Abstract
Cbf5 is the catalytic subunit of the H/ACA small nucleolar/Cajal body ribonucleoprotein particles (RNPs) responsible for site specific isomerization of uridine in ribosomal and small nuclear RNA. Recent evidence from studies on archaeal Cbf5 suggests its second functional role in modifying tRNA U55 independent of guide RNA. In order to act both as a stand-alone and a RNP pseudouridine synthase, Cbf5 must differentiate features in H/ACA RNA from those in tRNA or rRNA. Most H/ACA RNAs contain a hallmark ACA trinucleotide downstream of the H/ACA motif. Here we challenged an archaeal Cbf5 (in the form of a ternary complex with its accessory proteins Nop10 and Gar1) with T-stem–loop RNAs with or without ACA trinucleotide in the stem. Although these substrates were previously shown to be substrates for the bacterial stand-alone pseudouridine synthase TruB, the Cbf5-Nop10-Gar1 complex was only able to modify those without ACA trinucleotide. A crystal structure of Cbf5-Nop10-Gar1 trimer bound with an ACA-containing T-stem–loop revealed that the ACA trinucleotide detracted Cbf5 from the stand-alone binding mode, thereby suggesting that the H/ACA RNP-associated function of Cbf5 likely supersedes its stand-alone function.
Keywords: pseudouridylase, ribosome biogenesis, RNA–protein interactions, X-ray crystal structure
INTRODUCTION
In eukaryotic cells, small nucleolar and small Cajal body RNAs (snoRNAs and scaRNAs) are abundant noncoding RNAs found in nucleolus and Cajal bodies, respectively, that participate in ribosome and spliceosome maturation (Maxwell and Fournier 1995; Balakin et al. 1996; Kiss 2002). Regardless of their nuclear location, sno/caRNAs are classified into two types based on their conserved sequence features and the maturation processes they involve. Box C/D RNAs guide protein enzymes to carry out site-specific 2′-O-methylation while box H/ACA RNAs guide site-specific pseudouridylation of ribosomal and spliceosomal RNAs (Maxwell and Fournier 1995; Balakin et al. 1996; Ganot et al. 1997; Kiss 2002). In this remarkable process, sno(ca)RNAs form multi-subunit ribonucleoprotein particles (RNPs) that function as RNA modification enzymes in the classical sense where the substrate is captured through Watson-Crick base-pairing with the sno(ca)RNA. Thus, the sno(ca)RNAs must be accurately positioned in the protein complex in order to dock the substrate RNA into the enzyme active site.
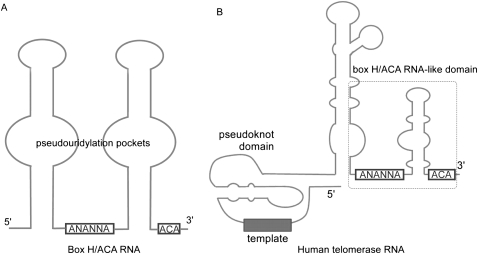
Box H/ACA RNAs may form one, two, or three “hairpin-hinge-hairpin tail” secondary structure motifs and each motif contains a conserved AC/UA trinucleotide at its 3′ end (Fig. 1A; Balakin et al. 1996; Ganot et al. 1997; Ni et al. 1997; Muller et al. 2008). There are four protein components associated with box H/ACA RNAs: Cbf5, Nop10, Gar1, and Nhp2 (Girard et al. 1992; Bousquet-Antonelli et al. 1997; Henras et al. 1998; Lafontaine et al. 1998; Watkins et al. 1998; Dragon et al. 2000; Watanabe and Gray 2000; Rozhdestvensky et al. 2003). Homologs of box H/ACA RNA and the associated proteins have also been identified in Archaea where the nomenclature of protein subunits is maintained with the exception that L7Ae in Archaea corresponds to Nhp2 (Tang et al. 2002; Baker et al. 2005; Charpentier et al. 2005; Muller et al. 2008). Cbf5 is the largest subunit of all and contains the catalytic site of the RNP pseudouridine synthase. Previous structural studies on archaeal box H/ACA ribonucleoprotein particles show that Cbf5 interacts directly and specifically with the H/ACA RNA