Abstract
Cytosine DNA methylation is evolutionarily ancient, and in eukaryotes this epigenetic modification is associated with gene silencing. Proteins with SRA (SET- or RING-associated) methyl-binding domains are required for the establishment and/or maintenance of DNA methylation in both plants and mammals. The 5-methyl-cytosine (5mC)-binding specificity of several SRA domains have been characterized, and each one has a preference for DNA methylation in different sequence contexts. Here we demonstrate through mobility shift assays and calorimetric measurements that the SU(VAR)3-9 HOMOLOG 5 (SUVH5) SRA domain differs from other SRA domains in that it can bind methylated DNA in all contexts to similar extents. Crystal structures of the SUVH5 SRA domain bound to 5mC-containing DNA in either the fully or hemimethylated CG context or the methylated CHH context revealed a dual flip-out mechanism where both the 5mC and a base (5mC, C, or G, respectively) from the partner strand are simultaneously extruded from the DNA duplex and positioned within binding pockets of individual SRA domains. Our structure-based in vivo studies suggest that a functional SUVH5 SRA domain is required for both DNA methylation and accumulation of the H3K9 dimethyl modification in vivo, suggesting a role for the SRA domain in recruitment of SUVH5 to genomic loci.
Keywords: DNA methylation, dual-base flip out, epigenetics, H3K9 methylation, SET domain, 5mC-binding pocket
DNA methylation is associated with gene silencing in most eukaryotic organisms. In mammals, DNA methylation is found predominantly in the symmetric CG context (Ehrlich et al. 1982; Lister et al. 2009), while in plants DNA methylation commonly occurs in all sequence contexts: the symmetric CG and CHG contexts (where H = A, T, or C) and the asymmetric CHH context (Zhang et al. 2006; Henderson and Jacobsen 2007; Law and Jacobsen 2010). In mammals, DNA methylation is established by the DNA methyltransferase 3 (DNMT3) family of de novo methyltransferases and is maintained by the DNMT1 methyltransferase (Goll and Bestor 2005; Cheng and Blumenthal 2008; Kim et al. 2009). In plants, DNA methylation in all sequence contexts is established by DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2), a homolog of the DNMT3 family, and is maintained by largely distinct pathways that use three different DNMTs (Law and Jacobsen 2010). DNA METHYLTRANSFERASE 1 (MET1), a homolog of DNMT1, maintains CG methylation; CHROMOMETHYLASE 3 (CMT3), a plant specific methyltransferase, maintains CHG methylation; and DRM2 maintains CHH methylation through persistent de novo methylation (Henderson and Jacobsen 2007; Law and Jacobsen 2010).
Roles for proteins that contain SET- and RING-associated (SRA) domains have been demonstrated at the level of establishment and/or maintenance of DNA methylation in both plants and animals. In Arabidopsis, de novo DNA methylation requires two (SET-associated) SRA domain proteins, SU(VAR)3-9 HOMOLOG 2 (SUVH2) and SUVH9, which function in a partially redundant manner and preferentially bind methylated DNA in the CG and CHH contexts, respectively (Johnson et al. 2008). These proteins function late in the DRM2 pathway and may aid in the recruitment or retention of DRM2 to methylated loci. Another family of plant SRA domain proteins, the RING-associated VARIANT IN METHYLATION (VIM)/ORTHRUS (ORTHUS) family, is required to maintain DNA methylation predominantly in the CG context (Woo et al. 2007, 2008; Kraft et al. 2008; Feng et al. 2010), and the SRA domains of VIM1/ORTH2 and VIM3/ORTH1 have been shown to bind methylated CG sites (Johnson et al. 2007; Woo et al. 2007). The mammalian homolog of the VIM proteins, Ubiquitin-like PHD and RING finger domain (UHRF1), is also required to maintain DNA methylation in the CG context (Bostick et al. 2007; Sharif et al. 2007). The SRA domain of UHRF1 specifically binds hemimethylated CG sites (Bostick et al. 2007; Sharif et al. 2007; Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008; Qian et al. 2008) and is necessary for the association of DNMT1 at chromatin (Bostick et al. 2007; Sharif et al. 2007), leading to a model in which UHRF1 recruits DNMT1 to sites of hemimethylated DNA and thus facilitates the restoration of hemimethylated DNA to the fully methylated state. In Arabidopsis, three other SRA domain proteins—SUVH4/KRYPTONITE, SUVH5, and SUVH6—are required to maintain DNA methylation in the CHG context (Jackson et al. 2002; Malagnac et al. 2002; Ebbs et al. 2005; Ebbs and Bender 2006). The SRA domains of SUVH4 and SUVH6 have different binding preferences, with SUVH4 strongly preferring CHG methylation over both CG and CHH methylation and SUVH6 preferring both CHG and CHH methylation strongly over CG methylation (Johnson et al. 2007).
In addition to their SRA domains, SUVH4, SUVH5, and SUVH6 also contain histone methyltransferase (HMTase) domains and catalyze histone 3 Lys 9 dimethylation (H3K9me2) in vitro (Jackson et al. 2002, 2004; Ebbs and Bender 2006), and are required for H3K9me2 methylation in vivo (Johnson et al. 2002, 2007; Jackson et al. 2004; Ebbs et al. 2005; Ebbs and Bender 2006). Genome-wide, the repressive H3K9me2 and CHG methylation modifications are strongly correlated (Bernatavichute et al. 2008), and CHG methylation is thought to be maintained by a reinforcing loop of DNA methylation by CMT3 and histone methylation by SUVH4, SUVH5, and SUVH6. However, these three HMTases do not contribute equally to the overall levels of H3K9me2 and DNA methylation in vivo. SUVH4 is the predominant H3K9me2 HMTase, and mutation of this gene significantly reduces levels of DNA methylation (Jackson et al. 2002, 2004; Malagnac et al. 2002). Nonetheless, the levels of H3K9me2 and DNA methylation are further reduced in suvh4 suvh5 or suvh4 suvh6 double and suvh4 suvh5 suvh6 triple mutants at specific genomic loci (Ebbs et al. 2005; Ebbs and Bender 2006). The mechanism governing the observed locus-specific contributions of SUVH4, SUVH5, and SUVH6 are poorly understood. For SUVH4, a mutation in the SRA domain exhibited significantly reduced binding to methylated DNA in vitro, and reduced both DNA methylation and H3K9me2 levels in vivo, suggesting a role for the SRA domain in the recruitment or retention of SUVH4 to silenced loci (Johnson et al. 2007).
Despite the conservation in the SRA domain, the binding specificities of previously characterized SRA domains vary greatly. Mechanistic insight into the ability of the SRA domain to recognize methylated DNA was revealed by the crystal structure of UHRF1 bound to hemimethylated CG DNA (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008). In this structure, the UHRF1 SRA domain is described as a hand grasping the DNA duplex, with two loops, termed the thumb and NKR finger (Avvakumov et al. 2008). The thumb loop contacts the DNA through the minor groove and the NKR finger enters the DNA duplex through the major groove and provides the arginine (Arg) residue that base-pairs with the orphaned guanine base (Avvakumov et al. 2008). Consistent with the binding of UHRF1 to hemimethylated DNA, an asparagine (Asn) residue within the finger loop of the UHRF1 structure is predicted to clash sterically if a methylated cytosine base is present on the opposite strand of DNA (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008). However, there is no structural data available for the other known SRA domain proteins.
In order to better understand the function and specificity of SRA domains in general, and of SUVH5 in particular, the binding preferences of the SUVH5 SRA domain for DNA methylation in all contexts were determined, and the structure of this domain bound to methylated DNA oligomers in several sequence contexts was determined by X-ray crystallography. Our studies establish for the first time a dual flip-out mechanism on partner strands for 5-methyl-cytosine (5mC) recognition by the SUVH5 SRA domain that is independent of DNA sequence context. The structural research was complemented by studying the impact of SUVH5 SRA mutants (involved in protein–DNA recognition) on the patterns of DNA methylation and H3K9 dimethylation in vivo.
Results
Electrophoretic mobility shift assay (EMSA) studies of the 5mC-binding specificity of the SUVH5 SRA domain
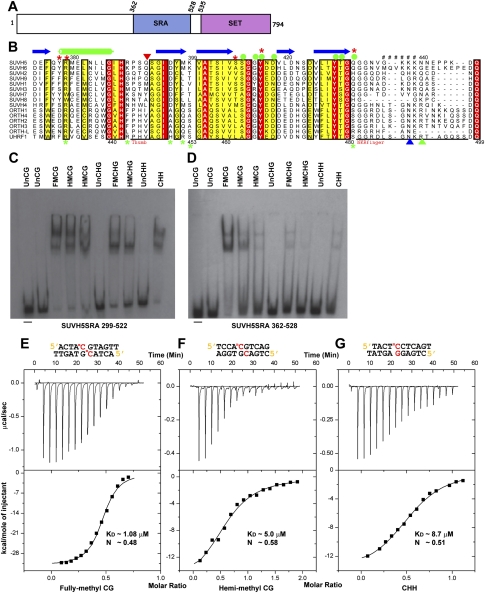
The C-terminal half of SUVH5 contains the SRA and SET domains (Fig. 1A). Sequence alignments of the SRA domains of the SUVH and ORTH proteins, as well as of UHRF1, are shown in Figure 1B and Supplemental Figure S1. Two GST fusion constructs corresponding to amino acids 299–522 and 362–528 of SUVH5 (Fig. 1A; Supplemental Fig. S1) were used to determine the in vitro specificity of its SRA domain for 5mC sites using EMSAs. DNA oligomers that contain multiple 5mC bases within a single sequence context were used to assess the overall binding preferences of these SRA proteins. Both the longer and shorter SRA proteins bound DNA oligomers containing methylation in all sequence contexts (CG, CHG, and CHH) and in both the hemimethylated and fully methylated states preferentially over the corresponding unmethylated oligomers (Fig. 1C,D). Untagged SRA proteins of both sizes that were used in the crystallographic analyses showed a similar specificity on these DNA substrates (Supplemental Fig. S2A). To further quantify the binding preference of the GST-SRA fusion proteins, binding curves were generated from EMSA experiments using the same DNA oligomers and titrations of each GST-SRA protein (Supplemental Fig. S2B–D). The larger GST-SRA protein, which corresponds to the previously characterized SRA domains of SUVH2, SUVH6, SUVH9, and UHRF1 (Bostick et al. 2007; Johnson et al. 2007, 2008), shows only minor preferences for DNA methylation in different sequence contexts (Fig. 1C; Supplemental Fig. S2). The smaller GST-SRA protein shows a preference for methylation in the CG context, but still binds methylation in other contexts over unmethylated DNA, and shows the same order of preference as the longer construct: fully or hemimethylated CG, then fully methylated CHG and methylated CHH, followed by hemimethylated CHG (Fig. 1C,D; Supplemental Fig. S2). These findings demonstrate that the SRA domain of SUVH5 exhibits a different specificity from that shown for other SRA domains (Bostick et al. 2007; Johnson et al. 2007, 2008; Woo et al. 2007). Furthermore, our finding that the SRA domain of SUVH5 binds to methylation in all contexts and has little preference for a fully over a hemimethylated state suggests that this SRA domain might either be recognizing only the methylated cytosine or be able to accommodate both a methylated and an unmethylated residue on the complementary DNA strand.
