Abstract
The active centers of the hairpin and VS ribozymes are both generated by the interaction of two internal loops, and both ribozymes use guanine and adenine nucleobases to accelerate cleavage and ligation reactions. The centers are topologically equivalent and the relative positioning of key elements the same. There is good evidence that the cleavage reaction of the VS ribozyme is catalyzed by the guanine (G638) acting as general base and the adenine (A756) as general acid. We now critically evaluate the experimental mechanistic evidence for the hairpin ribozyme. We conclude that all the available data are fully consistent with a major contribution to catalysis by general acid–base catalysis involving the adenine (A38) and guanine (G8). It appears that the two ribozymes are mechanistically equivalent.
Keywords: RNA catalysis, general acid–base catalysis, nucleobase
INTRODUCTION
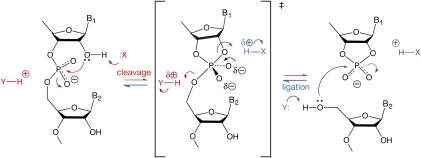
The hairpin and VS ribozymes are members of the class of nucleolytic ribozymes, which cleave or ligate RNA at a specific internal site (Lilley and Eckstein 2008). These ribozymes carry out cleavage by transesterification reactions in which the 2′-O attacks the 3′-phosphorus, with departure of the 5′-oxygen to leave a cyclic 2′-3′-phosphate (Fig. 1). In the ligation reaction the 5′-O attacks the phosphorus with departure of the 2′-O to open the cyclic phosphate. The reactions follow an SN2 mechanism, with inversion of configuration at the phosphate, and are accelerated by at least 105-fold when catalyzed by the ribozymes. While the overall structures of hairpin and VS ribozymes are quite different, their active sites have significant similarities. Herein we argue that these similarities extend to a common catalytic mechanism.
FIGURE 1.
The generally accepted chemical mechanism of cleavage and ligation in the nucleolytic ribozymes, and the possible rate acceleration by general acid–base catalysis. In the cleavage reaction (red) the 2′-O attacks the 3′-P in an SN2 process leading to a trigonal bipyramidal phosphorane that is probably close to the transition state. This is consistent with observed inversion of configuration, although it should be noted that a constrained stepwise mechanism could conceivably result in inversion. Simultaneous breakage of the bond to the 5′-O leads to the cyclic 2′3′ phosphate and 5′-O products. In the ligation reaction (blue), the 5′-O nucleophile attacks the P of the cyclic phosphate. The cleavage reaction can be potentially catalyzed by a general base (X) assisting in the removal of the proton from the 2′-OH, and a general acid (Y) that protonates the 5′-O− oxyanion leaving group. By the principle of microscopic reversibility, X and Y will act as general acid and base, respectively, in the ligation reaction. They have arbitrarily been shown as neutral in the unprotonated states, although this is not necessarily the case.
THE STRUCTURE AND ACTIVE SITE OF THE HAIRPIN RIBOZYME
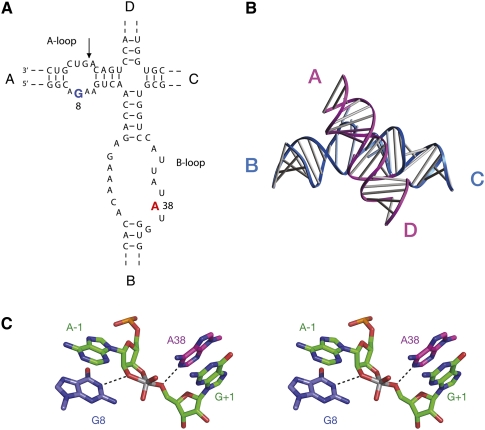
The hairpin ribozyme is centered on a four-way helical junction, with arms sequentially labeled A, B, C, and D (Fig. 2A; Hampel and Tritz 1989). Adjacent arms A and B contain loops that include all but one of the nucleotides shown to be essential for catalytic activity, and the site of cleavage/ligation is located in arm A. Crystal structures of the hairpin ribozyme show that the active conformation is generated by the intimate interaction of the A and B loops (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002; Grum-Tokars et al. 2003), as observed in the earlier FRET studies (Fig. 2B; Murchie et al. 1998; Walter et al. 1999; Wilson and Lilley 2002). The four-way junction is not required for catalytic activity, but acts as an auxiliary element that assists the folding of the ribozyme under physiological conditions (Walter et al. 1998a,b, 1999; Tan et al. 2003), akin to the loops of the hammerhead ribozyme (Khvorova et al. 2003; Penedo et al. 2004).
FIGURE 2.
The sequence and structure of the hairpin ribozyme. (A) The hairpin ribozyme secondary structure. The natural ribozyme is built on a scaffold of a four-way helical junction, with the arms labeled A through D. The important nucleotides for ribozyme activity are all contained within the internal loops in helices A and B. The key nucleotides A38 and G8 are highlighted. (Arrow) The position of cleavage and ligation. (B) Crystal structure of the hairpin ribozyme in its four-way junction form (Rupert and Ferré-D'Amaré 2001). (C) Parallel-eye stereoscopic view of the active site of a transition state analog of the hairpin ribozyme in which the pentacoordinate phosphorane is substituted by a vanadium atom (Rupert et al. 2002).
The first functional nucleobase identified in the hairpin ribozyme was G8, located in the internal loop of the A helix, on the opposite strand from the scissile phosphate. Substitution by other nucleotides led to rates of cleavage being reduced by two orders of magnitude in both the hinged form (Grasby et al. 1995; Shippy et al. 1998; Pinard et al. 2001) and the junction form of the ribozyme (Wilson et al. 2001), without affecting significantly ion-induced folding in the latter. Complete ablation of the nucleobase to leave an abasic site also led to a major loss of ribozyme activity (Kuzmin et al. 2004). The cleavage rate was found to decrease with pH when 2,6-diaminopurine was substituted at this position (whereas the rate of the natural ribozyme increases with pH up to ∼7), showing that the catalysis was now dependent on proton transfer involving a group with a pKa of 6.9 (Pinard et al. 2001), and replacement by an imidazole nucleoside analog gave cleavage and ligation rates 10-fold faster than that for a G8U variant (Wilson et al. 2006).
