of certain later-born cells, suggesting that different cell polarity mechanisms are likely to operate in these cells. Despite these differences, the internalizing cells that we examined accumulated myosin at their apical surface and activated it there, and most, if not all, cells depended on myosin for internalization. Our results suggest that the same cytoskeletal mechanism may be used reiteratively in cells, associated with different patterning mechanisms (Fig. 9).
Fig 9. Distinct cell fate and cell polarity regulators are associated with the same cytoskeletal mechanisms during the internalization of different cell lineages.
END-1 and END-3 are required for efficient internalization of the endodermal cells, whereas the T-box protein TBX-35 and the NK-2 homeodomain protein CEH-51 are required for timely internalization of the MS descendants, and the receptor tyrosine kinase-like protein MES-1 is required for timely internalization of the germ line. Endodermal cells are polarized by the PAR proteins, but these are not required for the timely internalization of multiple other lineages. These distinct patterning regulators appear to be associated with a common cytoskeletal mechanism in multiple lineages.
Our initial results further completed a description of the cells that gastrulate in C. elegans, allowing us to map cell identities onto the C. elegans embryonic cell lineage. Among the sixty-six cells found to gastrulate in C. elegans, we found precursors of much of the ring ganglion, sometimes referred to as the C. elegans brain (Thomas and Lockery, 1999). The internalization of a concentration of nervous system precursors might reasonably be referred to as the beginning of C. elegans neurulation, with ventral epidermal enclosure representing much of the rest of neurulation. However, because we found that many of these AB-derived cells internalized as a ventral patch of cells that formed portions of a larger, continuous, ventral stripe of gastrulating cells (Fig. 2B), for convenience, we refer instead to all of these cells as gastrulating cells. It will be interesting to learn how commonly gastrulation and early neurulation occur in concert in other systems.
Cells contributing to a wide variety of tissues internalize by apical constriction in other systems, although not necessarily in a coherent stripe of cells as in C. elegans. In Drosophila, apical constriction plays important roles in the internalization of cells of the ventral furrow, the posterior midgut, the amnioserosa, the trachea and the salivary glands (Chung and Andrew, 2008; Sawyer et al., 2010). Cell fate and polarity regulators have been identified as important for internalization in these tissues. The specific regulators used can vary from tissue to tissue. For example, apical constriction in tracheal tube formation is regulated by the cell fate regulator Trachealess, a transcription factor, through an EGF receptor signaling pathway and the RhoGAP Crossveinless (Brodu and Casanova, 2006). Mesodermal cells also invaginate by apical constriction downstream of different cell fate regulators, the transcription factors Twist and Snail (Leptin and Grunewald, 1990). Still, some commonalities do exist among regulators between tissues. For example the secreted protein fog serves as an apical determinant in ventral furrow formation and also functions in salivary gland morphogenesis (Dawes-Hoang et al., 2005; Kolesnikov and Beckendorf, 2007).
Given the spatial coherence of the ventral stripe of gastrulating cells in C. elegans, it seems plausible that unknown secreted signals could coordinate internalization. In Drosophila, the secreted protein fog is known to coordinate apical constriction between cells (Costa et al., 1994). Consistent with this in C. elegans, Nance and Priess (2002) found that if they genetically changed the fate of the E lineage and also laser ablated the fate-transformed cell, this could affect MS lineage internalization timing. On the other hand, they found that changing the fate of the E lineage without ablating cells failed to affect MS lineage internalization timing. Furthermore, we found that internalization timing can be altered in the germ line lineage without affecting internalization timing of neighboring lineages, in a mes-1 mutant. This raises an alternative possibility, that a coherent stripe may be cobbled together by relatively autonomous internalization behaviors among cells of diverse lineages.
Our data suggested that PAR-3 and PAR-6 do not provide significant, nonredundant polarity inputs to apical constriction in some lineages. Paralogs of other known polarity regulators exist in C. elegans, including members of the Crumbs/Pals/Patj and Scribble/Dlg/Lgl complexes (Assémat et al., 2008). Preliminary experiments targeting homologs of Crumbs and Scribble in wild-type and par-3(ZF1) embryos have failed to reveal roles for these complexes in cell internalization (JH, unpublished). It will be interesting to determine if other polarity regulators play a role instead, either in concert with or independent of the PAR proteins. In the endodermal lineage, loss of PAR-3 or PAR-6 delays cell internalization, rather than completely preventing internalization (Nance and Priess, 2002), suggesting that partially redundant polarity regulators may exist even at this early stage. Epithelial cells that lack PAR-6 are still polarized (Totong et al., 2007) and other polarity regulators function in other cells in C. elegans (St Johnston and Ahringer, 2010.) Together, these results suggest that different cells rely on different polarity regulators, although it remains possible that an overlapping set of polarity regulators functions in different cells but with variations in the balance of redundancy among specific cell polarity regulators.