Figure 1.
Binding of the SUVH5 SRA domain to 5mC-containing DNA in different sequence contexts. (A) Schematic representation of the domain architecture of SUVH5, with SRA and SET methyltransferase domains colored in blue and purple, respectively. (B) Multiple sequence alignment of the SRA domains from the SET domain-associated SUVH family and from the ORTH1–4, ORTH-L, and UHRF1 RING domain-associated proteins in the region flanking the thumb and NKR finger. Sequence numbering of the SUVH family proteins is based on SUVH5, whereas, for the RING domain-associated proteins, it is based on UHRF1. The secondary structural elements of the SUVH5 SRA are indicated above the sequence (α helices are in green cylinders, β-strands are in blue arrows, and disordered regions are represented by ##). Residues highlighted in a background color code correspond to conservation levels: fully conserved in red, and conservative substitutions in yellow. The thumb and NKR finger corresponding to the UHRF1 are underlined in black at the bottom. The residue that inserts into the duplex and displaces the 5mC residue in the SUVH5 SRA complex is indicated as an inverted red triangle. Green and blue upright triangles correspond to residues that replace the looped-out 5mC and mask the unmodified C in the UHRF1 SRA complex, respectively. Filled green circles designate residues that interact with the 5mC in the binding pocket of the SUVH5 SRA and UHRF1 SRA complexes. Red and green stars designate DNA backbone-interacting residues in the SUVH5 SRA and UHRF1 SRA complexes, respectively. (C) EMSAs using a GST-299–522 SUVH5 SRA domain fusion protein. The context and methylation state of each radiolabeled dsDNA oligonucleotide is indicated above. (Un) Unmethylated; (FM) fully methylated; (HM) hemimethylated; (−) no protein. (D) EMSA experiments using a GST-362–528 SUVH5 SRA domain fusion protein. In C and D, the first and second HMCG oligonucleotides harbor methylated cytosines on the sense and antisense DNA strand, respectively. (E) ITC measurements of the binding of the SUVH5 SRA domain to fully methylated CG DNA (diagram above). Experimental details are provided in the Materials and Methods. The measured binding parameters are KD = 1.08 μM and N = 0.48. (F) ITC measurements of the binding of the SUVH5 SRA domain to hemimethylated CG DNA. The measured binding parameters are KD = 5.0 μM and N = 0.58. (G) ITC measurements of the binding of the SUVH5 SRA domain to methylated CHH DNA. The measured binding parameters are KD = 8.7 μM and N = 0.51.
Calorimetric studies of the 5mC-binding specificity of the SUVH5 SRA domain
Although the substrate specificity of the SRA domains of several SUVH proteins have been reported (Johnson et al. 2007, 2008), no data on the affinity or stoichiometry of binding are available. Here, we used the isothermal titration calorimetric (ITC) approach to investigate these parameters for SUVH5 using the shorter SRA construct (362–528) and DNA oligomers containing methylation in the fully and hemimethylated CG sequence contexts, the methylated CHH context, and the fully methylated CHG context. The SUVH5 SRA domain binds to fully methylated CG with a dissociation constant (KD) of 1.08 μM (Fig. 1E). Consistent with the EMSA data using this SRA construct, the binding constant decreases by a factor of 4.6 (KD = 5.0 μM) (Fig. 1F) for hemimethylated CG, a factor of 8.1 (KD = 8.7 μM) (Fig. 1G) for methylated CHH, and a factor of 6.0 (KD = 6.6 μM) (Supplemental Fig. S3) for fully methylated CHG sequence contexts.
Together, these binding studies demonstrate that the SUVH5 SRA domain recognizes 5mC in all sequence contexts. Notably, the SRA domain of SUVH5 exhibits a 4.6-fold preference for fully methylated over hemimethylated CG sequences, which is in sharp contrast to the preference of the UHRF1 SRA domain, which exhibits the reverse preference; namely, a sevenfold preference for hemimethylated over fully methylated CG sequences (Bostick et al. 2007). Additionally, in the case of the SUVH5 SRA domain, the stoichiometry of binding is ∼0.5, which reflects the ratio of 0.5 duplex bound per SRA or, equivalently, two SRA domains bound per DNA duplex.
Structure of the SUVH5 SRA domain bound to fully methylated CG DNA
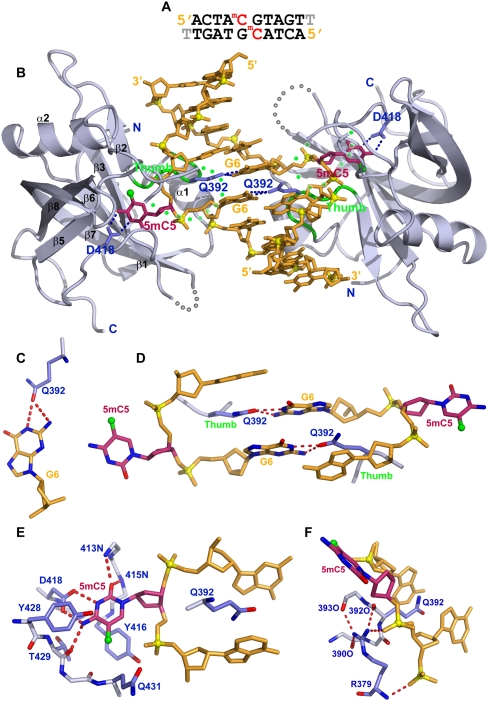
We grew diffraction-quality crystals and solved the structure of the SUVH5 SRA domain bound to a self-complementary 10-base-pair (bp) duplex (with a 3′ thymine overhang) containing a central fully methylated CG step (Fig. 2A). The crystals of this complex (space group P42212) diffracted to 2.20 Å resolution, and the structure was solved using phases derived from multiwavelength anomalous diffraction data collected from a crystal of the complex containing seleno–methionine-labeled protein. The asymmetric unit is composed of one SRA domain bound to one strand of the fully methylated DNA (X-ray statistics listed in Supplemental Table 1). A second structure of this complex was solved in the space group P6122 at 2.65 Å resolution using the molecular replacement method (Supplemental Table 1). The present study focuses on the higher-resolution 2.20 Å structure.
Figure 2.
Crystal structure of the SUVH5 SRA domain bound to fully methylated CG DNA. (A) Sequence of the self-complementary fully methylated CG 10-mer (with a 3′-T overhang) containing 5mC-G steps on partner strands in the center of the duplex. (B) Stick (DNA) and ribbon (protein) representation of the 2.2 Å crystal structure of the 2:1 SUVH5 SRA–fully methylated CG DNA duplex complex. The DNA is colored in orange, except for 5mC5, which is colored in purple. The 5-methyl group is shown as a small green sphere. Backbone phosphorus atoms are shown as yellow balls, and the 5′ and 3′ ends of the DNA are labeled. The SRA domain is colored in blue, with its secondary structural elements labeled with the same α/β numbering scheme as for the SRA domain of UHRF1 (Avvakumov et al. 2008). The thumb loop and disordered NKR loop segments are colored in green and dotted green, respectively. The 5mC5 residues on partner strands flip out through the minor groove and are positioned in binding pockets on individual SRA domains. The Watson-Crick edge of 5mC5 is hydrogen-bonded with the side chain of Asp418. The side chain of Gln392 inserts into and fills the gap created by the flipped-out base and pairs with the Watson-Crick edge of the G6 base. (C) Hydrogen bonding between the Watson-Crick edge of G6 and the carbonyl group of the Gln392 side chain. (D) Relative alignments of stacked G6–Gln392 interactions in the structure of the complex. Note that the side chain of Gln392 is sandwiched between bases. (E) Interaction of the flipped-out 5mC5 with residues lining the binding pocket. The 5mC5 base is positioned between the aromatic rings of Tyr416 and Tyr428, with its Watson-Crick edge hydrogen-bonded to the protein backbone and side chain (Asp418) residues. (F) The side chain of Arg379 forms a network of intramolecular hydrogen bonds with the backbone of the thumb segment and intermolecular hydrogen bonds with the phosphate backbone that buttresses the fold of the binding pocket.