A crystal structure of the ribozyme in which the scissile phosphate was replaced by vanadate as a model of a pentacoordinate phosphorane transition state (Rupert et al. 2002) showed the close proximity of G8 to the catalytic center, hydrogen bonded to the 2′-O and the proS non-bridging O of the scissile phosphate and thus positioned to participate in the catalytic chemistry (Fig. 2C). This proximity has been demonstrated functionally by the transfer to G8 of the alkyl group from bromoacetamide attached at the position of the 2′-OH nucleophile (Thomas and Perrin 2006). The crystal structure also revealed the presence of a second nucleotide juxtaposed with the scissile phosphate. The nucleobase of A38 (contributed by loop B) was found to form hydrogen bonds to the 5′-O and the proR O. Removal of this nucleobase resulted in a 10,000-fold loss of activity (Kuzmin et al. 2005), while NAIM experiments showed that ligation activity was sensitive to functional group changes at this position (Ryder et al. 2001).
THE STRUCTURE AND ACTIVE SITE OF THE VS RIBOZYME
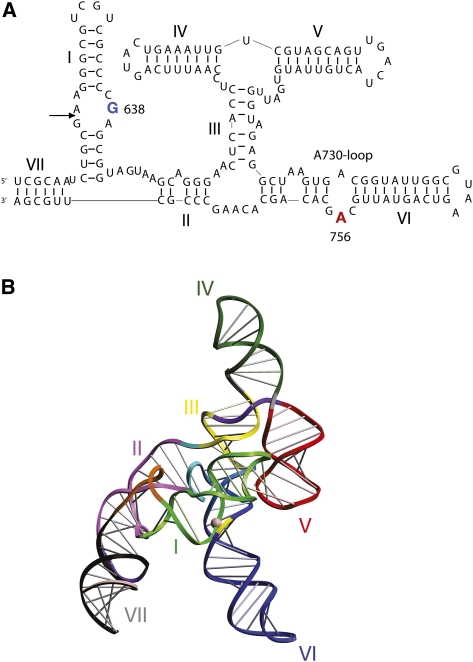
The sequence and deduced secondary structure of the VS ribozyme (Beattie et al. 1995; Jones et al. 2001) indicates that it comprises seven helical segments that are connected by three different three-way junctions (Fig. 3A). A trans-acting core of the ribozyme can be released, formed by five helices (II through VI) connected by two of the junctions to form a nominal H shape. Cleavage and ligation reactions occur within the internal loop of stem–loop I. Collins and coworkers showed that the terminal loop of helix I interacts with that of helix V (Rastogi et al. 1996).
FIGURE 3.
The sequence and structure of the VS ribozyme. (A) The VS ribozyme secondary structure. This consists of seven helical segments labeled I through VII that are associated through three three-way junctions. Cleavage occurs within the internal loop of helix I ([arrow] the position of cleavage and ligation) and requires the A730 loop within helix VI. The key nucleotides A756 and G638 are highlighted. (B) A model of the complete VS ribozyme derived from analysis of SAXS data (Lipfert et al. 2008). This is also in agreement with earlier studies based on analysis of the conformations of the individual component helical junctions (Lafontaine et al. 2002a). (Yellow) The A730 loop; (magenta sphere) the scissile phosphate.
Unlike the hairpin ribozyme, there is no crystal structure for the VS ribozyme so far. Nevertheless, biophysical studies have provided a good idea of the general fold of the ribozyme at low resolution. A model of the trans-acting core of the ribozyme was developed from studies of the individual three-way junctions, in which helices II and V were directed laterally from a central stem generated by the coaxial alignment of helices IV, III, and VI (Lafontaine et al. 2001a, 2002a). It was proposed that the probable location of helix I was in the cleft formed between helices II and VI, so that it could be connected to helix II and make the loop–loop interaction with helix V (Lafontaine et al. 2002a). More recently we have used data from small-angle X-ray scattering (SAXS) in solution to propose a model for the complete ribozyme comprising helices I through VII (Fig. 3B; Lipfert et al. 2008) that is broadly in agreement with the earlier model.
The key catalytic components of the VS ribozyme have been identified by nucleotide substitution. The A730 loop within helix VI had been found to be sensitive to ethylation interference and contained sites of interference by phosphorothioate incorporation and suppression by thiophilic metal ions (Sood et al. 1998). Moreover, sequence variants in this loop led to large effects on cleavage (Lafontaine et al. 2001b) and ligation (McLeod and Lilley 2004) activity without concomitant changes to the structure. Within the A730 loop, substitution of A756 resulted in loss of activity by three orders of magnitude (Lafontaine et al. 2002b). NAIM experiments revealed that this position was the most sensitive nucleotide to substitution by a range of analogs (Jones and Strobel 2003), and UV cross-linking data placed A756 physically close to the cleavage site in the substrate (Hiley et al. 2002). Functional group substitutions indicated that the Watson-Crick edge of the nucleobase of A756 is important for catalytic activity (Lafontaine et al. 2002b), and it was later shown that some activity could be retained when A756 was replaced by an imidazole nucleoside (Zhao et al. 2005). A second key nucleobase was found in the internal loop of the substrate helix I. Changes to G638 resulted in loss of activity by four orders of magnitude, while not affecting the binding affinity of the substrate to the ribozyme (Wilson et al. 2007). Thus as with the hairpin ribozyme, adenine and guanine nucleobases seem to be implicated as the key players in catalysis.
The low-resolution structure of the VS ribozyme (Lipfert et al. 2008) and cross-linking data (Hiley et al. 2002) suggest a close association of the substrate internal loop and the A730 loop. This interaction potentially brings together the scissile phosphate and G638 (in the substrate loop) and A756 (in the A730 loop). The association of these elements has been demonstrated by a restoration of activity in a mixture of two VS ribozymes that are mutated in one or other nucleobases (Ouellet et al. 2009).
