Abstract
Nonsense-mediated mRNA decay (NMD) is a eukaryotic surveillance pathway that regulates the degradation of mRNAs harboring premature translation termination codons. NMD also influences the expression of many physiological transcripts. SMG-1 is a large kinase essential to NMD that phosphorylates Upf1, which seems to be the definitive signal triggering mRNA decay. However, the regulation of the kinase activity of SMG-1 remains poorly understood. Here, we reveal the three-dimensional architecture of SMG-1 in complex with SMG-8 and SMG-9, and the structural mechanisms regulating SMG-1 kinase. A bent arm comprising a long region of HEAT (huntington, elongation factor 3, a subunit of PP2A and TOR1) repeats at the N terminus of SMG-1 functions as a scaffold for SMG-8 and SMG-9, and projects from the C-terminal core containing the phosphatidylinositol 3-kinase domain. SMG-9 seems to control the activity of SMG-1 indirectly through the recruitment of SMG-8 to the N-terminal HEAT repeat region of SMG-1. Notably, SMG-8 binding to the SMG-1:SMG-9 complex specifically down-regulates the kinase activity of SMG-1 on Upf1 without contacting the catalytic domain. Assembly of the SMG-1:SMG-8:SMG-9 complex induces a significant motion of the HEAT repeats that is signaled to the kinase domain. Thus, large-scale conformational changes induced by SMG-8 after SMG-9-mediated recruitment tune SMG-1 kinase activity to modulate NMD.
Keywords: nonsense-mediated mRNA decay, NMD, SMG-1, SMG-8, SMG-9, cryo-EM
Eukaryotic gene expression is exquisitely regulated at several steps from gene transcription to the final synthesis of proteins to ensure fidelity in the conversion of genetic information into biological functions. A post-transcriptional surveillance pathway known as nonsense-mediated mRNA decay (NMD) is responsible for the recognition and degradation of mRNAs containing premature translation termination codons (PTCs). The main function of this pathway is to ensure that mutated mRNAs harboring a PTC, which could generate potentially harmful truncated proteins, are removed before translation (Isken and Maquat 2008; Shyu et al. 2008). NMD also regulates the abundance of many physiological mRNAs contributing to the regulation of global gen expression (Mendell et al. 2004; Stalder and Muhlemann 2008).
Seven conserved core proteins have been identified in most metazoans as required for NMD. These are SMG-1, Upf1, Upf2, Upf3, SMG-5, SMG-6, and SMG-7 (Yamashita et al. 2005; Isken and Maquat 2008), a list that has been enlarged recently by novel components of the NMD machinery; namely, SMG-8, SMG-9, Ruvbl1, Ruvbl2, RPB5, and SMG-10 (Yamashita et al. 2009; Izumi et al. 2010). Recent advances in the field have provided significant insights into the mechanisms of NMD, but our understanding is still incomplete (Rebbapragada and Lykke-Andersen 2009; Hwang et al. 2010; Nicholson et al. 2010). SMG-1 is one of the essential players triggering an NMD response, since it is the kinase that phosphorylates the Upf1 protein. Upf1 phosphorylation is believed to be the definitive signal triggering NMD.
SMG-1 is a very large protein (∼410 kDa) belonging to the phosphatidylinositol 3-kinase-related protein kinase (PIKK) family of serine–threonine kinases (Abraham 2004; Yamashita et al. 2005). In humans, the PIKK family comprises six members: ATM, ATR, DNA-PKcs, mTOR, TRRAP, and SMG-1 (Abraham 2004; Lempiainen and Halazonetis 2009). The primary structure of all PIKKs contains a conserved C terminus preceded by a long stretch of helical, mostly HEAT (huntington, elongation factor 3, a subunit of PP2A and TOR1), repeats (Fig. 1A; Lempiainen and Halazonetis 2009). HEAT repeats are units of two anti-parallel α helices connected by flexible loops, and many of these units organize large, superhelical, frequently twisted structures that in many proteins function as a scaffold for protein–protein interactions (Bono et al. 2010). The required amino acid composition and length of the helices and loops connecting these helices in HEAT repeats are very flexible, allowing for significant sequence divergence within the PIKK family while maintaining the structural characteristics common to all HEAT repeats (Perry and Kleckner 2003). The extension of these HEAT regions, always N-terminal to the conserved C-terminal domains, varies among these kinases, and is significantly shorter in SMG-1 compared with DNA-PKcs, one of the larger PIKKs (Fig. 1A). These HEAT repeats are followed by a conserved C terminus, comprising an ∼600-amino-acid FAT (FRAP/TOR, ATM, and TRRAP) domain, also thought to be assembled from multiple helical repeats; the catalytic kinase domain with homology with the PI3K (∼370 residues); and a short (∼70-residue) C-terminal FATC (FAT C-terminal) domain. The FATC is a short, highly conserved α-helical domain that is critical for kinase activity and has been suggested to be important in protein–protein interactions (Lempiainen and Halazonetis 2009). Detailed information about sequence conservation has been reviewed elsewhere (Lempiainen and Halazonetis 2009). SMG-1 shares these structural features common to all PIKKs, but a unique >1000-amino-acid insertion of poorly characterized function is located between the kinase domain and the FATC (Fig. 1A). A C-terminal fragment of SMG-1 encompassing the catalytic domain, the FATC, and the insertion between these two domains has been implicated in Upf2 recognition (Kashima et al. 2006). For simplicity, in this study we use the terms N-terminal and C-terminal domains of SMG-1 when referring to the HEAT repeat region and the conserved C terminus (FAT, kinase, insertion, and FATC domains), respectively (Fig. 1A).
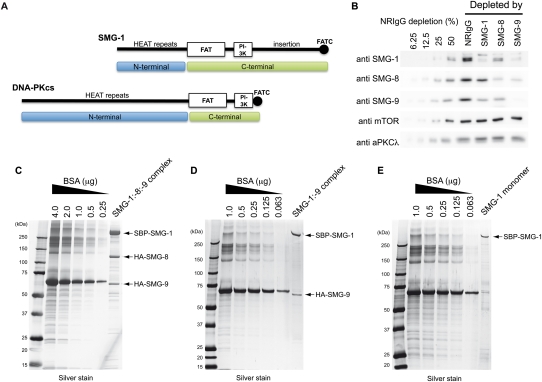
Figure 1.
Purification of SMG-1, SMG-1:SMG-9, and SMG1C. (A) SMG-1 primary structure and comparison with DNA-PKcs. In this study, the terms N-terminal and C-terminal are used to refer to the helical region and the conserved C-terminal domains, respectively. (B) Western blots of HeLa TetOff cell lysates immunodepleted with anti-SMG-1, anti-SMG-8, or anti-SMG-9 antibody, or NRIgG (normal rabbit immunoglobulin G). Immunodepleted lysates were analyzed using the antibodies indicated. Dilutions corresponding to 50%, 25%, 12.5%, or 6.25 of the NRIgG-depleted protein amount from the NRIgG-depleted lysate were loaded to assess the efficiency of the immunodepletion. (C–E) SDS gel after silver staining of the fractions eluted from the affinity column for the purification of the SMG1C complex (SMG-1:SMG-8:SMG-9) (C), the SMG-1:SMG-9 complex (D), or SMG-1 (E), all expressed in 293T cells.
The structural analysis of the PIKK kinases has been found to be extremely challenging due to their large size, their interaction with multiple partners, and the difficulty of their purification. One of the best structurally characterized members of the PIKK family is DNA-PKcs. The three-dimensional (3D) structure of DNA-PKcs and that of its complexes with DNA and Ku70/Ku80 have been studied by 3D electron microscopy (3D-EM) (Rivera-Calzada et al. 2005; Spagnolo et al. 2006; Williams et al. 2008) and SAXS (Hammel et al. 2010). These structures defined two main structural regions in the protein: the “head” (bulky and containing the kinase domain), and several arms containing regions of HEAT repeats. Recently, a model based on crystallographic data at 6.6 Å resolution was proposed that unambiguously confirmed the location of the kinase domain at the top of the head region and suggested that the head possibly comprised most of the conserved C-terminal domains (Sibanda et al. 2010). Interestingly, two arms of HEAT repeats project from the head, forming an open ring that might participate in DNA binding (Supplemental Fig. S1; Rivera-Calzada et al. 2005; Sibanda et al. 2010).