The structure of the complex shown in Figure 2B contains two SRA molecules bound per DNA duplex, with the 5mC residues on adjacent base pairs flipped out of the DNA helix and positioned in the binding pockets of two individual SRA domains. There is a crystallographic twofold axis perpendicular to the DNA helical axis, and the two SRA molecules form no contacts with each other in the complex. The observed stoichiometry of two SRA molecules bound per duplex is consistent with an N-value of 0.5 (DNA duplex:SRA domain ratio) observed in the ITC binding curve for complex formation of the SUVH5 SRA domain with fully methylated DNA (Fig. 1E). These findings support a model in which the stoichiometry represents the binding of two SRA domains to individual 5mC residues rather than the dimerization of the two SRA domains with each other.
The flipping out of the 5mC residues in the fully methylated CG crystal structure introduces a gap in the DNA duplex that is filled by the side chain of Gln392, which resides within the thumb loop (residues 390–395) of the SUVH5 SRA domain. Gln392 is inserted through the minor groove and is intercalated between flanking bases, forming intermolecular hydrogen bonds with the Watson-Crick edge of the orphan guanine G6 (Fig. 2C). The arrangement of the two G6–Gln392 pairs within the complex, which occur on both DNA strands, are shown in Figure 2D. The NKR finger loop (residues 433–444), which plays a critical role in the UHRF1 SRA–hemimethylated DNA complex (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008), is disordered in the SUVH5 SRA–fully methylated DNA complex. Despite the presence of two flipped-out 5mC residues on partner strands, the remaining bases in the DNA duplex are undistorted and essentially retain the regular B-form conformation.
The flipped out 5mC is positioned in a pocket within the SRA domain such that it is anchored in place via stacking interactions with Tyr416 and Tyr428 and by intermolecular hydrogen bonds between its Watson-Crick edge and the side chain of Asp418, the backbone amide nitrogens, and a carbonyl group (Fig. 2E). The 5mC adopts an anti alignment around the glycosidic bond (χ = −119°), and there is sufficient room to accommodate the methyl group at the cytosine 5 position, which is stabilized by van der Waals contacts, most prominently with the Cα and Cβ atoms of Gln431 (Fig. 2E). It should be noted that the N3 position of looped-out 5mC must be protonated in order to pair with the carboxylate group of Asp418 (Fig. 2E), which in turn would imply an unusual ionization constant (pKa) for the looped-out 5mC. The same alignment of the Asp residue and the 5mC, characteristic of an unusual pKa for the looped-out 5mC, was also observed in the published structures of the UHRF1 SRA–DNA complexes (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008).
Finally, Arg379 plays an important role in buttressing the thumb loop, which provides the Gln392 residue that inserts into the DNA helix, as well as in anchoring the DNA phosphodiester backbone flanking the flipped-out 5mC. These functions are mediated through the intra- and intermolecular hydrogen bond interactions depicted in Figure 2F.
The intermolecular contacts in the SUVH5 SRA complex are summarized in Supplemental Figure S4A. In essence, the key amino acids in the SUVH5 SRA domain that contribute to the molecular recognition of the flipped-out 5mC and stabilization of the binding pocket are Gln392, Asp418, Tyr416, Tyr428, and Arg379. Hence, these residues constitute appropriate candidates for mutational studies.
Structure of the SUVH5 SRA domain bound to hemimethylated CG DNA
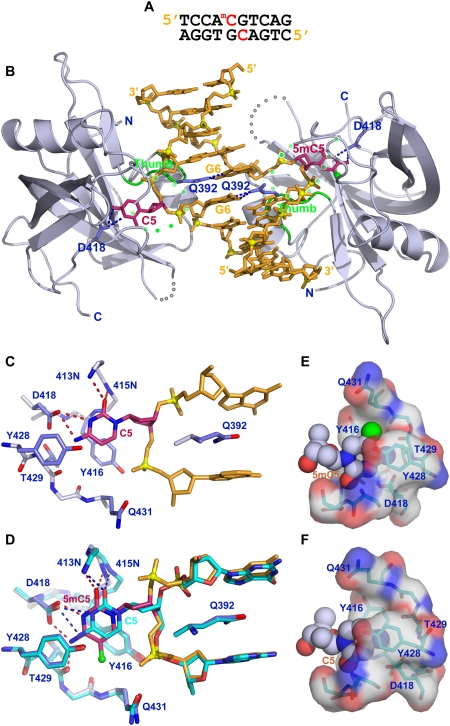
We next solved the crystal structure of the SUVH5 SRA domain bound to a complementary 10-bp duplex containing a central hemimethylated CG step (Fig. 3A). The crystal belongs to the P42 space group, diffracts to 2.37 Å resolution, and contains two SRA molecules and one DNA duplex in the asymmetric unit (crystallographic statistics listed in Supplemental Table 1).
Figure 3.
Crystal structure of the SUVH5 SRA domain bound to hemimethylated CG DNA. (A) Sequence of the complementary hemimethylated CG 10-mer containing a 5mC-G step on the top strand and an unmodified C-G step on the bottom strand in the center of the duplex. (B) Stick (DNA) and ribbon (protein) representation of the 2.37 Å crystal structure of the 2:1 SUVH5 SRA–hemimethylated CG DNA duplex complex. The color coding is the same as in Figure 2B. Note that the 5mC5 and the C5 from adjacent base pairs on partner strands flip out through the minor groove and are positioned in binding pockets of individual SRA domains. (C) Interaction of the looped-out C5 with residues lining the binding pocket in the structure of the complex. (D) Superposition of the flipped-out C5 (in green) and the flipped-out 5mC5 (in purple) within their respective binding pockets in the structure of the complex. (E) Insertion of anti 5mC5 (space-filling representation) into the binding pocket of the SRA domain (electrostatic surface presentation). (F) Insertion of anti C5 (space-filling representation) into the binding pocket of the SRA domain (electrostatic surface presentation).
The structure of the complex is shown in Figure 3B, and it contains two SUVH5 SRA domains bound per DNA duplex, consistent with the stoichiometry elucidated from ITC binding data (Fig. 1F). Unexpectedly, both the 5mC and the unmodified C from adjacent pairs on partner strands simultaneously flip out of the duplex and are positioned within binding pockets of individual SRA domains.
The alignment and intermolecular contacts of the flipped-out unmodified C within its binding pocket are shown in Figure 3C, and they compare favorably with that of the flipped-out 5mC, as can be seen in the superposition of the flipped-out C and 5mC residues (Fig. 3D). Alternate representations showing the flipped-out base in a space-filling representation and the binding pocket in a surface representation are shown accommodating the flipped-out 5mC and C residues in Figure 3, E and F, respectively. Importantly, both the flipped-out C (χ = −118°) and 5mC (χ = −112°) residues adopt anti conformations about their glycosidic bonds (Fig. 3E,F).
Structure of the SUVH5 SRA domain bound to methylated CHH DNA
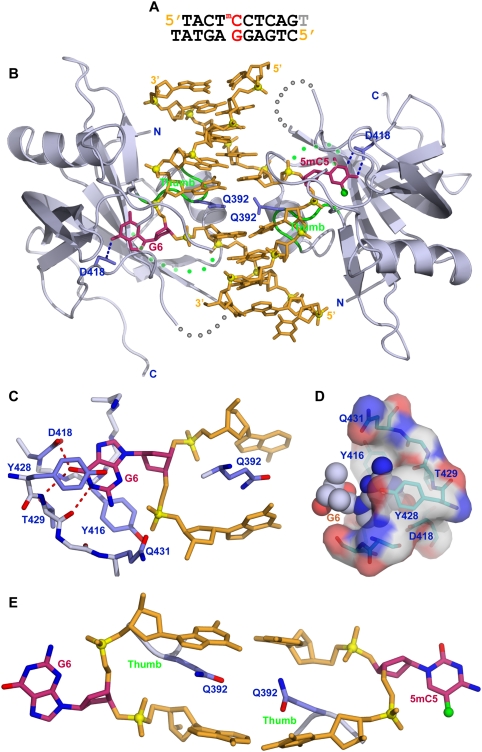
We also solved the crystal structure of the SUVH5 SRA domain bound to a complementary 10-bp duplex with a 3′ thymine overhang that contains a centrally positioned methylated CHH (where H = C, T, or A) step (Fig. 4A). The crystals belong to the P6122 space group, diffract to 2.75 Å resolution, and contain two SRA molecules and one DNA duplex in the asymmetric unit (crystallographic statistics listed in Supplemental Table 1).
Figure 4.
Crystal structure of the SUVH5 SRA domain bound to methylated CHH DNA. (A) Sequence of the complementary methylated CHH 10-mer (with a 3′-T overhang) containing a 5′-5mCCT step on the top strand and a 5′-AGG step opposite it on the bottom strand toward the center of the duplex. In essence, a 5mC is positioned opposite a G. (B) Stick (DNA) and ribbon (protein) representation of the 2.75 Å crystal structure of the 2:1 SUVH5 SRA–methylated CHH DNA duplex complex. The color coding is the same as in Figure 2B. Note that the 5mC5 and the G6 from the same base pair simultaneously flip out of the DNA duplex through the minor groove and are positioned in binding pockets of individual SRA domains. (C) Interaction of the flipped-out G6 in a syn conformation with residues lining the binding pocket in the structure of the complex. (D) Insertion of the syn G6 (space-filling representation) into the binding pocket of the SRA domain (electrostatic surface presentation). (E) Relative alignments of the inserted Gln392 residues that are positioned opposite each other in the structure of the complex. Note that the side chain of the Gln392 residue is sandwiched between two bases.
