Abstract
Metabolic flux analysis is a vital tool used to determine the ultimate output of cellular metabolism and thus detect biotechnologically relevant bottlenecks in productivity. 13C-based metabolic flux analysis (13C-MFA) and flux balance analysis (FBA) have many potential applications in biotechnology. However, noteworthy hurdles in fluxomics study are still present. First, several technical difficulties in both 13C-MFA and FBA severely limit the scope of fluxomics findings and the applicability of obtained metabolic information. Second, the complexity of metabolic regulation poses a great challenge for precise prediction and analysis of metabolic networks, as there are gaps between fluxomics results and other omics studies. Third, despite identified metabolic bottlenecks or sources of host stress from product synthesis, it remains difficult to overcome inherent metabolic robustness or to efficiently import and express nonnative pathways. Fourth, product yields often decrease as the number of enzymatic steps increases. Such decrease in yield may not be caused by rate-limiting enzymes, but rather is accumulated through each enzymatic reaction. Fifth, a high-throughput fluxomics tool hasnot been developed for characterizing nonmodel microorganisms and maximizing their application in industrial biotechnology. Refining fluxomics tools and understanding these obstacles will improve our ability to engineer highlyefficient metabolic pathways in microbial hosts.
1. Introduction
Numerous chemical compounds, ranging from the antimalaria drug artemisinin [1] to the “biofuel” butanol [2, 3], have been produced with the aid of synthetic biology tools. The ability to efficiently synthesize natural or unnatural products requires a systems-level understanding of metabolism. Functional genomics tools such as genome sequencing, profiling of mRNA transcripts, and proteomics, are widely used to attain a comprehensive knowledge of how metabolic components (genes, proteins and metabolites) are regulated. In contrast to traditional omics tools, flux analysis (measurement of metabolite turnover rates) has become instrumental for physiological prediction and enzymatic rate quantification in metabolic networks [4]. This technology also allows for the identification of metabolic interactions and the knowledge-based design of cellular functions. As such, one can utilize this tool to rationally modify biological hosts and analyze global physiological changes resulting from genetic modifications.
Fluxomics, the cell-wide quantification of intracellular metabolite turnover rates, was first performed via flux balance analysis (FBA). This method uses the stoichiometry of the metabolic reactions in addition to a series of physical, chemical and biological characteristics (thermodynamics, energy balance, gene regulation, etc.) to constrain the feasible fluxes under a given objective function (e.g., maximal biomass production). FBA is an underdetermined model (the number of constraints is smaller than the number of reactions in the metabolic network), which may give unrealistic metabolic readout. In spite of this limitation, FBA provides a useful framework for predicting a wide variety of cellular metabolisms. A complementary approach, 13C-based metabolic flux analysis (13C-MFA) allows for precise determinations of metabolic status under a particular growth condition. The key to 13C-MFA is isotopic labeling, whereby microbes are cultured using a carbon source with a known distribution of 13C. By tracing the transition path of the labeled atoms between metabolites in the biochemical network, one can quantitatively determine intracellular fluxes.
Flux analysis can not only provide genetic engineers with strategies for “rationally optimizing” a biological system, but also reveal novel enzymes useful for biotechnology applications [4]. However, flux analysis platforms are still not routinely established in biotechnology companies. This review paper addresses current developments and challenges in the field of fluxomics, which may guide future study to bridge the gap between systems analysis of cellular metabolism and application in biotechnology.
2. Advances and Limitations in Metabolic Flux Analysis
2.1. Steady-State Flux Model
FBA and 13C-MFA concentrate on the stoichiometric (rather than kinetic) properties of metabolic networks. FBA has been widely applied to predict cell growth rate, product yield using different feedstocks, lethality of gene knockouts, and advantageous pathway modifications [5]. Such a model provides general guidelines for metabolic engineering and thus is a viable first step towards improving biosynthetic yield [6]. The hallmark of large scale FBA is the constraint-based reconstruction and analysis toolbox (COBRA) [7], which provides a general platform for fluxomics studies.
A number of optimization algorithms and computational strategies for resolving in silico and in vivo inconsistencies have been proposed to improve the applicability of FBA [6, 8]. For example, incorporation of thermodynamic principles into FBA can constrain solution space (i.e., energy balance analysis) and obtain both stoichiometrically and thermodynamically feasible fluxes [9]. To describe the “nonoptimal” metabolic behaviors, FBA can use a bilevel optimization approach to estimate the potential trade-off between biomass accumulation and the yield of a desired product [10]. FBA can also relax the objective function for maximization of the biomass and apply a Minimization of Metabolic Adjustment Algorithm to solve fluxes in mutant strains [9]. Such an algorithm calculates fluxes by minimizing the difference between the wild-type flux distributions and the knockout-strain fluxes. Furthermore, FBA can be integrated with metabolic pathway analysis (MPA). MPA is a qualitative technique that examines functional routes existing in a metabolic network without requiring knowledge of reaction rates [11]. Combining MPA with FBA can quantitatively trace the plausible paths for optimal product synthesis, calculate cellular metabolism, and predict phenotypes under genetic manipulations or culture conditions [12]. One main advantage of FBA is its capability for genome-scale modeling (including thousands of reactions), which bridge genomic annotation and functional metabolic output. Accordingly, the number of FBA models has increased exponentially since 1999 [13].
13C-MFA aims to rigorously quantify pathway activities in intracellular metabolism by using both the isotopic labeling approach and in silico computation. 13C-MFA is accomplished by feeding microbes a 13C-labeled carbon source, measuring the enrichment pattern of the isotopomer in metabolites (e.g., amino acids), and deciphering the fluxes via computational routines [14]. Since carbon fluxes through a metabolic network generate unique labeling patterns in metabolites, the overall flux distributions can be determined using isotopomer information. Advances in 13C-MFA, including mass spectrometry-based metabolomics and isotopomer modeling approaches (such as novel algorithm using elementary metabolite units), have been discussed in recent papers [4, 15, 16].
Furthermore, open-source software has recently been published that facilitates in silico modelling. For example, WEbcoli is web-based software for flux balance analysis of E.coli [17]. In addition, OpenFLUX is computationally efficient software for 13C-MFA [15], which incorporates the elementary metabolite unit (EMU) framework for calculation of isotopomer balances [18]. User-friendly software such as this allows biologists to perform fluxomics studies with little programming knowledge.
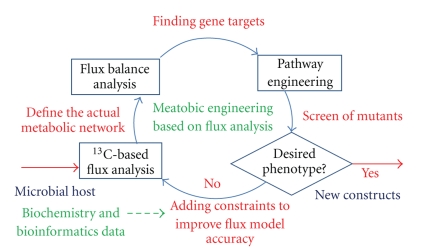
Methodologies for FBA and 13C-MFA share two key characteristics: the use of a metabolic network and the assumption of a steady metabolic state (for internal metabolites). However, the two techniques have different purposes. FBA profiles the “optimal” metabolism for the desired performance; 13C-MFA measures in vivo operation of a metabolic network. The two approaches to flux analysis are complementary when developing a rational metabolic engineering strategy. By comparing existent metabolic fluxes which were empirically determined via 13C-MFA to the optimal metabolisms predicted by both FBA and other “omics” tools (such as transcription analysis), one can deduce gene targets for solving biotechnologically relevant productivity bottlenecks [19]. Figure 1 shows that iterative flux analysis and genetic engineering of microbial hosts can remove competitive pathways or toxic byproducts, amplify genes encoding key metabolites, and balance energy metabolism [6].
Figure 1.
An iterative approach of fluxomic analysis and rational metabolic engineering.
2.2. Metabolic Control and Dynamic Flux Analysis
FBA and 13C-MFA disregard dynamic intracellular behavior. This avoids the difficulties in developing kinetic models and performing intracellular experimental measurements. However, many biological systems may not maintain a meaningful metabolic (or isotopic) steady state during the fermentation process [20–22]. The description of metabolic perturbation and regulatory mechanisms requires kinetic modeling and control theories. For example, metabolic control analysis (MCA) couples local enzyme kinetics with systematic behavior to predict the control exerted on the targeted pathways by different components (e.g., transcription, enzymes) [23]. Although MCA is not a quantitative measurement of flux, MCA can pinpoint bottle-neck enzymes (enzymes having the largest effect on the desired flux) in a pathway and allow the analysis of steady-state metabolism in response to changes in the cellular environment [24]. In addition to MCA, the cybernetic approach (a model based on process dynamics and control) has been introduced for study of multienzyme systems and metabolic regulation [25]. By incorporating both the enzyme kinetics in pathways and the enzyme synthesis kinetics, the cybernetic approach emphasizes microbial process dynamics and control during complicated fermentations [26].
