Abstract
The purpose of this study was to compare the sampling efficacy of rayon swabs and nylon flocked swabs, and of oropharyngeal and nasopharyngeal specimens for the detection of respiratory viruses in elderly patients. Samples were obtained from patients 60 years of age or above who were newly admitted to Sorlandet Hospital Arendal, Norway. The patients were interviewed for current symptoms of a respiratory tract infection. Using rayon swabs and nylon flocked swabs, comparable sets of mucosal samples were harvested from the nasopharynx and the oropharynx. The samples were analysed using real-time polymerase chain reaction (PCR) methods. A total of 223 patients (mean age 74.9 years, standard deviation [SD] 9.0 years) were swabbed and a virus was recovered from 11% of the symptomatic patients. Regardless of the sampling site, a calculated 4.8 times higher viral load (95% confidence interval [CI] 1.3–17, p = 0.017) was obtained using the nylon flocked swabs as compared to the rayon swabs. Also, regardless of the type of swab, a calculated 19 times higher viral load was found in the samples from the nasopharynx as compared to the oropharynx (95% CI 5.4–67.4, p < 0.001). When swabbing for respiratory viruses in elderly patients, nasopharyngeal rather than oropharyngeal samples should be obtained. Nylon flocked swabs appear to be more efficient than rayon swabs.
Keywords: Respiratory Syncytial Virus, Polymerase Chain Reaction Assay, Respiratory Syncytial Virus Infection, Respiratory Virus, Bordetella Pertussis
Introduction
In recent years, respiratory viruses have been established as significant causes of mortality and morbidity in the older population [1–5]. However, as respiratory pathogens may cause similar clinical pictures, the aetiological diagnosis depends on laboratory confirmation [6].
As demonstrated with respiratory syncytial virus (RSV), viral detection is more demanding in the elderly than in young individuals, as older people tend to shed less virus and have shorter viral-shedding periods [7]. Hence, rapid viral antigen tests that are useful in children may fail in the elderly [8]. Due to its high sensitivity and specificity, real-time polymerase chain reaction (PCR) on respiratory viral samples represents an important diagnostic opportunity also in the older age groups.
Samples for the diagnosis of a respiratory viral infection can be obtained by swabbing the oropharynx, the nasal cavity, the nasopharynx or alternatively, by nasopharyngeal aspiration (NPA) or nasopharyngeal washings (NPW). Carefully conducted NPA and NPW usually provide sufficient diagnostic material in children and young adults. However, in frail elderly, NPA and NPW are less well tolerated [9, 10]. For this reason, nasopharyngeal swabbing (NPS) is the preferred sampling method in older adults [11], although slightly fewer epithelial cells are recovered by NPS as compared to NPA and NPW [12].
Two structurally different swabs are available for microbial sampling in the upper airways: rayon swabs and nylon flocked swabs. A few studies comparing the respective efficacies of these two swabs in providing diagnostic specimens have been performed in children, but, to our knowledge, no such studies have been conducted in the elderly.
The aim of this study was to compare the respective efficacies of rayon swabs and nylon flocked swabs in providing material for direct respiratory virus detection by real-time PCR in adults above 60 years of age. We also compared the diagnostic yield of samples recovered from the oropharynx and the nasopharynx.
Materials and methods
Ethical committee
The study design was approved by the Norwegian Regional Committee of Research Ethics.
Study design and eligibility criteria
The study took place from 13 February 2008 until 3 February 2009 at the Department of Internal Medicine, Sorlandet Hospital Arendal, Norway. Twice a week, all patients born in 1948 or earlier and admitted to the hospital the previous day were interviewed by two team members for symptoms as described in Table 1. During the first and the last four weeks of the study, patients were asked to take part regardless of symptoms, whereas during the rest of the study period, only patients with symptoms were asked to participate.
Table 1.
Inclusion criteria
| Inclusion criteria |
|---|
| Patients born in 1948 or earlier |
| AND at least one of the following current symptoms with debut less than three weeks ago: |
| • Nasal congestion or runny nose |
| • Throat pain |
| • Fever |
| • Malaise |
| • Muscle pain |
| • Self-diagnosis of “the common cold” |
| • Diarrhoea or eye infection combined with laboratory values supporting an infection |
If a patient was unable to supply the information needed, the next of kin and/or the concerned hospital staff were interviewed. If a patient was not able to give informed consent, the next of kin was asked for permission.
