Abstract
Inflammation involves the activation of a highly coordinated gene expression program that is specific for the initial stimulus and occurs in a different manner in bystander parenchymal cells and professional immune system cells recruited to the inflamed site. Recent data demonstrate that developmental transcription factors like the macrophage fate-determining Pu.1 set the stage for the activity of ubiquitous transcription factors activated by inflammatory stimuli, like NF-kB, AP-1, and interferon regulatory factors (IRFs). The intersection of lineage-determining and stimulus-activated transcription factors at enhancers explains cell type specificity in inflammatory responses.
Keywords: ChIP-seq, NF-kB, Pu.1, chromatin, inflammation, macrophages
Living organisms are constantly exposed to an innumerable variety of internal and external stimuli. Some of them can be classified as danger signals that either represent a direct consequence of tissue damage (like the release of proteins and metabolites normally sequestered within cells) or indicate the presence of harmful agents that may threaten tissue and even organism integrity (like invading microbes) (Baccala et al. 2009; Schroder and Tschopp 2010). When such signals are detected, a complex response is set in motion that is aimed at eliminating the danger signals and eventually restoring tissue and organism homeostasis (Medzhitov 2008). This response is generically referred to as inflammation, and its overall blueprint has been determined early in the evolution of metazoa, as indicated by the presence of a typical inflammatory response to wounds in invertebrates like the starfish (the organism in which Elly Metchnikov [1887] discovered phagocytosis >100 years ago). Moreover, some of the molecules and the domains involved in inflammation can be traced back to the point where the evolution of the primordial animal and vegetal cells diverged, ∼2 billion years ago (Kimbrell and Beutler 2001). Inflammatory molecules and responses evolved at a fast pace, reflecting the different environmental challenges different animals are exposed to: In fact, genes involved in environmental and inflammatory responses (e.g., genes encoding chemo-attractants and scavenger receptors) display an unusually high rate of duplications and losses during evolution (Ponting 2008).
We now know that inflammatory responses are both essential for homeostasis and potentially dangerous, being involved in diseases as different as cancer, rheumatoid arthritis, and septic shock. The realization that inflammation is a causative agent of many diseases has definitely contributed to a strong increase in the general interest toward this response, which in turn has resulted in an exponential increase in our mechanistic knowledge of normal and pathologic inflammatory reactions.
Recent technological advancements, particularly the combination of chromatin immunoprecipitation (ChIP) with high-throughput sequencing (ChIP-seq) and the development of refined computational tools for the deconvolution of large data sets, are providing an increasingly clear picture of the basic functional and organizational principles that underlie inflammatory gene expression. This review is specifically focused on the interplay between transcription factors (TFs) and chromatin organization in orchestrating highly specialized inflammatory gene expression programs.
General features of the inflammatory gene expression programs
Independently of the initial stimulus, inflammatory responses show characteristic properties that reflect defined mutual interactions between sequence-specific TFs, pre-existing chromatin organization, and enzymes that modify chromatin (Amit et al. 2009).
Kinetic complexity
A large number of genes (several hundreds) are activated in response to inflammatory stimuli in a kinetically complex fashion—some immediately after the stimulus, and some after many hours. Evidence has accumulated suggesting that inducible recruitment to target genes of some TFs (like NF-kB) is influenced by the pre-existing chromatin state (Saccani et al. 2001; Smale 2010). In turn, nucleosomal organization at promoters seems to be largely dependent on simple sequence features (particularly GC content) that impact on nucleosomal stability (Ramirez-Carrozzi et al. 2009). A comparatively low GC content enables the assembly of thermodynamically stable nucleosomes, which are conversely incompatible with the high GC content characteristic of CpG islands. In turn, stable nucleosomes impart an absolute dependence on Swi/Snf chromatin remodeling complexes for both NF-kB recruitment and gene activation (Ramirez-Carrozzi et al. 2006, 2009; Kayama et al. 2008; Hargreaves et al. 2009). In general, the requirement for a chromatin remodeling step at inflammatory genes whose promoters have a low GC content has two types of consequences: First, it tends to cause slower kinetics of activation; and second, it imposes the requirement for additional TFs that promote the initial remodeling step. Some of the TFs that control remodeling (see below) are selectively induced by specific stimuli, which lays the grounds for stimulus specificity in inflammatory gene expression (Smale 2010).