THE ACTIVE SITES OF THE HAIRPIN AND VS RIBOZYMES ARE TOPOLOGICALLY SIMILAR
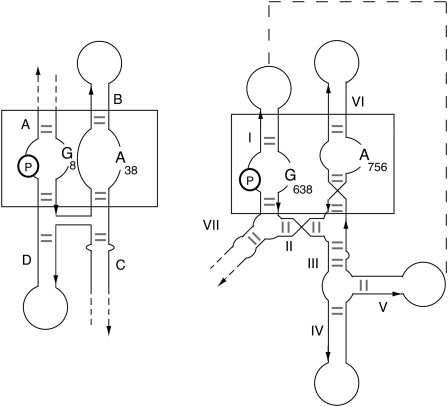
The active sites of both ribozymes appear to be generated by loop–loop interaction, which brings together the scissile phosphate with adenine and guanine nucleobases. This similarity goes further. We can draw the secondary structures of the two ribozymes to include the loop–loop interactions (Fig. 4), whereupon it becomes clear that the polarity of the strands and the relative positioning of the adenine, guanine, and sissile phosphate are the same in both cases. This suggests a fundamental similarity between the active sites of the two ribozymes, hinting at a deeper similarity in catalytic mechanism.
FIGURE 4.
Schematic of the structures of the hairpin and VS ribozyme drawn to show the equivalence of the two active sites. The scissile phosphate (P circled) and the important adenine and guanine nucleobases are highlighted for each ribozyme. The secondary structures are arranged to bring the interacting internal loops side by side, the A and B loops for the hairpin ribozyme and the substrate and A730 loops for the VS ribozyme. These regions are boxed in both ribozymes. Note that these are drawn so that the strand polarities (arrows point in the 3′ direction) are the same for both. When this is done, the relative positionings of the key functionalities occupy the same positions with respect to the equivalent loops.
THE CATALYTIC MECHANISM OF THE VS RIBOZYME
Like any functional RNA species, the VS ribozyme requires the presence of metal ions to fold into its active conformation. But the ribozyme is active in high concentrations of monovalent metal ions (Murray et al. 1998), so that the direct participation of a site-bound metal ion as a Lewis acid or in general acid–base catalysis is unlikely. An electrostatic role of non-site-bound metal ions, however, remains possible, perhaps even likely. We have established that the nucleobases of A756 and G638 are key players in the generation of the catalytic rate enhancement. They might stabilize the transition state by hydrogen-bonding the phosphorane, or a positively charged protonated adenine base might provide electrostatic stabilization of the dianionic transition state. The currently available evidence points toward an important role for general acid–base catalysis by these nucleobases, although this would not exclude an additional role in transition-state stabilization.
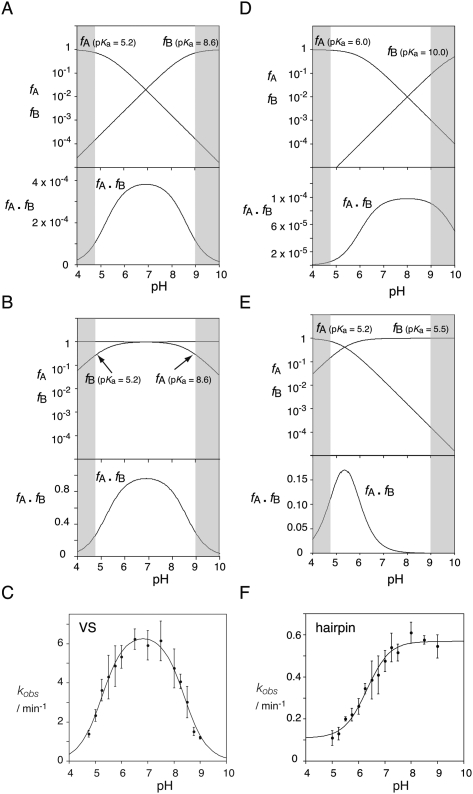
This would clearly require that the acid be protonated and the base unprotonated at the outset of the reaction (Fig. 1). The observed rate of reaction (kobs) will be given by the product of the rate of cleavage catalyzed by the ribozyme in the correct state of protonation (kcat) with the fractions of protonated acid and unprotonated base (fA and fB, respectively), i.e.,
We may simulate the pH dependence of cleavage rate by assuming probable pKa values, thereby calculating fA and fB (Bevilacqua 2003). Taking the example of an acid of pKa = 5.2 and a base of pKa = 8.6, a bell-shaped profile results (Fig. 5A). At low pH the rate rises because the acid is fully protonated (fA = 1) and the base is steadily deprotonating (fB increasing in a log-linear manner), but then the rate increase levels off as the acid begins to deprotonate (fA reducing) and the two fractions balance. Then the rate falls as the fraction of protonated acid steadily falls (fA continues to reduce log-linearly), while that of the base begins to saturate (approaches fB = 1).
FIGURE 5.
pH dependence of the rate of cleavage of the VS and hairpin ribozymes, and simulations of the pH dependence of reaction rates catalyzed by a general acid and a general base. The fractions of protonated acid (fA) and unprotonated base (fB) have been calculated as a function of pH and plotted on a logarithmic scale (upper panels). The product fA · fB should simulate the dependence of cleavage rate with pH if general acid–base catalysis is a major contributor to the catalytic rate enhancement. This is plotted on a linear scale. The gray areas at the sides are not accessible to experimental investigations. (A) Simulation of the pH dependence of reaction rate calculated for pKa (acid) = 5.2 and pKa (base) = 8.6, which gives a bell-shaped pH dependence of fA · fB. This corresponds to VS ribozyme cleavage. (B) The inversion of the pKa values for acid and base from case A. The simulation has been calculated for pKa (acid) = 8.6 and pKa (base) = 5.2. Although the absolute magnitude of the product fA · fB is higher in the second case, the profile with pH is identical to that in A. This corresponds to VS ribozyme ligation. (C) The experimental pH dependence of the cleavage reaction for the VS ribozyme in trans, in the presence of 200 mM Mg2+ ions (Wilson et al. 2007). (D) Simulation for a general base with a higher pKa. The simulation has been calculated for pKa (acid) = 6.0 and pKa (base) = 10.0. In this case, the reduction in fA · fB at high pH occurs mostly in the region pH > 9.0, where experimental investigation is difficult for RNA. This corresponds to the mechanism proposed for hairpin ribozyme cleavage. (E) Simulation for a general base with a lower pKa. The simulation has been calculated for pKa (acid) = 5.2 and pKa (base) = 5.5. This might correspond to the situation where a guanine general base has been substituted by diaminopurine, or exogenous cytosine. (F) The experimental pH dependence of the cleavage reaction for the hairpin ribozyme (in its full junction form) in the presence of 10 mM Mg2+ ions (Nahas et al. 2004).