containing a single hairpin motif via both the overall elongated body and the highly conserved 3′ ACA trinucleotide of the RNA (Li and Ye 2006; Duan et al. 2009; Liang et al. 2009). The ACA trinucleotide is recognized by the unique PUA (PseudoUridine synthase and Archaeosine transglycosylase) domain of Cbf5 in a sequence specific manner (Li and Ye 2006; Duan et al. 2009; Liang et al. 2009). The structural data corroborate previous biochemical observations. Mutational analysis showed that mutation or deletion of the ACA trinucleotide is detrimental to both guide RNA binding and in vitro pseudouridylation activities (Baker et al. 2005; Charpentier et al. 2005; Li and Ye 2006; Liang et al. 2009). Similarly, deletion of the PUA domain of Cbf5 was deleterious to H/ACA RNP mediated pseudouridylation activity (Baker et al. 2005; Manival et al. 2006). The ACA trinucleotide is also a hallmark motif in the 3′ end of the human, and other vertebrate, telomerase RNA (hTR) (Mitchell et al. 1999; Chen et al. 2000) and is responsible for recruitment of the four box H/ACA core proteins to the telomerase complex (Fig. 1B). The box H/ACA RNA domain of hTR has been shown to be essential for hTR accumulation, hTR 3′-end processing, and telemere elongation in vivo (Mitchell and Collins 2000; Fu and Collins 2003). Importantly, mutations in human Cbf5 are correlated with the genetic disease dyskeratosis congenita (DC) and many of these mutations occur near or on the PUA domain that interacts with the ACA trinucleotide (Li and Ye 2006; Rashid et al. 2006). In addition, DC mutants are found in the box H/ACA domain-like motif of hTR (Vulliamy et al. 2001). These mutations on both protein and RNA components could potentially disrupt the function of either telomerase or the H/ACA ribonucleoprotein pseuoduridine synthase.
FIGURE 1.
Conserved secondary structure features of the box H/ACA small nucleolar and Cajal body RNA (A) and the human telomerase RNA (B). The conserved H-box (hinge) and ACA trinucleotide are indicated. Letter “N” indicates any sequence.
Recent studies found that archaeal Cbf5 or its complex with Nop10 and Gar1 can act as a stand-alone pseudouridine synthase that isomerizes U55 of Pyrococcus abyssi and Saccharomyces cerevisiae tRNAAsp or U2603 of P. abyssi 23S rRNA in an H/ACA RNA-independent manner similar to its bacterial homolog TruB (Roovers et al. 2006; Muller et al. 2007). Previously, biochemical and structural data show that other stand-alone pseudouridine synthases can bind and modify small hairpin RNA mimicking the TΨC-stem–loop or anticodon stem-loop (Hoang and Ferre-D'Amare 2001; Pan et al. 2003; Spedaliere and Mueller 2004; Phannachet et al. 2005; Hamilton et al. 2006; Hoang et al. 2006). Two hairpin RNA substrates were co-crystallized with their respective stand-alone enzymes and their target uridine analog, 5-fluorouridine, was indeed rearranged in its crystal complexes (Hoang and Ferre-D'Amare 2001; Hoang et al. 2006). These data indicate that stand-alone pseudouridine synthases primarily recognize the hairpin motif of the RNA substrates. In TruB, the hairpin RNA is stabilized by two long insertions called insertion 1 and insertion 2 (thumb loop). The thumb loop reaches deep into the major groove of the hairpin and the insertion 1 loop interacts directly with the flipped out +1 nucleotide (Hoang and Ferre-D'Amare 2001). The PUA domain of TruB is hypothesized to bind the acceptor stem and the 3′ CCA terminus of tRNA, which further enhances but is not necessary for modification of the T-stem.