Two novel components of the NMD machinery have been described recently as forming a complex with SMG-1. These two proteins, SMG-8 (991 amino acids) and SMG-9 (520 amino acids), appear to be tightly bound to SMG-1 to form the so-called SMG1C complex (Yamashita et al. 2009). We recently described that SMG-9 comprises a C-terminal putative nucleotide-triphosphatase domain preceded by an intrinsically disordered N-terminal region (Yamashita et al. 2009; Fernandez et al. 2010). Both domains are required to interact with SMG-1, whereas the C terminus of SMG-9 interacts with SMG-8, showing also that SMG-8 and SMG-9 can form a complex in the absence of SMG-1 (Fernandez et al. 2010). The association between SMG-8 and SMG-9 with SMG-1 appears to be an absolute requirement for the regulation of the NMD response (Yamashita et al. 2009). Here, we provide the molecular architecture of the SMG1C complex using 3D-EM and the structural basis for the regulation of SMG-1 by SMG-8 and SMG-9.
Results
SMG-9 plays a central role in the assembly of several complexes in vivo
We performed a series of depletion experiments from HeLa TetOff cell extracts to reveal the complexes formed among endogenous SMG-1, SMG-8, and SMG-9 in vivo. The extracts were depleted of either protein by specific antibodies, and the remaining material was tested for the presence of SMG-1, SMG-8, and SMG-9 by Western blotting (Supplemental Fig. S2; Yamashita et al. 2009). Under the conditions tested, a significant reduction of the specific target protein was observed (Fig. 1B), but, interestingly, although SMG1C seemed to be an abundant complex, other subcomplexes were detected in vivo. When SMG-1 was immunodepleted, the remaining extract revealed significant amounts of SMG-8 and SMG-9 that were not bound to SMG-1. Depletion of SMG-8 revealed the existence of significant amounts of SMG-1 and SMG-9 molecules that were not bound to SMG-8, consistent with the formation of complexes between SMG-1 and only SMG-9, as later confirmed (see below). In addition, SMG-9 depletion removed most SMG-1 and a large proportion of SMG-8, suggesting that the majority of SMG-1 and SMG-8 are bound to SMG-9. In this experiment, nondepleted SMG-8 would correspond to free SMG-8 and/or putative SMG-1:SMG-8 complexes, but the small amount of SMG-1 remaining after SMG-9 depletion indicated that SMG-1 does not significantly interact with SMG-8 independently of SMG-9. Overall, these data revealed that SMG-1, SMG-8, and SMG-9 assemble several complexes in vivo besides SMG1C. Importantly, SMG-9 seems to play a major role in the assembly of these complexes, and SMG-1 was not detected bound to SMG-8 in the absence of SMG-9 in a significant proportion (Fig. 1B; Yamashita et al. 2009). This finding is consistent with the ability of SMG-8 and SMG-9 to interact in the absence of SMG-1 described previously (Fernandez et al. 2010).
These findings led us to purify SMG-1 as well as the SMG-1:SMG-9 and SMG-1:SMG-8:SMG-9 complexes after transfection of human 293T cells with tagged versions of the proteins (Fig. 1C,E). An extraordinary effort was required to solve all of the challenges encountered during the expression and purification of these complexes, and key to our success was the combination of siRNA, antibody depletion, and imaging in the electron microscope during purification. The SMG1C complex was purified from cells expressing SBP-SMG-1, HA-SMG-8, and HA-SMG-9 using the SBP tag (Fig. 1C). SMG-1 copurified with a significant amount of SMG-8 and SMG-9, suggesting that the SMG1C complex comprised the majority of the purified protein, as later confirmed by EM. Similar strategies were performed to purify SMG-1 and SMG-1:SMG-9. However, the purification of SMG-1 or SMG-1:SMG-9 revealed the presence of trace amounts of “contaminant” endogenous SMG-8 bound to exogenously expressed SMG-1:SMG:9, and endogenous SMG-8 and SMG-9 bound to exogenously expressed SMG-1 (Supplemental Fig. S3). The SMG-1:SMG-9 complex was successfully purified from cell extracts expressing SBP-SMG-1 and HA-SMG-9 together with the cotransfection of siRNA expression plasmid targeting SMG-8 (Fig. 1D; Supplemental Fig. S3). SMG-1 was purified from human cells after removing endogenous SMG-8 and SMG-9 using a sequential protocol where contaminant SMG-8 and SMG-9 were removed by immune depletion with specific antibodies (Fig. 1E; Supplemental Fig. S3).
3D structure of SMG-1:SMG-9
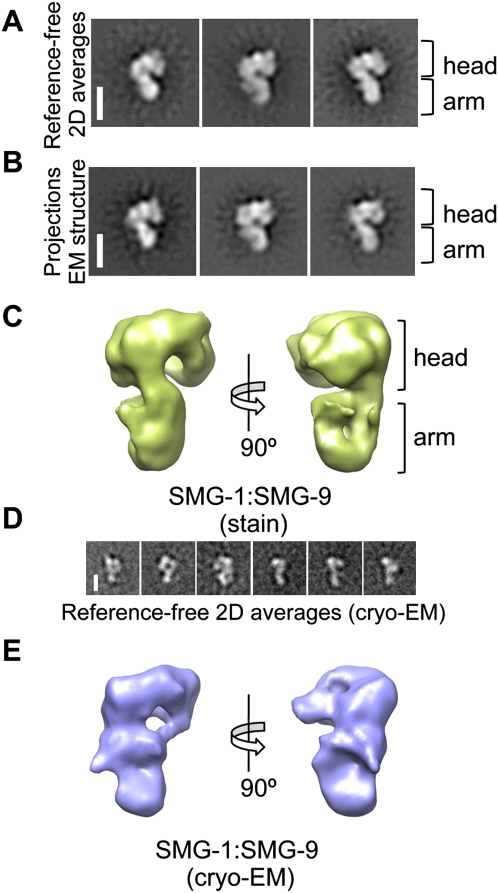
The SMG-1:SMG-9 complex was the best behaved of all of our preparations, and it was thus the target of our primary characterization of SMG-1 complexes. Images (15,608) of SMG-1:SMG-9 after negative staining were collected, and reference-free two-dimensional (2D) classification and averaging of these images revealed an elongated molecule with rough dimensions of 10 × 12 × 16 nm (Fig. 2A; Supplemental Fig. S4). A characteristic view of the complex showed a shape evocative of a question mark (Fig. 2A). Several ab initio reconstructions of SMG-1:SMG-9 were performed by the random conical tilt (RCT) method, where pairs of tilted and untilted images for each molecule were used to obtain structures of those molecules bound to the support film in the same orientation (Supplemental Fig. S5). We then refined the total data set using angular refinement methods and several initial templates for refinement, including the RCT reconstructions and random Gaussian blobs to discard a significant bias by the initial 3D reference (Supplemental Fig. S6). The data set consistently refined to a 24 Å structure of SMG-1:SMG-9 comprising a voluminous head connected to a thinner extension, the “arm” (Fig. 2C), and, importantly, the projections of the structure obtained matched the reference-free averages (Fig. 2A,B). We also performed EM experiments using unstained specimens preserved at liquid-nitrogen temperature (cryo-EM). Collecting cryo-EM data was complicated by the size of the complex and the low concentration of the sample. Cryo-EM images (15,500) of SMG-1:SMG-9 were collected, and reference-free 2D averages revealed a molecule with a prominent head and a thin arm (Fig. 2D; Supplemental Fig. S4). The 30 Å resolution cryo-EM structure of SMG-1:SMG-9 agreed well with the stained reconstructions (Fig. 2E).
Figure 2.
3D-EM structure of SMG-1:SMG-9. (A,B) Reference-free 2D averages of SMG-1:SMG-9 (A) compared with projections of the 3D structure (B) obtained after angular refinement.Bar, 10 nm. (C) Two views of the 3D structure of the SMG-1:SMG-9 complex obtained by angular refinement methods. (D) Reference-free 2D averages obtained from unstained specimens of the SMG-1:SMG-9 complex. Bar, 10 nm. (E) Cryo-EM structure of SMG-1:SMG-9.