We have obtained a bell-shaped pH dependence for the cleavage reaction in the presence of a high concentration of Mg2+ ions (Fig. 5C), which was fitted to a double-ionization model with apparent pKa values of 5.2 and 8.4 (Wilson et al. 2007). The lower value is very much in line with an adenine in an electronegative environment, while the upper value would be consistent with a guanine base if the pKa were reduced by proximity to metal ions. A bell-shaped pH dependence for cleavage, with pKa values of 5.8 and 8.3, was also observed using the fast-cleaving cis-acting form of the ribozyme (Smith and Collins 2007). The pH dependence of a reaction may reflect a change in the rate-limiting step rather than the protonation state of reactants, but several lines of evidence suggest that this is not the case for the VS ribozyme. Kinetic isotope effects in the fast, cis-acting form show that proton transfer occurs in the transition state of the cleavage reaction (Smith and Collins 2007). In the trans form of the ribozyme, the central conversion of substrate to product is rate limiting, with rapid and pH-independent substrate binding (Wilson et al. 2007). Lastly, the correlation between the pKa of the nucleobase at position 638 and the observed pKa of the cleavage reaction strongly suggests that the rate of reaction depends on the protonation state of the nucleobase (Wilson et al. 2007).
By themselves the pH profiles do not allow us to determine which nucleobase is the acid and which the base in the cleavage reaction. Comparison of Figure 5, A and B, shows that exchange of the acid and base result in identical simulated pH profiles of cleavage rate. This was resolved for the HDV ribozyme by using a 5′-phosphorothiolate (5′-PS) substitution at the scissile phosphate (Das and Piccirilli 2005). The 5′ sulfur atom is a much better leaving group than the oxygen it replaces, so does not require protonation. Substitutions in the ribozyme that impair the function of the general acid, lowering the activity of the oxy substrate, should have little effect on cleavage of a 5′-PS-containing substrate. We therefore made a 5′-phosphorothiolate substitution at the scissile phosphate of the VS ribozyme (Wilson et al. 2010). We found that the cleavage activity of VS A756G was impaired 1000-fold on the oxy (5′-PO) substrate, but the activity was completely restored for the 5′-PS-containing substrate. Thus the cleavage of the 5′-PS substrate is insensitive to substitution by guanine at position 756, and we conclude that A756 is the general acid for the cleavage reaction. In contrast, the rate of cleavage of a 5′-PS substrate with diaminopurine (DAP) at position 638 was similar to that observed for a 5′-PO substrate with G638DAP, and both were significantly slower than the natural sequence. The pH profile of cleavage rate for the G638DAP plus 5′-PO substrate was bell-shaped, corresponding to pKa values of 4.8 and 5.6 (Wilson et al. 2007), consistent with general acid–base catalysis by A756 and DAP at position 638. In contrast, with the 5′-PS substitution the reaction rate increased to pH ∼6 and remained at a plateau at higher pH (Wilson et al. 2010). A constant rate is to be expected at higher pH once the base is fully deprotonated since deprotonation of the acid is no longer relevant. These data were fitted to a single ionization, with a pKa = 5.3, consistent with general base catalysis by the diaminopurine at position 638. Thus all the available data are consistent with a catalytic mechanism for the VS ribozyme cleavage reaction in which G638 acts as general base to deprotonate the 2′-O nucleophile, and A756 is the general acid protonating the 5′-oxyanion leaving group. And by the principle of microscopic reversibility, in the ligation reaction protonated G638 should act as the general acid protonating the 2′-oxyanion leaving group, and unprotonated A756 as general base deprotonating the 5′-O nucleophile that attacks the cyclic phosphate.
DOES THE HAIRPIN RIBOZYME SHARE THE SAME CATALYTIC MECHANISM?
A striking similarity has emerged between the active sites of the VS and hairpin ribozymes in our current view. Both are formed through the interaction of two internal loops. In both cases, an active guanine lies on the opposing strand of the internal loop harboring the scissile phosphate, while an active adenine is provided by the second loop (Rupert and Ferré-D'Amaré 2001; Wilson et al. 2010). In the crystal structure of the hairpin ribozyme the positions of G8 and A38 are consistent with roles of base and acid, respectively, in the cleavage reaction, corresponding to the proposed functions of G638 and A756 in the VS ribozyme. Moreover, the topology of the two ribozymes is seen to be identical when the polarity of the strands is considered (Fig. 4). So do these ribozymes share a common mechanism using general acid–base catalysis?
It has been long recognized that G8 and A38 of the hairpin ribozyme might participate in general acid–base catalysis (Pinard et al. 2001; Rupert and Ferré-D'Amaré 2001; Lebruska et al. 2002; Rupert et al. 2002). In a very illuminating paper, Bevilacqua (2003) showed that the observed pH dependence of the reaction was consistent with general acid–base catalysis by an adenine and a guanine, and that the observed rates of reaction could be achieved despite having pKa values significantly far from neutrality. However, the function of G8 has been controversial, with some favoring the hypothesis that G8 contributes to positioning the nucleophile and stabilizing the transition state by electrostatics but not to proton transfer (Lebruska et al. 2002; Rupert et al. 2002; Kuzmin et al. 2004; Salter et al. 2006; Nam et al. 2008). These doubts have arisen from three observations.