The structure of the complex is shown in Figure 4B and contains two SUVH5 SRA domains bound per DNA duplex, consistent with the stoichiometry elucidated from ITC binding data (Fig. 1G). To our surprise, both the 5mC and the G positioned opposite it on the complementary strand are flipped out of the DNA duplex and are positioned in binding pockets within individual SRA domains. The flipped-out G adopts a syn conformation (χ = 30°) about its glycosidic bond (Fig. 4C,D), allowing the flipped-out G to form the same distribution of intermolecular hydrogen bonds (Fig. 4C) as are formed by the flipped-out anti 5mC (Fig. 2E). Since the flipped-out bases originate from the same base pair in the methylated CHH complex, the inserted Gln392 side chains are roughly in the same plane and are sandwiched between flanking base pairs (Fig. 4E). The stark difference in the arrangement of the two Gln392 residues in this complex compared with that observed in the fully methylated CG complex, where the flipped-out bases on partner strands originate from flanking base pairs, can be appreciated by comparing Figures 2D and 4E.
Overall, the structures reveal that the SUVH5 SRA complexes with fully methylated CG, hemimethylated CG, and methylated CHH DNAs all involve the flipping out of both the 5mC and a base—either a 5mC, a C, or a G, respectively—on the partner strand. The intermolecular contacts in the above three SUVH5 SRA–DNA complexes are summarized in Supplemental Figure S4, A–C, respectively. One notable difference between the structures is that the relative orientations of the two SRA domains are altered in the different complexes such that, in the fully methylated CG DNA and hemimethylated CG DNA complexes the two Gln392 residues lie in adjacent planes, while in the methylated CHH DNA complex the Gln392 residues lie in the same plane (Supplemental Fig. S4D,E).
Effects of SRA domain mutations on methyl-DNA binding in vitro
To test the importance of specific residues within the SUVH5 SRA domain for recognition of the flipped-out base, we engineered point mutations within the SUVH5 SRA domain that we predicated might be important based on the crystallographic structures of the complexes. These mutant SRA domain proteins were assessed for their ability to bind a fully methylated CG site by ITC experiments in vitro (Supplemental Fig. S5). Mutation of Gln392 (the residue that base-pairs with the orphaned guanine by inserting into the duplex and taking the place of the flipped-out base) to an alanine resulted in what appears to be a complete loss of binding affinity (Supplemental Fig. S5D). This finding strongly supports the crystallographic finding that the thumb loop rather than the NKR finger facilitates the base-flipping mechanism in the SUVH5 SRA domain. Mutation of both Tyr residues (Tyr416Ala/Tyr428Ala) that sandwich the flipped-out base also abrogates binding (Supplemental Fig. S5E). However, a single Tyr416Ala mutation only reduced the binding affinity (Supplemental Fig. S5C). Thus, both Tyr residues are important for positioning the 5mC in the binding pocket. The Asp418Ala mutant, which is predicted to disrupt the hydrogen bonds between the Asp side chain and the Watson-Crick edge of the flipped-out 5mC in the structure of the complex (Fig. 2E), exhibits a calculated KD = 21 μM (Supplemental Fig. S5A), reflecting a 20-fold reduction in binding affinity compared with wild type (Fig. 1E). This structure–function analysis supports the underlying structure-based intermolecular interactions observed in the crystal structure of the complex, where a combination of specific hydrogen bonding, stacking, and hydrophobic interactions contribute to the anchoring of the flipped-out 5mC within its binding pocket in the SUVH5 SRA domain.
Impact of guanine to inosine substitutions and the presence of abasic sites on binding to methylated DNA
To further investigate the interactions required for the binding of the SUVH5 SRA domain to methylated DNA, specifically those involving the DNA surrounding the flipped-out 5mC, we carried out in vitro binding assays using modified DNA duplexes. The guanine base present opposite the 5mC forms hydrogen bonds with the Gln392 residue of the SUVH5 SRA domain in both the fully and hemimethylated CG complexes, and mutation of Gln392 within SUVH5 ablates DNA binding, suggesting this interaction may be important. However, such an interaction is not observed in the complex containing methylation in the CHH context, suggesting it may not be required.
Thus, to investigate the relative importance of these guanine residues, binding assays were conducted using substrates in which the guanine bases were replaced with inosine bases. Replacement of the guanine bases with inosine bases in the fully methylated and hemimethylated CG duplexes had no impact on either the KD or N-values following complex formation with the SUVH5 SRA domain (Supplemental Fig. S5F,G). Thus, given that the GC and IC pairs contain three and two hydrogen bonds, respectively, which are disrupted upon complex formation, this finding demonstrates that reducing the capacity of Gln392 to hydrogen-bond with the Watson-Crick edge of guanine (Fig. 2E) does not have an impact on the binding affinity.
To investigate whether a dual base-flipping mechanism is required for binding to DNA methylation in the asymmetric CHH context, we replaced the guanine opposite the 5mC with an abasic site (designated dS-mCHH) and monitored binding by the SUVH5 SRA domain by ITC measurements. The observed KD of 2.7 μM and an N-value of 1.1 (Supplemental Fig. S5H) imply that the SRA domain still interacts with this type of substrate, and that flipping out of the base opposing the 5mC is not required for complex formation. Equally important, the N-value of 1.1 implies that a single SRA domain is bound to the dS-mCHH DNA. These findings further support the hypothesis that the SUVH5 SRA domain relies on the 5mC in the absence of complementary guanine.
Effects of SRA domain mutations on DNA methylation in vivo
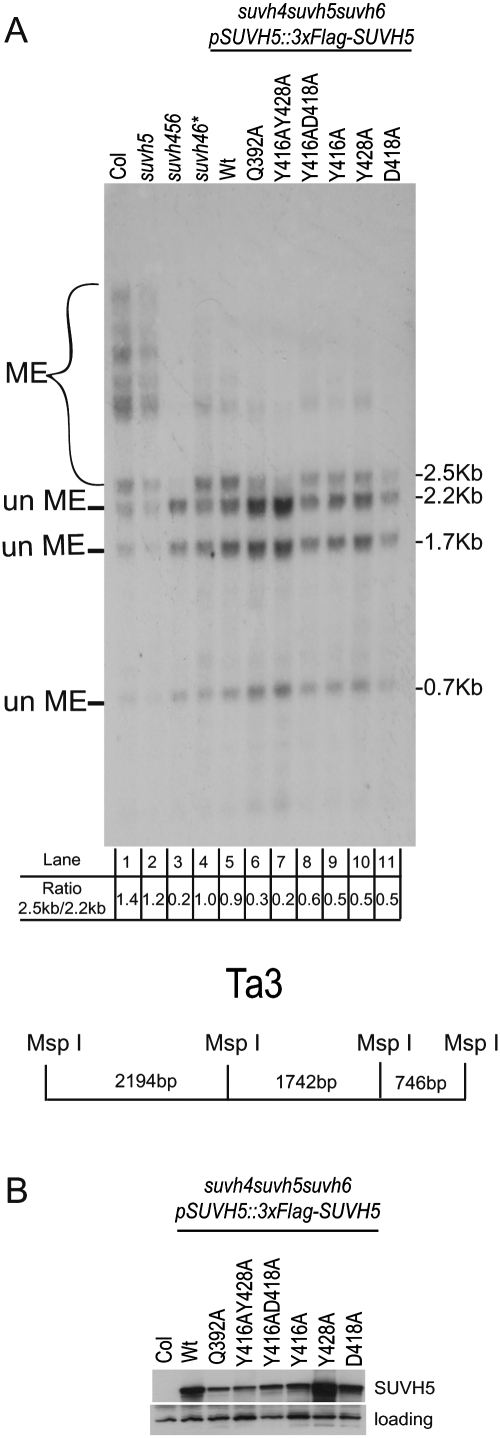
To determine the in vivo significance of key residues within the 5mC-binding pocket, the ability of an epitope-tagged SUVH5 transgene (pSUVH5∷3xFlag-SUVH5) carrying specific point mutations to restore methylation at the Ta3 locus was assessed by Southern blotting following digestion of genomic DNA with the methylation-sensitive restriction enzyme MspI. DNA methylation at Ta3 is redundantly controlled by SUVH4, SUVH5, and SUVH6 such that, in a suvh4 suvh5 suvh6 triple mutant, this locus is almost completely unmethylated (Ebbs and Bender 2006), resulting in the presence of three main bands (2.2 kb, 1.7 kb, and 0.7 kb in size) after Southern blotting (Fig. 5A, lane 3). In a suvh5 single mutant, methylation at this locus is indistinguishable from that of wild-type plants (Fig. 5A, cf. lanes 1 and 2; Ebbs and Bender 2006). However, in the sensitized suvh4 suvh5 suvh6 triple-mutant background, the addition of a functional SUVH5 wild-type transgene results in an increase in DNA methylation, which is visualized as the accumulation of a 2.5-kb band to a similar intensity as observed for the 2.2-kb and 1.7-kb bands (Fig. 5A, lane 5). This pattern is comparable with that observed when mutations in SUVH4 and SUVH6 are present (Fig. 5A, lane 4).
Figure 5.