Molecular architecture of SMG-1 and the SMG-1:SMG-9 complex
The structure of SMG-1:SMG-9 was compared with the structural models of DNA-PKcs, since this is the only PIKK for which high-resolution structural information is available (Supplemental Fig. 1; Sibanda et al. 2010). EM and crystallographic studies have shown that DNA-PKs contains a bulky region (head) containing the catalytic domain and likely all domains corresponding to the conserved C-terminal region, and two arms extending outward from the C-terminal region—one of them proposed to contain the N-terminal end of the molecule (Rivera-Calzada et al. 2005; Sibanda et al. 2010). These arms comprised regions of HEAT repeats arranged as superhelical structures. The limited resolution of those studies prevented a definition of the arrangement of the polypeptide chain in the structure, and how the HEAT repeats at the N-terminal side of the conserved C terminus can organize two arms projecting from the C-terminal end of the molecule is unclear (Supplemental Fig. S1). Except for a unique C-terminal insertion, the primary structure of SMG-1 is similar to other PIKK members and DNA-PKcs, but the extensive HEAT repeat region is significantly shorter in SMG-1 (Perry and Kleckner 2003). This similarity allowed the comparison between SMG-1 and DNA-PKcs, both showing a “bulky” head likely corresponding to the C-terminal portion of the molecule containing the kinase domain (Fig. 3D; Supplemental Fig. S1). Interestingly, SMG-1:SMG-9 revealed just one arm projecting from the head region, which would be consistent with the shorter HEAT repeat region present in the primary structure of SMG-1 compared with DNA-PKcs (Fig. 1A). This superhelical HEAT repeat region in SMG-1 formed a bent arm, as in DNA-PKcs (Rivera-Calzada et al. 2005; Sibanda et al. 2010).
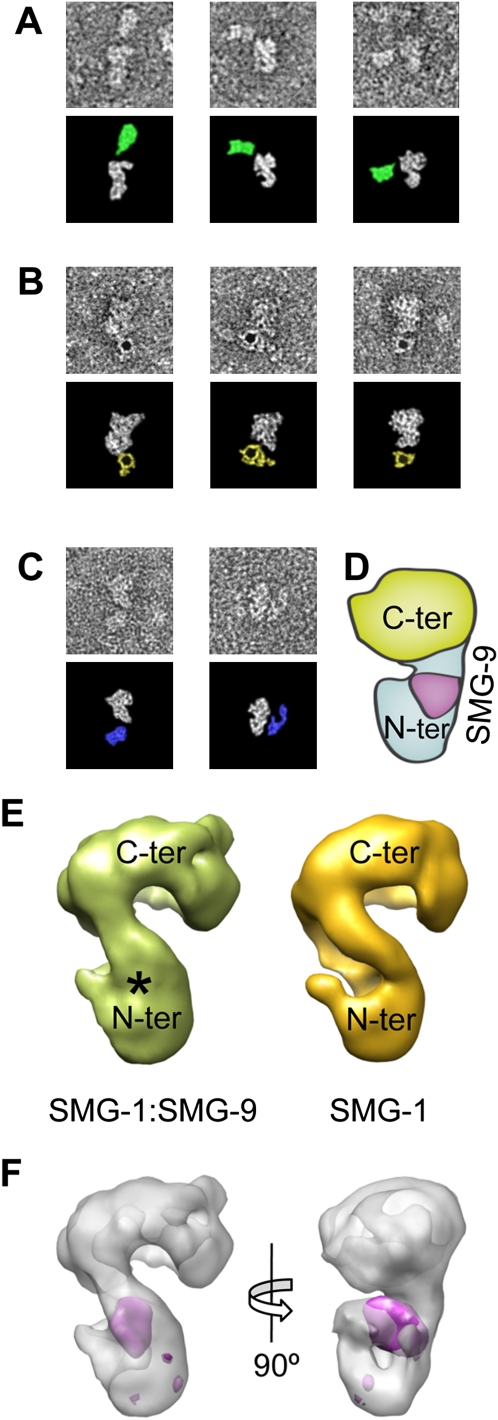
Figure 3.
3D architecture of SMG-1 and the SMG-1:SMG-9 complex. (A) Immunolabeling of SMG-1:SMG-9 complexes using a monoclonal antibody targeting residues 3547–3657 at the C terminus of SMG-1. (B) Labeling of SMG-1:SMG-9 complexes using a 5-nm gold particle coupled to streptavidin binding the SBP tag at the N terminus of SMG-1. (C) Immunolabeling of SMG-1:SMG-9 complexes using a monoclonal antibody targeting the SBP tag at the N terminus of SMG-1. The bottom panels of A–C highlight the position of the protein and the antibody in the single images by removing the surrounding background. Antibodies targeting the C terminus are colored in green, whereas those labeling the N terminus are colored in blue. (D) Cartoon model of SMG-1:SMG-9 with the proposed location for the N-terminal and C-terminal domains of SMG-1 and SMG-9. (E) Structure of SMG-1 and its comparison with the structure of SMG-1:SMG-9. (*) The region of density in SMG-1:SMG-9 that is not present in SMG-1. (F) Difference map of SMG-1:SMG-9 and SMG-1 (purple) superimposed on the structure of SMG-1:SMG-9, shown as a transparency.
Several labeling experiments supported the structural model of SMG-1 based on the comparison with DNA-PKcs (Fig. 3). The SMG-1:SMG-9 complex was incubated with an excess of different antibodies, and the immune complexes were visualized in the electron microscope (see the Materials and Methods). Images of SMG-1:SMG-9 incubated with a monoclonal antibody against residues 3547–3657 at the C terminus of SMG-1 consistently revealed an extra density, the putative antibody, which, in those favorable cases where the view of SMG-1:SMG-9 suggested the location of the head and arm regions, was found mapping the head region, consistent with the assignment of the head to the C terminus (Fig. 3A). The N terminus of SMG-1 was mapped by labeling the SBP tag using streptavidin coupled to a gold particle 5 nm in diameter (Fig. 3B) or an anti-SBP antibody (Fig. 3C). In both cases, the majority of images obtained revealed the label in proximity to the narrower end of SMG-1:SMG-9, compatible with the proposed location of the N terminus of SMG-1 at the arm region (Fig. 3D).
SMG-9 in SMG-1:SMG-9 was located by difference mapping between the 3D structures of SMG-1 and SMG-1:SMG-9. Our preparation of purified SMG-1 was confirmed to be free of contamination by SMG-8 and SMG-9 by Western blotting (Fig. 1D; Supplemental Fig. S3). Unfortunately, the amount and concentration of this material was very low, and SMG-1 was apparently unstable, since images collected in the electron microscope showed poor quality, and many reference-free averages appeared blurred and featureless (data not shown). Nonetheless, we collected 8032 particles from the best of our preparations, and distorted particles were removed from the data set by computationally selecting 3750 images showing better correlation with projections of SMG-1:SMG-9 (see the Materials and Methods; Supplemental Fig. S4F). These images, refined using the structure of SMG-1:SMG-9 as the initial template for refinement and parameters in EMAN that reduce model bias (classiter = 8) (Ludtke et al. 1999), consistently converged to a structure of SMG-1 at 27 Å resolution (Fig. 3E). As a control, an independent refinement was performed with 11,358 particles from a SMG-1 sample containing residual traces of SMG-1:SMG-8:SMG-9 complexes (Supplemental Fig. S3C) after the images from these contaminants were computationally eliminated from the data set by removing those images that showed a high correlation with the structure of SMG-1:SMG-8:SMG-9 (described below). This independent data set generated an identical structure of SMG-1, supporting the consistency of the differences found between SMG-1 and SMG-1:SMG-9 (data not shown).
SMG-1 showed an overall similarity with SMG-1:SMG-9, except that the head region tilts slightly and approaches the arm region. Interestingly, a significant density was found to disappear at the level of the arm (Fig. 3E, labeled with an asterisk in SMG-1:SMG-9). We calculated a difference map between SMG-1:SMG-9 and SMG-1 after alignment of the two structures (Fig. 3F). The head and arm regions of both molecules were aligned separately to account for the minor conformational change observed between the two molecules. Differences were found only at a precise location, which unequivocally mapped SMG-9 as a density bound to the arm region containing the HEAT repeats (Fig. 3F, purple). This finding would be consistent with the biochemical experiments performed (see below). The volume occupied by SMG-9 in SMG-1:SMG-9 was roughly consistent with a 40- to 60-kDa globular protein, based on the number of voxels occupied and the average density of proteins.