Specificity for stimulus
Specificity for stimulus largely reflects different signaling properties of receptors for inflammatory stimuli, which leads to the activation of a distinct panel of TFs and, ultimately, to a stimulus-specific gene expression program (Nau et al. 2002; Smale 2010). The paradigmatic case in the field is provided by Toll-like receptor 2 (TLR-2) on the one hand, and TLR-3/TLR-4 on the other, which recognize distinct classes of microbial molecules (Takeuchi and Akira 2010). TLR-3 and TLR-4, but not TLR-2, are able to trigger the phosphorylation and activation of IRF3 (interferon regulatory factor 3) (Doyle et al. 2002), which is required for the activation of a subset of Swi/Snf-dependent inflammatory genes, possibly by directly promoting recruitment of Swi/Snf (Ramirez-Carrozzi et al. 2009). In this case, a different wiring of the signal transduction cascade emanating from related receptors results in a different profile of activated TFs and a different ability of alternative stimuli to overcome the nucleosomal barrier.
Specificity for cell type (context dependence)
Specificity for cell type (context dependence) indicates that identical stimuli provoke different transcriptional outputs in different cell types. Based on recent genomic data (Ghisletti et al. 2010; Heinz et al. 2010), an essential point we argue here (see below) is that cell type specificity in inflammatory responses reflects the intersection of developmental inputs (cell type-specific and lineage-determining TFs) and environmental inputs (which act through non-cell type-specific TFs responsive to stimulation, like NF-kB, AP-1, and some IRFs) at specific cis-regulatory elements (particularly enhancers).
Requirement for precise tuning
An excessive reaction to inflammatory stimuli is an obvious cause of morbidity and mortality, as exemplified by septic shock and inflammatory diseases (e.g., rheumatoid arthritis) in humans. In mouse models, genetic ablation of negative regulators of inflammatory pathways (e.g., IkBa/Nfkbia and A20/Tnfaip3, which down-regulate NF-kB activation at different levels) leads to fatal inflammatory diseases (Beg et al. 1995; Lee et al. 2000). Recent genomic and genetic data point to a specific role of a sequence-specific and signal-responsive transcriptional repressor (Bcl-6) in limiting the activity of enhancers that control the inflammatory gene expression program in macrophages, thus preventing a hyperreaction to microbial stimuli (Barish et al. 2010).
The cis-regulatory repertoire of professional inflammatory cells
As discussed above, one of the most obvious features of inflammatory responses is that genes activated by identical stimuli differ extensively among cell types, even though induction of these genes depends on inflammatory TFs (like NF-kB and AP-1 family members) that are ubiquitously expressed. Recent genomic data (Ghisletti et al. 2010; Heinz et al. 2010) shed light on the precise molecular nature of such “context dependence,” which seems to depend mainly on the existence of a cell type-specific repertoire of functional cis-regulatory elements, particularly enhancers.
A firmly established concept in transcriptional control is that, although promoters in some cases contain cis-regulatory information that enables tissue-specific expression (like binding sites for a cell type-restricted TF), complex patterns of gene expression always require the combined activity of distant enhancers, each of them able to selectively function in a given cell type or tissue (Pennacchio et al. 2006). The genomic counterpart of the notion that enhancers function in a tissue-specific manner is the observation that the regions of the genome active as enhancers in different cell types show little overlap (Heintzman et al. 2009). This was definitively demonstrated by the genome-wide mapping in multiple cell types of (potential) enhancer regions, identified by the binding of histone acetyltransferases (like p300) as well as high levels of monomethylation of histone H3 Lys 4 (H3K4me1) in the absence of H3K4me3 (a modification associated with promoters and transcription start sites) (Heintzman et al. 2009).
Therefore, the chromatinized genome of different cell types shows a largely unique repertoire of active cis-regulatory regions. In turn, this unique repertoire reflects the underlying activity of cell type-specific activators of enhancer function. Such activators are nothing other than TFs responsible and required for terminal differentiation and constantly expressed in differentiated cells, where their presence is essential for maintenance of cell identity (Natoli 2010).