Our experiments on myosin localization and function suggest that many cells internalize by actomyosin-dependent apical constriction. It is possible that other mechanisms could contribute to their internalization as well. For example, forces could be produced by neighboring cells in a myosin-dependent cinching of neighboring cell contacts, as seen in wound healing (Wood et al., 2002). However, we consider this unlikely, as we detected little myosin enrichment in the rings of neighboring cells surrounding internalizing cells (Fig. 6). It will be interesting to determine exactly how specific fate and polarity regulators might impact the cytoskeletal force-producing mechanisms that move cells. It is possible, for example, that key cytoskeletal regulators could be transcriptional targets of the MS lineage fate regulators TBX-35 and CEH-51.
Many cell fate and cell polarity regulators identified first in invertebrate model systems have turned out to have mammalian homologs with similar functions (Erwin and Davidson, 2002; Goldstein and Macara, 2007). The variety of such regulators that can be associated with apical constriction between and within organisms suggests a large degree of evolutionary lability in the patterning mechanisms that can function upstream of apical constriction-dependent cell movements. For this reason, we would predict that the specific cell fate and cell polarity regulators found to be associated with apical constriction in any one tissue of a single animal system might not necessarily be expected to perform precisely the same function in vertebrate neural tube closure, a morphogenetic process with important implications for human health. This suggests casting a wide net when considering which cell fate and cell polarity regulators from invertebrate model systems might have vertebrate paralogs that regulate a specific morphogenetic process. It is possible that more direct regulators of cytoskeletal motility will more often have evolutionarily conserved functions in morphogenesis.
It will be interesting to dissect further the direct links between patterning mechanisms and cytoskeletal force-producing mechanisms in this system. Such links are not well understood in general in animal development. Establishing direct links will likely require uncovering new molecular players, which might shed further light on the regulation of morphogenesis in other systems.
Supplementary Material
MSpppa (top white asterisk) and MSappa (bottom white asterisk), which are the grandparents of somatic gonadal precursors Z1 and Z4 respectively, lie close to the germ line precursors Z2 and Z3 (black asterisks). To see the cell movements, see Supplemental Movies 1 and 2.
Lineages are drawn from wild-type (A) and individual tbx-35;ceh-51 embryos (B). Blue lines indicate cells in the MS lineage that internalized. (A) MS lineage in a wild-type embryo. Gray lines on lineage are standard deviations. White stars on the image of the embryo indicate cells that internalize in wild-type embryos. (B) MS lineages in four tbx-35;ceh-51 mutant embryos. Lineage lines without blue indicate cells that did not internalize during the time the embryos were filmed. Red asterisks are abnormalities in the internalization of cells. Question marks indicate cells for which we could not determine whether they internalized.
Lineages are drawn from wild-type (A) and individual mes-1 embryos (B). Orange lines indicate germ line cells that internalized and red lines indicate cells with a D cell fate that internalized. (A) Germ line and D cell lineages in wild-type embryos. Gray lines on lineage are standard deviations. White stars on the image of the embryo indicate cells of the relevant lineages that internalize in wild-type embryos. (B) Germ line and D lineages in four mes-1 embryos. Gray lines indicate cells that were born internalized by a cell division that left one cell. Lineage lines without colors to mark internalization indicate cells that did not internalize during the time the embryos were filmed. Red asterisks are abnormalities in the internalization of cells. Question marks indicate cells for which we could not determine whether they internalized.
Cell lineage color code is the same as in Fig 2. Renderings of cell outlines were generated from a membrane-marked embryo filmed by spinning disk confocal microscopy at a plane corresponding to the middle of the top layer of cells at each stage, i.e. halfway through the depth at the four cell stage, and rising to match the middle of the layer of cells nearest the objective lens as cell divisions resulted in smaller and smaller cells. View is initially a lateral view, becoming a ventral view as the embryo rotates after E lineage (green) internalization. E, MS, P4, D and all of their descendants are colored from the time they are born. In the AB and C lineages, only some descendants gastrulate. For these lineages, we have colored the AB cells (in purple) only during the cell cycle at which each cell internalization occurs, and we have colored the C lineage (in yellow) at the birth of C, with yellow later marking only those C lineage cells that internalize. 50 of the 66 gastrulating cells are shown here. The remaining 16 gastrulating cells (all from the AB lineage) internalized from a site other than the ventral side of the embryo. Frames were acquired 1 minute apart.
