Abstract
Using RNA-seq technology, we found that the majority of microRNAs (miRNAs) present in CFU-E erythroid progenitors are down-regulated during terminal erythroid differentiation. Of the developmentally down-regulated miRNAs, ectopic overexpression of miR-191 blocks erythroid enucleation but has minor effects on proliferation and differentiation. We identified two erythroid-enriched and developmentally up-regulated genes, Riok3 and Mxi1, as direct targets of miR-191. Knockdown of either Riok3 or Mxi1 blocks enucleation, and either physiological overexpression of miR-191 or knockdown of Riok3 or Mxi1 blocks chromatin condensation. Thus, down-regulation of miR-191 is essential for erythroid chromatin condensation and enucleation by allowing up-regulation of Riok3 and Mxi1.
Keywords: erythropoiesis, enucleation, RNA-seq, miR-191, Riok3, Mxi1
In mammals, terminal erythroid differentiation is a coordinated process that involves four to five cell divisions, induction of many erythroid-important genes, chromatin condensation, and enucleation. The first two cell divisions—from the committed erythroid progenitor CFU-E to the late basophilic erythroblast—are Erythropoietin (Epo)-dependent (Lodish et al. 2010). Epo regulates this process by binding to EpoR and triggering the activation of several downstream signaling pathways (Richmond et al. 2005). Further differentiation beyond this stage is no longer Epo-dependent, but requires the adhesion of erythroid progenitors to a fibronectin matrix (Eshghi et al. 2007). In addition to extracellular hormones, several key transcriptional factors, such as Gata1, are indispensable for erythropoiesis, regulating the expression of many genes (Cantor and Orkin 2002). In addition to the accumulation of erythroid lineage-specific gene products such as hemoglobin and chromatin condensation, in mammals, enucleation occurs, a process by which the condensed nucleus surrounded by the plasma membrane is budded off of the nascent reticulocyte (Ji et al. 2008).
MicroRNAs (miRNAs) are a class of recently identified regulatory small RNAs that down-regulate expression of their target genes by either mRNA degradation or translational inhibition (Bushati and Cohen 2007; Bartel 2009; Guo et al. 2010). So far, only a few miRNAs have been reported to regulate erythropoiesis (Zhao et al. 2010). Among these miRNAs, it was shown that miR-144 and miR-451 are Gata1 target genes and are required for erythropoiesis by targeting Klfd and Gata2, respectively (Dore et al. 2008; Fu et al. 2009; Pase et al. 2009). Recently, further research suggested that the miR-144–451 cluster is required for erythroid homeostasis (Rasmussen et al. 2010). Erythroid differentiation is defective in miR-451 mutant mice, due to the abnormal up-regulation of the miR-451 target gene 14-3-3ζ (Patrick et al. 2010). Additionally, miR-451 protects against erythroid oxidant stress by targeting 14-3-3ζ (Yu et al. 2010).
Here, using RNA-seq technology, we quantitatively determined the expression profile of miRNAs during terminal erythroid differentiation. Consistent with nuclear condensation and global gene expression shutdown during terminal erythroid differentiation, we found that the majority of miRNAs are down-regulated in more mature Ter119+ erythroblasts compared with CFU-E erythroid progenitors. We found that the down-regulation of one erythroid-abundant miRNA, miR-191, is required for the enucleation process. Our further experiments suggested that the erythroid-enriched genes Riok3 and Mxi1 are direct targets of miR-191. Both Riok3 and Mxi1 are up-regulated during terminal erythroid differentiation, and knockdown of either Riok3 or Mxi1 impaired chromatin condensation and enucleation.
Results and Discussion
The majority of late erythroblast CFU-E-abundant miRNAs are down-regulated during terminal erythroid differentiation
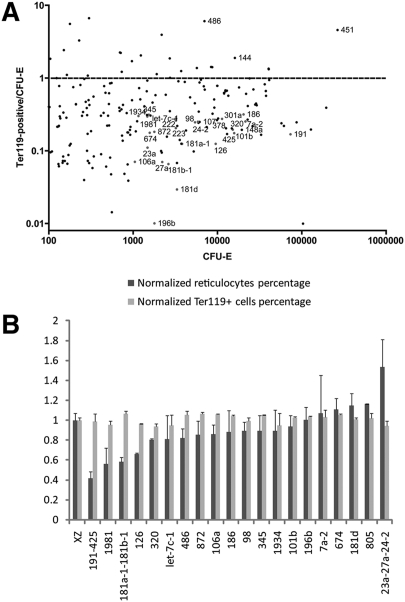
To understand quantitatively the global miRNA expression pattern during terminal erythroid differentiation, we carried out RNA-seq to profile miRNA expression patterns of Epo-dependent CFU-E erythroid progenitors and Ter119+ mature erythroblast cells. As shown in Figure 1A, one erythroid-abundant miRNA, miR-451, is sharply up-regulated during terminal differentiation and accounts for 12% of the total miRNAs at the CFU-E stage. This percentage increases to 58% at the Ter119+ stage. As a result of the sharp increase of miR-451 expression, the majority of other miRNAs are down-regulated in Ter119+ cells compared with CFU-Es, assuming the total amount of miRNA stays the same.