The enhancer repertoire specific to macrophages, a cell type with a central role in most types of normal and pathologic inflammatory responses, has been revealed recently by genome-wide ChIP studies. Like most other cell types analyzed to date, macrophages contain at least 35,000–45,000 identifiable genomic regions that may be classified as enhancers on the basis of the H3K4me1/H3K4me3 chromatin signature, the binding of p300, and the presence of conserved TF-binding sites (De Santa et al. 2010; Ghisletti et al. 2010). The unifying property of these enhancers is the almost invariable association with the essential macrophage fate-determining TF: the Ets protein Pu.1/Sfpi1 (Nerlov and Graf 1998; DeKoter and Singh 2000). Pu.1 and its paralogs, Spib and Spic, recognize both in vitro and in vivo a highly specific sequence that differs at a few critical positions from high-affinity binding sites for other subclasses of Ets proteins (Wei et al. 2010). A canonical Pu.1 site can be identified at >95% of genomic locations contacted by Pu.1 in vivo. Canonical Pu.1 sites can also be computationally identified at hundreds of thousands of genomic sites that are not contacted in cells (I Barozzi and G Natoli, unpubl.). As a whole, contacted sites show higher affinity for Pu.1 than the group of noncontacted ones, but, on an individual basis, some contacted sites show similar or lower affinity as compared with sites that are not bound in vivo; therefore, it is still unclear how Pu.1 selects sites to bind. One possibility is that Pu.1 cannot overcome some repressive chromatin configurations, which therefore efficiently prevent its binding; alternatively, Pu.1 recruitment may require cooperative interactions with other lineage-determining TFs, thus selecting restricted subsets among all possible sites.
In B cells, where Pu.1 concentration is estimated to be ∼10 times lower than in macrophages (DeKoter and Singh 2000), recruitment to target sites is strongly dependent on partner TFs (like E2a and Ebf), possibly through cooperative DNA binding: As a result, Pu.1 distribution as well as the repertoire of enhancers in B cells display little overlap with those in macrophages (Heinz et al. 2010).
It remains to be determined whether and to what extent genomic recruitment of Pu.1 in macrophages depends on partner TFs, or whether, alternatively, its high concentration makes cooperative interactions not required for binding to genomic sites.
Importantly, Pu.1 is not a simple marker of enhancers, but, conversely, it is directly involved in their activation. Indeed, Pu.1 expression in nonmyeloid cells (like fibroblasts) (Ghisletti et al. 2010) as well as Pu.1 re-expression in Pu.1-negative myeloid progenitors (Heinz et al. 2010) are both sufficient to induce nucleosome-free DNA stretches bracketed by nucleosomes marked by H3K4me1 at genomic regions that extensively overlap macrophage-specific enhancers. Therefore, Pu.1 provides a clear and paradigmatic example of how lineage-determining TFs directly organize a cell type-specific cis-regulatory repertoire.
A canonical and minimal enhancer module that controls inflammatory gene expression in macrophages
Genome-wide ChIP data combined with perturbation experiments have provided an initial interpretative framework of the minimal components of enhancers that control the inflammatory gene expression program in macrophages.