This movie includes Movie 1 in the lower right, the colored cells overlain on the original film, the drawn cell boundaries, and the raw movie of a plasma membrane-tagged embryo. Frames were acquired 1 minute apart.
Acknowledgments
We thank Morris Maduro, Jeremy Nance, Bob Waterston and the Caenorhabditis Genetics Center for strains, the Developmental Studies Hybridoma Bank for the PAR-3 antibody, and Jennifer Sallee and members of our lab for discussion and comments. We thank Janet Iwasa for her expertise in making the supplementary movies, from which we derived parts of Figure 2. This work was supported by funds from the UNC Cell and Developmental Biology Department to J.H. and by National Institutes of Health grant R01-GM083071.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson DC, Gill JS, Cinalli RM, Nance J. Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science. 2008;320:1771–1774. doi: 10.1126/science.1156063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assemat E, Bazellieres E, Pallesi-Pocachard E, Le Bivic A, Massey-Harroche D. Polarity complex proteins. Biochim Biophys Acta. 2008;1778:614–630. doi: 10.1016/j.bbamem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- Barrett K, Leptin M, Settleman J. The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell. 1997;91:905–915. doi: 10.1016/s0092-8674(00)80482-1. [DOI] [PubMed] [Google Scholar]
- Berkowitz LA, Strome S. MES-1, a protein required for unequal divisions of the germline in early C. elegans embryos, resembles receptor tyrosine kinases and is localized to the boundary between the germline and gut cells. Development. 2000;127:4419–4431. doi: 10.1242/dev.127.20.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodu V, Casanova J. The RhoGAP crossveinless-c links trachealess and EGFR signaling to cell shape remodeling in Drosophila tracheal invagination. Genes Dev. 2006;20:1817–1828. doi: 10.1101/gad.375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broitman-Maduro G, Owraghi M, Hung WW, Kuntz S, Sternberg PW, Maduro MF. The NK-2 class homeodomain factor CEH-51 and the T-box factor TBX-35 have overlapping function in C. elegans mesoderm development. Development. 2009;136:2735–2746. doi: 10.1242/dev.038307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capowski EE, Martin P, Garvin C, Strome S. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics. 1991;129:1061–1072. doi: 10.1093/genetics/129.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm AD, Hardin J. Epidermal morphogenesis. The C. elegans Research Community. WormBook. 2005 doi: 10.1895/wormbook.1.35.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Chung S, Andrew DJ. The formation of epithelial tubes. J Cell Sci. 2008;121:3501–3504. doi: 10.1242/jcs.037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp AJ, Greene ND. Genetics and development of neural tube defects. J Pathol. 2010;220:217–230. doi: 10.1002/path.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Wilson ET, Wieschaus E. A putative cell signal encoded by the folded gastrulation gene coordinates cell shape changes during Drosophila gastrulation. Cell. 1994;76:1075–1089. doi: 10.1016/0092-8674(94)90384-0. [DOI] [PubMed] [Google Scholar]
- Dawes-Hoang RE, Parmar KM, Christiansen AE, Phelps CB, Brand AH, Wieschaus EF. Folded gastrulation, cell shape change and the control of myosin localization. Development. 2005;132:4165–4178. doi: 10.1242/dev.01938. [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Erwin DH, Davidson EH. The last common bilaterain ancestor. Development. 2002;129:3021–3032. doi: 10.1242/dev.129.13.3021. [DOI] [PubMed] [Google Scholar]
- Fodor A, Riddle DL, Nelson FK, Golden JW. Comparison of a new wild-type Caenorhabditis briggsae with laboratory strains of C. briggsae and C. elegans. Nematologica. 1983;29:203–217. [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigo SL, Hildebrand JD, Harland RM, Wallingford JB. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr Biol. 2003;13:2125–2137. doi: 10.1016/j.cub.2003.11.054. [DOI] [PubMed] [Google Scholar]
- Harris TJ, Sawyer JK, Peifer M. How the cytoskeleton helps build the embryonic body plan: models of morphogenesis from Drosophila. Curr Top Dev Biol. 2009;89:55–85. doi: 10.1016/S0070-2153(09)89003-0. [DOI] [PubMed] [Google Scholar]
- Hildebrand JD. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J Cell Sci. 2005;118:5191–5203. doi: 10.1242/jcs.02626. [DOI] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell. 1999;99:485–497. doi: 10.1016/s0092-8674(00)81537-8. [DOI] [PubMed] [Google Scholar]