Figure 1.
miRNA expression profile, as determined by RNA-seq deep sequencing. (A) CFU-Es and Ter119+ cells are high-purity late erythroid progenitors and mature erythroblasts sorted from E14.5 mouse fetal liver (fsee the Supplemental Material for details of sorting procedures). RNA-seq deep sequencing specific for small RNAs was carried out for CFU-E and Ter119+ cells. Data from the RNA-seq were normalized so that the total number of reads from the Ter119+ cells is equivalent to the total number of reads from the CFU-Es. On the X-axis is the RPKM (reads per kilobase of exon per million mapped reads) of the CFU-Es. On the Y-axis is the ratio of the RPKM of the Ter119+ cells relative to the RPKM of the CFU-Es. (B) Twenty miRNAs or miRNA clusters were overexpressed in E14.5 mouse fetal Ter119− erythroid progenitors, followed by 2 d in vitro culture. Day 2 cells were stained with Ter119-APC, CD71-PE, and Hoechst. FACS analysis was carried out to determine the differentiation and enucleation statuses of these cells. The percentages of Ter119+Hochest− cells and Ter119+ cells were plotted normalized to a value of 1.0 representing the corresponding value for control XZ vector-infected cells.
Since the majority of miRNAs present in CFU-E cells are down-regulated, to uncover the functions of these miRNAs in erythropoiesis, we carried out a miRNA overexpression functional screen. Twenty abundant and developmentally down-regulated miRNAs or miRNA clusters were cloned into the retrovirus vector XZ-IRES-GFP. Retroviral vectors for these miRNAs or miRNA clusters were transduced into mouse embryonic day 14.5 (E14.5) fetal liver erythroid progenitors and cultured in vitro for 2 d to allow terminal erythroid cell proliferation and differentiation (Zhang et al. 2003). To determine the differentiation and enucleation statuses of miRNA-infected cells, day 2 cultured cells were labeled for the erythroid lineage-specific membrane protein Ter119, the transferrin receptor CD71, and the cell-permeable DNA dye Hoechst 33342, followed by FACS analysis. As shown previously, the cells first display induction of CD71 (CD71highTer119med), then induction of Ter119 and down-regulation of CD71 (CD71medTer119high) during 2 d in vitro culture. This mimics the normal in vivo erythroid progenitor differentiation processes (Zhang et al. 2003). At day 2, the HoechstlowTer119high population represents enucleated reticulocytes (Ji et al. 2008). As shown in Figure 1B, overexpression of the majority of these miRNAs had no or subtle effects on terminal erythroid differentiation or enucleation. Among the miRNAs we tested, overexpression of the miR-191–425 cluster, miR-1981, and the miR-181a-1–181b-1 cluster inhibited enucleation, whereas overexpression of the miR-23a–27a–24–2 cluster promoted enucleation. Since miR-191 is the most abundant miRNA among these miRNAs tested, we chose the miR-191–425 cluster for further analysis.
miR-191 modulates erythroblast enucleation
Further analysis showed that miR-191 but not miR-425 (data not shown) mediated the miR-191–425 overexpression phenotype. According to our RNA-seq data, miR-191 is an abundant miRNA that accounts for 3.5% of all miRNAs present in CFU-Es, while the most abundant miRNA, miR-451, accounts for 12.7%. As shown in Figure 1, miR-191 is down-regulated approximately sixfold from the CFU-E to the Ter119+ stage.
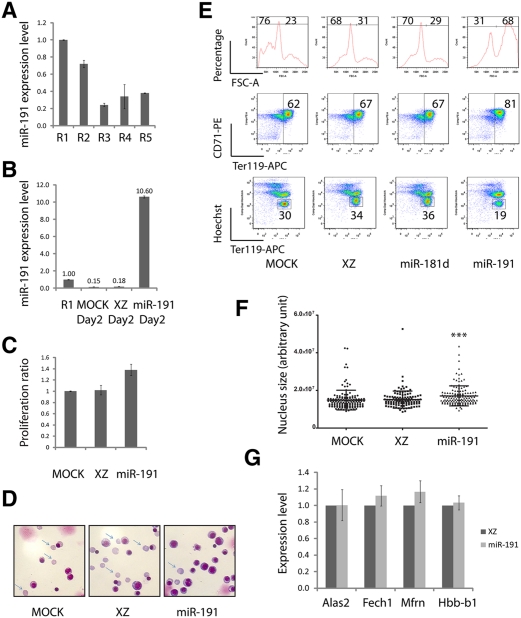
E14.5 fetal livers contain at least five distinct populations of cells, R1–R5, which are defined by their characteristic Ter119 and CD71 double-staining patterns. CFU-E cells and proerythroblasts make up the R1 population; R2 consists of proerythroblasts and early basophilic erythroblasts; R3 includes early and late basophilic erythroblasts; R4 is mostly chromatophilic and orthochromatophilic erythroblasts; and R5 is comprised of late orthochromatophilic erythroblasts and reticulocytes (Zhang et al. 2003). As shown in Figure 2A, miR-191 is down-regulated around fivefold from the R1 stage to the R3 stage.
