Abstract
Controlling for sample site is considered to be an important aspect of chronic wound microbiological investigations; yet, macro-scale spatial variation in wound microbiota has not been well characterized. A total of 31 curette samples were collected at the leading edge, opposing leading edge and/or center of 13 chronic wounds. Bacterial community composition was characterized using a combination of 16S rRNA gene based pyrosequencing; heat map display; hierarchical clustering; non-metric multidimensional scaling; and permutation multivariate analysis of variance. A total of 58 bacterial families and 91 bacterial genera were characterized among the 13 wounds. While substantial macro-scale spatial variation was observed among the wounds, bacterial communities at different sites within individual wounds were significantly more similar than those in different wounds (p = 0.001). Our results support the prevalent opinion that controlling for sample site may improve the quality of wound microbiota studies; however, the significant similarity in bacterial communities from different sites within individual wounds indicates that studies failing to control for sampling site should not be disregarded based solely on this criterion. A composite sample from multiple sites across the surface of individual wounds may provide the most robust characterization of wound microbiota.
Keywords: chronic wound, microbiota, bacteria, infection, spatial variation, pyrosequencing, 16S rRNA gene
Introduction
There is a continued need to define the role of microbiota in wound healing to insure that appropriate therapy is utilized. Such studies are particularly relevant pertaining to chronic wounds as microbial colonization is considered to be one of the key factors leading to delayed healing (1). Each year, hundreds of thousands of Americans are treated for chronic wounds, resulting in billions of dollars in healthcare expenditures (2-3). Fully characterizing the role of microbiota in wound healing will require studies using a combination of robust molecular analyses and appropriate epidemiological design that considers all relevant factors such as macro-scale spatial variation (i.e., variation across the surface of the wound) in chronic wound microbiota. Studies that do not consider these variables are likely to present an incomplete or biased characterization of the chronic wound microbiota.
Previous studies using culture-independent molecular-based methods to characterize chronic wound microbiota indicate that standard culture-based methods frequently fail to capture the full scope of bacteria present within a wound (4-9). The combination of incorrect growth conditions (media formulation, temperature, gas mixture, time, etc.) and fastidious or non-culturable bacteria has proven difficult to overcome using culture-based methods; whereas, DNA- and RNA-based methods are unaffected by these challenges. Moreover, in contrast to culture-based methods, molecular methods—aided by next-generation sequencing technologies—can be used to characterize complex bacterial communities with relative ease.
Understanding macro-scale, spatial variation in chronic wound microbiota is critical to evaluating the relative importance of controlling for sample site in chronic wound investigations; however, such variation has yet to be thoroughly evaluated. Physiological conditions can vary spatially within a wound and create diverse microenvironments that may support different microbial communities. For example, the surface of a wound may be better oxygenated than the deeper wound center and favor more aerobic bacterial species. A review of the wound literature over the past three decades reveals few studies on spatial variation in chronic wound microbiota. Two recent studies used fluorescent in situ hybridization-based methods to reveal non-random, micro-scale spatial variation in chronic microbiota (10-11). To date, however, only one study has applied culture-independent methods to evaluate macro-scale, spatial variation within chronic wounds (12). In this study, Wolcott et al. examined topological variation in four chronic venous leg ulcers by sampling from multiple sites at the leading edge and center of each wound and concluded that there was substantial variation across the surface of the four chronic wounds.
In the current study, we characterized the bacterial microbiota of 13 chronic wounds using 16S rRNA-based pyrosequencing analysis.
Methods
Ethics Statement
Participants were provided written informed consent for enrollment. The protocol was reviewed and approved by the Johns Hopkins University IRB and Western IRB (for TGen). Each patient included in the current analyses was assigned a code beginning with WS or TR followed by a two-digit number.
Sample collection
Chronic wound tissue samples (n = 31) were collected from 12 patients enrolled and consented at the Johns Hopkins Wound Center, a tertiary wound center in Baltimore, MD. Briefly, after consent and topical anesthesia with xylocain, tissue was collected from the wound base with a 3 or 7 mm curette. Samples were taken from two to three sites from each wound: site A, the leading edge of the wound; site B, the opposing leading edge; and/or site C, the wound center (Figure S1). Each specimen was placed in nonbacteriostatic saline, immediately transported to the laboratory, minced, and divided for molecular- and culture-based analyses.
Culture-based assessment
For each sample, 20 to 40 mg of tissue was transferred to a sterile 50 ml disposable tissue grinder containing 5 ml of sterile saline and hand-homogenized for 30 seconds. The tissue homogenate was then serially diluted and plated on non-selective media including sheep blood agar, MacConkey and chocolate agar plates and incubated overnight at 35°C in 5% CO2. Colonies were counted and used to determine the Colony Forming Units per gram of tissue (CFU/g) for each species. Speciation was carried out on the MicroScan Walk-Away® system (Dade Behring Inc., West Sacramento, CA).