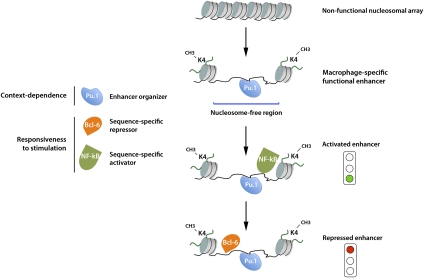
First of all, as discussed above, enhancers seem to be almost universally bound by Pu.1 in macrophages (Ghisletti et al. 2010; Heinz et al. 2010). Although the underlying mechanistic bases are still unclear, it is likely that Pu.1 attracts chromatin remodelers able to displace or remodel nucleosomes, thus leading to the formation of small accessible regions centered on the Pu.1-binding site (Fig. 1). In simple words, Pu.1 acts as an organizer responsible for the activation of selected genomic regions as enhancers (Natoli 2010). Such organization activity entails both nucleosome displacement (or remodeling) and the deposition of histone marks characteristic of enhancers (like H3K4me1) at adjacent nucleosomes. An inference of this model, which still awaits experimental validation, is that Pu.1 must be able to bind nucleosomal sites. While the relevance of H3K4me1 (and other histone modifications) for enhancer function is still unclear, the creation of nucleosome-free regions may be essential for the recruitment of TFs like NF-kB, which may not be able to associate with binding sites embedded in a nucleosomal context (Natoli 2009). In this light, functional cooperation between two or more TFs can be seen as mediated by a repressive nucleosomal organization that must be overcome by one TF (Pu.1) in order to allow the binding of the other (e.g., NF-kB), which is ultimately responsible for the recruitment of the transcriptional machinery.
Figure 1.
Organization and function of macrophage-specific enhancers responsive to inflammatory stimuli. The essential macrophage fate-determining TF Pu.1 activates as enhancers the genomic regions it binds. Enhancer organization entails both the formation of a nucleosome-free region centered on the Pu.1-binding site and the deposition of histone modifications like H3K4me1. A fraction of the macrophage-specific enhancers contains binding sites for TFs activated by inflammatory stimuli (like NF-kB), which are recruited in response to stimulation. Binding sites for sequence-specific transcriptional repressors (e.g., Bcl-6) are involved in negative regulation of enhancer activity. Bcl-6 association with most genomic regions it binds is attenuated or completely eliminated in response to inflammatory stimuli.
The notion that TFs involved in inducible gene expression in macrophages rely on Pu.1 for their recruitment to chromatin is supported by the behavior of Liver X Receptor-β (Lxrβ). LXRs are nuclear receptors that act mainly as intranuclear cholesterol sensors and activate genes involved in cholesterol handling and metabolism (Zelcer and Tontonoz 2006). At the same time, they negatively regulate inflammatory genes in macrophages. A large fraction of Lxrβ-binding sites detected by ChIP-seq are adjacent to Pu.1 sites; these sites, but not those distal to a Pu.1-binding site, disappear in cells lacking Pu.1 (Heinz et al. 2010). Clearly, these data are also compatible with the possibility that cooperative binding (rather than nucleosome displacement) is required for recruitment, and additional targeted experiments will be needed to discriminate between these different mechanisms. However, a notable observation is that combined genetic ablation of Lxrβ and Lxrα does not impair Pu.1 recruitment at Lxr-bound regions, thus indicating an obvious hierarchy in which prior Pu.1 binding prepares the ground for subsequent Lxrβ recruitment (Heinz et al. 2010).
Irrespective of the precise underlying mechanisms, the convergence at enhancers of a lineage-determining TF (like Pu.1), stimulus-responsive TFs (like Lxrβ), and the classic inflammatory TFs (like NF-kB and IRFs) provides a simple and unifying explanation of the role of the cellular context in modulating the response to inflammatory triggers and, more generally, environmental stimuli. In other words, enhancers are the genomic locations where developmental and environmental cues intersect, and the minimal inflammatory enhancer module in macrophages includes two binding sites: one for Pu.1 (the cell type-specific organizer) and one for a TF responsive to stimulation (the non-cell type-specific responder; e.g., NF-kB, AP1, IRFs, and LXRs) (Fig. 1).