Figure 2.
miR-191 regulates cultured mouse fetal erythroblast enucleation. (A) miR-191 expression levels from R1 to R5 as determined by qRT–PCR. (B) miR-191 overexpression levels at day 2 of an in vitro cultured erythroid progenitor, as determined by qRT–PCR. (A,B) U6 was the loading control. Error bar is standard deviation (SD) (n = 3). (C) Mouse fetal erythroid progenitors were infected with indicated viruses. Twelve hours after infection, GFP+ cells were sorted and cultured for another 36 h. The numbers of GFP+ cells at 12 h and 48 h were determined by a cell-counting machine (BD). The normalized ratios of cell numbers at 48 h relative to 12 h were plotted. Error bar is SD (n = 3). (D) May-Grunwald Giemsa staining carried out on day 2 cultured cells infected with indicated viruses; representative images are shown (n = 3). Arrows indicate reticulocytes. (E) Day 2 in vitro cultured cells as in D stained with Ter119-APC, CD71-PE, and Hoechst. Representative FACS plots are shown (n = 3). (F) Day 2 in vitro cultured cells as in E stained with DAPI; nucleus sizes were quantified by image processing. (***) P < 0.001 in Student t-test. (G) qRT–PCR performed on day 2 cultured cells infected with indicated viruses. Error bar is SD (n = 3).
As shown in Figure 2B, using our retrovirus vector, miR-191 was overexpressed to a level 10-fold higher than control R1 stage cells. Overexpression of miR-191 significantly impaired enucleation; ∼19% reticulocytes were detected at day 2, while cells infected by the empty vector XZ or by a vector encoding a control miRNA, miR-181d, generated almost twice that number (34%) of reticulocytes (Fig. 2E). The blockage of enucleation was further supported by May-Grunwald Giemsa staining, which clearly showed that miR-191 overexpression decreased the percentage of enucleated cells (Fig. 2D). However, miR-191 overexpression did not affect the induction of Ter119, suggesting that overexpression of miR-191 had no influence on differentiation (Fig. 2E). In the Ter119-APC/Hoechst FACS plot and Ter119negative in theTer119-APC/CD71-PE FACS plot (Fig. 2E), the slight increase of the Ter119+ percentage in the miR-191 overexpression sample is due to the decrease in the percentage of extruded nuclei, the populations that are Ter119negativeHoechsthigh. Supporting this contention, Figure 2G shows that miR-191 overexpression had no effect on the expression levels of several erythroid-important genes tested (including Alas2, Fech1, and Mfrn) and Hbb-b1 (β-globin). Overexpression of miR-191 slightly enhanced the rate of cell division (Fig. 2C).
The blockage of enucleation might be due to the direct defects in cytokinesis (Ji et al. 2008) or in the preceding event, chromatin condensation. To determine whether chromatin condensation was impaired by miR-191 overexpression, we carried out a single-cell nucleus size quantification assay. As shown in Figure 2F, overexpression of miR-191 significantly blocked chromatin condensation. Furthermore, as indicated by the differences in the FACS forward scatter (FSC) parameters, overexpression of miR-191 inhibited the normal decrease in cell volume late in differentiation, consistent with a defect in chromatin/nuclear condensation (Fig. 2E).
The above results suggest that ectopic expression of miR-191 at a physiological level significantly inhibited chromatin condensation and enucleation; in other words, prevention of the normal down-regulation of miR-191 prevented chromatin condensation and enucleation. Since miR-191 is sharply down-regulated approximately sixfold from the CFU-E to the Ter119+ stage, further down-regulation of miR-191 by loss-of-function approaches likely would not generate any obvious morphological phenotype. To test this hypothesis, we carried out a miRNA sponge-mediated miR-191 knockdown experiment (Ebert et al. 2007). miR-191 was successfully knocked down by the miR-191-sponge, as shown by quantitative RT–PCR (qRT–PCR) (Supplemental Fig. 1A). Compared with a control sponge, only ∼18% of miR-191 was detected at day 2 of in vitro cultured erythroid cells following miR-191 sponge infection. No obvious differentiation, enucleation, or proliferation defects were observed (Supplemental Fig. 1B–D).
Two developmentally up-regulated and erythroid enriched genes, Riok3 and Mxi1, are direct targets of miR-191
To further understand how miR-191 modulates erythroblast enucleation, we used three criteria to pick potential miR-191 target genes: those predicted as miR-191 target genes by TargetScan, those developmentally up-regulated during terminal erythroid differentiation according to our mRNA RNA-seq data (J Flygare and P Wong, unpubl.), and those down-regulated upon miR-191 overexpression in our in vitro erythroid progenitor culture system. Among all of the genes tested, Riok3 and Mxi1 fulfilled all three criteria (Fig. 3).