Technical replicates
From each of two study participants (TR01 and TR02), we collected a single curette sample from the leading edge of their wound, and then divided the sample into three approximately equal portions. Each of the wound portions was then processed and analyzed separately in order to evaluate the reproducibility of our methods. Note: As performed, the technical replicates themselves were vulnerable to spatial variation within the individual curette sample.
DNA extraction and purification
Genomic DNA was extracted from curette samples using a bead-beating and enzymatic lysis protocol, followed by purification using a QIAamp DNA Mini Kit (Qiagen, Valencia, USA). Briefly, the frozen tissue samples were thawed on ice and 0.75 ml of TE50 (10mM Tris-HCl + 50mM EDTA, pH 8.0) solution added. 500 μl of the solution was transferred to a clean, sterile bead-beating tube (MP Biomedicals, Solon, USA) and kept on ice. A lytic enzyme cocktail was prepared at the time of extraction and added to each sample as follows: 50 μl Lysozyme (450 kU ml-1), 6 μl Mutanolysin (25 kU ml-1), 3 μl Lysostaphin (4 kU ml-1) and 41 μl TE50 for a final volume of 100 ml per sample. Samples were digested by incubating at 37°C for 60 min in a dry heat block before centrifugation at 1200 rpm for 1 min. To each digested sample, 750 mg of sterile 0.1 mm diameter zirconia silica beads (BioSpec Products Inc., Bartlesville, USA) were added. Bead-beating was performed for 1 min at 2100 rpm using a BioSpec Mini-Bead Beater-96. Following bead disruption, the tubes were centrifuged at 1200 rpm for 1 min. Two 200 ml aliquots of crude lysate from each sample were transferred to new, sterile microcentrifuge tubes. To each tube, 25 μl of Proteinase K (20 mg/ml (>600 mAU/ml)) and 200 μl of Qiagen buffer AL were added. Samples were mixed by pulse-vortexing for 15 sec and then incubated at 56°C for 10 min before being centrifuged at 1200 rpm for 1 min. For each 200 μl crude lysate, 20 μl of 3 M sodium acetate, pH 5.5 was added followed by 200 μl of molecular grade ethanol (96–99.5%). Vortexing was repeated for an additional 15 sec before being centrifuged at 1200 rpm for 1 min. From this point onward, purification was carried out using the QIAmp DNA Purification from Blood or Body Fluids as per manufacturer's instructions. Aliquots from the same sample were loaded onto the same column. Purified genomic DNA was stored at -80°C until analysis.
Pyrosequencing analysis of the 16S rRNA gene V3-V4 region on the 454® platform using fusion primers
The 16S rRNA gene was amplified in two replicate 15 μl reaction volumes. In each 15 μl reaction, 1 μl was added to 14 μl of PCR reaction mix containing 450 nM of each broad range forward (5′- CCATCTCATCCCTGCGTGTCTCCGACTCAGnnnnnnnnnn-CCTACGGGDGGCWGCA-3′) and reverse primer (5′-CCTATCCCCTGTGTGCCTTGGCAGTC-TCAGGGACTACHVGGGTMTCTAATC-3′), 1X PCR buffer without MgCl2 (Invitrogen, Carlsbad, USA), 3 mM MgCl2, 0.2 mM dNTP mix, 1 U platinum Taq (Invitrogen) using the following touch-down PCR condition: 90s at 95°C for initial denaturation and 20 cycles of 30s at 95°C for denaturation, 30s at 60°C for annealing, 30s at 72°C for extension with the annealing temperature decreasing by 0.5°C for each subsequent cycle for followed by 10 cycles of 30s at 95°C for denaturation, 30s at 45°C for annealing, 30s at 72°C for extension, and a final extension for 7 min at 72°C. The final tagged PCR products were then purified using the Agencourt® AMPure® Kit (Agencourt, Beverly, USA). Gel electrophoresis was performed using 5ul of the purified PCR product using the E-Gel® 96-well System (Invitrogen, Carlsbad, USA) for quality check and gel-based quantification. A second quantification was performed using an in-house 16S rRNA gene quantitative real-time PCR assay. The PCR products were then pooled in an equimolar fashion to generate the amplicon library for pyrosequencing. The emulsion PCR and pyrosequencing analysis using a Roche 454 XLR Titanium chemistry on the FLX pyrosequencer was performed according to the manufacturer instructions (454 Life Sciences, Branford, USA) at the Institute for Genome Sciences of University of Maryland.