Cbf5 differs from TruB in both the hairpin-binding motifs as well as the PUA domain. Both insertion loops are replaced by short peptides in Cbf5. Consistently, an archaeal Cbf5 was shown to have low stand-alone activity on the tRNA substrate lacking the CCA end, although Nop10 and Gar1 can enhance Ψ55 formation in this case (Muller et al. 2007). Surprisingly, when bound with Nop10 and Gar1, Cbf5 was found to also act on small hairpin substrates derived from rRNA (Muller et al. 2008). These findings reveal both similarities as well as differences between TruB and Cbf5 and, at the time, raised questions on how Cbf5 interacts differently with hairpin-like substrates than with H/ACA RNAs.
Here, we report functional and structural studies of an archaeal Cbf5-Nop10-Gar1 complex acting as a stand-alone pseudouridine synthase on small hairpin substrates. We tested the ability of the Cbf5-Nop10-Gar1 complex to catalyze pseudouridylation of two previously characterized small hairpin RNA substrates of TruB and obtained a co-crystal structure of the protein complex bound to one of the substrates. A 22-mer T-stem–loop (TSL 22 mer) (Phannachet and Huang 2004) and a TSL 17 mer containing ACA trinucleotide (TSL-ACA 17 mer) (Pan et al. 2003) were previously shown to be substrates for bacterial stand-alone enzyme TruB. We found that Cbf5-Nop10-Gar1 is able to modify the 22-mer hairpin substrate but not the 17-mer ACA-containing substrate. The crystal structure of the Cbf5-Nop10-Gar1 complex bound with the TSL-ACA 17-mer RNA revealed the structural basis for the discrimination of the ACA-containing substrate. Our results demonstrate the strong preference for ACA trinucleotide by the PUA domain of Cbf5 that is different from the PUA domain of other homologs such as TruB.
RESULTS
The impact of ACA trinucleotide on the stand-alone activity of the Cbf5-Nop10-Gar1 complex
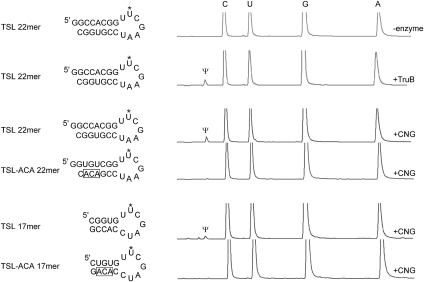
We first tested the ability of the Pyrococcus furiosus (Pf) Cbf5-Nop10-Gar1 complex to catalyze isomerization of the target uridine of the TSL 22-mer RNA and the TSL-ACA 17-mer RNA, respectively, by a mass spectrometry method similar to that used in studying TruB and RluA (Fig. 2A; Phannachet et al. 2005; Hamilton et al. 2006). These RNA were previously shown to be modified by TruB in vitro (Hoang and Ferre-D'Amare 2001; Pan et al. 2003; Phannachet and Huang 2004). We incubated TruB or the ternary protein complex with each hairpin RNA at 37°C (TruB) or 70°C (Cbf5 complex) for 2 h and isolated the reacted RNA by phenol extraction and ethanol precipitation. We then digested the isolated RNA by nuclease P1 and alkaline phosphatase in order to generate nucleosides that were analyzed by C18 chromatography (Varian, 100-5 250 × 4.6 mm) and an in-line ESI-TOF (JMS-T100LC “AccuTOF,” JEOL Ltd.). The positions of four natural nucleosides (A, U, G, and C) were identified using unreacted RNA or that incubated with a catalytically deficient TruB. The peak intensity ratios of the four nucleosides from the C18 chromatography matched their sequence composition and their detected masses were consistent with the theoretical molecular weights (data not shown). In enzyme-reacted TSL 22-mer RNA but not the TSL-ACA 17-mer RNA, a small peak preceding cytidine was detected that has the same molecular weight as uridine. The nucleoside was identified as Ψ. The ratio of Ψ to U (1:8) indicates that ∼37.5% of the substrate was modified by the enzyme (assuming the enzyme is specific for U13), suggesting that the Cbf5-Nop10-Gar1 complex is reasonably efficient in modifying these small hairpin substrates similar to what was previously observed for a small rRNA hairpin substrate (Muller et al. 2007).
FIGURE 2.