Both MCA and the cybernetics approach focus on a simplified pathway network. To perform cell-wide quantitative analysis of a dynamic system, it is necessary to integrate the kinetic modeling with FBA and 13C-MFA. Dynamic FBA (dFBA) has been developed to illuminate changing global enzyme activities [27, 28]. To avoid ordinary differential equations and dynamic optimization for describing intracellular metabolism, dFBA can use the Static Optimization Approach (SOA) [29] which divides the time-course into numerous small intervals. At each time interval, a steady-state flux is calculated under the assumption of fast intracellular dynamics. By combining stoichiometric FBA for intracellular metabolism with dynamic mass balances on extracellular substrates and products, it is possible to reconstruct dFBA model for genome-scale analysis of microbial metabolisms in industrial fermentations, where product synthesis is often under dynamic control [30, 31].
Recently, 13C-dMFA (dynamic metabolic flux analysis) has been developed for isotopically nonstationary cultures. To profile the flux distributions for fed-batch cultures (slow dynamic metabolism), isotopic pseudosteady state was assumed and two dilution parameters were introduced to account for isotopic transients [92]. Another approach (Kinetic Flux Profiling) for solving intracellular fluxes is to create a sudden increase of the portion of 13C in the substrate feed, then measure time-course samples as 13C moves from the substrate into the metabolites [33]. The fluxes can be calculated based on the rates of isotopic enrichment multiplied by the intracellular metabolite concentrations. A similar principle has been proposed for the flux analysis of photoautotrophic microorganisms [34] and E. coli in an isotopic transient phase [35]. If the culture is under both metabolic and isotopic nonstationary state, exploratory and sophisticated 13C-dMFA (dynamic 13C-MFA) models have to be used to calculate both metabolic and isotopic kinetics [20, 36, 37]. To solve the 13C-dMFA problem efficiently, a set of computational algorithms have been developed for tracing nonstationary isotopomer labeling in response to in vivo flux distributions [20, 36, 37]. The EMU (elementary metabolite unit) framework has also been applied in 13C-dMFA [18, 38], because such algorithm can significantly improve computational times for tracing the labeling information [39]. To avoid extensive simulation of dynamic isotopomer patterns, the SOA has to be applied by dividing the growth period into small time intervals (30~60 min), then the “mini” quasi-steady state 13C-MFA can be applied at each time interval based on constraints from simultaneous isotopomer analysis of the fast turnover metabolites [40]. By examining flux profiles over all time intervals, one can resolve the metabolic transients during the entire cultivation period.
2.3. Technical Limitations of Fluxomics
Cell-wide fluxomics tools (i.e., FBA and 13C-MFA) have technical limitations. In genome-scale FBA models, the number of constraints (i.e., the availability of quantitative metabolite data) is much smaller than the number of reactions in the metabolic network. The calculation of such underdetermined systems depends on objective functions where one assumes that the metabolism optimizes its native ‘‘goals” (such as biomass or cofactor production) [41]. This optimization principle has been questioned for several reasons. First, biological systems (e.g., Bacillus subtilis) seem to display suboptimal growth performance [42]. Second, a previous study examined 11 objective functions in E. coli and found no single objective function that can perfectly describe flux states under various growth conditions [43]. For example, unlimited aerobic growth on glucose is best described by a nonlinear maximization of the ATP yield per flux unit, but nutrient-limited continuous cultures favor biomass yield as the objective function. Third, some native cellular processes cannot be simply described by FBA. For example, cyanobacterial species (i.e., Cyanothece 51142) maintain their circadian rhythms (e.g., nitrogen fixation and light dependent reaction activities) under nutrient-sufficient and continuous light conditions [44, 45].
The application of 13C-MFA in industrial biotechnology also has several bottlenecks. The most prevalent constriction occurs because current techniques are insufficient for measuring large-scale metabolic networks. Obtaining labeling information of free metabolites rather than amino acids and solving large-scale nonlinear flux models pose two key challenges. As a result, most obtained flux information is limited to central metabolism. To date, only two large-scale 13C-MFA (>300 reactions) have been reported, but many fluxes in their reports cannot be precisely determined due to insufficient constraints [46, 47]. The genome-scale 13C-MFA is still in its infancy and requires further development of the relevant experimental techniques and computational tools [48]. A second issue is that 13C-dMFA is still poorly developed for determining dynamic metabolic behavior. It is difficult for rapid sampling and precise measurements of metabolites at short time intervals throughout the entire culture period. For example, to measure absolute intracellular metabolite concentrations, one has to grow cell in fully 13C-labeled medium, then the labeled cells are extracted with quenching solvent containing known concentrations of unlabeled internal standards (the concentrations of metabolites are calculated using the isotope ratio-based approach) [49]. Such measurement requires extremely high cost of analytical efforts including quick sampling, rapid metabolite extraction, and a high-resolution LC-MS instrument. Furthermore, the time-dependent model includes ordinary differential equations and significantly increases the computational complexity [20, 35]. Third, flux determination assumes that enzymatic reactions are homogenous inside the cell and that there are no transport limitations between metabolite pools. However, eukaryotes have organelles (compartments) that may have diffusion limitations or metabolite channeling [14, 50]. Compartmentalization of amino acid biosynthesis further clouds the obtained amino acid-based labeling information [51]. Therefore, confident 13C-MFA for eukaryotes not only requires the combination of different analytical tools (GC-MS, LC-MS and NMR) to obtain extensive labeling information [52], but also adequate sample processing and extraction methods (e.g., separation of compartments by ultracentrifugation). A fourth problem is that some industrial hosts and the great majority of environmental microbes resist cultivation in minimal media, and introducing other nutrient sources often significantly complicates metabolite labeling measurements and flux analyses [53]. Finally, a microbial community demonstrates complex metabolic interactions between species. To date, only a few FBA models have been developed for community studies [54, 55]. The exchange of metabolites among species is nearly impossible to unravel by 13C-MFA because complete separation and measurement of metabolites from a single species in a microbial community is impossible [4]. These technical limitations in both FBA and MFA models are responsible for the gap between fluxomics and its applicability in biotechnology.
3. Integration of Fluxomics with Other “Omics”
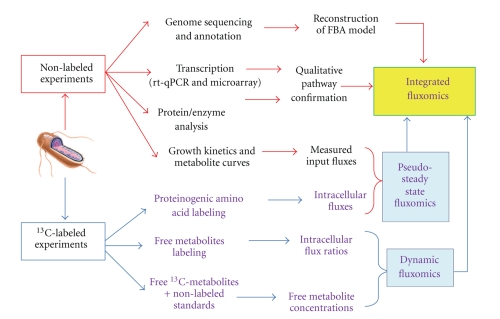
It is desirable to integrate the concepts of systems biology (which combines the readouts from transcription as well as protein/metabolite profiling) with fluxomics (Figure 2) [48]. For example, 13C-MFA, enzyme activity assays, and RT-PCR analysis can be used together to study E.coli mutants' metabolism [56]. Additionally, the responses of E. coli to genetic modification have been systematically examined by utilizing multiple high-throughput “omics” methods [57]. The results illuminate relatively small changes in mRNA and proteins in response to genetic disruptions, which allow the cell to maintain a stable metabolic state under changing growth conditions. A similar approach to the study of Synechocystis 6803 has shown that the regulation of some enzymes is sensitive to light conditions [58]. Many other regulatory mechanisms, however, still remain unknown. Furthermore, global regulators in industrial microorganisms have been successfully identified by correlating transcript/transduction levels and metabolic fluxes [59–62]. The discovery of functioning regulators provides insight to the entire regulation in metabolic network.
Figure 2.
13C-assisted cellular metabolism analysis.
On the other hand, challenges in integrated “omics” studies are also present. The lack of understanding of metabolic regulation at different metabolic levels complicates the rational design of biological systems, which is a major barrier in industrial biotechnology. For example, posttranscriptional regulation poses a significant challenge in integrating fluxomics with other “omics” studies. It is well known that transcript and protein data correlate relatively well for specific pathways, yet this correlation can be poor in cell-wide analyses [76]. Furthermore, most mRNA expression studies insufficiently predict enzyme activities or flux changes in many E. coli pathways [77]. In studies on the adaptation of E. coli to environmental perturbations, the tricarboxylic acid cycle is found to correlate well with molecular changes at the transcriptional level, but flux alterations in other central metabolic pathways seem to be uncorrelated to changes in the transcriptional network [78]. Because of the complexity of regulatory mechanisms spanning multiple cellular processes, fluxomics and other “omics” studies may have inconsistent observations which complicate systems-level analyses.
4. Fluxomics of Microbes for Industrial Biotechnology
FBA allows in silico simulations of metabolism in “industrial workhorses,” from which desired strains or targeted mutations can be identified. 13C-MFA can assess in vivo metabolism of engineered strains under specific growth conditions and validate FBA results. Here, we summarize recent applications of FBA and 13C-MFA for commonly-used industrial chassis (i.e., E. coli, B. subtilis and S. cerevisiae) and for nonmodel microorganisms (i.e., less-characterized or newly-discovered microorganisms).