Swabs and mucosal swabbing
Rayon swabs
Virocult swabs MW950 and MW975 (Virocult, Medical Wire & Equipment, Corsham, UK) were used in the oropharynx and nasopharynx, respectively. The rayon swabs were inserted into the Virocult medium immediately after sampling and the tip of the tube was squeezed as instructed by the manufacturer.
Nylon flocked swabs
Nylon flocked swabs 502CS01 regular and 503CS01 nasopharyngeal (Copan Italia, Brescia, Italy) were used in the oropharynx and nasopharynx respectively. The nylon flocked swabs were inserted into universal transport medium (UTM) tubes (1.5 ml medium, no beads, Copan Italia) and the tubes were shaken for 10 s.
Swabbing in the oropharynx
The sample was taken from the tonsils or posterior oropharyngeal area. The nylon flocked swab was used on one side and the rayon swab on the other. A tongue depressor was used during the procedure and care was taken to avoid touching the tongue with the swabs.
Swabbing in the nasopharynx
The swab was inserted through the nostril, pushed back as far as possible, then rotated, and withdrawn. The nylon flocked swab was used in one nostril and the rayon swab in the other.
Swabbing was conducted by two experienced team members only. The patients underwent mucosa swabbing of the oropharynx and nasopharynx the day after admittance to the hospital. After four weeks, all patients testing positive for a respiratory virus were retested with oropharyngeal and nasopharyngeal swabs. The swabbing was performed in the following order: right oropharyngeal area, left oropharyngeal area, left nasopharynx and right nasopharynx. The order of the two types of swabs used was decided by block randomisation. Only one swab was used per transport tube, regardless of the type of swab or the location swabbed. The tubes were stored at room temperature and transported to the microbiological laboratory within 48 h.
Sample processing and diagnostic procedures
Specimens were kept at room temperature until nucleic acid extraction, which usually took place within 24 h and always within 48 h. PCR analyses were performed within the next 24 h or else the nucleic acid eluates were stored at −70°C until testing. Using monoplex (RSV and human metapneumovirus) or multiplex (influenza A/B, adenovirus/internal control and parainfluenza virus 1–4) PCR methods, the specimens were examined for, in total, nine different respiratory viruses (Table 2).
Table 2.
Characteristics and sequences of primers and probes for the real-time polymerase chain reaction (PCR) assays
| Agents detected | Target gene | Oligonucleotide sequence (5’-3’) | Reference |
|---|---|---|---|
| Mycoplasma pneumoniae | P1 adhesin | F GAACCCTCGACCAAGCCAAC | [21] |
| R GTCCTGCGTGGTTAAACTATCC | |||
| P CCACACCAAGTTCACGAGCGCTACG | |||
| Chlamydophila pneumoniae | Pmp 4 | F GCTAGGCCATTGAGAGTGACC | [22] |
| R GCTGCAGGTGCCTTAGTATTG | |||
| P CGCTTGCCTCAAAAGTAGTCCCTCC | |||
| Bordetella pertussis | IS 481 | F CGGATGAACACCCATAAGCAT | [23] |
| R CGATCAATTGCTGGACCATTT | |||
| P CCCGATTGACCTTCCTACGTCGACTC | |||
| Influenza virus A | M | F CATGGAATGGCTAAAGACAAGACC | [24] |
| R CCATTTAGGGCATTTTGGACA | |||
| P TTTGTGTTYACGCTCACCGTGCCCA | |||
| Influenza virus B | HA | F AAATACGGTGGATTAAATAAAAGCAA | [25] |
| R CCAGCAATAGCTCCGAAGAAA | |||