In vivo DNA methylation analysis. (A) Southern blot showing the methylation status of the Ta3 locus. Genomic DNA was extracted from the indicated Arabidopsis genotypes and digested with the methylation-sensitive MspI restriction enzyme. pSUVH5∷3xFlag-SUVH5 indicates the epitope-tagged SUVH5 transgene, driven by its endogenous promoter, encoding either the wild-type SUVH5 protein (wild type [Wt]) or the indicated mutant protein; the asterisk (*) indicates suvh2 and suvh9 mutations are also present in this background. Previous studies have shown these mutations do not affect methylation at Ta3. (un Me) DNA fragments generated by cleavage of unmethylated DNA by MspI; (Me) DNA fragment generated when DNA methylation blocks cleavage by MspI. The ratio of the 2.5-kb band intensity to the 2.2-kb band intensity is shown below each lane number and was used to score the complementation level of each SUVH5 transgene (either the wild-type genomic sequence [lane 5] or the indicated point mutants [lanes 6–11]). A schematic diagram of the Ta3 locus showing the MspI restriction sites is shown below. (B) Western blot using an antibody against the Flag epitope showing the expression of the wild-type and mutant versions of the SUVH5 protein from the same T1 plants characterized in A. Protein extracted from the nontransgenic Colombia (Col) ecotype was used as a negative control. An unknown protein that cross-reacts with the Flag antibody and serves as an internal loading control is shown in the bottom panel.
Mutant versions of SUVH5 were tested for their ability to complement the suvh4 suvh5 suvh6 mutant phenotype, and the expression of each mutant protein in vivo was assessed by Western blotting using an antibody against the Flag epitope (Fig. 5B). A SUVH5 transgene encoding a protein with mutations in both of the tyrosine residues that form stacking interactions with the flipped-out 5mC (namely, Tyr416/Tyr428 in the crystal structure) to Ala failed to complement the methylation defect at Ta3 (Fig. 5A, lane 7), and a transgene encoding a mutation in the glutamine that base-pairs with the orphaned guanine base, Gln392, also significantly reduced the level of complementation (Fig. 5A, lane 6). Mutation of either stacking tyrosine alone (Tyr416 or Tyr428); of Asp418, another residue that forms interactions within the methyl-cytosine-binding pocket; or of both Tyr416 and Asp418 was able to partially restore methylation at Ta3. These data confirm that residues in the thumb loop of the SUVH5 SRA domain are important for the in vivo function of SUVH5, and further support the crystallographic data showing that the residue that replaces the flipped-out 5mC and base-pairs with the orphaned G in SUVH5 differs from that observed in UHRF1, where this role is filled by an arginine that resides in the NKR finger loop (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008).
Effects of SRA domain mutations on H3K9 dimethylation in vivo
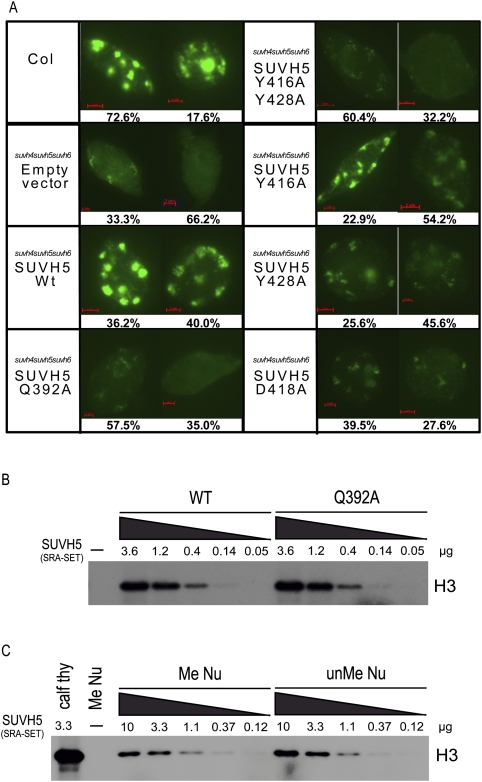
To determine the effect of mutations within the SRA domain of SUVH5 on H3K9 dimethylation levels in vivo, immunofluorescence experiments were conducted using nuclei isolated from the progeny of the transgenic SUVH5 lines characterized in Figure 5. Similar expression levels of the various SUVH5 transgenes in this generation were confirmed by Western blotting (Supplemental Fig. S6A). As expected based on previous immunofluorescence experiments in Arabidopsis (Johnson et al. 2008), nuclei isolated from wild-type (Col ecotype) plants showed an intense H3K9 dimethylation signal at chromocenters, and nuclei isolated from a suvh4 suvh5 suvh6 triple mutant transformed with an empty vector as a negative control showed a weak and largely diffuse signal (Fig. 6A; Supplemental Fig. S6). A strong H3K9 dimethylation signal at chromocenters was restored in nuclei isolated from suvh4 suvh5 suvh6 mutant plants transformed with a wild-type copy of SUVH5, demonstrating that this transgene is able to complement both the DNA methylation and the histone methylation defects (Fig. 6A). Consistent with the DNA methylation analysis at the Ta3 locus, suvh4 suvh5 suvh6 mutants transformed with a SUVH5 transgene carrying alanine mutations at either Gln392 or both Tyr416 and Tyr428 had the strongest effect on global levels of H3K9 dimethylation (Fig. 6A). Mutation of either tyrosine alone or of Asp418 had less severe effects on H3K9 levels, with a portion of the nuclei exhibiting a strong to moderate H3K9 dimethylation signal at chromocenters (Fig. 6A). Together, these finding suggest that, like what has been observed for SUVH4 (Johnson et al. 2007), a functional SRA domain is important for the activity of SUVH5 at the level of either recruitment and/or retention at genomic loci or HMTase activation.
Figure 6.
Histone methylation analysis. (A, left) Immunofluorescence detection of H3K9 dimethylation in nuclei isolated from the indicated genotype. Images representing the two most predominant classes of nuclei are shown, and percentages out of 200 nuclei are indicated below. See Supplemental Figure 6 for a full breakdown of classes and percentages for each mutant. (B) HMTase assays using a wild-type or a Q392A mutant SUVH5 SRA-SET protein (amino acids 362–794) and calf thymus histones as a substrate. 3H-radiolabeled SAM was supplied as the methyl donor. The concentration of SUVH5 protein used for each assay is indicated in micrograms, and the position of histone 3 (H3) is indicated (at the right). (C) HMTase assays using a wild-type SUVH5 SRA-SET protein (amino acids 362–794) and either calf thymus histones (Calf thy) or mononucleosomes assembled using methylated (Me Nu) or unmethylated (unMe Nu) DNA. The concentration of SUVH5 protein used for each assay is indicated in micrograms, and the position of histone 3 (H3) is indicated (at the right).
In order to differentiate between potential roles for the SUVH5 SRA domain in targeting verses stimulation of its H3K9 HMTase activity, the in vitro HMTase activity of SUVH5 proteins containing both the SRA and the SET HMTase domains was assessed. To confirm that a functional SRA domain is not required for the HMTase activity of SUVH5, and thus is unlikely to account for the decreased level of H3K9 methylation observed in vivo, the HMTase activity of a wild-type or Gln392Ala mutant SUVH5 SRA-SET protein was assessed using calf thymus histones as a substrate (Fig. 6B). To determine whether the binding of the SRA domain to methylated DNA has a stimulatory effect on the HMTase activity of SUVH5, the HMTase activity of wild-type SUVH5 was assessed using mononucleosomes assembled with CG-methylated or unmethylated DNA. If the presence of DNA methylation results in an increase in the level of HMTase activity, that may suggest that the binding of the SRA domain to methylated DNA causes conformational changes in the protein that allows more efficient HMTase activity. Alternatively, if the presence of DNA methylation does not have an effect on the level of HMTase activity, that may suggest that the function of the SUVH5 SRA domain is to recruit or retain SUVH5 at specific genomic loci and thereby allow deposition of the H3K9 dimethylation mark in a loci-specific manner. Under the conditions used, similar levels of HMTase activity were observed on methylated and unmethylated nucleosomes (Fig. 6C). Nucleosomes that contain a lysine-to-alanine mutation at position 9 of the H3 tail (H3K9A) were used as a control to confirm the H3K9 specificity of SUVH5 in vitro (Supplemental Fig. S6C). These findings support the hypothesis that the SRA domain of SUVH5 is important for the recruitment of SUVH5, rather than activation of its HMTase activity.
Discussion
SUVH5 is unusual among the SUVH family members in that its SRA domain efficiently binds methylated DNA in the fully and hemimethylated CG contexts (Fig. 1E,F, respectively), as well as in the methylated CHH and fully methylated CHG contexts (Fig. 1G; Supplemental Fig. S3, respectively). In an attempt to understand the principles underlying this broad specificity, we initiated a structure–function study aimed at solving the crystal structures of the SUVH5 SRA in a complex with methylated DNA in all of the above sequence contexts. We were successful in growing crystals and solving the structures of the first three complexes, but were unable to grow diffraction-quality crystals of SUVH5 SRA bound to fully methylated CHG DNA. These structures identified the intermolecular protein–DNA contacts within the complexes, allowing us to monitor the impact of specific SUVH5 SRA mutants on DNA methylation and H3K9 dimethylation in vivo.
Dual flipping out of 5mC/C positioned on adjacent pairs on partner strands
Base flipping was first identified in the structures of the bacterial M.HhaI (Klimasauskas et al. 1994) and HaeIII (Reinisch et al. 1995) methyltransferases bound to CG-containing DNA, and since then has been identified as a conserved mechanism that is widely used by DNA- and RNA-modifying and repair enzymes (Huffman et al. 2005). A central feature of these complexes involves the flipping out of a single base, which is then inserted into an active site pocket within the bound protein.
We anticipated that the structure of the SUVH5 SRA bound to fully methylated CG DNA (Fig. 2A) could involve flipping out of either one or both 5mC residues of the adjacent base pairs, depending on the number of SRA domains bound per DNA duplex. Both the ITC measurements (Fig. 1E) and the structure of the complex (Fig. 2B) established a stoichiometry of two SRA domains bound per fully methylated CG DNA duplex, with both 5mC residues flipping out and being positioned within their individual SRA-binding pockets.