Sequences and predicted secondary structures of the small hairpin RNA substrates used in the study (left) and their pseudouridylation activity assay results (right). “TSL” is the abbreviation for T-stem–loop and “ACA” denotes the presence of ACA trinucleotide in the RNA. “CNG” stands for the Cbf5-Nop10-Gar1 ternary complex. The U55 is indicated by a star. The four major peaks are eluted standard nucleosides and their identities are labeled at the top (C for cytidine, U for uridine, G for guanosine, and A for adenosine). Pseudouridine is the earliest eluted peak and is labeled by Ψ.
We noted that the TSL-ACA 17-mer RNA substrate contains an ACA trinucleotide near the 3′ end of the stem and hypothesized its ability to disrupt the hairpin structure when bound with Cbf5. We thus mutated the bases ACA and its complementary bases to create TSL 17mer RNA substrate that contains no ACA (TLS 17 mer, Fig. 2). Similarly, we also introduced an ACA trinucleotide in the reactive TSL 22-mer RNA (with compensatory mutations) substrate to create TSL-ACA 22-mer RNA substrate (Fig. 2). Consistent with our hypothesis, the two hairpin RNAs containing the ACA trinucleotide in their stems were not modified by the Cbf5-Nop10-Gar1 complex (Fig. 2). These results demonstrate the close resemblance between Cbf5 (with Nop10 and Gar1) and TruB but highlight the unique ability of Cbf5 to interact with ACA-containing RNAs.
The impact of ACA trinucleotide on interactions between hairpin RNA and Cbf5
In order to understand the molecular basis for Cbf5's ability to discriminate ACA-containing RNA substrates, we obtained a crystal structure of the Cbf5-Nop10-Gar1 complex bound to the the TSL-ACA 17-mer RNA (Fig. 3). The structure was solved by the molecular replacement method using a previously determined protein complex as the search probe (Rashid et al. 2006). The computed electron density map using refined protein coordinates had sufficiently high quality to allow tracing of the RNA molecule.
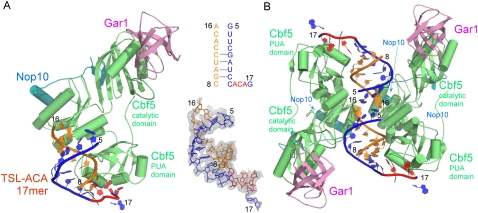
FIGURE 3.
Structure of the Cbf5-Nop10-Gar1 complex bound to TSL-ACA 17mer RNA. (A) The overall view of the Cbf5-Nop10-Gar1 complex. The 3Fo-2Fc map computed prior to modeling of the RNA is shown around the final RNA model. The ACA-binding strand is colored in blue (ACA is in red) and contains nucleotides 5–17. The complementary strand is colored in orange and contains nucleotides 8–16. (B) The packing of two symmetry related protein-RNA complexes indicates some weak interactions between the ends of the two RNA duplexes.
The TSL-ACA 17-mer RNA did not form the intended hairpin secondary structure. Instead, it forms a staggered and imperfect duplex with a symmetry-related TSL RNA (Fig. 3A). The protein complex interacts with more than half of the imperfect duplex and the rest is apparently disordered (Fig. 3B). The target uridine is not placed in the active site of the enzyme. This result explains why the two ACA-containing hairpin RNA were not the substrates for the protein complex as described previously.
Disruption of the hairpin secondary structure of the RNA is likely a result of strong interactions between the ACA trinucleotide and the PUA domain of Cbf5 (Fig. 4A). In previously observed H/ACA RNP structures (Li and Ye 2006; Duan et al. 2009; Liang et al. 2009), the ACA trinuleotide forms specific interactions with the PUA domain of Cbf5 (Fig. 4A). The two adenine nucleotides both are in syn conformation and are buried by a hybrid pocket formed by both RNA and protein surfaces (Fig. 4B). Although no specific side chains of Cbf5 residues contact the nucleobases of the adenine nucleotides, the backbone atoms are strategically placed around the edges of the bases to form sequence specific interactions (Li and Ye 2006; Duan et al. 2009; Liang et al. 2009). Here, the same interactions are observed (Fig. 4B), suggesting that Cbf5 can interact with the trinucleotide regardless of the context of the RNA.