First, the N1 of G8 donates a hydrogen bond to the O2′ nucleophile in crystal structures in which cleavage is prevented by a 2′-O-methyl group (Rupert and Ferré-D'Amaré 2001; Salter et al. 2006). It has been argued that such a hydrogen bond is not compatible with the removal of a proton from the nucleophile by G8. G8 N1 is also seen to donate a hydrogen bond to the 2′-bridging oxygen in the transition-state structure, and also to the 2′-bridging oxygen of the 2′-3′-cyclic phosphate in a product structure (Rupert et al. 2002). These structures are consistent with the proposition that G8 N1 remains protonated throughout the reaction. However, some caution is required in the interpretation of crystal structures that are inherently snapshots of ground-state structures, often with chemical modifications. In the structure of the cleavage product, G8 is positioned to act as a general acid in the ligation reaction, protonating the O2′ leaving group. If so, by microscopic reversibility it should act as a general base during cleavage. Furthermore, Wedekind and coworkers have demonstrated the sensitivity of the active-site structure to substitutions at position 8 and to modifications at the O2′ (MacElrevey et al. 2008). It seems reasonable to propose that in the active ribozyme the hydrogen bond donated by N1 is either not present or readily broken, allowing N1 to deprotonate and function as a base.
Second, the pH dependence of the ribozyme superficially appears to correspond to a single pKa for both cleavage and ligation (Pinard et al. 2001; Kuzmin et al. 2004; Nahas et al. 2004), with activity rising with pH until a plateau level is achieved around neutrality, corresponding to a pKa close to 6 (Fig. 5F). This is consistent with a single titratable group that could not plausibly be a guanine. Moreover, substitution of an abasic residue for G8 did not change the pH profile, although activity was reduced by three orders of magnitude (Lebruska et al. 2002; Kuzmin et al. 2004). Neither observation is inconsistent with general acid–base catalysis, but the absence of a significant lowering of rate at high pH would require that G8 has a pKa > 10. This is illustrated by the simulation shown in Figure 5D. If the higher pKa is 10, this results in very little loss of activity up to pH 9; the decline in fA is balanced by the increase in fB so there is no detectable effect within the experimentally observable range of pH. An elevated pKa is plausible in the vicinity of a phosphate. Removal of the nucleobase from position 8 would result in the same pH profile if an alternative base of high pKa acted in its place. Specific base catalysis or a Mg2+-bound hydroxide ion could be candidates for this role. It has been argued that the pH dependence of cleavage and ligation reactions should be the inverse of each other if rate acceleration occurs by general acid–base catalysis, whereas electrostatic catalysis would retain the same pH profile for both reactions (Lebruska et al. 2002; Kuzmin et al. 2004). While it is true that the pH dependence for an individual functional group will be inverted for the reverse reaction, where catalysis involves both a general acid and general base working together, the two inversions offset each other. Identical pH profiles result for cleavage and ligation, as demonstrated by comparison of Figure 5, A and B. Thus, ribozymes using general acid–base catalysis are expected to exhibit similar pH dependence for cleavage and ligation, as is observed experimentally for the hairpin ribozyme.
Third, data on the restoration of activity to a ribozyme with an abasic site at position 8 by addition of exogenous compounds were suggested to be inconsistent with general acid–base catalysis. In a detailed study, Fedor and coworkers (Lebruska et al. 2002; Kuzmin et al. 2004) found that each compound leading to increased activity shared an amidine group with guanine, but in contrast to guanine, each of the rescuing compounds carried a positive charge when protonated. The observed decrease in rate with increasing pH was taken to indicate that the exogenous bases were active in their protonated, cationic form. However, as illustrated in Figure 5, in general acid–base catalysis, the decline in activity at high pH is due to the deprotonation of the acid (i.e., a reduction in fA) in the presence of a fully deprotonated base. Thus, it is the neutral deprotonated forms of the exogenous bases that would be active in the cleavage reaction. The bell-shaped pH dependence and apparent pKa values of exogenous base rescue by cytosine and 2-aminopyridine (Kuzmin et al. 2004) are consistent with the hypothesis that the exogenous base is acting as a general base during cleavage, corresponding to the simulation shown in Figure 5E. The equivalent pH dependence observed for the reverse ligation reaction is also in agreement with the hypothesis. Thus the pH dependence is completely consistent with general acid–base catalysis and does not require an interpretation indicating a role in electrostatic stabilization, although electrostatic transition state stabilization could occur in parallel.
Replacing G8 with nucleobases of lower pKa provides direct support for the general base role of this nucleobase. Pinard et al. (2001) found that substitution of diaminopurine or 2-aminopurine at position 8 resulted in bell-shaped pH profiles centered near pH 6 and pH 5.5, respectively, consistent with general acid–base catalysis arising from two nucleobases of low pKa, much like the simulation shown in Figure 5E. These investigators discounted the decline in activity in low pH because it was reduced by an increased Mg2+ concentration, suggesting that this loss of activity was due to destabilization of the folded ribozyme. However, increasing the Mg2+ concentration is also expected to make the pKa values of the nucleobases more acidic, which could also explain the increased activity at low pH. Ribozymes substituted with imidazole nucleoside at position 8 also gave bell-shaped pH profiles for both cleavage (apparent pKa values of 6.2 and 7.8) and ligation (apparent pKa values of 6.6 and 7.0) (Wilson et al. 2006). The lower pKa values are in the range normally observed for the hairpin ribozyme. The upper pKa values are higher than generally observed for imidazole heterocycles substituted at position 4 but fall within the range observed for histidine in proteins. While other mechanisms cannot be discounted, these data are consistent with general acid–base catalysis involving A38 and imidazole at position 8.