Unexpectedly, ITC measurements established a stoichiometry of two SRA domains bound per hemimethylated CG DNA duplex (Figs. 1F, 3A), despite the presence of only a single 5mC base. The crystallographic analysis of this complex revealed that both the 5mC and the C from adjacent base pairs on the partner strands are looped out and positioned within individual SRA-binding pockets (Fig. 3B), consistent with the stoichiometry inferred from the ITC measurements. This contrasts strikingly with earlier structural studies of the UHRF1 SRA domain (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008), where UHRF1 was found to bind with a stoichiometry of one SRA domain bound per hemimethylated CG DNA duplex and only the 5mC was flipped out and positioned in the SRA-binding pocket, while the C on the adjacent base pair remained stacked within the duplex.
To our knowledge, the structures of the fully and hemimethylated complexes in Figures 2B and 3B, respectively, represent the first examples of a dual-base flipping from partner strands, thereby providing a novel mechanism for scanning both strands of the duplex simultaneously.
The versatility of SRA domains in binding DNA as either a single molecule (UHRF1 complexes) or two molecules (SUVH5 SRA complexes) is reminiscent of POU domains, which flex to fit the DNA duplex such that the two-part DNA-binding domain (POUH and POUS) partially encircles the DNA, without base flipping, and the subdomains adopt a DNA element-dependent range of conformations (Phillips and Luisi 2000).
Dual flipping out of the 5mC/G bases positioned opposite each other on partner strands
It was unclear what to expect as far as stoichiometry and the number of flipped-out residues for the SUVH5 SRA domain bound to the asymmetrically methylated CHH DNA duplex (Fig. 4A). However, the ITC measurements once again established that the stoichiometry was two SRA domains bound per duplex (Fig. 1G), as was also observed in the structure of the complex (Fig. 4B). In this structure, both the 5mC and the G from the same pair are flipping out of the DNA duplex and are positioned within individual SRA-binding pockets (Fig. 4B). Thus, the dual base-flipping mechanism exhibited by the SUVH5 SRA domain can involve bases located on either partner DNA strands that are positioned on adjacent pairs (fully and hemimethylated CG DNA) or within the same base pair (methylated CHH DNA).
Packing of the SUVH5 SRA domain complexes in the crystal lattice
We questioned whether some aspect of crystal packing could contribute to the dual-base flipping observed in all three SUVH5 SRA–DNA complexes. We observed the same crystal packing in all three complexes; namely, that the SRA domains from adjacent complexes (each of which contains two SUVH5 SRA domains bound per DNA duplex) in the crystal lattice pack against each other, as is shown for the complex of the SUVH5 SRA domain bound to fully methylated CG DNA (Supplemental Fig. S7A). There is an extensive interface (641 Å2 per monomer) involving hydrogen-bonding, hydrophobic, and salt bridge interactions formed between the interacting SRA domains (Supplemental Fig. S7B). It should be noted that the packing interaction occurs on the opposite SRA face (Supplemental Fig. S7A) from that which interacts with the DNA. It is unlikely that crystal packing alone contributes to the observed 2:1 (SUVH5 SRA:DNA duplex) stoichiometry, since the same stoichiometry is also observed from ITC studies (Fig. 1E–G; Supplemental Fig. S3) in solution. Furthermore, we demonstrate using both gel filtration (Supplemental Fig. S7C) and tandem gel filtration multiangle light scattering (MALS) (Supplemental Fig. S7D,E) analyses that the SUVH5 SRA domain migrates as a monomer in the absence of DNA and as a complex of two SRA molecules bound to DNA on complex formation. Together, these data, along with the crystallographic analyses, rule out protein-mediated dimerization for the SUVH5 SRA domain both in the free state and when bound to fully methylated CG DNA, and do not provide support for models where an SRA dimer facilitates looping of methylated DNA.
Comparison of the SUVH5 and UHRF1 SRA domains bound to hemimethylated CG DNA
We can directly compare the structure of the UHRF1 SRA domain bound to hemimethylated CG DNA (one SRA bound per duplex) (Supplemental Fig. S8), which has been published previously (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008), with our structure of the SUVH5 SRA domain bound to hemimethylated CG DNA (two SRAs bound per duplex) (Fig. 3B).
Overall, the structures of the SUVH5 and UHRF1 SRA domains in their DNA-bound complexes are similar (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008), with a root mean square deviation (r.m.s.d.) of 1.33 Å (superposed structures of complexes are shown in Supplemental Fig. S9). Nevertheless, we observe several conformational features associated with the 5mC and C recognition of the SUVH5 SRA domain that differ in significant aspects from that reported earlier for the UHRF1 SRA. Using the same secondary structural element designations previously assigned to the UHRF1 SRA domain structure, we observe that the loop that connects α2 and β6 is absent (Supplemental Fig. S1), and two additional loops—one that connects β6 and β7 (474–483), and another that connects β5 and α2 (435–441), termed the NKR finger—are disordered in the structure of the SUVH5 SRA–DNA complex (Fig. 2B). In the UHRF1 SRA complex, the NKR finger projects into the major groove of the hemimethylated CG DNA and provides two key residues: one that is involved in the flipping out of the 5mC, and another that is involved in discriminating against the presence of a methylated C on the complementary strand (Supplemental Fig. S8B, loops are shown in red; Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008).
In addition to using two SRA domains to recognize the DNA duplex in the SUVH5 SRA domain complex, the residues involved in the 5mC recognition also differ between the structures of the SUVH5 and UHRF1 SRA domain complexes. In the UHRF1 SRA complex, the thumb loop (444–449 residues) and the NKR finger loop (483–496 residues) have been described as a hand gasping the DNA such that the former enters the minor groove and the latter intrudes through the major groove (Avvakumov et al. 2008). Specifically, the NKR finger provides the Arg491 residue that replaces the 5mC in the DNA duplex and pairs with the Hoogsteen edge of the orphaned guanine (Supplemental Fig. S8C). The methylene side chain of this Arg also forms hydrophobic contacts with Val446 from the thumb domain (Supplemental Fig. S8D). Asn489 from the NKR finger appears to act as a selectivity filter by preventing the symmetry-related C5 base from flipping out of the DNA duplex (Supplemental Fig. S8D).
In the SUVH5 SRA domain complex, the NKR finger is disordered (in all three sequence contexts) and does not appear to play a role in DNA recognition. Rather, it is Gln392 from the thumb loop of each individual SRA domain that inserts into the DNA duplex from the minor groove and displaces the 5mC and C residues (Fig. 3B). Unlike Arg491, the Gln392 residue interacts with the Watson-Crick edge of G6 (Fig. 2C,D). Thus, the SUVH5 and UHRF1 SRA domains insert amino acids from different loops into the DNA duplex in order to base-pair with the orphan guanines: UHRF1 inserts a residue from its NKR finger loop into the major groove and recognizes the Hoogsteen edge of the guanine base, while SUVH5 inserts a residue from its thumb loop into the minor groove and recognizes the Watson-Crick edge of the guanine base. Despite these differences, the distribution of amino acids lining the binding pocket in the UHRF1 (Supplemental Fig. S8E) and SUVH5 (Fig. 3D) complexes is similar.
Implications for the SUVH5 and UHRF1 SRA domains bound to fully methylated CG DNA
Our structure of the SUVH5 SRA domain bound to fully methylated CG DNA established that two SRA domains can bind to the DNA duplex without generating steric clashes between the two bound SRA domains, allowing the two 5mC bases from the partner strands to be flipped out and positioned into the binding pockets of two individual SRA domains (Fig. 2B). This does not appear to be the case for the UHRF1 SRA domain, since our modeling of a second SRA domain onto the structure of the single SRA bound to a hemimethylated CG DNA duplex resulted in steric clashes between the NKR fingers of the individual SRA domains. Furthermore, it appears that Asn489 blocks access of the stacked C on the complementary strand from flipping out of the duplex in UHRF1 complexes, while no such block exists for the SUVH5 complexes, allowing for the dual base-flipping mechanism observed for the SUVH5 SRA domain.
Preferential recognition of the 5mC-containing DNA by the SUVH5 SRA domain
Our ITC and EMSA binding studies establish that the SUVH5 SRA domain binds to unmethylated CG-containing DNA very poorly. Thus, there is a requirement for a 5mC in all three DNA sequence (fully methylated CG, hemimethylated CG, and methylated CHH) contexts. This could reflect initial specific recognition of the methyl group of the 5mC during scanning by the SUVH5 SRA domain, similar to what has been proposed as a recognition mechanism for damaged DNA. Thus, although 5mC and C show only small differences in their positioning in the SUVH5 SRA-binding pocket (Fig. 3D), it appears that van der Waals contacts involving the methyl group must contribute to binding affinity.
It is conceivable that the SUVH5 SRA recognizes a looped-out 5mC and, in doing so, thermodynamically destabilizes the surrounding segment of the DNA duplex, thereby facilitating looping out of an adjacent 5mC or C on the partner strand for fully methylated and hemimethylated CG duplexes, or an opposing G on the partner strand in the methylated CHH duplex. There is some precedent for this in the literature in the structure of the Rad4 nucleotide excision repair protein bound to damaged DNA (Min and Pavletich 2007). In this example, Rad4 flips both the damaged cyclobutane pyrimidine dimer, and the two tandem mismatched thymine bases on the complementary strand out of the duplex.