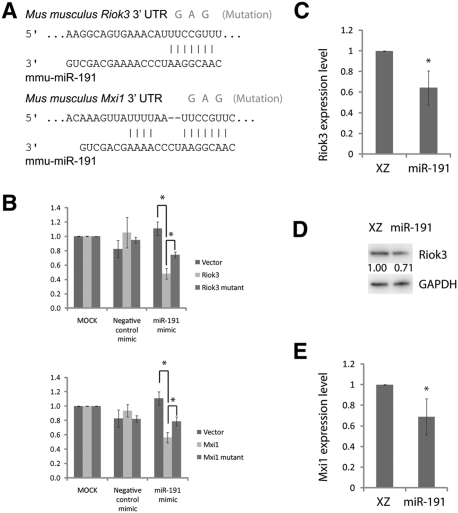
Figure 3.
Riok3 and Mxi1 are direct target genes of miR-191. (A) Mouse Riok3 and Mxi1 3′UTRs contain miR-191-binding sites; mutations introduced into the target sequences are shown. (B) Luciferase reporters together with miRNA mimics were cotransfected into 293T cells. The relative firefly/Renilla luciferase activity ratios were determined 48 h after transfection. Error bar is SD (n = 3). (C) qRT–PCR shows expression levels of Riok3 mRNA at day 2 of in vitro cultured erythroid progenitors following infection of indicated viruses. Error bar is SD (n = 3). (D) Western blot shows Riok3 protein levels at day 2 of in vitro cultured erythroid progenitors following infection of indicated viruses. Numbers below Riok3 bands are the relative Riok3/DAPDH band intensity ratios. Representative data are shown (n = 3). (E) qRT–PCR shows Mxi1 expression levels at day 2 of in vitro cultured erythroid progenitors following infection of indicated viruses. Error bar is SD (n = 3). (*) P < 0.05 in Student t-test (n = 3).
As shown in Figure 3A, both mouse Riok3 and Mxi1 3′ untranslated regions (UTRs) contain predicted miR-191-binding sites. To test whether these sites are under the regulation of miR-191, we performed luciferase reporter assays. Sequences from Riok3 and Mxi1 3′UTRs that are ∼500 base pairs (bp) in length and contain the miR-191-binding sites were cloned into the luciferase reporter vector psiCHECK2. The miR-191 mimic or the negative control mimic was cotransfected with luciferase reporters or control empty vectors into 293T cells, and the cells were lysed 48 h after transfection. As shown in Figure 3B, luciferase activities of reporters containing Riok3 or Mxi1 3′UTR sequences were significantly lower than the control empty vector when cotransfected with the miR-191 mimic but not with a negative control mimic. To further prove it was the miR-191-binding sites that mediated this regulation, three point mutations at a miR-191-binding site were introduced into the luciferase reporters (Fig. 3A). As shown in Figure 3B, the mutations significantly weakened the response of reporters to the miR-191 mimic but not to the control mimic.
To test whether Riok3 and Mxi1 are under the control of miR-191 during terminal erythroid differentiation, we measured Riok3 and Mxi1 expression levels in our day 2 in vitro cultured erythroid cells. As shown in Figure 3, C–E, overexpression of miR-191 down-regulated the expression of Riok3 and Mxi1. Collectively, these results suggest that Riok3 and Mxi1 are miR-191 target genes. Since little is known about the function of Riok3 and Mxi1 in erythropoiesis, we decided to continue our research to uncover the possible roles of these two genes in erythropoiesis.
Riok3 is required for erythroblast enucleation
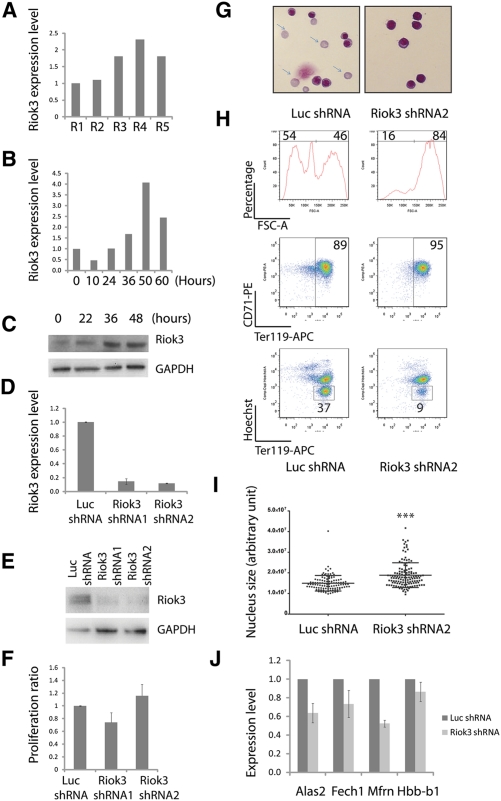
Riok3 belongs to the RIO family of atypical protein kinases (LaRonde-LeBlanc and Wlodawer 2005). As shown in Supplemental Figure 1A, Riok3 is enriched in the erythroid lineage compared with other tissues and cell types (Su et al. 2004). During terminal erythroid differentiation, Riok3 is continuously up-regulated from the R1 to the R5 stage (Fig. 4A), and, in our in vitro erythroid progenitor culture system, Riok3 showed a similar expression pattern (Fig. 4B,C).