| P CACCCATATTGGGCAATTTCCTATGGC | |||
| PIV type 1 | HN | F TGATTTAAACCCGGTAATTTCTCAT | [26] |
| R CCTTGTTCCTGCAGCTATTACAGA | |||
| P ACGACAACAGGAAATC-MGB | |||
| PIV type 2 | Pol | F TGCATGTTTTATAACTACTGATCTTGCTAA | [27] |
| R GTTCGAGCAAAATGGATTATGGT | |||
| P ACTGTCTTCAATGGAGATAT- LNA | |||
| PIV type 3 | M | F TGCTGTTCGATGCCAACAA | [27] |
| R ATTTTATGCTCCTATCTAGTGGAAGACA | |||
| P TTGCTCTTGCTCCTCA- MGB | |||
| PIV type 4 | P | F CCTGGAGTCCCATCAAAAGT | [28] and * |
| R GCATCTATACGAACACCTGCT | |||
| P ATCAAGACAATACAATTACACTTGA-LNA | |||
| hMPV | N | F CATATAAGCATGCTATATTAAAAGAGTCTC | [29] |
| R CCTATTTCTGCAGCATATTTGTAATCAG | |||
| P TGYAATGATGAGGGTGTCACTGCGGTTG | |||
| RSV (A/B) | N | F GATGGCTCTTAGCAAAGTCAAGTT | * |
| R TCTTTATAGTGTCTTCTCTTCCTAA | |||
| P CTGTCATCCAGCAAATACACCATCCAACG | |||
| AdV | LightMix® Kit Adenovirus TIB MOLBIOL | ||
| cat. no. 40-0303-16 | |||
| λ phage | F ATGCCACGTAAGCGAAACA | * | |
| R GCATAAACGAAGCAGTCGAGT | |||
| P ACCTTACCGAAATCGGTACGGATACCG |
*Designed by TIB MOLBIOL
Y = C/T; MGB minor groove binder; LNA locked nucleic acids
HA, hemagglutinin-neuraminidase; M, matrix; N, nucleocapsid; Pol, polymerase; P, phosphoprotein; hMPV, human metapneumovirus; RSV, respiratory syncytial virus; AdV, adenovirus; PIV, parainfluenza virus
To achieve a more complete diagnostic coverage of respiratory pathogens, PCR assays were added for the detection of Mycoplasma pneumonia, Chlamydophila pneumonia and Bordetella pertussis.
Extraction of total nucleic acids
The rayon swabs were incubated for 15 min in a TRIS-EDTA (TE) buffer. After thorough stirring, 200 μl of each sample was subjected to nucleic acid extraction in a MagNAPure LC instrument (Roche Diagnostics, Mannheim, Germany) using the MagNA Total Nucleic Acid Isolation kit (Roche ca. no. 03038505001). The samples were eluted in 100 μl buffer. To be able to detect inhibition of the PCR assay by specimen-specific inhibitors, a final concentration of 20,000 λ phage DNA copies/ml (TIB MOLBIOL cat. no. 80-5000-02) were added to the TE buffer (not done with the flocked swabs). A control sample with only λ phage DNA was included in all extraction runs, in order to ensure that negative results were not due to poor processing.
cDNA synthesis
cDNA synthesis was performed with the qScript cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, MD, USA; cat. no. 95047) following the manufacturer’s protocol. The reaction was incubated for 5 min at 25°C and for 30 min at 42°C, and then inactivated by 5 min at 95°C. The cDNA samples were subjected to amplification immediately.
Real-time PCR assay
Adenovirus real-time PCR assays were performed using LightMix® Kit TIB MOLBIOL (cat. no. 40-0303-16) according to the manufacturer’s protocol. The other real-time PCR was performed using 5 μl of (c)DNA in a 20-μl reaction mixture consisting of 5 mM MgCl2, 750 units uracil-DNA glycosylase (Eurogentec S.A., Seraing, Belgium), LightCycler FastStart DNA Master Mix (Roche cat. no. 12239272001), 0.5–1.0 μM of the primers and 0.2–0.4 μM of each of the probes. All primers and probes are listed in Table 2. The PCR cycling conditions were initiated with 2 min at 42°C, followed by 10 min at 95°C and 47 cycles of 15 s at 95°C, 30 s at 60°C and 20 s at 72°C.