Recently Liu et al. (2009) directly measured the ionization state of 8-azaguanine substituted at position 8 by means of the increased fluorescent emission when the base becomes deprotonated. Under standard conditions, the intrinsic pKa of 8-azaguanine at position 8 of an active hairpin ribozyme was 9.5. Since this is nearly 3 pH units higher than the apparent pKa of the activity of the hairpin ribozyme, the authors concluded that G8 functions in the protonated form. However, we believe that an inappropriate comparison was made. Applying the general acid–base hypothesis, inspection of Figure 5D shows that the pKa below pH 7 is due to the deprotonation of the acid (fA reducing; most probably A38), whereas the base of higher pKa (whether guanine or 8-azaguanine) would be steadily deprotonating in a log-linear manner over this range (fB increasing). In other words, the pKa measured from the pH dependence of the cleavage rate is expected to be that of A38, and it is not meaningful to compare this value with that of the 8-azaguanine measured by fluorescence. Furthermore, in contrast to the single apparent pKa observed for the pH dependence of cleavage by ribozymes with guanine at position 8 (pKa = 7.0 in that study), the pH dependence for the 8-azaguanine ribozyme activity exhibited a decline in activity above pH 9 with apparent pKa values of 6.8 and 9.9. The higher value agrees reasonably well with that measured by fluorescence. Thus, in contrast to Liu et al., we argue that there is, in fact, good agreement between the fluorescence and activity data for the 8-azaguanine ribozyme.
It is interesting to note that the pKa of 8-azaguanine is ∼1 pH unit lower than that of guanine (Liu et al. 2009). Substitution of 8-azaguanine for guanine would therefore be expected to increase fB at pH 8 since a greater proportion of molecules would have a deprotonated base. This should result in a corresponding increase in kobs, and a threefold increase in rate was, indeed, observed. Assuming that the substitution is non-perturbing and that the difference in activity is solely due to the different extent of base deprotonation, a pKa of 10.6 can be calculated for G8, in agreement with our assumption that G8 has a pKa > 10. Hence, the data of Liu et al. (2009) are entirely consistent with the hypothesis that G8 participates in general acid–base catalysis.
In contrast to G8, the importance of A38 has not been disputed, but the exact role it plays has been debated. Poisson-Boltzmann calculations (Tang et al. 2007) and Raman spectroscopy (Guo et al. 2009) have shown that A38 has an elevated pKa (pKa values of 5.9 and 5.46, respectively), consistent with the observed pH dependence of activity arising from proton transfer at this nucleobase. Exogenous base rescue and substitution with other nucleobases have demonstrated the importance of both the N1 and N6 of A38 for full activity (Kuzmin et al. 2005). Most such experiments have substituted adenine for a nucleobase with a similar pKa, resulting in a similar pH profile; however, two substitutions are particularly informative. Wedekind and colleagues (Spitale et al. 2009) substituted an N1-deazaadenine at position 38. X-Ray crystallography reveals that the ribozyme with this substitution folds into an apparently unperturbed active structure, yet one that has lost all measureable activity. This demonstrates that the N1 is essential for activity and suggests that A38 donates/accepts a proton. A ribozyme with an isoguanine substitution, which retains the N6 exocyclic amine of adenine but has a pKa similar to guanine, has negligible activity at neutral pH but is as active as the native ribozyme at high pH (Kuzmin et al. 2005). This observation is consistent with general acid–base catalysis by two nucleobases of high pKa. Isoguanine will be negatively charged when deprotonated, demonstrating that the assumption that a negative charge will not form in the electronegative environment of the active site, sometimes presented as an argument against G8 acting as a general base (Kuzmin et al. 2004; Salter et al. 2006; Nam et al. 2008), is unlikely to be valid. Lastly, in a recent study Strobel and coworkers (Suydam et al. 2010) carried out a NAIM study of ligation in the hairpin ribozyme using a series of adenosine analogs of varying pKa. They found strong interference using compounds of low pKa, consistent with ionization of N1 being required for the catalytic activity of the ribozyme.
CONCLUSION
It is practically impossible to elucidate catalytic mechanism with total certainty, either for enzymes or ribozymes, and we cannot claim to know the mechanism of either the VS or hairpin ribozymes for sure. The common features in the proposed active sites of these ribozymes suggest that their similarities could extend to their catalytic mechanisms. The evidence for an important contribution from general acid–base catalysis to the mechanism of the VS ribozyme is strong, and all the available evidence for the hairpin ribozyme is consistent with the same mechanism for that RNA. Moreover, we are unaware of any data that are not consistent with this mechanism for either ribozyme, which is presented in Figure 6. This does not exclude other contributions to the total rate enhancement. Stabilization by enhanced coordination of the TS was observed for the hairpin ribozyme (Rupert et al. 2002) and may also contribute to the VS ribozyme since the exocyclic amines of A756 and G638 are necessary for full activity (Lafontaine et al. 2002b; Wilson et al. 2007), and electrostatic effects may also contribute. But the pH dependence of a G638DAP-substituted VS ribozyme suggested that proton transfer contributes at least 100- to 1000-fold to the catalytic power of that ribozyme (Wilson et al. 2007), so general acid–base catalysis is likely to be a major source of catalytic rate enhancement. Indeed, evidence suggests that general acid–base catalysis is not restricted to the hairpin and VS ribozymes. Guanine nucleobases appear to be strong contenders for the role of general base in cleavage reactions catalyzed by the hammerhead (G12) (Han and Burke 2005; Martick et al. 2008) and GlmS (G33 or 40, depending on the source of the ribozyme) (Cochrane et al. 2007, 2009; Klein et al. 2007) ribozymes. However, the guanine plus adenine combination is restricted to the hairpin and VS ribozymes. The hammerhead ribozyme is proposed to employ a 2′-hydroxyl (Martick and Scott 2006), and the GlmS ribozyme uses an exogenous glucosamine-6-phosphate as general acid in cleavage reactions (Klein and Ferré-D'Amaré 2006; Cochrane et al. 2009), while the HDV ribozyme uses a Mg2+ ion to activate the nucleophile, either as general base or Lewis acid (Nakano et al. 2000; Chen et al. 2010), and a cytosine nucleobase as general acid (Ferré-D'Amaré et al. 1998; Nakano et al. 2000; Ke et al. 2004; Das and Piccirilli 2005). Thus it seems probable that general acid–base catalysis is a source of catalytic rate enhancement in all the nucleolytic ribozymes.
FIGURE 6.