Figure 4.
Riok3 is required for erythroblast enucleation. (A) Riok3 mRNA expression levels from R1 to R5, as determined by qRT–PCR. 18s rRNA was the loading control. (B) Riok3 expression levels of in vitro cultured erythroid progenitors, as determined by qRT–PCR. 18s was the loading control. Error bar is SD (n = 3). (C) Riok3 protein expression levels of in vitro cultured erythroid progenitors, as determined by Western blot. Representative data are shown (n = 3). (D) Riok3 mRNA levels at day 2 of culture following addition of shRNAs, as determined by qRT–PCR. 18s rRNA was the loading control. Error bar is SD (n = 3). (E) Riok3 protein levels at day 2 determined by Western blot following addition of shRNAs. Representative data are shown (n = 3). (F) Similar to Figure 2C; for samples with infection of indicated viruses, the normalized ratios of cell numbers at 48 h relative to 12 h were plotted. Error bar is SD (n = 3). (G) May-Grunwald Giemsa staining carried out on day 2 cultured cells infected with indicated viruses; representative images are shown (n = 3). Arrows indicate reticulocytes. (H) Day 2 in vitro cultured cells as in G stained with Ter119-APC, CD71-PE, and Hoechst. Representative FACS plots are shown (n = 3). (I) Day 2 in vitro cultured cells, as in H, stained with DAPI; nucleus sizes were quantified by image processing. (***) P < 0.001 in Student t-test. (J) qRT–PCR performed on day 2 cultured cells infected with indicated viruses. Error bar is SD (n = 3).
To uncover Riok3's function in erythropoiesis, two Riok3-specific shRNAs and a control Luc shRNA were introduced by retroviral infection into mouse E14.5 fetal liver erythroid progenitors, followed by 2 d of in vitro culture. As shown in Figure 4, D and E, two different and nonoverlapping Riok3 shRNAs, but not a control Luc shRNA, blocked Riok3 expression at both the mRNA and protein levels. Strikingly, knockdown of Riok3 almost completely blocked enucleation (Fig. 4H). May-Grunwald Giemsa staining further supported the blockage of enucleation, as almost no enucleated cells could be found in the Riok3 shRNA-infected sample (Fig. 4G). As shown in Figure 4H, Riok3 shRNA infection did not influence the induction of Ter119, which suggested that knockdown of Riok3 had little if any influence on differentiation. This contention was further supported by a qRT–PCR assay showing that knockdown of Riok3 had no effect on Hbb-b1 (β-globin) expression and only slightly inhibited the expression of the erythroid-important genes tested, including Alas2, Fech1, and Mfrn (Fig. 4J). Knockdown of Riok3 also had no influence on the cell division rate (Fig. 4F), and, by measuring the size of individual nuclei, we found that knockdown of Riok3 severely blocked chromatin condensation (Fig. 4I). This is consistent with a blockage of the normal decrease in cell volume, as indicated by the difference in the FSC parameter (Fig. 4H). Collectively, these results suggested that normal up-regulation of Riok3 is required for chromatin condensation and enucleation, and that its down-regulation by ectopic expression of miR-191 is one reason why chromatin condensation and enucleation are blocked.
Mxi1 is required for erythroblast enucleation
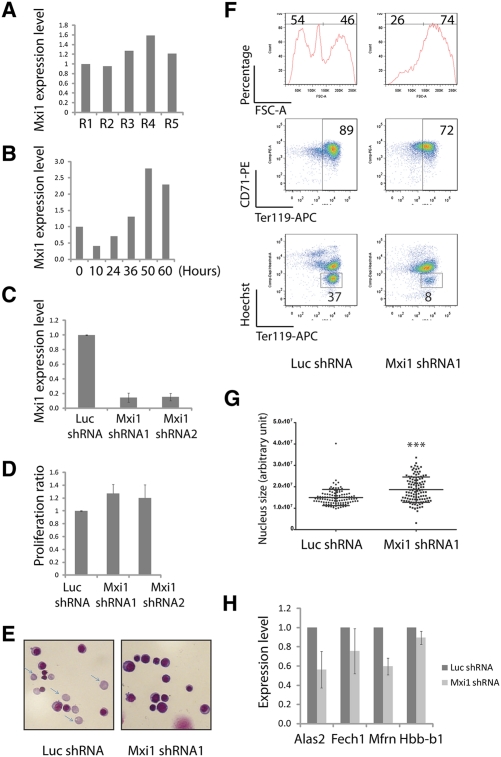
Mxi1 is a well-known c-Myc antagonist (Schreiber-Agus and DePinho 1998). Similar to Riok3, Mxi1 is enriched in the erythroid lineage (Supplemental Fig. 1B; Su et al. 2004). As shown in Figure 5A, Mxi1 is up-regulated from the R1 to the R5 stage during terminal erythroid differentiation (Fig. 5A), and this expression pattern was recapitulated in our in vitro erythroid progenitor culture system (Fig. 5B).