Negative regulation of inflammatory gene transcription at enhancers
While this minimal enhancer module ensures that a macrophage-specific inflammatory gene expression program is activated, it does not provide regulatory mechanisms to limit the intensity of the response. Because of the potential pathogenicity of inflammation, it can easily be imagined that such regulatory mechanisms must have been positively selected during evolution. Recent genomic and genetic data demonstrated that this is indeed the case, and that a central regulatory role is played by Bcl-6. Bcl-6 is a sequence-specific transcriptional repressor known for its role in B-cell differentiation (mainly in germinal centers) and B-cell lymphomas (Basso and Dalla-Favera 2010), but in fact is expressed across many tissues and cell types (Ravasi et al. 2010). Bcl-6−/− mice are unable to mount a germinal center reaction (Dent et al. 1997; Ye et al. 1997). Moreover, they develop a severe multiorgan inflammatory response that is dependent on nonlymphoid cells and has been at least in part attributed to the excessive production of inflammatory mediators by macrophages (Toney et al. 2000). New data greatly contribute to explain this phenotype. Bcl-6 was found to be responsible for both maintenance of a low basal level of transcription and prevention of exaggerated activation of a large fraction of lipopolysaccharide (LPS)-inducible genes (Barish et al. 2010). Since only 5% of Bcl-6-binding sites are located at promoters, negative regulation of inflammatory genes occurs mainly at distant sites that coincide extensively with Pu.1-bound enhancers. Although Bcl-6 is considered mainly a developmental regulator, it is responsive to extracellular cues (Bereshchenko et al. 2002; Pantano et al. 2006). In fact, LPS stimulation resulted in the loss of >90% of Bcl-6-binding events detected in basal cells, together with the appearance of novel sites. Importantly, nearly 2500 genomic sites contacted by Bcl-6 recruited the main inflammatory TF, NF-kB, in response to LPS stimulation. Overall, ∼18% of LPS-induced genes are associated with enhancers contacted by both Bcl-6 and NF-kB, suggesting a direct cross-talk between activator and repressor that takes place at a subset of macrophage-specific enhancers.
Although it is still unclear whether binding sites for other repressors coexist with binding sites for activators at other enhancers involved in inflammatory gene control, this seems to be a likely scenario that will be clarified by future genomic studies.
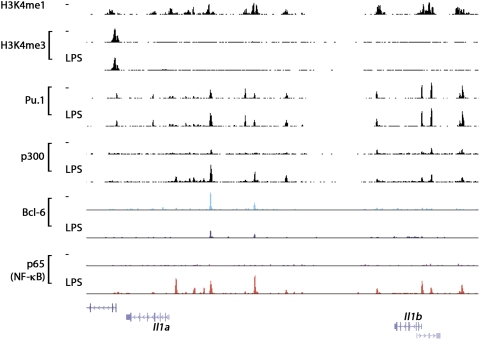
A typical landscape of a genomic region containing enhancer elements that confer responsiveness to microbial stimulation in macrophages is shown in Figure 2.
Figure 2.
A canonical genomic landscape of inflammation. A macrophage genomic region is shown that contains multiple potential enhancer elements marked by high constitutive H3K4me1 levels. These enhancers are constitutively associated with the macrophage lineage commitment and identity factor Pu.1, and recruit inflammatory TFs like NF-kB in response to microbial stimulation. In turn, inflammatory TFs promote the recruitment of the histone acetyltransferase p300. In some cases, enhancers are also constitutively bound by Bcl-6, a sequence-specific transcriptional repressor that is in part released upon microbial stimulation. Data sets used for the figure were from Barish et al. (2010), Ghisletti et al. (2010), and De Santa et al. (2010).
Conclusions and future perspectives
Combination of ChIP-seq data and genetic studies has provided an initial yet essential framework explaining the mechanistic bases of inflammatory gene regulation in professional innate immune system cells—macrophages in particular. However, it is important to realize that at this stage the whole complement of players involved in the response remains largely unknown. Specifically, the complete panel of TFs that contribute to the organization and maintenance of the specific cistrome characteristic of the different cells participating in inflammation is incompletely defined, and even more so the repertoire of coactivators and corepressors that contribute to positive and negative regulation of inflammatory gene expression in various cell types. Such a limited characterization of the players involved hinders a systems-level understanding of the inflammatory gene expression program and urges more systematic efforts to identify such players. It is also important to realize, as data obtained in macrophages unambiguously demonstrate, that responses to environmental stimuli are embedded in (and determined by) the specific differentiation program of each cell type. Therefore, the activity of inflammatory TFs will have to be studied and interpreted in the specific context created and maintained by lineage-determining TFs. This may create an exciting point of convergence between developmental biologists and scientists interested in environmental responses.