Pyrosequencing data processing
Experimental sequences were processed using a custom PERL script, which performed the following: the script filtered the sequence files and retained only sequences that were 200-nt or longer. Regular expressions were then applied to the remaining sequences to search for a single barcode sequence in each FASTA sequence, binned each sequence accordingly, and scanned each binned sequence for the 16S forward primer sequence. The script then trimmed off the forward primer sequence and oriented the remaining sequence such that all sequences begin with the 5′ end according to standard sense strand conventions. As a result of our processing, sequences that were shorter than 200-nt or had multiple barcode or primer motifs were excluded from the analysis.
Taxonomic assignment
We classified the 16S rRNA gene sequences at each taxonomic level (i.e., phylum, class, order, family, genus) at ≥95% bootstrap confidence level using a web service for the Naïve Bayesian Classifier made available by the Ribosomal Database Project (13). An SQL database was used to store and query the results. In order to control for potential errors due to pyrosequencing that may have created spurious sequences, we removed all taxonomic groups occurring only once (i.e., singletons). The taxonomic data was converted into a data matrix in R, which we converted to a proportional abundance data matrix calculated by dividing the bacterial family abundance by the total number of sequences assigned to the Bacterial domain from the 31 samples. To faciliate in silico evaluation of the robustness of using combined samples to evaluate the overall wound microbiota, we simulated a combined dataset from all sampled sites for each wound. This was performed by summing the number of sequences from each sampling site for each detected bacterial family and genus. Next, the proportional abundance was generated accordingly (i.e., the sum of each bacterial family/genus from all sampled sites divided by sum of total number of sequences from all sampled sites assigned to the domain of Bacteria). Subsequent microbiota analyses were performed using both the experimental wound data and the simulated combined wound data.
Visualization of the wound microbiota data
Subsequent data visualization and analyses were performed in R version 2.9.1 unless otherwise specified (14). We visualized the wound microbioa using two different approaches: 1) a heatmap display and 2) non-metric multi-dimensional scaling (nMDS). Heatmap display. We visually assessed the wound microbiota using a heatmap display, where the relative abundances of the different taxa are represented by different colors (Figure 1; Figure S2). Non-metric multidimensional scaling (nMDS) analysis. We utilized nMDS ordination to visually assess patterns microbiota compositional differences among our samples. All nMDS plots were generated in R using functions from ecodist (15), ellipse (16), and BiodiversityR (17). To generate each nMDS plot, we began by evaluating the number of dimensions required to appropriately present the bacterial communities using a stress plot. We applied a conventional cutoff of < 0.2 to determine the acceptable number of dimensions. Using the appropriate number of dimensions n = 50 iterations, Using the appropriate number of dimensions, the nMDS procedure was repeated with n = 50 iterations. The final nMDS plot was generated using the ordinated dataset. The distance between points in the plot can be interpreted as the relative difference in community composition; hence, points that are closer are more similar than points that are more distant.
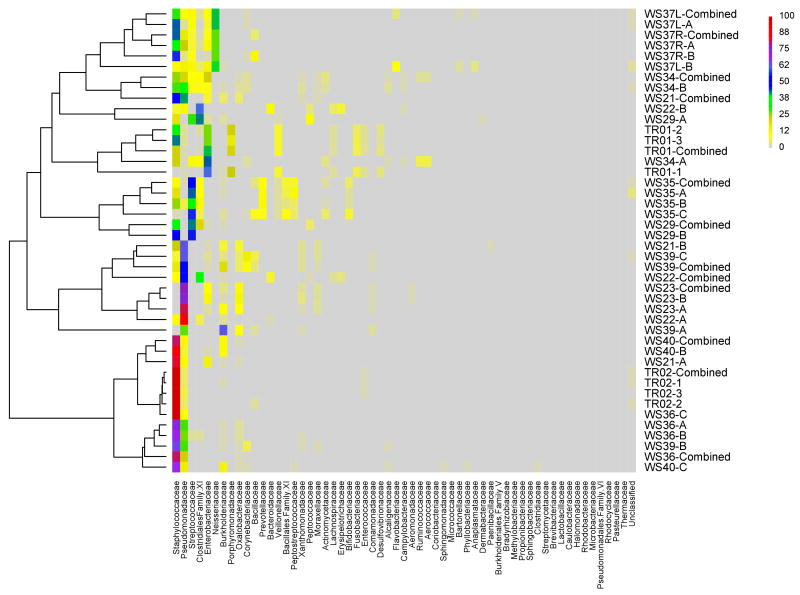
Figure 1.