Figure 5.
Mxi1 is required for erythroblast enucleation. (A) Mxi1 mRNA expression levels from R1 to R5, as determined by qRT–PCR. 18s rRNA was the loading control. (B) Mxi1 expression levels of in vitro cultured erythroid progenitors, as determined by qRT–PCR. 18s was the loading control. Error bar is SD (n = 3). (C) Mxi1 mRNA levels at day 2 of culture following addition of shRNAs, determined by qRT–PCR. 18s rRNA was the loading control. Error bar is SD (n = 3). (D) Similar to Figure 2C; for samples with infection of indicated viruses, the normalized ratios of cell numbers at 48 h relative to 12 h were plotted. Error bar is SD (n = 3). (E) May-Grunwald Giemsa staining carried out on day 2 cultured cells infected with indicated viruses; representative images are shown (n = 3). Arrows indicate reticulocytes. (F) Day 2 in vitro cultured cells, as in E, stained with Ter119-APC, CD71-PE, and Hoechst. Representative FACS plots are shown (n = 3). (G) Day 2 in vitro cultured cells, as in F, stained with DAPI; nucleus sizes were quantified by image processing. (***) P < 0.001 in Student t-test. (H) qRT–PCR performed on day 2 cultured cells infected with indicated viruses. Error bar is SD (n = 3).
To uncover Mxi1's function in erythropoiesis, shRNAs targeting Mxi1 and a control Luc shRNA were introduced by retroviral infection into mouse E14.5 fetal liver erythroid progenitors, followed by 2 d in vitro culture. Two different and nonoverlapping Mxi1 shRNAs, but not a control Luc shRNA, blocked Mxi1 expression (Fig. 5C). As shown in Figure 5F, knockdown of Mxi1 almost completely blocked enucleation. This was further supported by May-Grunwald Giemsa staining, which showed that almost no enucleated cells could be detected (Fig. 5E). As shown in Figure 5F, Mxi1 shRNA infection did not influence the induction of Ter119, suggesting that knockdown of Mxi1 had no influence on differentiation. This is further supported by the qRT–PCR assay shown in Figure 5H: Knockdown of Mxi1 had no effect on Hbb-b1 (β-globin) expression and only slightly inhibited the expression of the erythroid-important genes tested, including Alas2, Fech1, and Mfrn. In addition, knockdown of Mxi1 slightly increased the rate of cell division (Fig. 5D). Importantly, knockdown of Mxi1 severely blocked chromatin condensation (Fig. 5G), and concomitantly blocked the normal decrease in cell volume (Fig. 5F). These results suggest that normal up-regulation of Mxi1 is required for chromatin condensation and enucleation.
Knockdown of Riok3 or Mix1 or overexpression of miR-191 blocked Gcn5 down-regulation
Erythroblast enucleation occurs only in terminal mammalian erythroblasts: Late erythroblasts extrude their condensed nuclei surrounded by a plasma membrane, forming enucleated reticulocytes. Several cytoskeletal components are involved in enucleation (Koury et al. 1989). Preceding enucleation is chromatin condensation, which is accompanied by histone modifications (Grigoryev et al. 2006). Additionally, the requirement for histone deacetylases (HDACs) in erythroblast chromatin condensation was suggested by the observation that treatment with a pan-HDAC inhibitor blocked erythroblast chromatin condensation and enucleation (Popova et al. 2009). Popova et al. (2009), using a Friend virus-infected erythroblast as a model system, further suggested that HDAC5 is up-regulated during the terminal differentiation stages of erythroblast differentiation, and that expression of HDAC5 is correlated with chromatin condensation. In contrast, using our primary erythroid progenitor in vitro culture system and an shRNA-based knockdown approach, we showed that HDAC2, but not HDAC5, is required for chromatin condensation and enucleation (Ji et al. 2010). Additionally, c-Myc is strongly down-regulated during terminal erythroid differentiation, and ectopic expression of c-Myc blocks the down-regulation of the histone acetyltransferase (HAT) Gcn5. Ectopic expression of either c-Myc or Gcn5 blocks chromatin condensation and enucleation (Jayapal et al. 2010). Histone deacetylation is correlated with terminal erythroid chromatin condensation, but much remains to be learned about the mechanisms and proteins used in erythroid cells to condense their nucleus.