4.1. Escherichia coli Model
E. coli is the most commonly utilized species in fermentation industry. E. coli flux models were reported as early as the 1990s [79, 80]. For biotechnology applications, the Liao group first applied metabolic pathway analysis (MPA) to guide the genetic manipulation of E.coli strains and channel the metabolic fluxes from carbohydrate to the aromatic amino acid pathway [81]. The Maranas group has integrated cell growth and product synthesis in the OptKnock toolbox [10] and applied it to construct high-performance mutants. The computer-aided designs have shown improved lactic acid, succinate, and 1,3-propanediol production [82]. FBA can predict lethality in a metabolic network where deletions of more than one nonessential gene mutants may trigger the death of the organism. For example, the Maranas group [83] analyzed the gene/reaction essentiality in a genome-scale model of E. coli and systemically identified possible pairs of synthetic lethals: nonessential genes whose simultaneous knockouts would have a potentially lethal effect. Incorporating information about synthetic lethality into the new model will curb the construction of ill-designed biological systems for biotechnology. Furthermore, FBA can be used to find rate-limiting steps for product synthesis. For example, FBA revealed gene targets, and modification of those genes (i.e., knocking out the genes for pyruvate forming enzymes, overexpression of the glyoxylate shunt and glucose transport system) resulted in more than a ten-fold increase in succinate production [84–86]. FBA has also been used to improve genetic strategies for the overproduction of secondary metabolites, such as amino acids [87] and lycopene [88].
Besides genetic strategies, FBA can provide useful information for the design of optimal fermentation conditions. For example, an FBA model was used to identify nutrient limitations during recombinant interleukin-2 (IL-2) production in E. coli. By supplementing specific amino acids, IL-2 production increased two-fold in fed-batch fermentation [89]. Recently, a reactor-scale dFBA model was developed via a static optimization Approach to analyze E. coli metabolism for the production of a biopharmaceutical drug [27]. dFBA contains a steady state FBA model embedded within a dynamic kinetic model that describes the time evolution of fermentation process variables (e.g., biomass growth, glucose consumption and products synthesis). Such a model provided guidelines for the optimization of fermentations at the scale of a 1000L process.
The 13C-MFA model was first used to investigate metabolic regulation in E.coli under different genetic and environmental conditions [90]. 13C-MFA has also been used to examine various biotechnological processes involved in the production of pharmaceuticals, amino acids and polymers. A large scale 13C-MFA with over 300 reactions was successfully developed for amorphadiene (a precursor of the antimalaria drug) producing E. coli strains [46]. Another study revealed a growth phase-dependent metabolic shift in a lysine-producing E. coli strain [91]. This work was performed in a fed-batch culture with rich medium (containing yeast extract), and metabolic fluxes in both exponential growth and stationary phases were estimated by measuring free metabolites. Metabolic analysis of the stationary phase is important since many products are synthesized during a nongrowth phase. In a third example, 13C-MFA of a 1,3-propanediol producing E. coli strain was conducted in fed-batch fermentation [92]. The 13C-MFA results showed a decrease in the split ratio between glycolysis and the pentose-phosphate pathway over the time-course of the culture in response to increasing 1,3-propanediol fluxes.
4.2. Bacillus subtilis Model
B. subtilis is the industrial organism of choice for the production of vitamins, antibiotics, enzymes, and nucleosides. The FBA model for B. subtilis was constructed based on a combination of genomic, biochemical, and physiological information [93]. The FBA model was iteratively corrected and improved using information from high-throughput phenotypic screens of mutants, substrate utilization, gene essentiality, and sequence analyses. The B. subtilis flux model is mostly studied for riboflavin production, focusing on four aspects: investigating phenotypes of wild type and knock-out strains, assessing production capacity, identifying the impact of different carbon sources on biosynthesis, and characterizing the cellular response to different culture conditions. The Sauer group has extensively investigated riboflavin-producing strains. They first used an FBA model to quantify growth maintenance coefficients, the maximum growth yield, and the specific riboflavin production rate in continuous cultivation [94]. Later on, they applied 13C-MFA to the same strain and found that genetic manipulations should target the NADPH balance and riboflavin biosynthetic pathways [95]. In other studies on B. subtilis, they revealed several guidelines for high-yield riboflavin production (1) they compared the metabolic flux distributions and maintenance energy of eight Bacillus strains and discovered that B. licheniformis was the most suitable for industrial biotechnology [96], (2) they found that using malate as a substrate resulted in a suppressed respiratory TCA cycle and an enhanced overflow metabolism [97], and (3) they found the pentose precursors of riboflavin were mainly synthesized via the nonoxidative pentose-phosphate pathway, so any suggested genetic modification should decrease the activity of the oxidative pentose-phosphate pathway [98]. Recently, they developed a 13C-dMFA model for B. subtilis to identify the metabolic response of riboflavin overproduction under a glucose-limited fed-batch culture [40]. This dynamic flux analysis was obtained by recording changes in labeling patterns of intracellular amino acids under a metabolic pseudosteady state assumption.
4.3. Saccharomyces cerevisiae Model
S. cerevisiae is a robust eukaryotic chassis used for the expression of a wide range of products. For example, flux analysis revealed target genes in two native pathways for the overexpression of succinate: the TCA and glyoxylate cycles [99]. Another study showed the enhancement of sesquiterpene production via in silico driven metabolic engineering [100]. Additionally, flux analysis has been extensively applied for improving ethanol production. First, a number of strategies were developed for the metabolic engineering of redox processes in S. cerevisiae, resulting in a decrease in the yield of glycerol by 40% and an increase in ethanol production under both glucose and xylose/glucose growth conditions [101]. Second, Dikicioglu et al. [102] applied a genome-scale FBA model to analyze respiration-deficient mutants of S. cerevisiae for ethanol production. They found that many genetic manipulation strategies (e.g., the overexpression of the glutamate synthase gene) were unnecessary in a respiration-deficient metabolic background. This indicates that the rate limiting steps for ethanol production can change after the initial genetic manipulations of targeted genes. Third, a 13C-MFA model was used to screen ethanol production in 14 hemiascomycetous yeast strains [51]. This study suggests that S. cerevisiae is the ideal ethanol production candidate due to a strong NADPH-driven pentose-phosphate pathway. Other 13C-MFA studies characterized the metabolic shift between oxidative growth and fermentative growth with ethanol production [103], investigated alternative carbon substrate (xylose) metabolisms [104], revealed key factors influencing biomass growth on xylose [32], and examined the consumption of ethanol and other storage carbohydrates in a glucose-limited chemostat culture [105].
Furthermore, a genome-scale FBA indicates an apparent enzyme dispensability, that is, 80% of yeast genes seem to be nonessential for viability under laboratory conditions [106]. The FBA illustrated the influence of nonessential genes on metabolic robustness and environmental fitness due to genetic buffering through alternative genes, while a 13C-MFA (consisting of over 700 reactions) revealed a similar effect of metabolic network robustness on null mutations [47]. Understanding the role of these redundant genes is important for a valid and efficient genetic modification.
4.4. Nonmodel Microorganisms
Fluxomics is an important tool for the rigorous study of metabolism in less-characterized microbes that provides novel insights for application of these species to biotechnology. However, fluxomics have not been sufficiently applied to nonmodel microorganisms as compared to model microbial hosts. Table 1 summarizes some milestone papers in fluxomics studies on nonmodel species that are potentially useful for synthetic biology. Compared to the work done in the field of fluxomics for industrial workhorses, far fewer studies have been performed on nonmodel microorganisms. This is due to the complicated growth conditions, poorly-understood metabolic networks, and significant lack of genetic and molecular biology tools. However, nonmodel environmental microorganisms are also important for industrial biotechnology because they often possess native biochemical pathways for chemical synthesis or the ability to utilize cheap substrates [120]. Furthermore, flux analysis can be used to discover novel enzymes that can be cloned into industrial microbes to improve their capacity for product synthesis. For example, 13C-MFA revealed a citramalate pathway for isoleucine biosynthesis (independent of the common threonine ammonia-lyase pathway) [121, 122]. Citramalate synthase, which has also been detected in some environmental bacteria [123–125], can be engineered into E. coli for 1-propanol and 1-butanol production. The new pathway bypasses threonine biosynthesis and represents the shortest keto-acid-mediated pathway; as such, it improved biofuel yield 9 to 22-fold [126]. Currently, high-throughput genome sequencing methods are mapping genomes in novel microbes at a pace that far exceeds the pace of functional characterization of these species. Therefore, a high throughput 13C-MFA technique is required for screening nonmodel microorganisms for new enzymes and maximizing their application in industrial biotechnology [4].
Table 1.