SMG-8 induces large conformational changes in the SMG1C complex
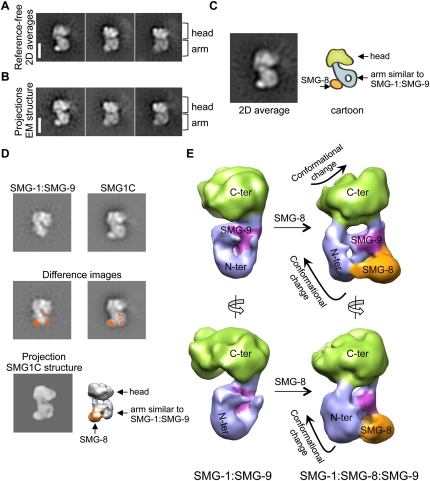
We analyzed the 3D structure of SMG1C by EM using similar methodology (Supplemental Figs. S4, S7), and the 2D averages revealed a molecule clearly distinct from SMG-1:SMG-9 (Fig. 4A). SMG1C showed longitudinal dimensions similar to SMG-1:SMG-9 and a bulky head, but the arm region now appeared to be embedded within a wider molecule. In these images, the outline of a thin and bent arm containing a region of lower density at its center was similar to the arm found in the structure of SMG-1:SMG-9 (Fig. 2), but a dot of density was now clearly attached (Fig. 4C). SMG-8 was mapped in SMG1C as this clear density by aligning reference-free averages of SMG1:SMG-9 and SMG1C and calculation of a difference image (Fig. 4D, middle panels). Images (13,853) of SMG1C were processed to obtain a 3D structure of the complex at a resolution of 24 Å (Fig. 4E). The consistency of the reconstruction was supported by the strong resemblance between the projections of the structure of the complex obtained after 3D refinement and 2D reference-free averages (Fig. 4A,B). Then, SMG-8 was located in the 3D structure by comparing reference-free averages of SMG1C—in which SMG-8 had been mapped as the difference image of SMG1C and SMG-1:SMG-9 (Fig. 4D, middle panels)—with all possible projections of the structure by cross-correlation. The best match placed the density of SMG-8 as a precise density in the 3D structure (Fig. 4D, bottom panels).
Figure 4.
3D architecture of the SMG1C complex. (A,B) Reference-free 2D averages of SMG1C (A) compared with projections of the 3D structure of SMG1C (B). Bar, 10 nm. (C) Cartoon of a 2D average of SMG1C. (D) Comparison and difference between 2D reference-free averages of SMG-1:SMG-9 (top left) and SMG-1:SMG-8:SMG-9 (top right). The difference image (orange) locates the position of SMG-8 in the SMG1C complex, and is represented superimposed on the average of SMG-1:SMG-9 (middle left) and SMG1C (middle right). Several 2D averages of SMG1C were cross-correlated with projections of the 3D structure of SMG1C, permitting the assignment of the mass detected by difference mapping, SMG-8, within the structure of SMG1C. All of the different averages used mapped the same density. (E) Views of the 3D structure of SMG1C compared with the compatible view in SMG-1:SMG-9. Color codes are as follows: The putative mapping of SMG-8 is colored in orange, the C-terminal head region is in green, the N-terminal arm domain is in blue, and SMG-9 is in purple.
SMG1C showed a prominent head region (Fig. 4E, green), whereas the arm revealed—in addition to the density mapped above as corresponding to SMG-8 (Fig. 4E, orange)—a region whose overall shape (roughly triangular and with lower density in its center) was similar to the structure of the bent arm in SMG-1:SMG-9, although with a different orientation (Fig. 4E, purple and blue). Based on this similarity, SMG-9 (Fig. 4E, purple) and the arm of SMG-1 (Fig. 4E, blue) could be placed within the structure of SMG1C, and, interestingly, SMG-8 was found contacting SMG-9. SMG-8 and SMG-9 appeared to form a complex within SMG1C, a finding that justifies the requirement of SMG-9 for the recognition between SMG-1 and SMG-8 (Fig. 1B; Yamashita et al. 2009), and previous findings indicating that SMG-8 and SMG-9 interact in the absence of SMG-1 (Fernandez et al. 2010). Remarkably, a motion of the arm of SMG1C when compared with SMG-1:SMG-9 was observed (Fig. 4E, blue), and the 3D structure of the C-terminal region of SMG1C showed manifest differences from that of SMG-1:SMG-9 (Fig. 4E, green), despite SMG-8 contacting only the N terminus of SMG-1:SMG-9.
The N terminus of SMG-1 recruits SMG-8 and SMG-9
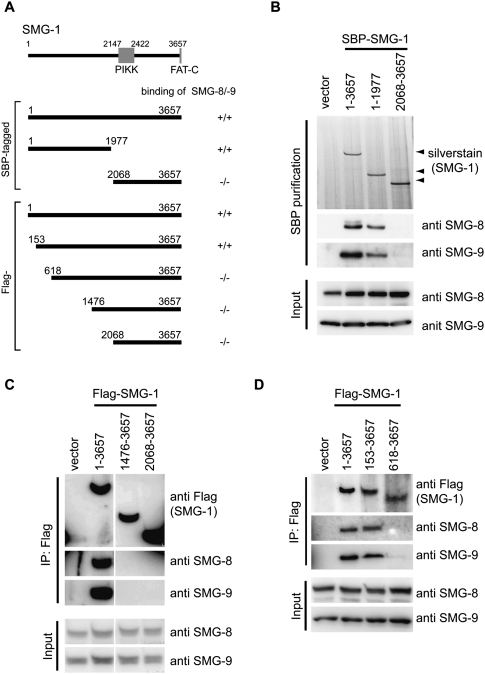
Several experiments were performed to test the structural model of SMG1C in the presence of RNaseA to remove RNA-mediated interactions. Initially, several SBP-tagged constructs of SMG-1 were expressed in 293T cells, and the presence of bound SMG-8 and SMG-9 was tested after affinity purification in the presence of RNaseA and Western blotting (Fig. 5A). Consistent with the structural model, whereas full-length SMG-1 (SMG-11–3657) and an N-terminal fragment (SMG-11–1977) were capable of binding both SMG-8 and SMG-9, a C-terminal fragment containing the kinase domain (SMG-12068–3657) did not bind detectible amounts of these two proteins (Fig. 5B). Similar experiments were performed with Flag-tagged fragments SMG-11–3657, SMG-1153–3657, SMG-1618–365, SMG-11476–3657, and SMG-12068–3657, which were designed based on the evolutionally conserved regions at the N-terminal side of SMG-1 (Fig. 5A; Morita et al. 2007). These experiments revealed that, whereas SMG-1153–3657 recognized SMG-8 and SMG-9 as efficiently as the full-length protein, SMG-1618–3657 showed a significant reduction in the amount of SMG-8 and SMG-9 bound to SMG-1 (Fig. 5C,D). As a whole, these results would be consistent with a role of the N-terminal side of SMG-1 (Fig. 1A) as the module implicated in the assembly of SMG-8 and SMG-9.
Figure 5.
SMG-8 and SMG-9 bind the N-terminal region of SMG-1. (A) SBP-tagged and Flag-tagged SMG-1 plasmids used. (B–D) 293T cells were transfected with SBP-tagged (B) and Flag-tagged (C,D) SMG-1 plasmids shown in A, together with a plasmid expressing the siRNA targeted to the 3′ untranslated region (UTR) of SMG-1. Cells were lysed and pulled down with the streptavidin sepharose (B) or anti-Flag (C,D) antibody in the presence of RNaseA. Pulled-down products or cell lysates (input) were then probed with the antibodies shown on the right.