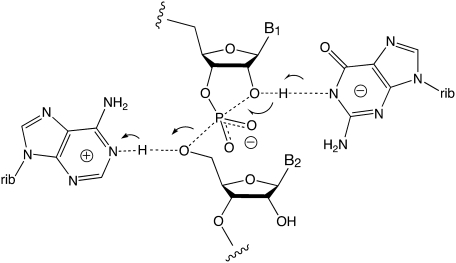
Proposed common catalytic mechanism of the VS and hairpin ribozymes. The cleavage reaction is depicted, in which a deprotonated guanine nucleobase acts as a general base to facilitate deprotonation of the 2′-O, and a protonated adenine nucleobase acts as a general acid to protonate the 5′-oxyanion leaving group.
ACKNOWLEDGMENTS
We thank our colleagues and Philip Bevilacqua for discussion, Joe Piccirilli and Shinya Harusawa for collaboration, and Cancer Research UK for funding ribozyme research in Dundee.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2473711.
REFERENCES
- Beattie TL, Olive JE, Collins RA 1995. A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc Natl Acad Sci 92: 4686–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua PC 2003. Mechanistic considerations for general acid–base catalysis by RNA: Revisiting the mechanism of the hairpin ribozyme. Biochemistry 42: 2259–2265 [DOI] [PubMed] [Google Scholar]
- Chen JH, Yajima R, Chadalavada DM, Chase E, Bevilacqua PC, Golden BL 2010. A 1.9 Å crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage. Biochemistry 49: 6508–6518 [DOI] [PubMed] [Google Scholar]
- Cochrane JC, Lipchock SV, Strobel SA 2007. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem Biol 14: 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane JC, Lipchock SV, Smith KD, Strobel SA 2009. Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme. Biochemistry 48: 3239–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SR, Piccirilli JA 2005. General acid catalysis by the hepatitis delta virus ribozyme. Nat Chem Biol 1: 45–52 [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Zhou K, Doudna JA 1998. Crystal structure of a hepatitis delta virus ribozyme. Nature 395: 567–574 [DOI] [PubMed] [Google Scholar]
- Grasby JA, Mersmann K, Singh M, Gait MJ 1995. Purine functional groups in essential residues of the hairpin ribozyme required for catalytic cleavage of RNA. Biochemistry 34: 4068–4076 [DOI] [PubMed] [Google Scholar]
- Grum-Tokars V, Milovanovic M, Wedekind JE 2003. Crystallization and X-ray diffraction analysis of an all-RNA U39C mutant of the minimal hairpin ribozyme. Acta Crystallogr D Biol Crystallogr 59: 142–145 [DOI] [PubMed] [Google Scholar]
- Guo M, Spitale RC, Volpini R, Krucinska J, Cristalli G, Carey PR, Wedekind JE 2009. Direct Raman measurement of an elevated base pKa in the active site of a small ribozyme in a precatalytic conformation. J Am Chem Soc 131: 12908–12909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel A, Tritz R 1989. RNA catalytic properties of the minimum (-)sTRSV sequence. Biochemistry 28: 4929–4933 [DOI] [PubMed] [Google Scholar]
- Han J, Burke JM 2005. Model for general acid–base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry 44: 7864–7870 [DOI] [PubMed] [Google Scholar]
- Hiley SL, Sood VD, Fan J, Collins RA 2002. 4-thio-U cross-linking identifies the active site of the VS ribozyme. EMBO J 21: 4691–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FD, Strobel SA 2003. Ionization of a critical adenosine residue in the Neurospora Varkud Satellite ribozyme active site. Biochemistry 42: 4265–4276 [DOI] [PubMed] [Google Scholar]
- Jones FD, Ryder SP, Strobel SA 2001. An efficient ligation reaction promoted by a Varkud Satellite ribozyme with extended 5′- and 3′-termini. Nucleic Acids Res 29: 5115–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke A, Zhou K, Ding F, Cate JH, Doudna JA 2004. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature 429: 201–205 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol 10: 1–5 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Ferré-D'Amaré AR 2006. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313: 1752–1756 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Been MD, Ferré-D'Amaré AR 2007. Essential role of an active-site guanine in glmS ribozyme catalysis. J Am Chem Soc 129: 14858–14859 [DOI] [PubMed] [Google Scholar]
- Kuzmin YI, Da Costa CP, Fedor MJ 2004. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J Mol Biol 340: 233–251 [DOI] [PubMed] [Google Scholar]
- Kuzmin YI, Da Costa CP, Cottrell JW, Fedor MJ 2005. Role of an active site adenine in hairpin ribozyme catalysis. J Mol Biol 349: 989–1010 [DOI] [PubMed] [Google Scholar]
- Lafontaine DA, Norman DG, Lilley DMJ 2001a. Structure, folding and activity of the VS ribozyme: importance of the 2-3-6 helical junction. EMBO J 20: 1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DA, Wilson TJ, Norman DG, Lilley DMJ 2001b. The A730 loop is an important component of the active site of the VS ribozyme. J Mol Biol 312: 663–674 [DOI] [PubMed] [Google Scholar]
- Lafontaine DA, Norman DG, Lilley DMJ 2002a. The global structure of the VS ribozyme. EMBO J 21: 2461–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DA, Wilson TJ, Zhao Z-Y, Lilley DMJ 2002b. Functional group requirements in the probable active site of the VS ribozyme. J Mol Biol 323: 23–34 [DOI] [PubMed] [Google Scholar]
- Lebruska LL, Kuzmine I, Fedor MJ 2002. Rescue of an abasic hairpin ribozyme by cationic nucleobases. Evidence for a novel mechanism of RNA catalysis. Chem Biol 9: 465–473 [DOI] [PubMed] [Google Scholar]
- Lilley DMJ, Eckstein F, ed. 2008. Ribozymes and RNA catalysis. RSC Publishing, Cambridge, UK [Google Scholar]
- Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley DMJ 2008. The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure 16: 1357–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cottrell JW, Scott LG, Fedor MJ 2009. Direct measurement of the ionization state of an essential guanine in the hairpin ribozyme. Nat Chem Biol 5: 351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacElrevey C, Salter JD, Krucinska J, Wedekind JE 2008. Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme. RNA 14: 1600–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Scott WG 2006. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126: 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Horan LH, Noller HF, Scott WG 2008. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 454: 899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod AC, Lilley DMJ 2004. Efficient, pH-dependent RNA ligation by the VS ribozyme in trans. Biochemistry 43: 1118–1125 [DOI] [PubMed] [Google Scholar]