Recent application of fluxomics of nonmodel microbes to bioproduct synthesis.
| Species | Product | Substrate | Model description | Results from study | Reference |
|---|---|---|---|---|---|
| Corynebacterium glutamicum | Lysine | Glucose (sucrose, fructose) | 13C-MFA | MFA models (combining transcriptome, metabolome analysis) have been developed to study fluxes under different cultivation modes (minibioreactor, batch, fed-batch) using various carbon sources. | [107] |
| Corynebacterium glutamicum | Methionine | Glucose | 13C-MFA only focuses on flux distribution in the methionine pathway. | The C. glutamicum mutant (mcbR) showed no overproduction of methionine, but accumulation of homolanthionine. | [108] |
| Corynebacterium glutamicum | Glutamate | Glucose | 13C-MFA (focus on anaplerotic pathways) | The flux from phosphoenolpyruvate to oxaloacetate catalyzed by phosphoenolpyruvate carboxylase (PEPc) was active in the growth phase, whereas pyruvate carboxylase was inactive. | [109] |
| Actinobacillus succinogenes | Succinate formate and acetate | Glucose NaHCO3 | 13C-MFA (via NMR and GC-MS) and enzyme assay | The model indicated (1) NADPH was produced primarily by transhydrogenase and/or by NADP-dependent malic enzyme (2) oxaloacetate and malate were converted to pyruvate (3) the effects of NaHCO3 and H2 on metabolic fluxes were quantified. | [110, 111] |
| Geobacillus thermoglucosidasius | Ethanol | Glucose | FBA and 13C-MFA | The model characterized the ethanol production under three oxygen conditions. The FBA analysis pointed out several gene targets for improving ethanol production. | [19] |
| Clostridium acetobutylicum | Butanol | Glucose | Genome-scale-FBA | The engineered strain was able to produce 154 mM butanol with 9.9 mM acetone at pH 5.5, resulting in a butanol selectivity (a molar ratio of butanol to total solvents) of 0.84. | [112] |
| Penicillium chrysogenum | Penicillin | Gluconate/glucose | 13C -MFA (focus on pentose phose phase pathway and glycolysis) | The model determined the pentose-phosphate pathway split ratio and estimated NADPH metabolism. | [113] |
| Synechocystis sp. PCC6803 | Hydrogen | CO2 | FBA | The results included H2 photoproduction, strategies to avoid oxygen inhibition, and analysis of hetero-, auto-, and mixotrophic metabolisms. | [114, 115] |
| Synechocystis sp. PCC6803 | Light energy & Biomass | Glucose/CO2 | 13C-MFA and dynamic 13C -MFA | The model analyzed heterotrophic, autotrophic and mixotrophic metabolisms. | [34, 58] |
| Chlamydomonas reinhardtii | Light energy & Biomass | CO2 | FBA model including three metabolically active compartments | The model indicated that heterotrophic growth had a low biomass yield on carbon, while mixotrophical and autotrophical growth had higher carbon utilization efficiency. | [116] |
| Zymomonas mobilis | Ethanol | Glucose/xylose | FBA with various biological objectives | Model analyzed the metabolic boundaries of Z. mobilis. The study indicated that ethanol and biomass production depend on anaerobic respiration stoichiometry and activity. | [117] |
| Zymomonas mobilis | Ethanol | Glucose/fructose/ xylose | 13C–MFA via 1H-NMR 31P-NMR spectroscopy | The model characterized the intracellular metabolic state during growth on glucose, fructose and xylose in defined continuous cultures. | [118] |
| Coculture (Desulfovibrio vulgaris and Methanococcus maripaludis) | CH4 | Lactate | FBA analysis of microbial consortia | The model predicted the ratio of D. vulgaris to M. maripaludis cells during growth. It was possible to eliminate formate as an interspecies electron shuttle, but H2 transfer was essential for syntrophic growth. | [55] |
| Community (oxygenic phototrophs, filamentous anoxygenic phototrophs, and sulfate-reducing bacteria). | Biomass and nitrogen fixation | CO2 | FBA and elementary mode analysis | The model predicted and described relative abundances of species, by-products, and the metabolic interactions. | [54] |
| Phaffia rhodozyma and Haematococcus pluvialis | Astaxanthin | Glucose with (peptone & yeast extract) | FBA analysis of mix culture | The two major astaxanthin-producing microorganisms exhibited elevated yields (2.8-fold) under mixed culture conditions compared to pure culture. | [119] |
5. Finding Bottlenecks for Industrial Biotechnology
One of the main goals of fluxomics is to indentify bottlenecks for industrial biotechnology and thereby assist in the creation of rational engineering strategies. Simple measurements of metabolism, however, are not enough to overcome unpredictable challenges in industrial biotechnology. Metabolic regulation is very complex, and systems biology tools are incapable of revealing a general strategy for synthetic biology [127].
Bottlenecks in industrial biotechnology can be explained from the view of fluxomics. First, metabolic robustness (the ability to maintain metabolic performance under genetic or environmental perturbations) is a long-recognized key property of microbial systems [128]. This basic mechanism is often responsible for the gap between computationally aided design and final experimental outcomes. For example, a 13C-MFA study indicates that E. coli shows remarkable robustness in the central carbon metabolism in the presence of genetic variation, and is even more flexible in response to altered environmental conditions (e.g., different nutrients or oxygen levels) [90]. Analyses of E. coli components at multiple “omics” levels also reveal unexpectedly small changes in messenger RNA, proteins and metabolite levels for most genetic disruptions. This is because E. coli actively regulates enzyme levels to maintain a stable metabolic state in the presence of perturbations [57, 78]. Similarly, B. subtilis shows rigidity and suboptimal performance for its flux regulation in over 137 genetic backgrounds [42]. Furthermore, gene essentiality and pairwise genetic interactions have been investigated in S. cerevisiae [106, 129]. It has been found that a gene's function is buffered by duplication in S. cerevisiae genomic DNA or by an alternative biochemical pathway. Although only 13% of genes were suggested to be essential by single knockout experiments, simultaneous deletion of pairs of nonessential genes (>70% of the total metabolic genes) were found to inhibit growth. Invariability of metabolic flux under mutagenic genotypes seems to be an important feature in many biological systems, and thus successful metabolic strategies highly depend on an understanding of robust cellular nature [130–132].
Metabolic engineering of industrial chassis is based on the premise that the yield of a desired product can be increased by identifying and overexpressing the enzymes that catalyze the rate-limiting steps in a given metabolic pathway. However, a method based on overexpressing rate-limiting enzymes will only work if these rate-limiting enzymes exist and remain rate-limiting when their activities are increased. Previous studies have shown that the commonly-believed “rate-limiting” enzymes may not exist in some industrial microbes and an increase in productivity has to be achieved by coordinated expression of entire pathways [133]. Furthermore, rate-limiting steps in a metabolic network often shift after initial targets have been engineered. For example, phenotypic data in S. cerevisiae mutants revealed that some FBA-predicted gene targets for ethanol production are invalid if the cell's respiratory genes have been knocked-out [102]. Another example of this phenomenon is highlighted by the metabolic consequences of the deletion of the methionine and cysteine biosynthesis repressor protein (McbR) in Corynebacterium glutamicum, which yielded no overproduction of methionine but drastic accumulation of homolanthionine [108]. The above evidence indicates that rate-limiting steps often shift after initial targets have been engineered. Additionally, simultaneous importation and expression of a few heterologous genes to improve the rate-limiting pathway may fail if the nonnative pathway is incompatible with the host. These efforts often lead to metabolic imbalance and accumulation of toxic metabolites [2, 3].
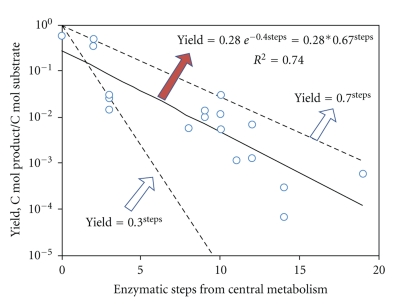
Based on the recent publications, we have constructed a linear regression model which shows that the yield of biosynthetic products decreases exponentially as a function of the steps away from central metabolism in S. cerevisiae (Figure 3). It is easier to achieve high carbon fluxes to the central metabolites, possibly because enzyme efficiency in central metabolism is usually high [134]. However, the yields of secondary metabolites are smaller because each additional enzymatic step may not be perfectly efficient (model regression shows an average of ~67% efficiency in each enzymatic step in secondary metabolisms). This loss of yield is unavoidable due to the metabolism channeling the intermediates away from the desired product. Potential solutions to this problem include (1) designing host-compatible enzymes with high product specificity [135], (2) feeding intermediates to the cell to reduce the number of enzymatic steps to final product [136], and (3) creating synthetic protein scaffolds, which significantly improve intermediate conversion efficiency and overall biosynthetic yield [137].
Figure 3.