SMG-8 regulates the kinase activity of SMG-1 on Upf1
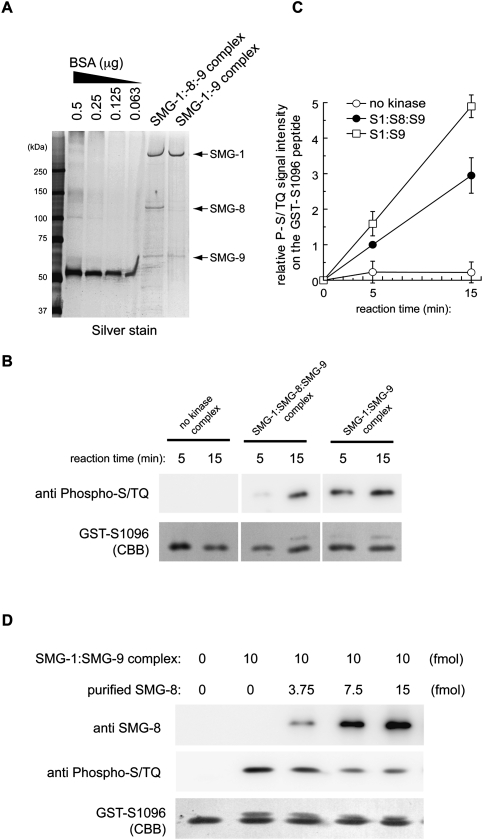
We evaluated the role of SMG-8 and SMG-9 in regulating the kinase activity of SMG-1 in vitro on a GST fusion peptide containing residue Ser 1096 of Upf1 (GST-Upf1-S1096), which can be phosphorylated by SMG-1, and this phosphorylation can be detected using the anti-phospho-ATM/ATR antibody (Fig. 6; Yamashita et al. 2009). We tested the activity of the recombinant SMG1C and SMG-1:SMG-9 complexes purified using methods identical to those used for the complexes visualized in the electron microscope. The purity of these preparations was analyzed by SDS electrophoresis, and we verified that SMG-8 was not a contaminant in the SMG-1:SMG-9 sample (Fig. 6A). SMG-1 was also purified, but these preparations appeared to be unstable, resulting in nonreproducible measurements of its kinase activity, in agreement with the behavior of SMG-1 when analyzed by EM. Thus, the kinase activity of SMG-1 was not further analyzed. These experiments revealed that the recruitment of SMG-8 down-regulated the kinase activity of the SMG-1:SMG-9 complex (Fig. 6B,C). The effect of SMG-8 on the kinase activity of SMG-1:SMG-9 complexes was further explored by the purification of recombinant SMG-8 (Supplemental Fig. S8) and the analysis of the kinase activity of recombinant SMG-1:SMG-9 in the presence of increasing amounts of SMG-8. SMG-8 was found to suppress the activity of the SMG-1:SMG-9 complex in a dose-dependent manner (Fig. 6D).
Figure 6.
Kinase activity of recombinant SMG-1:SMG-9 and SMG-1:SMG-8:SMG-9 complexes. (A) Fifty femtomoles of SMG-1 complexes was separated by SDS-PAGE and silver-stained. (B) In vitro kinase assay using 10 fmol of purified complexes and 30 pmol of GST-Upf1-S1096 peptide as the substrate. (C) Quantification of the phosphorylation level of GST-Upf1-S1096 in vitro. Relative values against the 5-min activity of the SMG-1:SMG-8:SMG-9 complex are shown. Mean values ± SD from three independent experiments are shown. (D) In vitro kinase assay using the indicated amount of purified SMG-8, 10 fmol of purified SMG-1:SMG-9 complex, and 30 pmol of GST-Upf1-S1096 peptide as a substrate for 10 min.
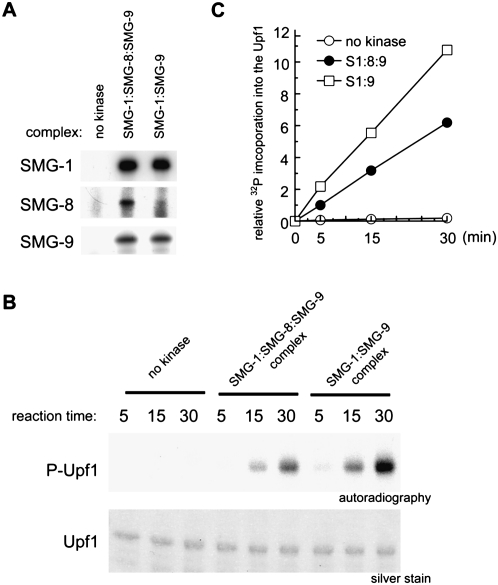
To assess the effect of SMG-8 under conditions closer to those found in vivo, we also purified endogenous complexes of the SMG-1 kinase by immunopurification, and its kinase activity was tested using its natural substrate, the Upf1 protein. We immunopurified SMG-1:SMG-8:SMG-9 and SMG-1:SMG-9 complexes from cell extracts using anti-SMG-1-C antiserum (Yamashita et al. 2001), and the presence or absence of each subunit in the complex was verified by Western blotting (Fig. 7A). To purify SMG-1:SMG-9, SMG-8 was knocked down by siRNA. The kinase activity of the endogenous SMG-1 complexes on purified recombinant Upf1 (Supplemental Fig. S9) showed that SMG-8 specifically reduced the amount of phosphorylation of SMG-1 on Upf1 (Fig. 7B,C).
Figure 7.
Kinase activity of endogenous SMG-1:SMG-9 and SMG-1:SMG-8:SMG-9 complexes. (A) HeLa TetOff cells were transfected with the nonsilencing siRNA (for those experiments labeled as “no kinase” and “SMG-1:SMG-8:SMG-9 complex”) or siRNA targeted to SMG-8 (for the SMG-1:SMG-9 complex). SMG-1 complexes were immunoprecipitated with preimmune serum (labeled “no kinase,” since it should not immunoprecipitate SMG-1) or anti-SMG-1-C antiserum for the SMG-1:SMG-8:SMG-9 complex and the SMG-1:SMG-9 complex, and were analyzed by Western blotting. (B) In vitro kinase assay with SMG-1 immunoprecipitates using and SBP-Upf1 as a substrate. (C) Quantification of the phosphorylation level of SBP-Upf1 in vitro. Relative values against a nonsilencing siRNA-treated control are shown. Mean values from two independent experiments are shown.
Discussion
Here, we reveal the 3D architecture of SMG-1 and the SMG1C complex (Yamashita et al. 2009; Fernandez et al. 2010). SMG-1 is organized in two major structurally distinct regions: a head and an arm projecting outward from the head. SMG-1, as well as all other members of the PIKK family of kinases, contains a conserved C-terminal region comprising the kinase domain as well as two conserved α-helical domains named FAT and FATC, whereas an extensive N-terminal domain is made of poorly conserved helical regions, mainly HEAT repeats, arranged as superhelical structures (Fig. 1A; Perry and Kleckner 2003). Atomic information on any PIKK kinase is lacking, but DNA-PKcs is best characterized by EM and a 6.6 Å resolution crystallographic model where the polypeptide chain was not traced (Rivera-Calzada et al. 2005; Sibanda et al. 2010). These studies reveled that the kinase, FAT, and FATC domains of DNA-PKcs are organized as a compact region (the head), whereas the long N-terminal HEAT repeat domain, accounting for 70% of the protein, appeared, at least a large part of it, as two twisted helical arms forming an open ring (Supplemental Fig. S1). In these studies, the kinase domain was unambiguously mapped at the top of the head with the catalytic site accessible, using antibody-labeling experiments and computational fitting in the EM structures (Rivera-Calzada et al. 2005; Sibanda et al. 2010) and by direct detection of the PI3K fold in the crystal structure (Sibanda et al. 2010). How the HEAT repeat regions at the N terminus of DNA-PKcs can organize in two arms projecting from the C-terminal head is uncertain given the lack of higher-resolution structures. The comparison of the structure of SMG-1 with that of DNA-PKcs, as well as the antibody-labeling performed in this study, strongly suggests that the head domain in SMG-1 comprises the catalytic kinase domain and probably most of the conserved C-terminal domains, including a C-terminal insertion unique to SMG-1. The function of this C-terminal insertion is unknown, and this region is missing in the SMG-1 ortholog in Caenorhabditis elegans, suggesting that it might be an adaptation to a more complex regulation acquired during evolution. Biochemical mapping has implicated a C-terminal fragment of SMG-1 containing the kinase domain, this insertion, and the FATC domain in recognition of Upf2 (Kashima et al. 2006). Since the interaction between SMG-1 and the Upf1:Upf2:Upf3 complex regulates the activation of SMG-1 and Upf1 phosphorylation, a tight structural association among the C-terminal domains in SMG-1 could facilitate complex regulatory interplays. For instance, interactions of the C-terminal FATC and the unique insertion with other proteins (Upf2, for instance) could contribute to regulation by inducing conformational changes in the kinase domain nearby. The kinase domain in the PIKK family is homologous to PI3Ks; thus, these mechanisms could be similar to the regulation described for PI3K, where a helical domain tightly packs with the catalytic domain, and interactions of regulatory subunits with this helical domain suppress the kinase activity (Lempiainen and Halazonetis 2009).