Acknowledgments
Work related to the topic of this review in our laboratory was supported by EC (FP6 grant Trans-Tar and FP7 grant Model-In) and the Italian Association for Research on Cancer (AIRC).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2018811.
References
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. 2009. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326: 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccala R, Gonzalez-Quintial R, Lawson BR, Stern ME, Kono DH, Beutler B, Theofilopoulos AN 2009. Sensors of the innate immune system: Their mode of action. Nat Rev Rheumatol 5: 448–456 [DOI] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM 2010. Bcl-6 and NF-κB cistromes mediate opposing regulation of the innate immune response. Genes Dev 24: 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Dalla-Favera R 2010. BCL6: Master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv Immunol 105: 193–210 [DOI] [PubMed] [Google Scholar]
- Beg AA, Sha WC, Bronson RT, Baltimore D 1995. Constitutive NF-κ B activation, enhanced granulopoiesis, and neonatal lethality in IκBα-deficient mice. Genes Dev 9: 2736–2746 [DOI] [PubMed] [Google Scholar]
- Bereshchenko OR, Gu W, Dalla-Favera R 2002. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet 32: 606–613 [DOI] [PubMed] [Google Scholar]
- DeKoter RP, Singh H 2000. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 288: 1439–1441 [DOI] [PubMed] [Google Scholar]
- Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM 1997. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 276: 589–592 [DOI] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384 doi: 10.1371/journal.pbio.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, et al. 2002. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17: 251–263 [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138: 129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayama H, Ramirez-Carrozzi VR, Yamamoto M, Mizutani T, Kuwata H, Iba H, Matsumoto M, Honda K, Smale ST, Takeda K 2008. Class-specific regulation of pro-inflammatory genes by MyD88 pathways and IκBζ. J Biol Chem 283: 12468–12477 [DOI] [PubMed] [Google Scholar]
- Kimbrell DA, Beutler B 2001. The evolution and genetics of innate immunity. Nat Rev Genet 2: 256–267 [DOI] [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A 2000. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science 289: 2350–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R 2008. Origin and physiological roles of inflammation. Nature 454: 428–435 [DOI] [PubMed] [Google Scholar]
- Metchnikov E 1887. Sur la lutte des cellules de l'organisme contre l'invasion des microbes. Annales de l'Institut Pasteur 1: 321–336 [Google Scholar]
- Natoli G 2009. Control of NF-κB-dependent transcriptional responses by chromatin organization. Cold Spring Harb Perspect Biol 1: a000224 doi: 10.1101/cshperspect.a000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G 2010. Maintaining cell identity through global control of genomic organization. Immunity 33: 12–24 [DOI] [PubMed] [Google Scholar]
- Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA 2002. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci 99: 1503–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C, Graf T 1998. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev 12: 2403–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantano S, Jarrossay D, Saccani S, Bosisio D, Natoli G 2006. Plastic downregulation of the transcriptional repressor BCL6 during maturation of human dendritic cells. Exp Cell Res 312: 1312–1322 [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444: 499–502 [DOI] [PubMed] [Google Scholar]
- Ponting C 2008. The functional repertoires of metazoan genomes. Nat Rev Genet 9: 689–698 [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST 2006. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev 20: 282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. 2010. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140: 744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G 2001. Two waves of nuclear factor κB recruitment to target promoters. J Exp Med 193: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Tschopp J 2010. The inflammasomes. Cell 140: 821–832 [DOI] [PubMed] [Google Scholar]
- Smale ST 2010. Selective transcription in response to an inflammatory stimulus. Cell 140: 833–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S 2010. Pattern recognition receptors and inflammation. Cell 140: 805–820 [DOI] [PubMed] [Google Scholar]
- Toney LM, Cattoretti G, Graf JA, Merghoub T, Pandolfi PP, Dalla-Favera R, Ye BH, Dent AL 2000. BCL-6 regulates chemokine gene transcription in macrophages. Nat Immunol 1: 214–220 [DOI] [PubMed] [Google Scholar]
- Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al. 2010. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J 29: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS, et al. 1997. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet 16: 161–170 [DOI] [PubMed] [Google Scholar]
- Zelcer N, Tontonoz P 2006. Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest 116: 607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]