During PCR amplification, the amount of DNA is doubled in each cycle until the components in the reaction reaches a critical level. There is an exponential relationship between the initial amount of template DNA and the cycle threshold (CT) values. A high CT value represents a low microbial load in the specimen. Samples that differ by a factor of two in the original DNA concentration would be expected to be one cycle apart in the run, whereas samples that differ by a factor of ten would be approximately 3.3 cycles apart. The differences in CT values obtained from the same experimental run demonstrate a relative difference in the concentration between samples, and is used for calculating differences in viral load. The amplification efficiency for all of the PCR assays used in this study was measured to be above 94%.
Statistics
Descriptive statistics were used when describing the population. A linear mixed model was applied to the data when comparing the two types of swabs or the origin of the samples with a random effect of patient. The statistical analysis was based on CT values. A CT value of 38 was designated as the cut-off value for positive results. Thus, a lower CT value in a sample corresponds to a higher microbial load in the sample. The CT values were analysed as continuous data. All statistical analyses were performed using SPSS 16.0 (SPSS Inc., Chicago, IL, USA).
Results
A total of 922 patients were interviewed for current symptoms of a respiratory tract infection (Table 1). Of these, 191 (21%) patients reported at least one such symptom and 168 agreed to be tested according to the study protocol. Twenty-six patients refused to participate (14 male, 12 female, mean age 74.8 years, standard deviation [SD] 7.9 years). During the first and last four weeks of the study, 55 patients without symptoms agreed to be swabbed as well. Altogether, 223 patients (134 males, 89 females, mean age 74.9 years, SD 9.0 years) were swabbed and 845 swabs were used. A total of 183 patients were swabbed with all four swabs, 34 patients with three, five patients with two, and one patient with one swab only.
A virus was recovered from at least one swab in 19 (11%) of the symptomatic patients (Table 3), all of whom had reported symptoms of respiratory tract infection. No patients tested positive for more than one virus and no virus was found when reswabbing previously positive patients after four weeks. One patient tested positive for Bordetella pertussis and none for Chlamydophila pneumonia and Mycoplasma pneumonia. As bacterial infections were not the focus of the current study, these results were excluded from the following analysis.
Table 3.
Types of viruses found and number of positive swabs
| Virus | Number of positive patients | Oropharynx | Nasopharynx | ||
|---|---|---|---|---|---|
| Rayon swab | Flocked swab | Rayon swab | Flocked swab | ||
| Positive swabs/total swabs used | Positive swabs/total swabs used | Positive swabs/total swabs used | Positive swabs/total swabs used | ||
| Influenza A | 7 | 3/7 | 4/7 | 6/7 | 7/7 |
| RSV | 3 | 3/3 | 2/2 | 1/3 | 2/2 |
| hMPV | 3 | 1/3 | 2/3 | 1/3 | 3/3 |
| AdV | 2 | 1/2 | 1/2 | 1/2 | 1/2 |
| PIV type 3 | 2 | 2/2 | 2/2 | 1/1 | 1/2 |
| Influenza B | 1 | 1/1 | 1/1 | 1/1 | 1/1 |
| PIV type 4 | 1 | 1/1 | 0/1 | 0/1 | 0/1 |
| Total | 19 | 12/19 | 12/18 | 11/18 | 15/18 |
RSV, respiratory syncytial virus; hMPV, human metapneumovirus; AdV, adenovirus; PIV, parainfluenza virus
Nylon flocked swabs, regardless of sample origin, were positive at a lower CT value compared to rayon swabs (mean difference CT 2.25, 95% confidence interval [CI] 0.42–4.09, p = 0.017), representing a calculated 4.8 times higher (95% CI 1.3–17) viral load found on the flocked swabs compared to the rayon swabs.
In general, nasopharyngeal swabs were positive at a lower CT value compared to oropharyngeal swabs (mean difference CT 4.25, 95% CI 2.43–6.07, p < 0.001), regardless of the sample type (Fig. 1). The calculated viral load was 19 times higher (95% CI 5.4–67.2) than in the oropharynx. Influenza A virus-positive samples showed a lower CT value in the nasopharynx than in the oropharynx, with a mean difference in CT value of 8.16 (95% CI 6.98–10.35, p < 0.001), which represents a calculated 286 times higher (95% CI 126–1,305) viral load in the nasopharynx.