Figure 1A. Heat map display of bacterial families comprising chronic wound microbiota (samples are grouped based on hierarchical clustering of bacterial communities).
The color key for the number of sequences from each bacterial family is shown on the right. A = leading edge; B = apposing leading edge; C = center; Combined = simulated combined wound; TR01 and TR02 (1, 2, & 3) are technical replicates created by dividing a single curette sample into three equal parts prior to processing.
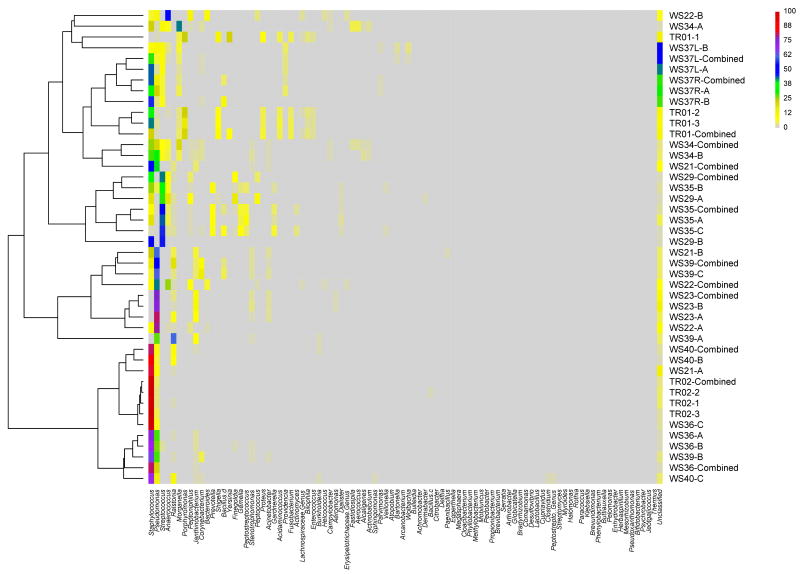
Figure 1B. Heat map display of bacterial genera comprising chronic wound microbiota (samples are grouped based on hierarchical clustering of bacterial communities).
The color key for the number of sequences from each bacterial family is shown on the right. (A = leading edge; B = apposing leading edge; C = center; Combined = simulated combined wound; TR01 and TR02 (1, 2, & 3) are technical replicates).
Multivariate ecological analyses
In order to compare the microbiota at different sites within individual wounds and among the different wounds, we applied the multivariate analysis method called Permutation Multivariate Analysis of Variance (PerMANOVA), which is often used for ecological community data. We used PerMANOVA to test the null-hypothesis of no-difference between the bacterial communities found within a single wound and those in different wounds (α = 0.05). PerMANOVA is a permutation-based version of the multivariate analysis of variance (18). PerMANOVA uses the distances between samples to partition variance and randomizations or permutations of the data to produce the p-value for the hypothesis test. It is non-parametric (or semi-parametric for multi-factor models) and, therefore, robust to the assumption of multivariate normality making it less prone to Type I errors. PerMANOVA analyses were performed in R using the “adonis” function from the vegan package (19).
Culture-based data was compared in a pair-wise fashion between different sites within individual wounds using Kendall's tau (τ) correlation coefficient.
Results
We enrolled 12 study participants at the Johns Hopkins Wound Center in Baltimore, MD and collected 31 curette samples from 13 wounds (one participant was sampled from venous ulcers on both legs) (Table 1). For each wound, we collected a curette sample from two to three sites: 1) site A, the leading edge of the wound; 2) site B, the opposing leading edge; and/or 3) site C, the center of the wound (Figure S1).
Table 1. Wound and patient characteristics.
| Wound | Patient Sex | Patient Age | Primary Diagnosis | DM1 | Systemic ABx2 within 14 days | Systemic ABx2 Name | Topical Treatment within 24 h |
|---|---|---|---|---|---|---|---|
| TR01 | F | 86 | Decubitus | 0 | 0 | ||
| TR02 | M | 71 | Decubitus | 0 | 1 | Unknown | |
| WS21 | M | 39 | Decubitus | 0 | 0 | ||
| WS22 | M | 76 | Venous Stasis | 0 | Silvadene® | ||
| WS23 | F | 48 | Post surgical | 1 | 0 | Aquacel® Ag | |
| WS29 | F | 57 | Neuropathic | 1 | 0 | Prisma™ (collagen dressing) | |
| WS34 | F | 54 | Other | 1 | 0 | ||
| WS35 | F | 59 | Neuropathic | 1 | 0 | ||
| WS36 | M | 42 | Other | 0 | 1 | Cipro | Mesalt |
| WS37L | F | 54 | Venous Stasis | 1 | 0 | ||
| WS37R | F | 54 | Venous Stasis | 1 | 0 | ||
| WS39 | F | 57 | Neuropathic | 1 | 1 | Bactrim DS | |
| WS40 | M | 30 | Decubitus | 0 | 0 | Unknown gel |
DM, diabetes mellitus;
ABx, antibiotic
Aerobic bacteria at different sites within the same wounds were well correlated as assessed by qualitative culture-based methods. Agreement was assessed using Kendall's tau (τ) correlation coefficient. None of the sample pairs were significantly different from one another. All tau p-values were greater than or equal to 0.719. As reported previously, culture-based methods revealed much fewer species among wound samples as compared to culture-independent 16S rRNA based methods (9).