Since Mxi1 is a well-known c-Myc antagonist (Schreiber-Agus and DePinho 1998), it is possible that the up-regulation of Mxi1 during terminal erythroid differentiation is required for the down-modulation of c-Myc activity, the down-regulation of Gcn5 expression, and proper chromatin condensation and enucleation. As shown in Supplemental Figure 3A, in our in vitro culture system, Gcn5 expression normally is down-regulated approximately fivefold at day 2 compared with day 0. Importantly, we found that knockdown of either Mxi1 or Riok3 blocked Gcn5 down-regulation, as did overexpression of miR-191 (Supplemental Fig. 3B). At day 2 of our in vitro cultures, the Gcn5 expression level in either Mxi1 or Riok3 knockdown cells is approximately twofold that of control cells, which is comparable with the reported expression level of ectopic Gcn5 (Jayapal et al. 2010) that blocks chromatin condensation and enucleation. These data suggest that blockage of Gcn5 down-regulation is at least part of the molecular explanation of how Mxi1 and Riok3 are required for normal chromatin condensation and enucleation. In addition, we found that overexpression of miR-191 also blocked Gcn5 down-regulation (Supplemental Fig. 3B), which further supports the notion that Riok3 and Mxi1 are target genes of miR-191.
According to our RNA-seq data, there are, in addition to miR-191, many other miRNAs that are also abundant in CFU-E progenitors and developmentally down-regulated during terminal erythroid differentiation. In our miRNA functional screen, we found that ectopic expression of the majority of miRNAs tested had a subtle or no effect on erythroid differentiation or enucleation. However, our functional screen may underestimate the complexity of these biological systems as well as the important roles that miRNAs play as gene expression modulators, such as buffering environment perturbations and maintaining the robustness of biological systems (Herranz and Cohen 2010). One such important lesson comes from recent research focusing on miR-451's function in erythropoiesis: miR-451 is strongly up-regulated during terminal erythroid differentiation and is dispensable for erythropoiesis under normal conditions. However, mice lacking miR-451 display a reduction in hematrocrit, an erythroid differentiation defect, and ineffective erythropoiesis in response to oxidative stress (Patrick et al. 2010; Rasmussen et al. 2010; Yu et al. 2010). Hence, it is possible that other miRNAs without obvious phenotypes in our screen may indeed be important for regulating cellular functions, such as responses to stress or other environmental perturbations not analyzed in our developmental screen.
Despite the fact that mammalian Riok3 was originally cloned as the homolog of the Aspergillus nidulans sudD gene >10 years ago (Anaya et al. 1998), the biological function of Riok3 is largely uncharacterized. Here we showed that Riok3 is up-regulated during terminal erythroid differentiation, and that knockdown of Riok3 blocked chromatin condensation and enucleation. Our data further suggest that Riok3 may be involved in regulating Gcn5 expression. Mxi1 is a well-known c-Myc activities antagonist. Our data suggested that Mxi1, c-Myc, and Gcn5 may form a cascade regulating histone deacetylation and chromatin condensation during terminal erythroid differentiation.
Altogether, our results illustrate and extend the increasing evidence that miRNAs play an important and necessary role in erythropoiesis, and how the quieting of a specific miRNA may be crucial at a very specific critical stage of erythroid development.
Materials and methods
RNA-seq
RNA-seq was carried out for high-purity CFU-Es and Ter119+ cells by using the Small RNA Sample Prep kit and genome analyzer (Illumina). RNA-seq data were deposited into the Gene Expression Omnibus (GEO) database with accession number GSE25602.
Mouse fetal liver erythroid progenitor purification, retrovirus infection, and in vitro culture
Erythroid progenitors were purified from mouse E14.5 fetal liver by depleting mature erythrocytes and differentiated nonerythroid cells. After viral infection, the erythroid cells were in vitro cultured for 2 d at 37°C.
Flow cytometry analysis and sorting
Flow cytometry analysis and sorting were carried out using a BD LSR flow cytometer and BD FACSAria cell sorter, respectively.
Detailed information of experimental procedures is provided in the Supplemental Material.
Acknowledgments
We thank Lodish laboratory colleagues Peng Ji, Karly Burke, Shilpa Hattangadi, Yutong Sun, Prakash Rao, Lei Sun, Violeta Rayon Estrada, Jing Wang, Song Chou, and Marina Bousquet for discussions and technical support, and Drs. David Bartel and Steven Cohen for discussions. We thank Patti Wisniewski and Chad Araneo for assistance with FACS sorting and analysis. L.Z. was supported by a graduate fellowship from the Singapore-Massachusetts Institute of Technology Alliance. This work was supported by SMA grant C-382-641-001-091 (to B.L. and H.F.L.), NIH grants DK047636 and AI054973 (to B.L.), and NIH grants DK047618, DK068348, and 5P01 HL066105 (to H.F.L.). L.Z. designed and performed the experiments, analyzed the data, and wrote the paper. J.F. performed the miRNA RNA-seq experiments. P.W. provided training in erythroid progenitor purification and culture and FACS analysis. B.L. and H.F.L. supervised the experimental design and wrote the paper.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1998711.