Product yields as a function of enzymatic steps from central metabolism. The solid line is the regression of published product yields by S. cerevisiae as a function of reaction steps from intermediate metabolites in central metabolism (including glycolysis, TCA cycle and pentose-phosphate pathways). The yield declines exponentially as the number of reaction steps increases. The dotted lines are boundary curves with yield efficiencies of 30% and 70% respectively. All yield data from initial carbon sources are estimated from recent papers using our best judgment. The synthesized products and reaction steps are: Poly(R-3-hydroxybutyrate) [63] (steps = 3); Glycerol [64] (steps = 2); Artemisinic acid [1] (steps = 10); Amorphadiene [65] (steps = 9); Pyruvate [66] (steps = 0); Geranylgeraniol [67] (steps = 10); Hydrocortisone [68] (steps = 19); Squalene [69] (steps = 9); β-carotene [70] (steps = 12); Lycopene [70] (steps = 11); Phytoene [70] (steps = 10); p-hydroxycinnamic acid [71] (steps = 12); Naringenin [72] (steps = 14); Pinocembrin [72] (steps = 14); Xylitol and Ribitol [73] (steps = 3); Ethanol [74] (steps = 2); L-ascorbic acid [75] (steps = 8).
In conclusion, fluxomics studies enable the quantification of intracellular metabolism. However, this tool is not fully developed, and it remains difficult to deduce cell-wide pathway bottlenecks and to provide effective strategies for biotechnology applications. Numerous technical difficulties in developing flux analysis methods and complicated metabolic regulatory mechanisms have severely limited the scope of fluxomics in industrial biotechnology. It is necessary for the future development of flux analysis to combine other advanced “omics” analysis and molecular biology techniques to resolve challenges in the fluxomics fields.
Acknowledgments
This study was supported in part by an NSF Career Grant (MCB0954016) to Y. J. Tang and in part by a DOE Bioenergy Research Grant (DEFG0208ER64694) to HBP. The authors are grateful to all of the students (Bing Wu, Arul Varman, Yin Wang, Stephanie Suen, Craig Jacobson, and Wenying Liu) in the 2010 Metabolic Engineering class (ChE596) at Washington University for their useful input during class discussions.
References
- 1.Ro DK, Paradise EM, Quellet M, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 2.Atsumi S, Higashide W, Liao JC. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nature Biotechnology. 2009;27(12):1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- 3.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 4.Zamboni N, Sauer U. Novel biological insights through metabolomics and 13C-flux analysis. Current Opinion in Microbiology. 2009;12(5):553–558. doi: 10.1016/j.mib.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Feist AM, Zielinski DC, Orth JD, Schellenberger J, Herrgard MJ, Palsson BØ. Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metabolic Engineering. 2010;12(3):173–186. doi: 10.1016/j.ymben.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blazeck J, Alper H. Systems metabolic engineering: genome-scale models and beyond. Biotechnology Journal. 2010;5(7):647–659. doi: 10.1002/biot.200900247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker SA, Feist AM, Mo ML, Hannum G, Palsson BØ, Herrgard MJ. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA toolbox. Nature Protocols. 2007;2(3):727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 8.Kumar VS, Maranas CD. GrowMatch: an automated method for reconciling in silico/in vivo growth predictions. PLoS Computational Biology. 2009;5(3) doi: 10.1371/journal.pcbi.1000308. Article ID e1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boghigian BA, Seth G, Kiss R, Pfeifer BA. Metabolic flux analysis and pharmaceutical production. Metabolic Engineering. 2010;12(2):81–95. doi: 10.1016/j.ymben.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Burgard AP, Pharkya P, Maranas CD. OptKnock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnology and Bioengineering. 2003;84(6):647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 11.Trinh CT, Wlaschin A, Srienc F. Elementary mode analysis: a useful metabolic pathway analysis tool for characterizing cellular metabolism. Applied Microbiology and Biotechnology. 2009;81(5):813–826. doi: 10.1007/s00253-008-1770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schilling CH, Edwards JS, Letscher D, Palsson BO. Combining pathway analysis with flux balance analysis for the comprehensive study of metabolic systems. Biotechnology and Bioengineering. 2000;71(4):286–306. [PubMed] [Google Scholar]
- 13.Suthers PF, Dasika MS, Kumar VS, Denisov G, Glass JI, Maranas CD. Genome-scale metabolic reconstruction of mycoplasma genitalium, iPS189. PLoS Computational Biology. 2009;5(2) doi: 10.1371/journal.pcbi.1000285. Article ID e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zamboni N, Fendt SM, Rühl M, Sauer U. C-based metabolic flux analysis. Nature Protocols. 2009;4(6):878–892. doi: 10.1038/nprot.2009.58. [DOI] [PubMed] [Google Scholar]
- 15.Quek LE, Wittmann C, Nielsen LK, Krömer JO. OpenFLUX: efficient modelling software for 13C-based metabolic flux analysis. Microbial Cell Factories. 2009;8, article 25 doi: 10.1186/1475-2859-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang YJ, Martin HG, Myers S, Rodriguez S, Baidoo EEK, Keasling JD. Advances in analysis of microbial Metabolic fluxes via 13C isotopic labeling. Mass Spectrometry Reviews. 2009;28(2):362–375. doi: 10.1002/mas.20191. [DOI] [PubMed] [Google Scholar]
- 17.Jung TS, Yeo HC, Reddy SG, Cho WS, Lee DY. WEbcoli: an interactive and asynchronous web application for in silico design and analysis of genome-scale E. coli model. Bioinformatics. 2009;25(21):2850–2852. doi: 10.1093/bioinformatics/btp496. [DOI] [PubMed] [Google Scholar]
- 18.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metabolic Engineering. 2007;9(1):68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang YJ, Sapra R, Joyner D, et al. Analysis of metabolic pathways and fluxes in a newly discovered thermophilic and ethanol-tolerant geobacillus strain. Biotechnology and Bioengineering. 2009;102(5):1377–1386. doi: 10.1002/bit.22181. [DOI] [PubMed] [Google Scholar]
- 20.Nöh K, Grönke K, Luo B, Takors R, Oldiges M, Wiechert W. Metabolic flux analysis at ultra short time scale: isotopically non-stationary 13C labeling experiments. Journal of Biotechnology. 2007;129(2):249–267. doi: 10.1016/j.jbiotec.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 21.Gianchandani EP, Chavali AK, Papin JA. The application of flux balance analysis in systems biology. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2010;2(3):372–382. doi: 10.1002/wsbm.60. [DOI] [PubMed] [Google Scholar]
- 22.Tang YJ, Meadows AL, Keasling JD. A kinetic model describing Shewanella oneidensis MR-1 growth, substrate consumption, and product secretion. Biotechnology and Bioengineering. 2007;96(1):125–133. doi: 10.1002/bit.21101. [DOI] [PubMed] [Google Scholar]
- 23.Wildermuth MC. Metabolic control analysis: biological applications and insights. Genome Biology. 2000;1(6) doi: 10.1186/gb-2000-1-6-reviews1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoefnagel MHN, Starrenburg MJC, Martens DE, et al. Metabolic engineering of lactic acid bacteria, the combined approach: kinetic modelling, metabolic control and experimental analysis. Microbiology. 2002;148(4):1003–1013. doi: 10.1099/00221287-148-4-1003. [DOI] [PubMed] [Google Scholar]
- 25.Namjoshi AA, Hu WS, Ramkrishna D. Unveiling steady-state multiplicity in hybridoma cultures: the cybernetic approach. Biotechnology and Bioengineering. 2003;81(1):80–91. doi: 10.1002/bit.10447. [DOI] [PubMed] [Google Scholar]
- 26.Song HS, Morgan JA, Ramkrishna D. Systematic development of hybrid cybernetic models: application to recombinant yeast co-consuming glucose and xylose. Biotechnology and Bioengineering. 2009;103(5):984–1002. doi: 10.1002/bit.22332. [DOI] [PubMed] [Google Scholar]
- 27.Meadows AL, Karnik R, Lam H, Forestell S, Snedecor B. Application of dynamic flux balance analysis to an industrial Escherichia coli fermentation. Metabolic Engineering. 2010;12(2):150–160. doi: 10.1016/j.ymben.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Gianchandani EP, Chavali AK, Papin JA. The application of flux balance analysis in systems biology. Systems Biology and Medicine. 2010;2(3):372–382. doi: 10.1002/wsbm.60. [DOI] [PubMed] [Google Scholar]
- 29.Mahadevan R, Edwards JS, Doyle FJ. Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophysical Journal. 2002;83(3):1331–1340. doi: 10.1016/S0006-3495(02)73903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hjersted JL, Henson MA, Mahadevan R. Genome-scale analysis of Saccharomyces cerevisiae metabolism and ethanol production in fed-batch culture. Biotechnology and Bioengineering. 2007;97(5):1190–1204. doi: 10.1002/bit.21332. [DOI] [PubMed] [Google Scholar]