Fig. 1.
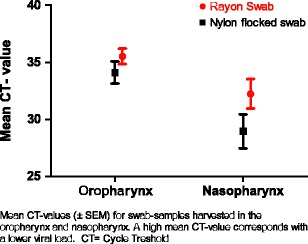
Comparison of mean cycle threshold (CT) values for samples harvested in the oropharynx and the nasopharynx. The data show the mean CT values (± standard error of the mean [SEM]) for swab samples harvested in the oropharynx and the nasopharynx. A high mean CT value corresponds to a lower viral load
Discussion
To our knowledge, no comparison between nylon flocked swabs and rayon swabs has been conducted in the elderly population. The present study favours nasopharyngeal sampling with nylon flocked swabs: a calculated 4.8 times higher viral DNA concentration was found on the nylon flocked swabs, regardless of the sample origin. Nylon flocked swabs seem to adhere more epithelial cells than rayon swabs [13] and this might, at least in part, explain the lower CT values obtained by using flocked swabs. Nasopharyngeal aspirate, which has been considered as a gold standard material for the diagnosis of respiratory viruses in children, has a sensitivity at the level of, or slightly above, nasopharyngeal nylon flocked swabs [14, 15]. A comparison between nylon flocked swabs and nasopharyngeal aspirate in the elderly population has not been published.
In terms of CT, we found nasopharyngeal samples to be superior to oropharyngeal samples, yielding a calculated 19 times higher concentration of viral nucleic acids. This corresponds to the results found in a study by Lieberman et al. [16], with a much broader variation in patient age. Our subgroup analysis of influenza A virus reveals a calculated 286 times higher viral load in samples from the nasopharynx. As shown in Table 3, several samples were negative for influenza A virus in the oropharynx and positive in the nasopharynx.
Nasopharyngeal swabs are easy to use after proper instruction, but swabbing for microbiological agents is often left with the least trained medical staff. In such cases, suboptimal swabbing may be relatively common. The sample collection in our study was performed by two experienced team members, thus, minimising the risk of suboptimal sampling. Whereas usually, considerable efforts are made to optimise the diagnostic procedures in the laboratory, less emphasis tends to be placed on the preceding procedures of sample collection. We believe that there is a lot to be gained from an increased awareness of proper sampling techniques and sampling tools.
For semi-quantitative assessments of respiratory pathogens in children, immunofluorescent assays or viral culture have been widely applied [14, 17–19], whereas in the current study, CT values were obtained for this purpose. Most studies applying real-time PCR for the detection of respiratory agents simply determine whether there is an infection or not. However, within each PCR experiment, there is a close relationship between the achieved CT value and the initial amount of specific nucleic acids in the sample. In this study, only samples taken at the same time from the same patient are compared and the CT values used for the calculations are collected from the same experiment. The calculations of differences in viral load relied upon the use of PCR assays with high amplification efficiencies. Factors other than the ones we have been examining are then minimised. In this manner, the method provides valid relative quantitative data when comparing distinct sample materials.
We found respiratory viruses in 11% of our study population. Corresponding numbers reported by other authors range from 10 to 43% [12, 20], depending on which viruses were included in the PCR analysis. We chose to include only microbial agents responsible for significant illness, thus, excluding frequently occurring causes of upper respiratory tract infections, such as rhinovirus and coronavirus. The number of patients diagnosed with influenza A and RSV infections were lower in our study than those reported by others, probably due to annual variations. We did not find any respiratory viruses in non-symptomatic patients. During the two periods when all patients regardless of symptoms were swabbed, respiratory viruses were found in 8.6% (6 patients) of the total study population.
Some authors have concluded that the combination of oropharyngeal sampling, nasopharyngeal sampling and NPA represents the ideal sampling method for the diagnosis of respiratory tract viruses [16]. However, in a busy clinical setting, it may be hard enough to ensure one properly collected sample from each of the concerned patients. Our conclusion is that, when a respiratory tract infection requires aetiological clarification, a rational approach to the virological part of the investigation is to obtain a nasopharyngeal sample, preferably collected by trained personnel and, at least where costs are comparable, using nylon flocked swabs rather than rayon swabs.