Pyrosequencing analysis of V3-V4 segment of the 16S rRNA gene generated a total of 61,422 sequences from the 31 curette samples (Per sample mean, 1982; SD, 941; min, 540; max, 4762). We performed taxonomic classification of sequences using the Ribosomal Database Project (RDP) Naïve Bayesian Classifier (13). Greater than 99% of the sequences analyzed were identified to the phylum, class, order and family levels at ≥95% confidence; however, the proportion of sequences assigned to the genus level decreased to 91.0% (genus level taxonomic assignment varied from 0 to 100% among the different bacterial families). Table 1 shows the 25 most common families and genera identified among the wounds.
We detected a total of 58 bacterial families (Average per wound,18.9; SD, 4.1; range, 13 to 29) from the 31 samples analyzed. Among these, Staphylococcaceae was the most common, followed by Pseudomonadaceae, Streptococcaceae, Clostridiales Family XI and Enterobacteriaceae (Table 2). We identified 91 bacterial genera among the wounds (Average per wound, 20.9; SD, 6.2; range 11 to 34). The ten most common genera included: Staphylococcus, Pseudomonas, Streptococcus, Anaerococcus, Ralstonia, Morganella, Porphyromonas, Peptoniphilus, Janthinobacterium and Corynebacterium.
Table 2. The 25 most common bacterial families found within the wounds.
| Family | Number of Sequences | Percent Total | Genus | Number of Sequences | Percent Family |
|---|---|---|---|---|---|
| Staphylococcaceae | 19565 | 32.10 | |||
| Staphylococcus | 19561 | ||||
| Jeotgalicoccus | 2 | ||||
| sum | 19563 | 99.99 | |||
| Pseudomonadaceae | 13077 | 21.45 | |||
| Pseudomonas | 12408 | 94.88 | |||
| Streptococcaceae | 5725 | 9.39 | |||
| Streptococcus | 5722 | 99.95 | |||
| Clostridiales Incertae Sedis XI | 4701 | 7.71 | |||
| Anaerococcus | 3036 | ||||
| Peptoniphilus | 1025 | ||||
| Finegoldia | 434 | ||||
| Helcococcus | 126 | ||||
| Parvimonas | 64 | ||||
| sum | 4685 | 99.66 | |||
| Enterobacteriaceae | 3826 | 6.28 | |||
| Morganella | 1162 | ||||
| Shigella | 533 | ||||
| Yersinia | 440 | ||||
| Proteus | 332 | ||||
| Providencia | 244 | ||||
| Citrobacter | 28 | ||||
| Serratia | 10 | ||||
| Klebsiella | 4 | ||||
| Buttiauxella | 3 | ||||
| sum | 2756 | 72.03 | |||
| Neisseriaceae | 2364 | 3.88 | |||
| Neisseria | 1 | 0.04 | |||
| Burkholderiaceae | 1668 | 2.74 | |||
| Ralstonia | 1526 | ||||
| Burkholderia | 129 | ||||
| Cupriavidus | 7 | ||||
| sum | 1662 | 99.64 | |||
| Porphyromonadaceae | 1035 | 1.70 | |||
| Porphyromonas | 1033 | ||||
| Dysgonomonas | 1 | ||||
| sum | 1034 | 99.90 | |||
| Oxalobacteraceae | 1004 | 1.65 | |||
| Janthinobacterium | 996 | ||||
| Herbaspirillum | 3 | ||||
| sum | 999 | 99.50 | |||
| Corynebacteriaceae | 628 | 1.03 | |||
| Corynebacterium | 628 | 100.00 | |||
| Bacillaceae | 598 | 0.98 | |||
| Bacillus d | 529 | ||||
| Bacillus c | 29 | ||||
| Bacillus f | 1 | ||||
| sum | 559 | 93.48 | |||
| Prevotellaceae | 564 | 0.93 | |||
| Prevotella | 534 | 94.68 | |||
| Bacteroidaceae | 546 | 0.90 | |||
| Bacteroides | 546 | 100.00 | |||
| Veillonellaceae | 491 | 0.81 | |||
| Acidaminococcus | 259 | ||||
| Dialister | 116 | ||||
| Veillonella | 60 | ||||
| Megasphaera | 20 | ||||
| sum | 455 | 92.67 | |||
| Bacillales Incertae Sedis XI | 433 | 0.71 | |||
| Gemella | 433 | 100.00 | |||
| Peptostreptococcaceae | 396 | 0.65 | |||
| Peptostreptococcus | 389 | ||||
| Peptostreptococcaceae Incertae Sedis | 7 | ||||
| sum | 396 | 100.00 | |||
| Xanthomonadaceae | 359 | 0.59 | |||
| Stenotrophomonas | 356 | ||||
| Pseudoxanthomonas | 3 | ||||
| sum | 359 | 100.00 | |||