- 31.Oddone GM, Mills DA, Block DE. A dynamic, genome-scale flux model of Lactococcus lactis to increase specific recombinant protein expression. Metabolic Engineering. 2009;11(6):367–381. doi: 10.1016/j.ymben.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Sonderegger M, Jeppsson M, Hahn-Hägerdal B, Sauer U. Molecular basis for anaerobic growth of Saccharomyces cerevisiae on xylose, investigated by global gene expression and metabolic flux analysis. Applied and Environmental Microbiology. 2004;70(4):2307–2317. doi: 10.1128/AEM.70.4.2307-2317.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan J, Bennett BD, Rabinowitz JD. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nature Protocols. 2008;3(8):1328–1340. doi: 10.1038/nprot.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shastri AA, Morgan JA. A transient isotopic labeling methodology for 13C metabolic flux analysis of photoautotrophic microorganisms. Phytochemistry. 2007;68(16–18):2302–2312. doi: 10.1016/j.phytochem.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 35.Schaub J, Mauch K, Reuss M. Metabolic flux analysis in Escherichia coli by integrating isotopic dynamic and isotopic stationary 13C labeling data. Biotechnology and Bioengineering. 2008;99(5):1170–1185. doi: 10.1002/bit.21675. [DOI] [PubMed] [Google Scholar]
- 36.Nöh K, Wahl A, Wiechert W. Computational tools for isotopically instationary 13C labeling experiments under metabolic steady state conditions. Metabolic Engineering. 2006;8(6):554–577. doi: 10.1016/j.ymben.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Wahl SA, Nöh K, Wiechert W. 13C labeling experiments at metabolic nonstationary conditions: an exploratory study. BMC Bioinformatics. 2008;9, article 152 doi: 10.1186/1471-2105-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young JD, Walther JL, Antoniewicz MR, Yoo H, Stephanopoulos G. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnology and Bioengineering. 2008;99(3):686–699. doi: 10.1002/bit.21632. [DOI] [PubMed] [Google Scholar]
- 39.Suthers PF, Chang YJ, Maranas CD. Improved computational performance of MFA using elementary metabolite units and flux coupling. Metabolic Engineering. 2010;12(2):123–128. doi: 10.1016/j.ymben.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Rühl M, Zamboni N, Sauer U. Dynamic flux responses in riboflavin overproducing Bacillus subtilis to increasing glucose limitation in fed-batch culture. Biotechnology and Bioengineering. 2010;105(4):795–804. doi: 10.1002/bit.22591. [DOI] [PubMed] [Google Scholar]
- 41.Stephanopoulos GN, Aristidou AA, Nielsen J. Metabolic Engineering Principles and Methodologies. San Diego, Calif, USA: Academic Press; 1998. [Google Scholar]
- 42.Fischer E, Sauer U. Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nature Genetics. 2005;37(6):636–640. doi: 10.1038/ng1555. [DOI] [PubMed] [Google Scholar]
- 43.Schuetz R, Kuepfer L, Sauer U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Molecular Systems Biology. 2007;3:p. 119. doi: 10.1038/msb4100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colón-López MS, Sherman DM, Sherman LA. Transcriptional and translational regulation of nitrogenase in light- dark- and continuous-light-grown cultures of the unicellular cyanobacterium Cyanothece sp. strain ATCC 51142. Journal of Bacteriology. 1997;179(13):4319–4327. doi: 10.1128/jb.179.13.4319-4327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toepel J, Welsh E, Summerfield TC, Pakrasi HB, Sherman LA. Differential transcriptional analysis of the cyanobacterium Cyanothece sp. strain ATCC 51142 during light-dark and continuous-light growth. Journal of Bacteriology. 2008;190(11):3904–3913. doi: 10.1128/JB.00206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suthers PF, Burgard AP, Dasika MS, et al. Metabolic flux elucidation for large-scale models using 13C labeled isotopes. Metabolic Engineering. 2007;9(5-6):387–405. doi: 10.1016/j.ymben.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blank LM, Kuepfer L, Sauer U. Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biology. 2005;6(6):p. R49. doi: 10.1186/gb-2005-6-6-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dauner M. From fluxes and isotope labeling patterns towards in silico cells. Current Opinion in Biotechnology. 2010;21(1):55–62. doi: 10.1016/j.copbio.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 49.Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nature Protocols. 2008;3(8):1299–1311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malaisse WJ, Zhang Y, Jijakli H, Courtois P, Sener A. Enzyme-to-enzyme channelling in the early steps of glycolysis in rat pancreatic islets. International Journal of Biochemistry and Cell Biology. 2004;36(8):1510–1520. doi: 10.1016/j.biocel.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 51.Blank LM, Lehmbeck F, Sauer U. Metabolic-flux and network analysis in fourteen hemiascomycetous yeasts. FEMS Yeast Research. 2005;5(6-7):545–558. doi: 10.1016/j.femsyr.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Kleijn RJ, Geertman JMA, Nfor BK, et al. Metabolic flux analysis of a glycerol-overproducing Saccharomyces cerevisiae strain based on GC-MS, LC-MS and NMR-derived C-labelling data. FEMS Yeast Research. 2007;7(2):216–231. doi: 10.1111/j.1567-1364.2006.00180.x. [DOI] [PubMed] [Google Scholar]
- 53.Kaeberlein T, Lewis K, Epstein SS. Isolating "uncultivabte" microorganisms in pure culture in a simulated natural environment. Science. 2002;296(5570):1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 54.Taffs R, Aston JE, Brileya K, et al. In Silico approaches to study mass and energy flows in microbial consortia: a syntrophic case study. BMC Systems Biology. 2009;3, article 114 doi: 10.1186/1752-0509-3-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stolyar S, Van Dien S, Hillesland KL, et al. Metabolic modeling of a mutualistic microbial community. Molecular Systems Biology. 2007;3:p. 92. doi: 10.1038/msb4100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimizu K. Metabolic flux analysis based on 13C-labeling experiments and integration of the information with gene and protein expression patterns. Advances in Biochemical Engineering/Biotechnology. 2004;91:1–49. doi: 10.1007/b94204. [DOI] [PubMed] [Google Scholar]
- 57.Ishii N, Nakahigashi K, Baba T, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316(5824):593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 58.Yang C, Hua Q, Shimizu K. Metabolic flux analysis in Synechocystis using isotope distribution from 13C-labeled glucose. Metabolic Engineering. 2002;4(3):202–216. doi: 10.1006/mben.2002.0226. [DOI] [PubMed] [Google Scholar]
- 59.Moxley JF, Jewett MC, Antoniewicz MR, et al. Linking high-resolution metabolic flux phenotypes and transcriptional regulation in yeast modulated by the global regulator Gcn4p. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(16):6477–6482. doi: 10.1073/pnas.0811091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lemuth K, Hardiman T, Winter S, et al. Global transcription and metabolic flux analysis of Escherichia coli in glucose-limited fed-batch cultivations. Applied and Environmental Microbiology. 2008;74(22):7002–7015. doi: 10.1128/AEM.01327-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tännler S, Fischer E, Le Coq D, et al. CcpN controls central carbon fluxes in Bacillus subtilis. Journal of Bacteriology. 2008;190(18):6178–6187. doi: 10.1128/JB.00552-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nanchen A, Schicker A, Revelles O, Sauer U. Cyclic AMP-dependent catabolite repression is the dominant control mechanism of metabolic fluxes under glucose limitation in Escherichia coli. Journal of Bacteriology. 2008;190(7):2323–2330. doi: 10.1128/JB.01353-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carlson R, Srienc F. Effects of recombinant precursor pathway variations on poly[(R)-3-hydroxybutyrate] synthesis in Saccharomyces cerevisiae. Journal of Biotechnology. 2006;124(3):561–573. doi: 10.1016/j.jbiotec.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 64.Overkamp KM, Bakker BM, Kötter P, Luttik MAH, Van Dijken JP, Pronk JT. Metabolic engineering of glycerol production in Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2002;68(6):2814–2821. doi: 10.1128/AEM.68.6.2814-2821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shiba Y, Paradise EM, Kirby J, Ro DK, Keasling JD. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metabolic Engineering. 2007;9(2):160–168. doi: 10.1016/j.ymben.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 66.Van Maris AJA, Geertman JMA, Vermeulen A, et al. Directed evolution of pyruvate decarboxylase-negative Saccharomyces cerevisiae, yielding a C2-independent, glucose-tolerant, and pyruvate-hyperproducing yeast. Applied and Environmental Microbiology. 2004;70(1):159–166. doi: 10.1128/AEM.70.1.159-166.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tokuhiro K, Muramatsu M, Ohto C, et al. Overproduction of geranylgeraniol by metabolically engineered Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2009;75(17):5536–5543. doi: 10.1128/AEM.00277-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szczebara FM, Chandelier C, Villeret C, et al. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nature Biotechnology. 2003;21(2):143–149. doi: 10.1038/nbt775. [DOI] [PubMed] [Google Scholar]