Acknowledgements
This project was financed with the aid of EXTRA funds from the Norwegian Foundation for Health and Rehabilitation in association with the Norwegian Heart and Lung Patient Organisation, from the South-Eastern Norway Regional Health Authority and from Sorlandet Hospital HF.
Conflict of interest
The nylon flocked swabs and UTM transport tubes were donated by Copan Italia, Brescia, Italy, whereas the rayon swabs and transport medium were donated by Chemi-Teknik A/S, Oslo, Norway. Both suppliers delivered the swabs free of charge. The suppliers had no role in the planning, running, evaluation or reporting of this trial. The authors declare that they have no conflict of interest related to this report.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Boivin G, de Serres G, Hamelin ME, Côté S, Argouin M, Tremblay G, et al. An outbreak of severe respiratory tract infection due to human metapneumovirus in a long-term care facility. Clin Infect Dis. 2007;44:1152–1158. doi: 10.1086/513204. [DOI] [PubMed] [Google Scholar]
- 2.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. 2005;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 3.Han LL, Alexander JP, Anderson LJ. Respiratory syncytial virus pneumonia among the elderly: an assessment of disease burden. J Infect Dis. 1999;179:25–30. doi: 10.1086/314567. [DOI] [PubMed] [Google Scholar]
- 4.Louie JK, Schnurr DP, Pan CY, Kiang D, Carter C, Tougaw S, et al. A summer outbreak of human metapneumovirus infection in a long-term-care facility. J Infect Dis. 2007;196:705–708. doi: 10.1086/519846. [DOI] [PubMed] [Google Scholar]
- 5.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 6.Walsh EE, Peterson DR, Falsey AR. Is clinical recognition of respiratory syncytial virus infection in hospitalized elderly and high-risk adults possible? J Infect Dis. 2007;195:1046–1051. doi: 10.1086/511986. [DOI] [PubMed] [Google Scholar]
- 7.Falsey AR, McCann RM, Hall WJ, Criddle MM. Evaluation of four methods for the diagnosis of respiratory syncytial virus infection in older adults. J Am Geriatr Soc. 1996;44:71–73. doi: 10.1111/j.1532-5415.1996.tb05641.x. [DOI] [PubMed] [Google Scholar]
- 8.Casiano-Colón AE, Hulbert BB, Mayer TK, Walsh EE, Falsey AR. Lack of sensitivity of rapid antigen tests for the diagnosis of respiratory syncytial virus infection in adults. J Clin Virol. 2003;28:169–174. doi: 10.1016/S1386-6532(03)00002-7. [DOI] [PubMed] [Google Scholar]
- 9.Gooskens J, Swaan CM, Claas EC, Kroes AC. Rapid molecular detection of influenza outbreaks in nursing homes. J Clin Virol. 2008;41:7–12. doi: 10.1016/j.jcv.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 10.Falsey AR, Walsh EE. Viral pneumonia in older adults. Clin Infect Dis. 2006;42:518–524. doi: 10.1086/499955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh EE, Falsey AR. A simple and reproducible method for collecting nasal secretions in frail elderly adults, for measurement of virus-specific IgA. J Infect Dis. 1999;179:1268–1273. doi: 10.1086/314726. [DOI] [PubMed] [Google Scholar]
- 12.Rezza G, Valdarchi C, Puzelli S, Ciotti M, Farchi F, Fabiani C, et al. Respiratory viruses and influenza-like illness: a survey in the area of Rome, winter 2004–2005. Euro Surveill. 2006;11:251–253. [PubMed] [Google Scholar]
- 13.Daley P, Castriciano S, Chernesky M, Smieja M. Comparison of flocked and rayon swabs for collection of respiratory epithelial cells from uninfected volunteers and symptomatic patients. J Clin Microbiol. 2006;44:2265–2267. doi: 10.1128/JCM.02055-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan KH, Peiris JS, Lim W, Nicholls JM, Chiu SS. Comparison of nasopharyngeal flocked swabs and aspirates for rapid diagnosis of respiratory viruses in children. J Clin Virol. 2008;42:65–69. doi: 10.1016/j.jcv.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Walsh P, Overmyer CL, Pham K, Michaelson S, Gofman L, DeSalvia L, et al. Comparison of respiratory virus detection rates for infants and toddlers by use of flocked swabs, saline aspirates, and saline aspirates mixed in universal transport medium for room temperature storage and shipping. J Clin Microbiol. 