The region of SMG-1 defined as the arm would comprise the HEAT repeats placed N-terminal to the conserved head. This arm domain is bent, similar to what it is found in DNA-PKcs (Supplemental Fig. S1) and other HEAT repeat-containing proteins. Interestingly, SMG-1 is composed of one rather than the two arm regions found in DNA-PKcs. Approximately 70% of the residues in DNA-PKcs primary structure (∼2800 amino acids out of 4127) correspond to the helical repeats placed N-terminal to the conserved FAT, kinase, and FATC domains, suggesting that this longer HEAT repeat region is accommodated as a duplication of the arm rather than a longer single-helical structure.
We found that the N-terminal region of SMG-1 functions as a scaffold for SMG-9. The results obtained in the interaction assays, combined with the structure of the SMG-1:SMG-9 complex and its comparison with DNA-PKcs, indicate that the HEAT repeats of SMG-1 recognize SMG-9. This finding agrees with the general role of HEAT repeats as protein scaffolds. The interaction of SMG-1 and SMG-9 enhances the stability of SMG-1, which, in our hands, damages frequently during purification and/or storage. SMG-8 interacts with SMG-9 when it is a part of SMG1C, and the evidence supports that SMG-9 recruits SMG-8 to the complex: (1) SMG-8 and SMG-9 form complexes in solution devoid of SMG-1 (Fernandez et al. 2010), and this interaction would likely be maintained when bound to SMG-1. (2) The C-terminal domain of SMG-9 binds SMG-8 (Fernandez et al. 2010). (3) Whereas SMG-1 and SMG-9 interact to form the SMG-1:SMG-9 complex, SMG-8 complexes with SMG-1 mostly contain SMG-9 (Fig. 1B). (4) Both proteins interact in vivo with the same region of SMG-1 (Fig. 5). Thus, we propose that SMG-9 possibly functions as a bridge between SMG-1 and SMG-8. Given the central role of SMG-9 in the assembly of SMG1C, it is possible that the modulation of SMG-1 might be controlled via a regulated assembly pathway of SMG1C directed by SMG-9. Although SMG-9 has been found capable of forming SMG-9:SMG-9 homodimers (Fernandez et al. 2010), the region occupied by SMG-9 in the structure of the SMG-1:SMG-9 complex is more compatible with a monomer of SMG-9 bound to SMG-1, and it could hardly accommodate a 120-kDa dimer.
Regions composed of adjacent HEAT repeats are present in many proteins acting as scaffolds for protein–protein interactions, usually adopting a superhelical structure. These structures are prone to conformational changes induced by the interaction with a partner, as revealed by the structure of importin 13 bound to Mago-Y14 (Bono et al. 2010). In the SMG1C complex, an apparent motion of the HEAT repeat-containing arm region is detected after SMG-8 binding to SMG-1:SMG-9. Importantly, whereas biochemical and structural data show that SMG-8 contacts SMG-1 far away from the location of the kinase domain, inferred from its location in DNA-PKcs (Supplemental Fig. S1), SMG-8 is found to strongly down-regulate the catalytic activity of SMG-1 (Figs. 6, 7). This modulation correlates with the conformational changes we observed in the C-terminal head domain in SMG1C containing the catalytic domain (Fig. 4), thus suggesting that conformational changes in the N-terminal region would be transmitted to the head. Consistent with this idea, the N-terminal region of SMG-1 has been found to be essential for its kinase activity (Morita et al. 2007).
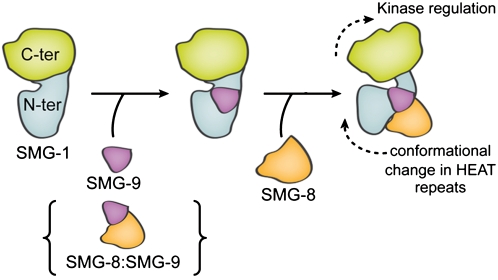
In conclusion, SMG-8-induced conformational changes modulate SMG-1 activation, and SMG-9 seems to be a major controller of SMG1C assembly by recruiting SMG-8 to SMG-1 (Fig. 8). SMG-9 could bind SMG-1 first to recruit SMG-8, or, alternatively, SMG-8 could be recruited as part of a preassembled SMG-8:SMG-9 complex, since SMG-8:SMG-9 complexes that do not contain SMG-1 have been detected previously (Fernandez et al. 2010). Then, SMG-8 recruits the inactive SMG1C complex to the PTC recognition complex (Yamashita et al. 2009). The N-terminal and C-terminal domains of SMG-9 are essential for binding to SMG-1, whereas only the C terminus of SMG-9 is required to recruit SMG-8 to the complex (Fernandez et al. 2010). SMG-8 and SMG-9 bind to the HEAT repeat region of SMG-1, inducing large-scale conformational changes that modify the conformation of the catalytic domain, and tuning the kinase activity of SMG-1 to the requirements of NMD machinery.
Figure 8.
Model of the regulation of the kinase activity of SMG-1 through the assembly of the SMG1C complex. SMG-8 could be recruited to a preformed SMG-1:SMG-9 complex or as part of a pre-existing SMG-8:SMG-9 complex.
Materials and methods
Plasmids
pEF_Flag-HA-SBP-SMG-1 (1–3657), pEF_Flag-HA-SBP-SMG-1 (126–3657), pSR_Strep-HA-SMG-9 (2–520), pSR_HA-SMG-8 (2–991), and pEF_Flag-HA-SBP-SMG-8 (2–991) plasmids were constructed by cloning each cDNA fragment using standard methods. siLentGene-puro-siSMG8cds, siLentGene-puro-siSMG9cds, or siLentGene-puro-siSMG1cds, U6 promoter-driven shRNA expression vectors (siLentGene-puro; Promega) used for SMG-8 (siRNA sequence: siGENOME duplex D-016480-02 [Dharmacon]), SMG-9 (siRNA sequence: siGENOME duplexD-020561-01 [Dharmacon]), or SMG-1 (siRNA sequence: GTGTATGTGCGCCAAAGTA), respectively, were constructed as described in the manual.
pNTAP-SMG-1-N (amino acids–1977), pNTAP-SMG-1-C (amino acids 2068–3657), pSR_Flag-SMG-1 full (1–3657), pSR_Flag-SMG-1153–3657, pSR_Flag-SMG-1618–3657, pSR_Flag-SMG-11476–3657, pSR_Flag-SMG-12068–3657, and siLentGene-puro-siSMG-1UTR (siRNA sequence targeted to the 3′ untranslated region [UTR] of the SMG-1 mRNA; target sequence: GGAAGATTTGATGCATTCA) were described previously (Morita et al. 2007; Izumi et al. 2010).
Antibodies
SMG-1 and SMG-8 antibodies used in immunodepletion experiments were generated against recombinant human SMG-1 (amino acids 1827–1925) or human SMG-8 (amino acids 621–734) proteins fused to maltose-binding protein (MBP). Affinity purification of antibodies was performed using standard procedures. Anti-SMG-1, anti-SMG-8, and anti-SMG-9 antibodies used in Western blotting have been described previously (Yamashita et al. 2009). Anti-HA-HRP (clone 3F10) (Roche), anti-Flag (clone M2) (Sigma), anti-mTOR (Cell Signaling Technology), and anti-aPKCλ (C-20) (SantaCruz Biotechnology) were obtained commercially.