- Murchie AIH, Thomson JB, Walter F, Lilley DMJ 1998. Folding of the hairpin ribozyme in its natural conformation achieves close physical proximity of the loops. Mol Cell 1: 873–881 [DOI] [PubMed] [Google Scholar]
- Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG 1998. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol 5: 587–595 [DOI] [PubMed] [Google Scholar]
- Nahas MK, Wilson TJ, Hohng S, Jarvie K, Lilley DMJ, Ha T 2004. Observation of internal cleavage and ligation reactions of a ribozyme. Nat Struct Mol Biol 11: 1107–1113 [DOI] [PubMed] [Google Scholar]
- Nakano S, Chadalavada DM, Bevilacqua PC 2000. General acid–base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science 287: 1493–1497 [DOI] [PubMed] [Google Scholar]
- Nam K, Gao J, York DM 2008. Electrostatic interactions in the hairpin ribozyme account for the majority of the rate acceleration without chemical participation by nucleobases. RNA 14: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet J, Byrne M, Lilley DMJ 2009. Formation of an active site in trans by interaction of two complete Varkud Satellite ribozymes. RNA 15: 1822–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo JC, Wilson TJ, Jayasena SD, Khvorova A, Lilley DMJ 2004. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA 10: 880–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinard R, Hampel KJ, Heckman JE, Lambert D, Chan PA, Major F, Burke JM 2001. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J 20: 6434–6442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi T, Beattie TL, Olive JE, Collins RA 1996. A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J 15: 2820–2825 [PMC free article] [PubMed] [Google Scholar]
- Rupert PB, Ferré-D'Amaré AR 2001. Crystal structure of a hairpin ribozyme–inhibitor complex with implications for catalysis. Nature 410: 780–786 [DOI] [PubMed] [Google Scholar]
- Rupert PB, Massey AP, Sigurdsson ST, Ferré-D'Amaré AR 2002. Transition state stabilization by a catalytic RNA. Science 298: 1421–1424 [DOI] [PubMed] [Google Scholar]
- Ryder SP, Oyelere AK, Padilla JL, Klostermeier D, Millar DP, Strobel SA 2001. Investigation of adenosine base ionization in the hairpin ribozyme by nucleotide analog interference mapping. RNA 7: 1454–1463 [PMC free article] [PubMed] [Google Scholar]
- Salter J, Krucinska J, Alam S, Grum-Tokars V, Wedekind JE 2006. Water in the active site of an all-RNA hairpin ribozyme and effects of Gua8 base variants on the geometry of phosphoryl transfer. Biochemistry 45: 686–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy R, Siwkowski A, Hampel A 1998. Mutational analysis of loops 1 and 5 of the hairpin ribozyme. Biochemistry 37: 564–570 [DOI] [PubMed] [Google Scholar]
- Smith MD, Collins RA 2007. Evidence for proton transfer in the rate-limiting step of a fast-cleaving Varkud satellite ribozyme. Proc Natl Acad Sci 104: 5818–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood VD, Beattie TL, Collins RA 1998. Identification of phosphate groups involved in metal binding and tertiary interactions in the core of the Neurospora VS ribozyme. J Mol Biol 282: 741–750 [DOI] [PubMed] [Google Scholar]
- Spitale RC, Volpini R, Heller MG, Krucinska J, Cristalli G, Wedekind JE 2009. Identification of an imino group indispensable for cleavage by a small ribozyme. J Am Chem Soc 131: 6093–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suydam IT, Levandoski SD, Strobel SA 2010. Catalytic importance of a protonated adenosine in the hairpin ribozyme active site. Biochemistry 49: 3723–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E, Wilson TJ, Nahas MK, Clegg RM, Lilley DMJ, Ha T 2003. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc Natl Acad Sci 100: 9308–9313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CL, Alexov E, Pyle AM, Honig B 2007. Calculation of pKas in RNA: On the structural origins and functional roles of protonated nucleotides. J Mol Biol 366: 1475–1496 [DOI] [PubMed] [Google Scholar]
- Thomas JM, Perrin DM 2006. Active site labeling of G8 in the hairpin ribozyme: Implications for structure and mechanism. J Am Chem Soc 128: 16540–16545 [DOI] [PubMed] [Google Scholar]
- Walter F, Murchie AIH, Lilley DMJ 1998a. The folding of the four-way RNA junction of the hairpin ribozyme. Biochemistry 37: 17629–17636 [DOI] [PubMed] [Google Scholar]
- Walter F, Murchie AIH, Thomson JB, Lilley DMJ 1998b. Structure and activity of the hairpin ribozyme in its natural junction conformation; effect of metal ions. Biochemistry 37: 14195–14203 [DOI] [PubMed] [Google Scholar]
- Walter NG, Burke JM, Millar DP 1999. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat Struct Biol 6: 544–549 [DOI] [PubMed] [Google Scholar]
- Wilson TJ, Lilley DMJ 2002. Metal ion binding and the folding of the hairpin ribozyme. RNA 8: 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TJ, Zhao Z-Y, Maxwell K, Kontogiannis L, Lilley DMJ 2001. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry 40: 2291–2302 [DOI] [PubMed] [Google Scholar]
- Wilson TJ, Ouellet J, Zhao ZY, Harusawa S, Araki L, Kurihara T, Lilley DMJ 2006. Nucleobase catalysis in the hairpin ribozyme. RNA 12: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TJ, McLeod AC, Lilley DMJ 2007. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J 26: 2489–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TJ, Li N-S, Lu J, Frederiksen JK, Piccirilli JA, Lilley DMJ 2010. Nucleobase-mediated general acid–base catalysis in the Varkud satellite ribozyme. Proc Natl Acad Sci 107: 11751–11756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, McLeod A, Harusawa S, Araki L, Yamaguchi M, Kurihara T, Lilley DMJ 2005. Nucleobase participation in ribozyme catalysis. J Am Chem Soc 127: 5026–5027 [DOI] [PubMed] [Google Scholar]