Comparison with other proteins that recognize 5mC bases present in DNA duplexes
The structures of the SUVH5 SRA domain bound to methylated DNA reported in this study mark the first structures of a SET-associated SRA domain bound to 5mC-containing DNA in different sequence contexts. In addition to the RING-associated SRA domain protein UHRF1 (Supplemental Fig. S8B; Avvakumov et al. 2008), structures have also been reported for the 5mC-binding domains (MBD) of human MBD1 (Ohki et al. 2001) and MeCP2 (Ho et al. 2008) in complexes with methylated DNA in the fully methylated CG context. Both MBD1 and MeCP2 recognize fully methylated CG DNA with a stoichiometry of one protein bound per duplex, and this recognition does not require base flipping on either strand (structure of MeCP2–DNA complex shown in Supplemental Fig. S10A). In the MBD1–DNA complex, recognition of the methyl group of 5mC is facilitated by a hydrophobic patch (Ohki et al. 2001), and, in the MeCP2–DNA, it is facilitated by a predominantly hydrophilic surface that includes tightly bound water molecules (Supplemental Fig. S10B; Ho et al. 2008). Thus, for the UHRF1 SRA domain, as well as the MBD domains of MBD1 and MeCP2, recognition of the 5mC-containing DNA involves a single domain that is able to recognize both strands of the methylated CG DNA, whereas, in the SUVH5 complex, two SRA domains bind independently to each strand of the DNA duplex at either a fully or hemimethylated CG site (Figs. 2B, 3B) or a methylated CHH site (Fig. 4B).
Plasticity in substrate recognition by the SUVH family of SRA domain proteins
Cytosine and H3K9 methylation marks play a decisive role in determining the epigenetic repression of eukaryotic genes. Animals contain only RING-associated SRA domain proteins, while plants—including Arabidopsis, Oryza sativa, and Zea mays—contain both SET (SUVH)- and RING (VIM)-associated SRA domain proteins. The SRA domains of the SUVH family of proteins show a range of plasticity in 5mC recognition in different sequence contexts: SUVH9 preferentially binds methylated CHH sites (Johnson et al. 2008), SUVH2 prefers methylated CG sites (Johnson et al. 2008), SUVH4 prefers CHG methylation over both CG and CHH methylation (Johnson et al. 2007), and SUVH6 prefers methylated CHG and CHH over CG sites (Johnson et al. 2007). Finally, this study shows that the SUVH5 can bind to methylated DNA in all sequence contexts to similar extents. Such plasticity makes it difficult to predict the sequence specificity of specific SRA domains on the basis of either primary sequence or their unliganded structure. However, based on the structure of the SUVH5 and UHRF1 SRA domains and their sequence comparisons, it is conceivable that the different specificities observed for the Arabidopsis SET-associated SUVH SRA domains might be attributed to the high degree of sequence and length variation within their thumb and NKR finger loop motifs (Fig. 1B; Supplemental Fig. S1).
The SRA domains of SUVH5, SUVH6, SUVH2, and SUVH9—all of which have been shown to bind methylated DNA—possess residues within their predicted thumb loops (like Gln) that could perform a base replacement function analogous to the role observed for Gln392 of the SUVH5 SRA domain. For SUVH1, SUVH3, and SUVH8, there is no obvious residue in the predicted thumb loop to perform the base replacement function, and the NKR finger loops are much shorter in comparison with the UHRF1 structure, suggesting that, if these proteins actively bind 5mC sites, the base replacement residue might come from the shorter NKR finger loop or elsewhere in the molecule.
The SUVH5 SRA exhibits plasticity in its substrate recognition such that it can recognize and flip out noncognate cytosine and guanine bases, which is reminiscent of HhaI cytosine methyltransferase. HhaI binds to noncognate mismatch target bases (G • A, G • U, G • T, and G • A) in the presence of the complementary 5′-GCGC-3′ recognition sequence, and even exhibits methyltransferase activity by transferring a methyl group as long as a G • U mismatch is present (Klimasauskas and Roberts 1995). The structure of HhaI in a complex with a mismatched recognition sequence has clearly demonstrated that noncognate base (G • A, G • U, and G • abasic) flipping and recognition by the active site residues can occur in the presence of a cognate complementary recognition sequence (O'Gara et al. 1998). For the SUVH5 SRA domain, the noncognate base flipping occurs only when there is a cognate 5mC present on the complementary strand. However, the mechanism of the noncognate base flipping, its potential presence in vivo, and its biological role require further investigation.
In contrast to the SET-associated SUVH family of SRA domain proteins, the “GNKR” motif within the NKR finger is conserved in all RING-associated SRA domain proteins, including UHRF1/ICBP90 in mammals (Avvakumov et al. 2008; Hashimoto et al. 2008) and its paralogs, the VIM/ORTHUS family of proteins (except in uncharacterized ORTH-L) in Arabidopsis (Fig. 1B; Johnson et al. 2007; Woo et al. 2007, 2008). In UHRF1, the “GNKR” motif provides both the base replacement residue (Arg491) and the complementary strand C/5mC-masking residue (Asn489). These residues are conserved not only in the UHRF1 family of proteins in animals, but also in their paralogs from plants, suggesting that these residues may be a hallmark of proteins that recognize hemimethylated CG sites (Fig. 1B), and that, like UHRF1, the VIM family of proteins might also function to preferentially read hemimethylated CG sites.
The conserved NKR finger motif is also present in one of the SET-associated SUVH proteins, SUVH4, and, like UHRF1, SUVH4 has a strong preference for hemimethylated over fully methylated DNA. Furthermore, a Asn417Ala mutation in SUVH5 that corresponds to a null mutation in SUVH4 has little effect on SUVH5 activity in vivo (data not shown). In the structure of the SUVH5 SRA–DNA complex, this residue is present in the binding pocket next to the residue that participates in the stacking interaction with the flipped-out 5mC, but does not participate directly in methyl-binding pocket interactions. Together, these observations suggest that the binding of SUVH4 to methylated DNA might be more mechanistically similar to UHRF1 than to the other SUVH family members.
Structural basis of substrate specificity and in vivo function
Although the base-flipping mechanism is conserved in 5mC readers like SRA domain proteins (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008) and 5mC writers like DNMTs (Klimasauskas et al. 1994; Reinisch et al. 1995), no apparent structural or sequence similarity has been observed between them. However, all of these proteins use two or three loops to approach the DNA from both the major and minor grooves simultaneously, and they also all bind the flipped-out base using a concave surface in an analogous manner.
It is of note that, while the SUVH5 SRA domain structure has striking primary and tertiary structural similarity with that of the UHRF1 SRA domain structure, only the thumb loop segment projects through the minor groove and is involved in the base-flipping process in the SUVH5 structure, while the same thumb loop has a minimal role in the UHRF1 structure. The opposite is observed for the NKR loop segment, which is disordered and has no role in base flipping in the SUVH5 structure, while it projects through the major groove and plays a key a role in base flipping in the UHRF1 structure (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008).
These stark structural differences between the UHRF1 and SUVH5 SRA domains may account for the different biological roles hypothesized for these two proteins. A single SRA domain within UHRF1 recognizes both strands of the hemimethylated CG site, which allows differentiation between fully and hemimethylated sites. Recognition of a 5mC in the hemimethylated CG context by UHRF1 is proposed to aid in the recruitment of the maintenance DNMT, DNMT1, to replication foci, where it assists in the restoration of hemimethylated DNA, which is generated during replication, to the fully methylated state (Bostick et al. 2007; Sharif et al. 2007).
In contrast, SUVH5 is a SET domain H3K9 methyltransferase that, along with SUVH4 and SUVH6, is required for the maintenance of DNA methylation in the CHG context. Given that, in Arabidopsis, DNA methylation at transposons and other repeat elements is dense and occurs in all contexts, the ability of SUVH5 to recognize methylation in all contexts may aid the recruitment of this protein to silenced loci in order to ensure maintenance of both DNA and histone methylation levels. The finding that the SRA domains of SUVH4, SUVH5, and SUVH6 have different binding preferences, in combination with previous genetic data showing that SUVH4, SUVH5, and SUVH6 contribute to the level of DNA methylation to different extents in a locus-specific manner, suggests that the different specificities of the SRA domains within these proteins could contribute to the targeting or retention of these proteins at specific silenced loci. In addition to the methyl-binding specificity of the different SRA domains, the SUVH family members could interact with different proteins or protein complexes in vivo, and such interactions could modulate the methyl-binding activity of the SRA domain or impart locus specificity through other unknown mechanisms. Currently, the effects of single and combinatorial mutants of SUVH4, SUVH5, and SUVH6 have been assessed at only a few loci. However, further studies looking at the effects of various SUVH mutants on a genome-wide scale as well as identification of potential SUVH4-, SUVH5-, and SUVH6-interacting partners should aid in clarifying the roles of these SRA domain proteins.
Based on the crystal structure of the SUVH5 SRA domain, we hypothesize that if the NKR finger loop was involved in the base-flipping mechanism in a manner analogous to that observed for the UHRF1 SRA domain, then it would likely face steric clashes from its symmetrically related mate present on the opposite DNA strand. Such structural constraints might restrict the binding affinity of the SUVH5 SRA domain to a particular sequence context, which is inconsistent with the ITC and EMSA analyses showing that the SUVH5 SRA domain binds 5mC in all sequence contexts. Thus, nature might have used the same SRA fold with subtle sequence variations within the thumb and/or NKR finger loops to engineer SUVH proteins that recognize 5mC in a particular DNA sequence context to achieve proper targeting and/or retention of these proteins at a given silenced loci. For SUVH5, the shorter thumb loop projects Gln392, in contrast to Val at the same position in UHRF1, thereby allowing pairing with the orphaned guanine (Fig. 2C).
Our findings demonstrate an unanticipated level of versatility within the SRA domain of SUVH5 in recognizing 5mC in all sequence contexts. A structural comparison of the SUVH5 SRA domain complex with that of the UHRF1 SRA domain complex (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008) demonstrates that the likely basis for recognition of 5mC bases in all sequence contexts by SUVH5, and for the preferential recognition of hemimethylated CG by UHRF1, lies within different parts of the SRA domain and uses structurally and chemically diverse residues: Gln392 in the thumb loop for SUVH5, and Arg491 in the NKR finger for UHRF1.