| Peptococcaceae | 355 | 0.58 | |||
| Peptococcus | 355 | 100.00 | |||
| Moraxellaceae | 334 | 0.55 | |||
| Acinetobacter | 332 | ||||
| Psychrobacter | 2 | ||||
| sum | 334 | 100.00 | |||
| Actinomycetaceae | 332 | 0.54 | |||
| Actinomyces | 196 | ||||
| Actinobaculum | 80 | ||||
| Arcanobacterium | 44 | ||||
| Mobiluncus | 12 | ||||
| sum | 332 | 100.00 | |||
| Lachnospiraceae | 326 | 0.53 | |||
| Lachnospiraceae Incertae Sedis | 162 | 49.69 | |||
| Erysipelotrichaceae | 301 | 0.49 | |||
| Erysipelotrichaceae Incertae Sedis | 108 | ||||
| Bulleidia | 38 | ||||
| Erysipelothrix | 1 | ||||
| Turicibacter | 1 | ||||
| sum | 148 | 49.17 | |||
| Bifidobacteriaceae | 275 | 0.45 | |||
| Gardnerella | 273 | ||||
| Bifidobacterium | 2 | ||||
| sum | 275 | 100.00 | |||
| Fusobacteriaceae | 240 | 0.39 | |||
| Fusobacterium | 240 | 100.00 | |||
| Enterococcaceae | 227 | 0.37 | |||
| Enterococcus | 132 | 58.15 | |||
The heat map display of wound microbiota composition showed that while Staphylococcaceae was the most common family overall and dominated many samples, others were dominated by different families including: Pseudomonadaceae (e.g., WS21-B, WS22-A, WS23-A, WS23-B & WS39-C); Streptococcaceae (e.g., WS35-A & WS35-C); Clostridiales Family XI (e.g., WS22-B); Enterobacteriaceae (e.g., TR01-1 & WS34-A); Neisseriaceae (e.g., WS37L-B); and Porphyromonadaceae (e.g., WS39-A) (Figure S2). Two to three different organisms dominated some samples including: TR01-2, TR01-3, WS29-A, WS29-B, WS34-B, WS35-B, WS37L-A and WS37R-A.
Samples from different sites within individual wounds shared similarities in bacterial community compositions as shown with hierarchal clustering of bacterial families on the heat map display (Figure 1A). Similarities among communities from the same wound were maintained at the genus level as well (Figure 1B). Technical replicates from sample TR02 were highly similar; whereas, replicates from sample TR01 appeared to be as diverse as samples taken from different sites within a wound. The simulated combined wound data, as would be expected, closely clustered with the experimental wound microbiota data (Figure 1A-B).
Samples taken from within a single wound were more similar to one another than those taken from different wounds as was shown using a community analysis method commonly used in ecology research, termed non-metric multi-dimensional scaling (nMDS), an iterative, ordination-based method that reduces data complexity while retaining the most meaningful components (Figure 2). The significant similarity across a single wound was confirmed using a statistical test that compare the within-group variance with the between-group variance using a permutation-based approach, Permutational Multivariate Analysis of Variance (PerMANOVA), which revealed that the variation between communities in different wounds was significantly greater than the variation among different sites within the same wound (p = 0.001). Microbial community variation within individual wounds was similar whether comparing opposing leading edges or comparing leading edge samples with those taken from the center of the wound (Figure 2). Likewise, between our two technical replicate sets, one set formed a tight cluster, while the other set appeared to show dissimilarities akin to samples taken from different sites within a single wound. The combined wound microbiota appear to be representative of all replicates from each wound site, as the composite microbiota was consistently located near the centroid of the wound microbiota from different sites (Figure 2).