- 69.Asadollahi MA, Maury J, Schalk M, Clark A, Nielsen J. Enhancement of farnesyl diphosphate pool as direct precursor of sesquiterpenes through metabolic engineering of the mevalonate pathway in Saccharomyces cerevisiae. Biotechnology and Bioengineering. 2010;106(1):86–96. doi: 10.1002/bit.22668. [DOI] [PubMed] [Google Scholar]
- 70.Verwaal R, Wang J, Meijnen JP, et al. High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Applied and Environmental Microbiology. 2007;73(13):4342–4350. doi: 10.1128/AEM.02759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vannelli T, Wei Qi W, Sweigard J, Gatenby AA, Sariaslani FS. Production of p-hydroxycinnamic acid from glucose in Saccharomyces cerevisiae and Escherichia coli by expression of heterologous genes from plants and fungi. Metabolic Engineering. 2007;9(2):142–151. doi: 10.1016/j.ymben.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 72.Jiang H, Wood KV, Morgan JA. Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2005;71(6):2962–2969. doi: 10.1128/AEM.71.6.2962-2969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Toivari MH, Ruohonen L, Miasnikov AN, Richard P, Penttilä M. Metabolic engineering of Saccharomyces cerevisiae for conversion of D-glucose to xylitol and other five-carbon sugars and sugar alcohols. Applied and Environmental Microbiology. 2007;73(17):5471–5476. doi: 10.1128/AEM.02707-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314(5805):1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 75.Sauer M, Branduardi P, Valli M, Porro D. Production of L-ascorbic acid by metabolically engineered Saccharomyces cerevisiae and Zygosaccharomyces bailii. Applied and Environmental Microbiology. 2004;70(10):6086–6091. doi: 10.1128/AEM.70.10.6086-6091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mukhopadhyay A, Redding AM, Rutherford BJ, Keasling JD. Importance of systems biology in engineering microbes for biofuel production. Current Opinion in Biotechnology. 2008;19(3):228–234. doi: 10.1016/j.copbio.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 77.Hua Q, Joyce AR, Palsson BØ, Fong SS. Metabolic characterization of Escherichia coli strains adapted to growth on lactate. Applied and Environmental Microbiology. 2007;73(14):4639–4647. doi: 10.1128/AEM.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fong SS, Nanchen A, Palsson BO, Sauer U. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. Journal of Biological Chemistry. 2006;281(12):8024–8033. doi: 10.1074/jbc.M510016200. [DOI] [PubMed] [Google Scholar]
- 79.Varma A, Palsson BO. Metabolic flux balancing: basic concepts, scientific and practical use. Bio/Technology. 1994;12(10):994–998. [Google Scholar]
- 80.Varma A, Palsson BO. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Applied and Environmental Microbiology. 1994;60(10):3724–3731. doi: 10.1128/aem.60.10.3724-3731.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liao JC, Hou SY, Chao YP. Pathway analysis, engineering, and physiological considerations for redirecting central metabolism. Biotechnology and Bioengineering. 1996;52(1):129–140. doi: 10.1002/(SICI)1097-0290(19961005)52:1<129::AID-BIT13>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 82.Fong SS, Burgard AP, Herring CD, et al. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnology and Bioengineering. 2005;91(5):643–648. doi: 10.1002/bit.20542. [DOI] [PubMed] [Google Scholar]
- 83.Suthers PF, Zomorrodi A, Maranas CD. Genome-scale gene/reaction essentiality and synthetic lethality analysis. Molecular Systems Biology. 2009;5, article 301 doi: 10.1038/msb.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee SY, Hong SH, Moon SY. In silico metabolic pathway analysis and design: succinic acid production by metabolically engineered Escherichia coli as an example. Genome Informatics. 2002;13:214–223. [PubMed] [Google Scholar]
- 85.Lee SJ, Lee DY, Kim TY, Kim BH, Lee J, Lee SY. Metabolic engineering of Escherichia coli for enhanced production of succinic acid, based on genome comparison and in silico gene knockout simulation. Applied and Environmental Microbiology. 2005;71(12):7880–7887. doi: 10.1128/AEM.71.12.7880-7887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Q, Chen X, Yang Y, Zhao X. Genome-scale in silico aided metabolic analysis and flux comparisons of Escherichia coli to improve succinate production. Applied Microbiology and Biotechnology. 2006;73(4):887–894. doi: 10.1007/s00253-006-0535-y. [DOI] [PubMed] [Google Scholar]
- 87.Park JH, Lee SY. Towards systems metabolic engineering of microorganisms for amino acid production. Current Opinion in Biotechnology. 2008;19(5):454–460. doi: 10.1016/j.copbio.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 88.Alper H, Jin YS, Moxley JF, Stephanopoulos G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metabolic Engineering. 2005;7(3):155–164. doi: 10.1016/j.ymben.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 89.Yegane-Sarkandy S, Farnoud AM, Shojaosadati SA, et al. Overproduction of human interleukin-2 in recombinant Escherichia coli BL21 high-cell-density culture by the determination and optimization of essential amino acids using a simple stoichiometric model. Biotechnology and Applied Biochemistry. 2009;54(1):31–39. doi: 10.1042/BA20080300. [DOI] [PubMed] [Google Scholar]
- 90.Sauer U, Lasko DR, Fiaux J, et al. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. Journal of Bacteriology. 1999;181(21):6679–6688. doi: 10.1128/jb.181.21.6679-6688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Iwatani S, Van Dien S, Shimbo K, et al. Determination of metabolic flux changes during fed-batch cultivation from measurements of intracellular amino acids by LC-MS/MS. Journal of Biotechnology. 2007;128(1):93–111. doi: 10.1016/j.jbiotec.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 92.Antoniewicz MR, Kraynie DF, Laffend LA, González-Lergier J, Kelleher JK, Stephanopoulos G. Metabolic flux analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of E. coli producing 1,3-propanediol. Metabolic Engineering. 2007;9(3):277–292. doi: 10.1016/j.ymben.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oh YK, Palsson BO, Park SM, Schilling CH, Mahadevan R. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. Journal of Biological Chemistry. 2007;282(39):28791–28799. doi: 10.1074/jbc.M703759200. [DOI] [PubMed] [Google Scholar]
- 94.Sauer U, Hatzimanikatis V, Hohmann HP, Manneberg M, Van Loon APGM, Bailey JE. Physiology and metabolic fluxes of wild-type and riboflavin-producing Bacillus subtilis. Applied and Environmental Microbiology. 1996;62(10):3687–3696. doi: 10.1128/aem.62.10.3687-3696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sauer U, Hatzimanikatis V, Bailey JE, Hochuli M, Szyperski T, Wüthrich K. Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nature Biotechnology. 1997;15(5):448–452. doi: 10.1038/nbt0597-448. [DOI] [PubMed] [Google Scholar]
- 96.Tännler S, Decasper S, Sauer U. Maintenance metabolism and carbon fluxes in Bacillus species. Microbial Cell Factories. 2008;7, article 19 doi: 10.1186/1475-2859-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kleijn RJ, Buescher JM, Le Chat L, Jules M, Aymerich S, Sauer U. Metabolic fluxes during strong carbon catabolite repression by malate in Bacillus subtilis. Journal of Biological Chemistry. 2010;285(3):1587–1596. doi: 10.1074/jbc.M109.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zamboni N, Fischer E, Muffler A, Wyss M, Hohmann HP, Sauer U. Transient expression and flux changes during a shift from high to low riboflavin production in continuous cultures of Bacillus subtilis. Biotechnology and Bioengineering. 2005;89(2):219–232. doi: 10.1002/bit.20338. [DOI] [PubMed] [Google Scholar]
- 99.Otero JM, Olssona L, Nielsen J. Metabolic engineering of Saccharomyces cerevisiae microbial cell factories for succinic acid production. Journal of Biotechnology. 2007;131(2):p. 205. [Google Scholar]