- Junkersdorf B, Schierenberg E. Embryogenesis in C. elegans after elimination of individual blastomeres or induced alteration of the celldivision order. Roux’s Arch. Dev. Biology. 1992;202:17–22. doi: 10.1007/BF00364593. [DOI] [PubMed] [Google Scholar]
- Kachur TM, Audhya A, Pilgrim DB. UNC-45 is required for NMY-2 contractile function in early embryonic polarity establishment and germline cellularization in C. elegans. Dev Biol. 2008;314:287–299. doi: 10.1016/j.ydbio.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Kolesnikov T, Beckendorf SK. 18 wheeler regulates apical constriction of salivary gland cells via the Rho-GTPase signaling pathway. Dev Biol. 2007;307:53–61. doi: 10.1016/j.ydbio.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolsch V, Seher T, Fernandez-Ballester GJ, Serrano L, Leptin M. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 2007;315:384–386. doi: 10.1126/science.1134833. [DOI] [PubMed] [Google Scholar]
- Lee JY, Goldstein B. Mechanisms of cell positioning during C. elegans gastrulation. Development. 2003;130:307–320. doi: 10.1242/dev.00211. [DOI] [PubMed] [Google Scholar]
- Lee JY, Marston DJ, Walston T, Hardin J, Halberstadt A, Goldstein B. Wnt/Frizzled signaling controls C. elegans gastrulation by activating actomyosin contractility. Curr Biol. 2006;16:1986–1997. doi: 10.1016/j.cub.2006.08.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leptin M, Grunewald B. Cell shape changes during gastrulation in Drosophila. Development. 1990;110:73–84. doi: 10.1242/dev.110.1.73. [DOI] [PubMed] [Google Scholar]
- Liu J, Maduzia LL, Shirayama M, Mello CC. NMY-2 maintaings cellular assymmetry and cell boundaries, and promotes a SRC-dependent asymmetric cell division. Dev Biol. 2010;339:366–373. doi: 10.1016/j.ydbio.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Hill RJ, Heid PJ, Newman-Smith ED, Zhu J, Priess JR, Rothman JH. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev Biol. 2005;284:509–522. doi: 10.1016/j.ydbio.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Maduro MF. Endomesoderm specification in Caenorhabditis elegans and other nematodes. Bioessays. 2006;10:1010–1022. doi: 10.1002/bies.20480. [DOI] [PubMed] [Google Scholar]
- Nance J, Priess JR. Cell polarity and gastrulation in C. elegans. Development. 2002;129:387–397. doi: 10.1242/dev.129.2.387. [DOI] [PubMed] [Google Scholar]
- Nance J, Munro EM, Priess JR. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development. 2003;130:5339–5350. doi: 10.1242/dev.00735. [DOI] [PubMed] [Google Scholar]
- Nikolaidou KK, Barrett K. A Rho GTPase signaling pathway is used reiteratively in epithelial folding and potentially selects the out come of Rho activation. Curr Biol. 2004;14:1822–1826. doi: 10.1016/j.cub.2004.09.080. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Takeichi M. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development. 2008;135:1493–1502. doi: 10.1242/dev.019646. [DOI] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Hacker U, Turck C, Vale RD. Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr Biol. 2004;14:1827–1833. doi: 10.1016/j.cub.2004.09.078. [DOI] [PubMed] [Google Scholar]
- Roh-Johnson M, Goldstein B. In vivo roles for Arp2/3 in cortical actin organization during C. elegans gastrulation. J Cell Sci. 2009;122:3983–3993. doi: 10.1242/jcs.057562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JM, Harrell JR, Shemer G, Sullivan-Brown J, Roh-Johnson M, Goldstein B. Apical constriction: A cell shape change that can drive morphogenesis. Dev Biol. 2010;341:5–19. doi: 10.1016/j.ydbio.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somlyo AP, Somlyo AV. Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: modulated by G proteins, kinases, and myosin phosphatase. Physiol Rev. 2003;4:1325–1358. doi: 10.1152/physrev.00023.2003. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Strome S. Specification of the germ line. The C. elegans Research Community. WormBook. 2005 doi: 10.1895/wormbook.1.9.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Strome S, Martin P, Schierenberg E, Paulsen J. Transformation of the germ line into muscle in mes-1 mutant embryos of C. elegans. Development. 1995;121:2961–2972. doi: 10.1242/dev.121.9.2961. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tenlen JR, Molk JN, London N, Page BD, Priess JR. MEX-5 asymmetry in one-cell C. elegans embryos requires PAR-4- and PAR-1-dependent phosphorylation. Development. 2008;135:3665–3675. doi: 10.1242/dev.027060. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Lockery SR. Neurobiology. In: Hope I, editor. C. elegans: A Practical Approach. New York: Oxford University Press, Inc; 1999. pp. 143–180. [Google Scholar]