Purification of SMG-1, SMG-1:SMG-9, SMG-1:SMG-8:SMG-9, SMG-8, and Upf1
For the SMG1C (SMG-1:SMG-8:SMG-9) complex, 5 × 108 293T cells were cotransfected with pEF_Flag-HA-SBP-SMG-1 (1–3657), pSR_Strep-HA-SMG-8 (2–991), and pSR_Strep-HA-SMG-9 (2–520) plasmids at a 2:1:1 ratio using HEKfectin (Bio-Rad). For the SMG-1:SMG-9 complex, 5 × 108 293T cells were cotransfected with pEF_Flag-HA-SBP-SMG-1 (126–3657), pSR_Strep-HA-SMG-9 (2–520), and siLentGene-puro-siSMG-8cds at a 2:1:1 ratio using HEKfectin (Bio-Rad). For SMG-1, 5 × 108 293T cells were cotransfected with pEF_Flag-HA-SBP-SMG-1 (126–3657), siLentGene-puro-siSMG-9cds, and siLentGene-puro-siSMG-8cds at a 2:1:1 ratio using HEKfectin (Bio-Rad). For SMG-8, 1 × 108 293T cells were cotransfected with pEF_Flag-HA-SBP-SMG-8 (2–991), siLentGene-puro-siSMG-9cds, and siLentGene-puro-siSMG-1cds at a 1:1:1 ratio using HEKfectin (Bio-Rad). For Upf1, 1 × 108 293T cells were transfected with pSR_myc-SBP-Upf1. Two days after transfection, cells were lysed with a loose-fit Potter-Elvehjem homogenizer in F-buffer (10 mM HEPES-KOH at pH 7.5, 50 mM NaCl, 10% [w/v] glycerol, 10 mM MnCl2, 50 mM β-glycerophosphate, 1 mM DTT, protease inhibitor cocktail [Roche], phosphatise inhibitor cocktail [Roche]) with 100 μg/mL RNaseA. The soluble fractions were precleared with sepharose 4B (Sigma) and then incubated with streptavidin sepharose (GE Biotech) for 2 h at 4°C with gentle rotation. After washing with RNase(−) F-lysis buffer, the affinity-purified protein complexes were eluted by incubation for 30 min at 4°C with RNase(−) lysis buffer containing 2 mM biotin (Sigma). For the purification of SMG-1, the fractions containing SMG-1 were incubated with 4 μg of anti-SMG-8 and 2 μg of anti-SMG-9 antibodies for 1 h at 4°C, then incubated with Protein-G-Dynabeads for 1 h at 4°C to immunodeplete SMG-8 and SMG-9. The supernatant collected contained purified SMG-1.
Negatively stained EM and 3D reconstructions
SMG-1, the SMG-1:SMG-9 complex, and SMG1C were observed in a JEOL JEM-1230 electron microscope operated at 100 kV after staining with 2% uranyl formate. Images of single molecules were extracted from micrographs recorded at a nominal magnification of 50,000× under low-dose conditions and were scanned at a final 4.2 Å per pixel. Micrographs were taken at 0° and 20° tilting of the specimen holder to increase the number of views of the molecules. More than 20,000 images for each of the complexes—SMG-1:SMG-9 and SMG-1:SMG-8:SMG-9—were initially extracted.
Reference-free 2D methods implemented in EMAN (Ludtke et al. 1999) and 2D maximum-likelihood classification (Scheres et al. 2005) implemented in XMIPP (Scheres et al. 2008) were used to analyze the data from each of these samples. 2D classification was used to remove distorted particles from the data set to end up with 15,608 images of SMG-1:SMG-9 and 13,853 images of SMG-1:SMG-8:SMG-9 for subsequent analysis. 3D structures of SMG-1:SMG-9 and SMG-1:SMG-8:SMG-9 were obtained by angular refinement methods (Supplemental Fig. S4) using several starting templates, which converged to similar structures, supporting the consistency of the final structures (Supplemental Figs. S6, S7). Among these references, we used several unbiased Gaussian blobs of different shapes generated with EMAN. In addition, we performed an ab initio reconstruction of SMG-1:SMG-9 using the RCT method, with images of the same field taken at 0° and 50° tilting and using XMIPP (Supplemental Fig. S5; Scheres et al. 2008). Angular refinement was performed by projection matching combined with a maximum-likelihood algorithm using XMIPP (Scheres et al. 2008). The handedness of the reconstructions defined by the RCT models was the one that best matched the atomic model of DNA-PKcs, and this was used for rendering. The resolution of the reconstructions was estimated to correspond to 24 Å for SMG-1:SMG-9 and 24 Å for the SMG1C complex, using a cutoff of 0.5 of the Fourier shell correlation coefficient (Supplemental Fig. S4). Importantly, the consistency of the final structures of SMG-1:SMG-9 and SMG-1:SMG-8:SMG-9 was supported by the agreement between 2D averages obtained by reference-free methods and the 2D averages and projections after angular refinement (Figs. 2, 4).
Particles (8032) of purified SMG-1 (Fig. 1E) were collected. SMG-1 seemed prone to distortions, and reference-free 2D averages revealed particles of different quality. The best images of SMG-1 were selected computationally as those with the best cross-correlation with projections of the 3D structure of SMG-1:SMG-9. These images were extracted, which showed overall features similar to those of SMG-1:SMG-9 after reference-free averaging. The data set was refined using the structure of SMG-1:SMG-9 as a template and using parameters in EMAN to reduce model bias (classiter = 8) (Ludtke et al. 1999). From the first step of refinement, the resulting structure revealed differences from the template, which were further refined until convergence. During refinement, those class averages visually judged to be of poor quality were not used to build the new model. The final structure at a resolution of 27 Å was reconstructed using 3750 images. Images (11,358) of a different data set obtained from the sample shown in Supplemental Fig. S3 were also analyzed. Contamination by images of SMG1C complexes was computationally removed by supervised classification of the data set using as templates the structures of SMG-1:SMG-9 and SMG1C complexes. Particles correlating better with the structure of SMG-1:SMG-9 were split and processed independently, and a result similar to that obtained for the pure SMG-1 sample (Fig. 1E) was obtained.
Antibody labeling of SMG-1:SMG-9
The SMG-1:SMG-9 complex was incubated with a threefold molar excess of each antibody for 1 h at 25°C, and the immune complexes were observed in a JEOL 1230 microscope at a nominal magnification of 25,000× (36,250× at the camera) using a TemCam-F416 camera (TVIPS). The following antibodies were used: monoclonal antibody (M02), clone 1A8 against residues 3547–3657 of SMG-1 (Abnova), monoclonal antibody against the SBP tag (SantaCruz Biotechnology). Also, 5-nm gold particles coupled to streptavidin were used to label the SBP tag (BBInternational).
Cryo-EM and 3D reconstruction of SMG-1:SMG-9
The SMG-1:SMG-9 complex was vitrified using a FEI Vitrobot after deposition of the sample on Quantifoil R 1.2/1.3 grids (http://www.quantifoil.com). These grids were decorated previously with a thin layer of carbon on top to facilitate interaction of the sample, given the low concentration of the material purified. Cryo-EM images were recorded on film under low-dose conditions using a JEM-2200FS electron microscope (JEOL) operated at 200 kV and equipped with an energy filter to increase the signal to noise ratio of the images (Supplemental Fig. S4). Micrographs were scanned to 2.1 Å per pixel and averaged to 4.2 Å per pixel, and the 15,500 images were extracted using XMIPP (Scheres et al. 2008). Reference-free 2D averages were obtained by multivariate statistical analysis using SPIDER (Shaikh et al. 2008). The 3D structure of SMG-1:SMG-9 was refined using a combination of projection matching and maximum likelihood as implemented in XMIPP (Scheres et al. 2008), and using the 3D reconstruction of the complex obtained from stained specimens after low-pass filtering to lower resolution as the initial template. The cryo-EM structure of SMG-1:SMG-9 was solved at a resolution of 30 Å (cutoff of 0.5 of the Fourier shell correlation coefficient) (Supplemental Fig. S4).