- 100.Asadollahi MA, Maury J, Patil KR, Schalk M, Clark A, Nielsen J. Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metabolic Engineering. 2009;11(6):328–334. doi: 10.1016/j.ymben.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 101.Bro C, Regenberg B, Förster J, Nielsen J. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metabolic Engineering. 2006;8(2):102–111. doi: 10.1016/j.ymben.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 102.Dikicioglu D, Pir P, Onsan ZI, Ulgen KO, Kirdar B, Oliver SG. Integration of metabolic modeling and phenotypic data in evaluation and improvement of ethanol production using respiration-deficient mutants of Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2008;74(18):5809–5816. doi: 10.1128/AEM.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frick O, Wittmann C. Characterization of the metabolic shift between oxidative and fermentative growth in Saccharomyces cerevisiae by comparative 13C flux analysis. Microbial Cell Factories. 2005;4, article 30 doi: 10.1186/1475-2859-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grotkjær T, Christakopoulos P, Nielsen J, Olsson L. Comparative metabolic network analysis of two xylose fermenting recombinant Saccharomyces cerevisiae strains. Metabolic Engineering. 2005;7(5-6):437–444. doi: 10.1016/j.ymben.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 105.Van Winden WA, Van Dam JC, Ras C, et al. Metabolic-flux analysis of Saccharomyces cerevisiae CEN.PK113-7D based on mass isotopomer measurements of 13C-labeled primary metabolites. FEMS Yeast Research. 2005;5(6-7):559–568. doi: 10.1016/j.femsyr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 106.Papp B, Pál C, Hurst LD. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature. 2004;429(6992):661–664. doi: 10.1038/nature02636. [DOI] [PubMed] [Google Scholar]
- 107.Iwatani S, Yamada Y, Usuda Y. Metabolic flux analysis in biotechnology processes. Biotechnology Letters. 2008;30(5):791–799. doi: 10.1007/s10529-008-9633-5. [DOI] [PubMed] [Google Scholar]
- 108.Krömer JO, Heinzle E, Schröder H, Wittmann C. Accumulation of homolanthionine and activation of a novel pathway for isoleucine biosynthesis in Corynebacterium glutamicum McbR deletion strains. Journal of Bacteriology. 2006;188(2):609–618. doi: 10.1128/JB.188.2.609-618.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shirai T, Fujimura K, Furusawa C, Nagahisa K, Shioya S, Shimizu H. Study on roles of anaplerotic pathways in glutamate overproduction of Corynebacterium glutamicum by metabolic flux analysis. Microbial Cell Factories. 2007;6, article 19 doi: 10.1186/1475-2859-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McKinlay JB, Shachar-Hill Y, Zeikus JG, Vieille C. Determining Actinobacillus succinogenes metabolic pathways and fluxes by NMR and GC-MS analyses of 13C-labeled metabolic product isotopomers. Metabolic Engineering. 2007;9(2):177–192. doi: 10.1016/j.ymben.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 111.McKinlay JB, Vieille C. C-metabolic flux analysis of Actinobacillus succinogenes fermentative metabolism at different NaHCO3 and H2 concentrations. Metabolic Engineering. 2008;10(1):55–68. doi: 10.1016/j.ymben.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 112.Jin YL, Jang YS, Lee J, Papoutsakis ET, Lee SY. Metabolic engineering of Clostridium acetobutylicum M5 for highly selective butanol production. Biotechnology Journal. 2009;4(10):1432–1440. doi: 10.1002/biot.200900142. [DOI] [PubMed] [Google Scholar]
- 113.Kleijn RJ, Van Winden WA, Ras C, Van Gulik WM, Schipper D, Heijnen JJ. 13C-labeled gluconate tracing as a direct and accurate method for determining the pentose phosphate pathway split ratio in Penicillium chrysogenum. Applied and Environmental Microbiology. 2006;72(7):4743–4754. doi: 10.1128/AEM.02955-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Navarro E, Montagud A, Fernández de Córdoba P, Urchueguía JF. Metabolic flux analysis of the hydrogen production potential in Synechocystis sp. PCC6803. International Journal of Hydrogen Energy. 2009;34(21):8828–8838. [Google Scholar]
- 115.Shastri AA, Morgan JA. Flux balance analysis of photoautotrophic metabolism. Biotechnology Progress. 2005;21(6):1617–1626. doi: 10.1021/bp050246d. [DOI] [PubMed] [Google Scholar]
- 116.Boyle NR, Morgan JA. Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii. BMC Systems Biology. 2009;3, article 4 doi: 10.1186/1752-0509-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tsantili IC, Karim MN, Klapa MI. Quantifying the metabolic capabilities of engineered Zymomonas mobilis using linear programming analysis. Microbial Cell Factories. 2007;6, article 8 doi: 10.1186/1475-2859-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.De Graaf AA, Striegel K, Wittig RM, et al. Metabolic state of Zymomonas mobilis in glucose-, fructose-, and xylose- fed continuous cultures as analysed by 13C- and 31P-NMR spectroscopy. Archives of Microbiology. 1999;171(6):371–385. doi: 10.1007/s002030050724. [DOI] [PubMed] [Google Scholar]
- 119.Dong QL, Zhao XM, Ma HW, Xing XY, Sun NX. Metabolic flux analysis of the two astaxanthin-producing microorganisms Haematococcus pluvialis and Phaffia rhodozyma in the pure and mixed cultures. Biotechnology Journal. 2006;1(11):1283–1292. doi: 10.1002/biot.200600060. [DOI] [PubMed] [Google Scholar]
- 120.Alper H, Stephanopoulos G. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nature Reviews Microbiology. 2009;7(10):715–723. doi: 10.1038/nrmicro2186. [DOI] [PubMed] [Google Scholar]
- 121.Risso C, Van Dien SJ, Orloff A, Lovley DR, Coppi MV. Elucidation of an alternate isoleucine biosynthesis pathway in Geobacter sulfurreducens. Journal of Bacteriology. 2008;190(7):2266–2274. doi: 10.1128/JB.01841-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang YJ, Chakraborty R, Martín HG, Chu J, Hazen TC, Keasling JD. Flux analysis of central metabolic pathways in Geobacter metallireducens during reduction of soluble Fe(III)-nitrilotriacetic acid. Applied and Environmental Microbiology. 2007;73(12):3859–3864. doi: 10.1128/AEM.02986-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu B, Zhang B, Feng X, et al. Alternative isoleucine synthesis pathway in cyanobacterial species. Microbiology. 2010;156(2):596–602. doi: 10.1099/mic.0.031799-0. [DOI] [PubMed] [Google Scholar]
- 124.Feng X, Mouttaki H, Lin LU, et al. Characterization of the central metabolic pathways in Thermoanaerobacter sp. strain X514 via isotopomer-assisted metabolite analysis. Applied and Environmental Microbiology. 2009;75(15):5001–5008. doi: 10.1128/AEM.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Howell DM, Xu H, White RH. (R)-citramalate synthase in methanogenic archaea. Journal of Bacteriology. 1999;181(1):331–333. doi: 10.1128/jb.181.1.331-333.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Atsumi S, Liao JC. Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Applied and Environmental Microbiology. 2008;74(24):7802–7808. doi: 10.1128/AEM.02046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kwok R. Five hard truths for synthetic biology. Nature. 2010;463(7279):288–290. doi: 10.1038/463288a. [DOI] [PubMed] [Google Scholar]
- 128.Stelling J, Sauer U, Szallasi Z, Doyle FJ, Doyle J. Robustness of cellular functions. Cell. 2004;118(6):675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 129.Deutscher D, Meilijson I, Kupiec M, Ruppin E. Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nature Genetics. 2006;38(9):993–998. doi: 10.1038/ng1856. [DOI] [PubMed] [Google Scholar]
- 130.Kol S, Elena Merlo M, Scheltema RA, et al. Metabolomic characterization of the salt stress response in streptomyces coelicolor. Applied and Environmental Microbiology. 2010;76(8):2574–2581. doi: 10.1128/AEM.01992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tang YJ, Martin HG, Deutschbauer A, et al. Invariability of central metabolic flux distribution in shewanella oneidensis MR-1 under environmental or genetic perturbations. Biotechnology Progress. 2009;25(5):1254–1259. doi: 10.1002/btpr.227. [DOI] [PubMed] [Google Scholar]
- 132.Heinemann M, Sauer U. Systems biology of microbial metabolism. Current Opinion in Microbiology. 2010;13(3):337–343. doi: 10.1016/j.mib.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 133.Niederberger P, Prasad R, Miozzari G, Kacser H. A strategy for increasing an in vivo flux by genetic manipulations: the tryptophan system of yeast. Biochemical Journal. 1992;287(2):473–479. doi: 10.1042/bj2870473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Colletti P, Goyal Y, Varman A, Feng X, Wu B, Tang YJ. Evaluating factors that influence microbial synthesis yields by regression with numerical and categorical variables. doi: 10.1002/bit.22996. Biotechnology and Bioengineering. In press. [DOI] [PubMed] [Google Scholar]
- 135.Yoshikuni Y, Ferrin TE, Keasling JD. Designed divergent evolution of enzyme function. Nature. 2006;440(7087):1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 136.Eshkol N, Sendovski M, Bahalul M, Katz-Ezov T, Kashi Y, Fishman A. Production of 2-phenylethanol from L-phenylalanine by a stress tolerant Saccharomyces cerevisiae strain. Journal of Applied Microbiology. 2009;106(2):534–542. doi: 10.1111/j.1365-2672.2008.04023.x. [DOI] [PubMed] [Google Scholar]
- 137.Dueber JE, Wu GC, Malmirchegini GR, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nature Biotechnology. 2009;27(8):753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]