SUVH5 is not only a 5mC binder, but also an H3K9 dimethylation writer. Our structure-based in vivo studies suggest that a functional SUVH5 SRA domain is required for recruitment and/or retention of the SUVH5 H3K9 methyltransferase at genomic loci, and subsequently for the maintenance of both DNA and histone methylation. Given the variation in sequence within SRA domains and the observed plasticity in the 5mC-binding preferences of the different SUVH family members, it stands to reason that future structural analyses of the SRA domains present in other SUVH family members should aid in our understanding of their in vivo function.
Materials and methods
Protein expression and purification
Detailed procedures for vector construction of wild-type SUVH5 residues 362–528 or residues 362–794 in hexahistidine-sumo-tagged expression vector, expression conditions for selenomethionine-labeled and unlabeled protein, and sequential protein purification protocols are outlined in the Supplemental Material.
Generation of mutants
The site-directed mutagenesis kit (Stratagene) was used for creating several mutations on a plasmid carrying the cDNA of the SUVH5 SRA domain protein, and mutations were confirmed by DNA sequencing. Mutated proteins were expressed and purified as mentioned in “Protein Expression and Purification” in the Supplemental Material.
ITC measurements
Calorimetric experiments were performed using a VP-ITC calorimeter (MicroCal, LLC) at 25°C, and MicroCal Origin software was used for curve-fitting, equilibrium dissociation constant, and molar ratio calculations. For detailed procedures, see the Supplemental Material.
Crystallization, X-ray data collection, and structure determination
SUVH5 SRA–methylated DNA complex crystallization setup was conducted at 18°C by the sitting-drop-vapor diffusion method using the crystallization robot. Data sets of SUVH5 SRA complexed with fully methylated CG, hemimethylated CG, or methylated CHH were collected at APS NE-CAT 24ID-C or 24ID-E beamlines. The multiple wavelength anomalous dispersion (MAD) data set at the selenium edge was collected from the selenomethionine-labeled SUVH5 SRA–fully methylated CG complex crystal, and the structure was solved by the MAD technique. Subsequent structures of SUVH5 SRA complexed with a second fully methylated CG, hemimethylated CG, or methylated CHH were solved by the molecular replacement method using the SRA domain from a fully methylated CG complex structure as the search model. For detailed methods of crystallization, data collection, and structure calculation, see the Supplemental Material. The crystallographic statistics for all structures presented above are listed in Supplemental Table 1.
Protein constructs
SUVH5 SRA-GST fusion constructs were generated using the Gateway cloning system (Invitrogen) and a PCR product amplified from genomic DNA as outlined in the Supplemental Material. Proteins were expressed in BL21 AI cells and purified as described previously (Johnson et al. 2007, 2008). Epitope-tagged full-length SUVH5 constructs were generated using Gateway technology. A detailed account of the primers and vectors used is provided in the Supplemental Material.
EMSAs
The DNA probes and conditions were largely as described previously (Johnson et al. 2007). Briefly, 200 ng of each protein in binding buffer (50 mM Tris at pH 6.8, 80% glycerol, 1 mg/mL BSA, 14 mM βme, 2 mM DTT, 50 mM NaCl) and an excess of unlabeled λ DNA as a competitor were incubated with a 32P end-labeled probe, run on 6% acrylamide gels, fixed, dried, and exposed to film. For binding curves, the same conditions were used except the concentration of protein added was titrated. Autoradiographs were quantified using the ImageQuant TLv2005 software.
Western and Southern blotting
The Flag epitope tag was detected using the anti-Flag M2 monoclonal antibody-peroxidase conjugate (Sigma A 8592) at a dilution of 1:5000. Southern blots were conducted using MspI-digested genomic DNA and a probe specific to the Ta3 locus as described previously (Johnson et al. 2008).
Nuclei isolation and H3K9 immunofluorescence
Nuclei were isolated from seedlings as described previously (Jasencakova et al. 2003). A primary antibody to H3K9 dimethylation (Abcam 1220) was used at a dilution of 1:100, and anti-mouse Alexa Fluor 488 (Invitrogen) was used as a secondary antibody at a dilution of 1:150. Nuclei were visualized as described previously (Li et al. 2006).
Nucleosome preparation and HMTase assays
Histone methylation assays were done as described previously (Johnson et al. 2008). Activity was assessed using nucleosomes assembled as described in the Supplemental Material.
Database entries
The X-ray coordinates of SUVH5 SRA complex with fully methylated CG DNA in space group P42212 (accession no. 3Q0B) and space group P6122 (3Q0C); hemimethylated CG DNA (3Q0D); and methylated CHH DNA (3Q0F) have been deposited in the Protein Data Bank (PDB).
Acknowledgments
We thank members of the Jacobsen, Patel, and Reinberg laboratories for helpful discussion. E.R. acknowledges the assistance of Dr. Haitao Li in collecting ITC data, Professor Judith L. Bender for providing cDNA encoding full-length SUVH5 protein, and Dr. Qian Yin and Professor Hao Wu from Weill Medical College of Cornell University for providing access and user assistance to their MALS instrument. We thank the staff at beamlines 24ID-C and 24ID-E of the Advanced Photon Source at the Argonne National Laboratory for assistance with data collection. L.M.J. acknowledges the assistance of Emily Chien and Anuj Khattar in the generation of tagged SUVH5 constructs. The Jacobsen laboratory research was supported by U.S. National Institutes of Health grant GM60398. The Patel laboratory research was supported by the Abby Rockefeller Mauze Trust and the Maloris Foundation. J.A.L. was supported by the National Institutes of Health National Research Service Award (5F32GM820453). S.E.J. and D.R. are investigators of the Howard Hughes Medical Institute. Work in the Reinberg laboratory is also supported by the National Institutes of Health (grants GM064844 and R37GM037120). P.V. is supported by the Deutsche Akademie der Naturforscher Leopoldina (Leopoldina Fellowship Program, LPDS 2009-5).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1980311.
Supplemental material is available for this article.
References
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M 2008. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455: 818–821 [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S 2008. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455: 822–825 [DOI] [PubMed] [Google Scholar]
- Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE 2008. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One 3: e3156 doi: 10.1371/journal.pone.0003156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764 [DOI] [PubMed] [Google Scholar]
- Cheng X, Blumenthal RM 2008. Mammalian DNA methyltransferases: A structural perspective. Structure 16: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs ML, Bender J 2006. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18: 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs ML, Bartee L, Bender J 2005. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol 25: 10507–10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C 1982. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res 10: 2709–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH 2005. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514 [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X 2008. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455: 826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE 2007. Epigenetic inheritance in plants. Nature 447: 418–424 [DOI] [PubMed] [Google Scholar]
- Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD 2008. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol Cell 29: 525–531 [DOI] [PubMed] [Google Scholar]
- Huffman JL, Sundheim O, Tainer JA 2005. DNA base damage recognition and removal: New twists and grooves. Mutat Res 577: 55–76 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Johnson L, Jasencakova Z, Zhang X, PerezBurgos L, Singh PB, Cheng X, Schubert I, Jenuwein T, Jacobsen SE 2004. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112: 308–315 [DOI] [PubMed] [Google Scholar]
- Jasencakova Z, Soppe WJ, Meister A, Gernand D, Turner BM, Schubert I 2003. Histone modifications in Arabidopsis—High methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J 33: 471–480 [DOI] [PubMed] [Google Scholar]
- Johnson L, Cao X, Jacobsen S 2002. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12: 1360–1367 [DOI] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE 2007. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE 2008. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet 4: e1000280 doi: 10.1371/journal.pgen.1000280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Samaranayake M, Pradhan S 2009. Epigenetic mechanisms in mammals. Cell Mol Life Sci 66: 596–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S, Roberts RJ 1995. M.HhaI binds tightly to substrates containing mismatches at the target base. Nucleic Acids Res 23: 1388–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S, Kumar S, Roberts RJ, Cheng X 1994. HhaI methyltransferase flips its target base out of the DNA helix. Cell 76: 357–369 [DOI] [PubMed] [Google Scholar]
- Kraft E, Bostick M, Jacobsen SE, Callis J 2008. ORTH/VIM proteins that regulate DNA methylation are functional ubiquitin E3 ligases. Plant J 56: 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE 2006. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126: 93–106 [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J 2002. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J 21: 6842–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JH, Pavletich NP 2007. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature 449: 570–575 [DOI] [PubMed] [Google Scholar]
- O'Gara M, Horton JR, Roberts RJ, Cheng X 1998. Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base. Nat Struct Biol 5: 872–877 [DOI] [PubMed] [Google Scholar]
- Ohki I, Shimotake N, Fujita N, Jee J, Ikegami T, Nakao M, Shirakawa M 2001. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell 105: 487–497 [DOI] [PubMed] [Google Scholar]
- Phillips K, Luisi B 2000. The virtuoso of versatility: POU proteins that flex to fit. J Mol Biol 302: 1023–1039 [DOI] [PubMed] [Google Scholar]
- Qian C, Li S, Jakoncic J, Zeng L, Walsh MJ, Zhou MM 2008. Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. J Biol Chem 283: 34490–34494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch KM, Chen L, Verdine GL, Lipscomb WN 1995. The crystal structure of HaeIII methyltransferase convalently complexed to DNA: An extrahelical cytosine and rearranged base pairing. Cell 82: 143–153 [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912 [DOI] [PubMed] [Google Scholar]
- Woo HR, Pontes O, Pikaard CS, Richards EJ 2007. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev 21: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Dittmer TA, Richards EJ 2008. Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet 4: e1000156 doi: 10.1371/journal.pgen.1000156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]