Kinase activity of SMG-1
For those experiments using purified recombinant complexes, incubations were performed using 10 fmol of purified proteins in 20 μL of 1× kinase reaction mixture (10 mM Hepes-KOH at pH 7.5, 50 mM NaCl, 10% [w/v] glycerol, 10 mM MnCl2, 50 mM β-glycerophosphate, 1 mM DTT, 0.1%BSA [w/v], phosphatase inhibitor cocktail [Roche], protease inhibitor cocktail [Roche]) containing 50 μM ATP for 5 or 15 min at 30°C (Fig. 6B) or 10 min at 30°C (Fig. 6D), using 1 μg of purified GST-Upf1-S1096 as a substrate (Yamashita et al. 2009). After separation by SDS-PAGE, phosphorylated GST-Upf1-S1096 peptides, amino acids 1090–1113 (QIDVALSQDSTYQG) of Upf1 fused with GST, were detected by Western blotting using the anti-phospho-ATM/ATR substrate antibody (Cell Signaling Technology). Signals were quantified with LuminoImager LAS-3000 and Science Lab 2001 Image Gauge software (Fuji Photo Film).
For those experiments performed using endogenous SMG-1 complexes, cells were lysed in F-buffer. Endogenous SMG-1 was immunoprecipitated using anti-SMG-1-C antiserum (Yamashita et al. 2001). Immunoprecipitates were washed with F-buffer containing 0.25 M LiCl and 1× kinase reaction buffer. Kinase reactions were carried out in 50 μL of 1× kinase reaction buffer containing 50 μM ATP and 5 μCi [γ-32P]ATP for 5, 15, or 30 min at 30°C using 1 μg of purified wild-type SBP-Upf1 as substrate. After separation by SDS-PAGE, gels were silver-stained. Then, phospho-proteins were visualized by autoradiography. The extent of phosphorylation was evaluated using an imaging analyzer (BAS2500, Fuji Film).
Interactions assays
293T cells were transfected using HEKfectin (Bio-Rad) and were lysed with a loose-fit Potter-Elvehjem homogenizer in F-buffer (10 mM HEPES-KOH at pH 7.5, 50 mM NaCl, 10% [w/v] glycerol, 10 mM MnCl2, 50 mM β-glycerophosphate, 1 mM DTT, protease inhibitor cocktail [Roche], phosphatase inhibitor cocktail [Roche]) with 100 μg/mL RNaseA. The soluble fractions were precleared with sepharose 4B (Sigma) and then incubated with streptavidin sepharose (GE Biotech) for 2 h at 4°C with gentle rotation. Precleared lysates were incubated with streptavidin-sepharose or anti-Flag (M2) antibodies for 2 or 1 h at 4°C with gentle rotation. When using antibodies, the soluble fractions were subsequently incubated with 30 μL of protein G sepharose (GE Biotech) for an additional 1 h at 4°C with gentle rotation. After washing with RNase(−) F-lysis buffer, the affinity-purified protein complexes were eluted by incubation for 30 min at 4°C with RNase(−) lysis buffer containing 2 mM biotin (Sigma) or 3× Flag peptide (Sigma), respectively. All proteins in Western blot experiments were detected with an ECL Western blot detection kit (GE Biotechnology) or Lumi-Light (Roche). All experiments were performed two to three times, and typical results are shown.
For immunodepletion analysis, 4 × 105 HeLa TetOff cells were lysed with a loose-fit Potter-Elvehjem homogenizer in T-buffer (20 mM Hepes-NaOH at pH 7.5, 50 mM NaCl, 0.05% Tween 20, 2.5 mM MgCl2, 0.5 mM DTT, protease inhibitor cocktail [Roche], phosphatase inhibitor cocktail [Roche], 100 μg/mL RNaseA [Qiagen]). The lysates were incubated with 4 μg of the anti-SMG-1, anti-SMG-8, or anti-SMG-9 antibodies for 1 h at 4°C with gentle rotation. Subsequently, the soluble fractions were incubated with 1 mg of protein-G-Dynabease (Veritas) for an additional 1 h at 4°C with gentle rotation. Antibody–protein G-Dynabease complexes were removed by magnet and supernatants were stored as the “immunodepleted” fraction.
The 3D-EM structures of SMG-1:SMG-9 and SMG1C have been deposited in the Electron Microscopy Data Bank (EMDB).
Acknowledgments
We thank Reiko Muramatsu for her technical assistance. This work was funded by the Spanish Ministry of Science and Innovation (SAF2008-00451 to O.L.); the Red Temática de Investigación Cooperativa en Cáncer (RTICC) from the Instituto de Salud Carlos III (RD06/0020/1001 to O.L.); the Autonomous Region of Madrid (CAM S-BIO-0214-2006 to O.L.); the Human Frontiers Science Program (RGP39/2008 to O.L.); the Consejería de Educación de la Comunidad de Madrid y Fondo Social Europeo (to E.A.P.); the Japan Society for the Promotion of Science (to A.Y. and S.O.); the Japan Science and Technology Corporation (to A.Y. and S.O.); the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to S.O.); the Takeda Science Foundation (to S.O.); and the Yokohama Foundation for Advancement of Medical Science (to A.Y.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.606911.
Supplemental material is available for this article.
References
- Abraham RT 2004. PI 3-kinase related kinases: ‘Big’ players in stress-induced signaling pathways. DNA Repair (Amst) 3: 883–887 [DOI] [PubMed] [Google Scholar]
- Bono F, Cook AG, Grunwald M, Ebert J, Conti E 2010. Nuclear import mechanism of the EJC component Mago-Y14 revealed by structural studies of importin 13. Mol Cell 37: 211–222 [DOI] [PubMed] [Google Scholar]
- Fernandez IS, Yamashita A, Arias-Palomo E, Bamba Y, Bartolome RA, Canales MA, Teixido J, Ohno S, Llorca O 2010. Characterization of SMG-9, an essential component of the nonsense-mediated mRNA decay SMG1C complex. Nucleic Acids Res doi: 10.1093/nar/gkq749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, et al. 2010. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem 285: 1414–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE 2010. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell 39: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE 2008. The multiple lives of NMD factors: Balancing roles in gene and genome regulation. Nat Rev Genet 9: 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S 2010. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci Signal 3: ra27 doi: 10.1126/scisignal.2000468 [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S 2006. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiainen H, Halazonetis TD 2009. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J 28: 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W 1999. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Morita T, Yamashita A, Kashima I, Ogata K, Ishiura S, Ohno S 2007. Distant N- and C-terminal domains are required for intrinsic kinase activity of SMG-1, a critical component of nonsense-mediated mRNA decay. J Biol Chem 282: 7799–7808 [DOI] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O 2010. Nonsense-mediated mRNA decay in human cells: Mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci 67: 677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Kleckner N 2003. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112: 151–155 [DOI] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J 2009. Execution of nonsense-mediated mRNA decay: What defines a substrate? Curr Opin Cell Biol 21: 394–402 [DOI] [PubMed] [Google Scholar]
- Rivera-Calzada A, Maman JD, Spagnolo L, Pearl LH, Llorca O 2005. Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Structure 13: 243–255 [DOI] [PubMed] [Google Scholar]
- Scheres SH, Valle M, Nunez R, Sorzano CO, Marabini R, Herman GT, Carazo JM 2005. Maximum-likelihood multi-reference refinement for electron microscopy images. J Mol Biol 348: 139–149 [DOI] [PubMed] [Google Scholar]
- Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R 2008. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc 3: 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J 2008. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 3: 1941–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A 2008. Messenger RNA regulation: To translate or to degrade. EMBO J 27: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Chirgadze DY, Blundell TL 2010. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature 463: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O 2006. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell 22: 511–519 [DOI] [PubMed] [Google Scholar]
- Stalder L, Muhlemann O 2008. The meaning of nonsense. Trends Cell Biol 18: 315–321 [DOI] [PubMed] [Google Scholar]
- Williams DR, Lee KJ, Shi J, Chen DJ, Stewart PL 2008. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals α helices and insight into DNA binding. Structure 16: 468–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev 15: 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Kashima I, Ohno S 2005. The role of SMG-1 in nonsense-mediated mRNA decay. Biochim Biophys Acta 1754: 305–315 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. 2009. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 23: 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]