Supplemental material is available for this article.
References
- Anaya P, Evans S, Dai C, Lozano G, May G 1998. Isolation of the Aspergillus nidulans sudD gene and its human homologue. Gene 211: 323–329 [DOI] [PubMed] [Google Scholar]
- Bartel D 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen S 2007. microRNA functions. Annu Rev Cell Dev Biol 23: 175–205 [DOI] [PubMed] [Google Scholar]
- Cantor A, Orkin S 2002. Transcriptional regulation of erythropoiesis: An affair involving multiple partners. Oncogene 21: 3368–3376 [DOI] [PubMed] [Google Scholar]
- Dore L, Amigo J, Dos Santos C, Zhang Z, Gai X, Tobias J, Yu D, Klein A, Dorman C, Wu W, et al. 2008. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci 105: 3333–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M, Neilson J, Sharp P 2007. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4: 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshghi S, Vogelezang M, Hynes R, Griffith L, Lodish H 2007. α4β1 integrin and erythropoietin mediate temporally distinct steps in erythropoiesis: Integrins in red cell development. J Cell Biol 177: 871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Du T, Dong M, Zhu K, Jing C, Zhang Y, Wang L, Fan H, Chen Y, Jin Y, et al. 2009. mir-144 selectively regulates embryonic α-hemoglobin synthesis during primitive erythropoiesis. Blood 113: 1340–1349 [DOI] [PubMed] [Google Scholar]
- Grigoryev S, Bulynko Y, Popova E 2006. The end adjusts the means: Heterochromatin remodelling during terminal cell differentiation. Chromosome Res 14: 53–69 [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia N, Weissman J, Bartel D 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H, Cohen S 2010. MicroRNAs and gene regulatory networks: Managing the impact of noise in biological systems. Genes Dev 24: 1339–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayapal S, Lee K, Ji P, Kaldis P, Lim B, Lodish H 2010. Downregulation of MYC is essential for terminal erythroid maturation. J Biol Chem doi: 10.1074/jbc.M110.181073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Jayapal S, Lodish H 2008. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat Cell Biol 10: 314–321 [DOI] [PubMed] [Google Scholar]
- Ji P, Yeh V, Ramirez T, Murata-Hori M, Lodish H 2010. HDAC2 is required for chromatin condensation and subsequent enucleation of cultured mouse fetal erythroblasts. Haematologica 95: 2013–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koury S, Koury M, Bondurant M 1989. Cytoskeletal distribution and function during the maturation and enucleation of mammalian erythroblasts. J Cell Biol 109: 3005–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N, Wlodawer A 2005. The RIO kinases: An atypical protein kinase family required for ribosome biogenesis and cell cycle progression. Biochim Biophys Acta 1754: 14–24 [DOI] [PubMed] [Google Scholar]
- Lodish H, Flygare J, Chou S 2010. From stem cell to erythroblast: Regulation of red cell production at multiple levels by multiple hormones. IUBMB Life 62: 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pase L, Layton J, Kloosterman W, Carradice D, Waterhouse P, Lieschke G 2009. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood 113: 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick D, Zhang C, Tao Y, Yao H, Qi X, Schwartz R, Jun-Shen Huang L, Olson E 2010. Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3ζ. Genes Dev 24: 1614–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova E, Krauss S, Short S, Lee G, Villalobos J, Etzell J, Koury M, Ney P, Chasis J, Grigoryev S 2009. Chromatin condensation in terminally differentiating mouse erythroblasts does not involve special architectural proteins but depends on histone deacetylation. Chromosome Res 17: 47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen K, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, Horos R, Von Lindern M, Enright A, O'Carroll D 2010. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med 207: 1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T, Chohan M, Barber D 2005. Turning cells red: Signal transduction mediated by erythropoietin. Trends Cell Biol 15: 146–155 [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N, DePinho R 1998. Repression by the Mad(Mxi1)–Sin3 complex. Bioessays 20: 808–818 [DOI] [PubMed] [Google Scholar]
- Su A, Wiltshire T, Batalov S, Lapp H, Ching K, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci 101: 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, dos Santos C, Zhao G, Jiang J, Amigo J, Khandros E, Dore L, Yao Y, D'Souza J, Zhang Z, et al. 2010. miR-451 protects against erythroid oxidant stress by repressing 14-3-3ζ. Genes Dev 24: 1620–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross A, Lodish H 2003. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: Functional analysis by a flow cytometry-based novel culture system. Blood 102: 3938–3946 [DOI] [PubMed] [Google Scholar]
- Zhao G, Yu D, Weiss M 2010. MicroRNAs in erythropoiesis. Curr Opin Hematol 17: 155–162 [DOI] [PubMed] [Google Scholar]