- Totong R, Achillieos A, Nance J. PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development. 2007;134:1259–1268. doi: 10.1242/dev.02833. [DOI] [PubMed] [Google Scholar]
- Wieschaus EF. From Molecular Patterns to Morphogenesis: The Lessons from Drosophila. Nobel Lecture 1995 [Google Scholar]
- Williams-Masson EM, Malik AN, Hardin J. An actin-mediated two-step mechanism is required for ventral enclosure of the C. elegans hypodermis. Development. 1997;124:2889–28901. doi: 10.1242/dev.124.15.2889. [DOI] [PubMed] [Google Scholar]
- Wood W, Jacinto A, Grose R, Woolner S, Gale J, Wilson C, Martin P. Wound healing recapitulates morphogenesis in Drosophila embryos. Nat Cell Biol. 2002;4:907–912. doi: 10.1038/ncb875. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Boyle TJ, Bao Z, Murray JI, Mericle B, Waterston RH. Comparative analysis of embryonic cell lineage between Caenorhabditis briggsae and Caenorhabditis elegans. Dev Biol. 2008;314:93–99. doi: 10.1016/j.ydbio.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Hill RJ, Heid PJ, Fukuyama M, Sugimoto A, Priess JR, Rothman JH. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 1997;11:2883–2896. doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MSpppa (top white asterisk) and MSappa (bottom white asterisk), which are the grandparents of somatic gonadal precursors Z1 and Z4 respectively, lie close to the germ line precursors Z2 and Z3 (black asterisks). To see the cell movements, see Supplemental Movies 1 and 2.
Lineages are drawn from wild-type (A) and individual tbx-35;ceh-51 embryos (B). Blue lines indicate cells in the MS lineage that internalized. (A) MS lineage in a wild-type embryo. Gray lines on lineage are standard deviations. White stars on the image of the embryo indicate cells that internalize in wild-type embryos. (B) MS lineages in four tbx-35;ceh-51 mutant embryos. Lineage lines without blue indicate cells that did not internalize during the time the embryos were filmed. Red asterisks are abnormalities in the internalization of cells. Question marks indicate cells for which we could not determine whether they internalized.
Lineages are drawn from wild-type (A) and individual mes-1 embryos (B). Orange lines indicate germ line cells that internalized and red lines indicate cells with a D cell fate that internalized. (A) Germ line and D cell lineages in wild-type embryos. Gray lines on lineage are standard deviations. White stars on the image of the embryo indicate cells of the relevant lineages that internalize in wild-type embryos. (B) Germ line and D lineages in four mes-1 embryos. Gray lines indicate cells that were born internalized by a cell division that left one cell. Lineage lines without colors to mark internalization indicate cells that did not internalize during the time the embryos were filmed. Red asterisks are abnormalities in the internalization of cells. Question marks indicate cells for which we could not determine whether they internalized.
Cell lineage color code is the same as in Fig 2. Renderings of cell outlines were generated from a membrane-marked embryo filmed by spinning disk confocal microscopy at a plane corresponding to the middle of the top layer of cells at each stage, i.e. halfway through the depth at the four cell stage, and rising to match the middle of the layer of cells nearest the objective lens as cell divisions resulted in smaller and smaller cells. View is initially a lateral view, becoming a ventral view as the embryo rotates after E lineage (green) internalization. E, MS, P4, D and all of their descendants are colored from the time they are born. In the AB and C lineages, only some descendants gastrulate. For these lineages, we have colored the AB cells (in purple) only during the cell cycle at which each cell internalization occurs, and we have colored the C lineage (in yellow) at the birth of C, with yellow later marking only those C lineage cells that internalize. 50 of the 66 gastrulating cells are shown here. The remaining 16 gastrulating cells (all from the AB lineage) internalized from a site other than the ventral side of the embryo. Frames were acquired 1 minute apart.
This movie includes Movie 1 in the lower right, the colored cells overlain on the original film, the drawn cell boundaries, and the raw movie of a plasma membrane-tagged embryo. Frames were acquired 1 minute apart.