Figure 2.
Non-metric Multi-Dimensional Scaling (nMDS) plot of microbiota of wounds samples taken from different sites within the wounds.
(A = leading edge; B = apposing leading edge; C = center; star = simulated combined wound; TR01 and TR02 (1, 2, & 3) are technical replicates).
In the participant from whom we sampled venous leg ulcers from both legs (WS37R-A, WS37R-B, WS37L-A and WS37L-B), there was high concordance between the wounds, with both wounds showing abundant Staphylococcaceae and Neiserriaceae. This suggests that host factors—immunological and/or behavioral—may play an important role in determining the bacteria that colonize the host wounds, overcoming other environmental differences such as location of the wound on the body (Figures 1 and 2); however, the wound microbiota from additional patients with multiple wounds will have to be studied before any firm conclusions can be made.
Discussion
Our findings indicate that the microbiota at different sites on the leading edge of a wound may be as different from one another as they are from those taken from the center of the wound. At the time of submission, only one other study had been published reporting on the macro-scale spatial variation of microbiota across the surface of chronic wounds (12). In this previous study, four wounds were sampled at multiple sites of their leading edges and central regions. While the authors did not report statistical analyses, the tabular data revealed substantial variation in wound microbiota regardless of the regions from which the sample was taken. Thus, our data are consistent with these previous findings. Future studies, adjusting for wound type and size may further elucidate macro-scale wound microbiota variation. Likewise, a prospective analysis of macro-scale spatial variation may further reveal the relative importance and stability of such variation.
The variation in one of the technical replicates was somewhat disturbing; however, our previous experience with technical replication indicates that our molecular and statistical methods are highly reproducible (20). Therefore, we propose that the observed differences were most likely due to spatial variation within the single curette sample itself. The sample was not homogenized prior to dividing it into three equal parts; thus, the portions may have been from physiologically distinct regions of the sample site itself (e.g., one portion near the wound bed, another at the surface and the third in-between). In retrospect, it would have been more appropriate to isolate DNA from a single homogenized wound sample then split the DNA into three equal aliquots prior to starting replicate analyses. Unfortunately, technical replicates are rarely reported in the related scientific literature, so it is difficult to know how our data compare to those of other research groups.
Our findings suggest that a homogenized composite sample from multiple sites within a wound may provide a more robust picture of a wound's microbiota than simply sampling from a single site. Creating a post-analysis composite of DNA sequence data generated from multiple samples taken from the same wound may provide all the benefits of a physical homogenization while allowing for additional independent analyses of the different sites.
Our results support the prevalent opinion that controlling for sampling site within individual wounds can improve the quality of wound microbiota studies; however, the significant similarity in microbiota from different sites within individual wounds as compared to between different wounds suggests that studies failing to control for sampling site should not be considered invalid based solely on this criterion.
Supplementary Material
Figure S2. Heat map display of bacterial families comprising wound sample microbiota (samples are arranged by wound ID).
Figure S3. Heat map display of bacterial families comprising wound sample microbiota after subtracting the three most prevalent wound bacterial families (Staphylococcaceae, Pseudomonadaceae, and Streptococcaceae).
The color key for the number of sequences from each bacterial family is shown on the right. A = leading edge; B = apposing leading edge; C = center.
Figure S4. Non-metric Multi-Dimensional Scaling (nMDS) plot of wound sample microbiota subtracting the three most prevalent bacterial families (Staphylococcaceae, Pseudomonadaceae, and Streptococcaceae).
A = leading edge; B = apposing leading edge; C = center
Acknowledgments
The authors would like to acknowledge contributions by: Jolene Bowers; the Johns Hopkins Bayview Medical Center Clinical Microbiology Laboratory; and the Johns Hopkins Wound Center. The US Department of Defense, the National Institutes of Health, the Johns Hopkins Center for Innovative Medicine, and the Translational Genomics Research Institute provided financial support for this study.