2008;46:2374–2376. doi: 10.1128/JCM.00714-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieberman D, Lieberman D, Shimoni A, Keren-Naus A, Steinberg R, Shemer-Avni Y. Identification of respiratory viruses in adults: nasopharyngeal versus oropharyngeal sampling. J Clin Microbiol. 2009;47:3439–3443. doi: 10.1128/JCM.00886-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahluwalia G, Embree J, McNicol P, Law B, Hammond GW. Comparison of nasopharyngeal aspirate and nasopharyngeal swab specimens for respiratory syncytial virus diagnosis by cell culture, indirect immunofluorescence assay, and enzyme-linked immunosorbent assay. J Clin Microbiol. 1987;25:763–767. doi: 10.1128/jcm.25.5.763-767.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abu-Diab A, Azzeh M, Ghneim R, Ghneim R, Zoughbi M, Turkuman S, et al. Comparison between pernasal flocked swabs and nasopharyngeal aspirates for detection of common respiratory viruses in samples from children. J Clin Microbiol. 2008;46:2414–2417. doi: 10.1128/JCM.00369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heikkinen T, Marttila J, Salmi AA, Ruuskanen O. Nasal swab versus nasopharyngeal aspirate for isolation of respiratory viruses. J Clin Microbiol. 2002;40:4337–4339. doi: 10.1128/JCM.40.11.4337-4339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaye M, Skidmore S, Osman H, Weinbren M, Warren R. Surveillance of respiratory virus infections in adult hospital admissions using rapid methods. Epidemiol Infect. 2006;134:792–798. doi: 10.1017/S0950268805005364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waring AL, Halse TA, Csiza CK, Carlyn CJ, Arruda MK, Limberger RJ. Development of a genomics-based PCR assay for detection of Mycoplasma pneumoniae in a large outbreak in New York State. J Clin Microbiol. 2001;39:1385–1390. doi: 10.1128/JCM.39.4.1385-1390.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mygind T, Birkelund S, Falk E, Christiansen G. Evaluation of real-time quantitative PCR for identification and quantification of Chlamydia pneumoniae by comparison with immunohistochemistry. J Microbiol Methods. 2001;46:241–251. doi: 10.1016/S0167-7012(01)00282-2. [DOI] [PubMed] [Google Scholar]
- 23.Chan EL, Antonishyn N, McDonald R, et al. The use of TaqMan PCR assay for detection of Bordetella pertussis infection from clinical specimens. Arch Pathol Lab Med. 2002;126:173–176. doi: 10.5858/2002-126-0173-TUOTPA. [DOI] [PubMed] [Google Scholar]
- 24.Watzinger F, Suda M, Preuner S, et al. Real-time quantitative PCR assays for detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. J Clin Microbiol. 2004;42:5189–5198. doi: 10.1128/JCM.42.11.5189-5198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Elden LJ, van Essen GA, Boucher CA, et al. Clinical diagnosis of influenza virus infection: evaluation of diagnostic tools in general practice. Br J Gen Pract. 2001;51:630–634. [PMC free article] [PubMed] [Google Scholar]
- 26.van de Pol AC, van Loon AM, Wolfs TF, et al. Increased detection of respiratory syncytial virus, influenza viruses, parainfluenza viruses, and adenoviruses with real-time PCR in samples from patients with respiratory symptoms. J Clin Microbiol. 2007;45:2260–2262. doi: 10.1128/JCM.00848-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuypers J, Wright N, Ferrenberg J, et al. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J Clin Microbiol. 2006;44:2382–2388. doi: 10.1128/JCM.00216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Templeton KE, Scheltinga SA, Beersma MF, Kroes AC, Claas EC. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J Clin Microbiol. 2004;42:1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maertzdorf J, Wang CK, Brown JB, et al. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J Clin Microbiol. 2004;42:981–986. doi: 10.1128/JCM.42.3.981-986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