List of Abbreviations
- nMDS
non-Metric Multi-Dimensional Scaling
- PerMANOVA
Permutation Multivariate Analysis of Variance
- RDP
Ribosomal Database Project
References
- 1.Mustoe TA, O'Shaughnessy K, Kloeters O. Chronic wound pathogenesis and current treatment strategies: A unifying hypothesis. Plast Reconstr Surg. 2006;117(7 Suppl):35S–41S. doi: 10.1097/01.prs.0000225431.63010.1b. [DOI] [PubMed] [Google Scholar]
- 2.Fonder MA, Lazarus GS, Cowan DA, Aronson-Cook B, Kohli AR, Mamelak AJ. Treating the chronic wound: A practical approach to the care of nonhealing wounds and wound care dressings. J Am Acad Dermatol. 2008;158:185–206. doi: 10.1016/j.jaad.2007.08.048. [DOI] [PubMed] [Google Scholar]
- 3.Sen CK, Gordillo GM, Roy S, Kirsner R, Lambert L, Hunt TK, Gottrup F, Gurtner GC, Longaker MT. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 2009;17:763–71. doi: 10.1111/j.1524-475X.2009.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen A, Hill KE, Stephens P, Thomas DW, Jorgensen B, Krogfelt KA. Bacterial profiling using skin grafting, standard culture and molecular bacteriological methods. J Wound Care. 2007;16:171–5. doi: 10.12968/jowc.2007.16.4.27025. [DOI] [PubMed] [Google Scholar]
- 5.Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS One. 2008;3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank DN, Wysocki A, Specht-Glick DD, Rooney A, Feldman RA, St Amand AL, Pace NR, Trent JD. Microbial diversity in chronic open wounds. Wound Repair Regen. 2009;17:163–72. doi: 10.1111/j.1524-475X.2009.00472.x. [DOI] [PubMed] [Google Scholar]
- 8.Frankel YM, Melendez JH, Wang NY, Price LB, Zenilman JM, Lazarus GS. Defining wound microbial flora: molecular microbiology opening new horizons. Arch Dermatol. 2009;145:1193–5. doi: 10.1001/archdermatol.2009.246. [DOI] [PubMed] [Google Scholar]
- 9.Price LB, Liu CM, Melendez JH, Frankel YM, Engelthaler D, Aziz M, Bowers J, Rattray R, Ravel J, Kingsley C, Keim PS, Lazarus GS, Zenilman JM. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS One. 2009;4:e6462. doi: 10.1371/journal.pone.0006462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazli M, Bjarnsholt T, Kirketerp-Møller K, Jørgensen B, Andersen AS, Krogfelt KA, Givskov M, Tolker-Nielsen T. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J Clin Microbiol. 2009;47:4084–9. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malic S, Hill KE, Hayes A, Percival SL, Thomas DW, Williams DW. Detection and identification of specific bacteria in wound biofilms using peptide nucleic acid fluorescent in situ hybridization (PNA FISH) Microbiology. 2009;155:2603–11. doi: 10.1099/mic.0.028712-0. [DOI] [PubMed] [Google Scholar]
- 12.Wolcott RD, Gontcharova V, Sun Y, Dowd SE. Evaluation of the bacterial diversity among and within individual venous leg ulcers using bacterial tag-encoded FLX and titanium amplicon pyrosequencing and metagenomic approaches. BMC Microbiol. 2009;9:226. doi: 10.1186/1471-2180-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The ribosomal database project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R Development Core Team. R: A language and environment for statistical computing. 2009 [Google Scholar]
- 15.Goslee SC, Niering WA, Urban DL, Christensen NL. Influence of environment, history and vegetative interactions on stand dynamics in a connecticut forest. J Torrey Bot Soc. 2005;132:471–482. [Google Scholar]
- 16.Murdoch D, Chow ED. Ellipse: Functions for drawing ellipses and ellipse-like confidence regions. 2007 0.3-5. [Google Scholar]
- 17.Kindt R, Coe R. Tree diversity analysis. A manual and software for common statistical methods for ecological and biodiversity studies. Nairobi: World Agroforestry Centre (ICRAF); 2005. [Google Scholar]
- 18.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- 19.Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, et al. Vegan: Community ecology package. 2009 1.15–2. [Google Scholar]
- 20.Price LB, Liu CM, Johnson KE, Aziz M, Lau MK, Bowers J, Ravel J, Keim PS, Serwadda D, Wawer MJ, Gray RH. The Effects of Circumcision on the Penis Microbiome. PLoS One. 2010;5:e8422. doi: 10.1371/journal.pone.0008422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S2. Heat map display of bacterial families comprising wound sample microbiota (samples are arranged by wound ID).
Figure S3. Heat map display of bacterial families comprising wound sample microbiota after subtracting the three most prevalent wound bacterial families (Staphylococcaceae, Pseudomonadaceae, and Streptococcaceae).
The color key for the number of sequences from each bacterial family is shown on the right. A = leading edge; B = apposing leading edge; C = center.
Figure S4. Non-metric Multi-Dimensional Scaling (nMDS) plot of wound sample microbiota subtracting the three most prevalent bacterial families (Staphylococcaceae, Pseudomonadaceae, and Streptococcaceae).
A = leading edge; B = apposing leading edge; C = center