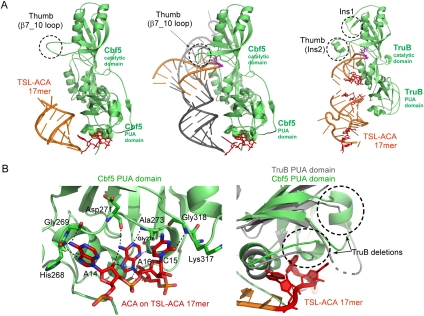
FIGURE 4.
(A) Comparison of the structures. (Left) Structure of the Cbf5-Nop10-Gar1 complex bound with TSL-ACA 17mer RNA; (middle) structure of the Cbf5-Nop10-L7Ae-H/ACA RNA-substrate RNA complex (PDBid: 3HJY); (right) structure of TruB bound with TSL-ACA 17mer RNA (PDBid: 1R3E). Cbf5 and TruB (green) are superimposed and the structures are shown in the same orientation. The substrate RNAs in all structures are in orange and the H/ACA RNA is in gray. Note the absence of substrate-interacting loops in Cbf5 (dashed circles). (B) Specific interactions observed between the PUA domain of Cbf5 and ACA (left) and the absence of ACA-interacting loops in TruB (dashed circles) (right).
In contrast, the same TSL-ACA 17-mer RNA binds to TruB in its predicted secondary structure and is correctly placed in the active site of TruB (Fig. 4A; Pan et al. 2003). The contrasting ability between TruB and Cbf5 to interact with the ACA trinucleotide demonstrates the extraordinary specificity of Cbf5 for ACA. Although Cbf5 and TruB have similar overall structures, their PUA domains differ in important regions that gives rise to their different abilities to interact with the ACA trinucleotide. The PUA domain of Cbf5 contains two insertions (residues 271–281, and 302–314), that are necessary for completion of the pocket for binding the two adenine nucleotides. The absence of the same loops from TruB is likely responsible for its lack of specificity for the ACA trinucleotide (Fig. 4B).
DISCUSSION
Cbf5 belongs to the TruB-family of pseudouridine synthases and is the catalytic subunit of the multi-subunit ribonucleoprotein particle pseudouridine synthases. In the RNA-guided pseudouridylation reaction, the guide RNA uses its internal loop to capture substrate RNA through base-pairing. The four protein subunits all participate in the process of substrate RNA placement (Liang et al. 2007, 2008, 2009). The protein subunits interact with the guide RNA in specific manners to tightly anchor it on the surface of Cbf5 such that the substrate RNA is correctly placed in the active site (Li and Ye 2006; Liang et al. 2007, 2009; Duan et al. 2009). In the case of TruB, its catalytic and PUA domains are sufficient to place the substrate RNA without the help of accessory proteins (Hoang and Ferre-D'Amare 2001; Pan et al. 2003; Phannachet and Huang 2004). Cbf5 differs from TruB in the specific elements required for binding hairpin RNA substrates. Surprisingly, Cbf5 (bound with Nop10 and Gar1) is able to catalyze small RNA hairpin substrates similarly as TruB. Our findings are consistent with previously discovered stand-alone activity of Cbf5 on tRNA or small hairpin rRNA substrates (Roovers et al. 2006; Muller et al. 2007, 2008) and suggest an inherent connection between Cbf5 and TruB at the most fundamental level of function and thus a common origin of evolution.
In eukaryotes, Cbf5 primarily functions as the catalytic subunit of the H/ACA sno(ca)RNPs through its specific association with H/ACA RNAs and partner proteins. The ACA trinucleotide is highly conserved in H/ACA sno(ca)RNAs and is required for formation of the Cbf5-containg RNPs (Baker et al. 2005; Charpentier et al. 2005). ACA trinucleotide is also conserved in human telomerase RNA and its interaction with Cbf5 plays an essential role in hTR accumulation, hTR 3′-end processing, and telomerase activity (Mitchell et al. 1999). However, some recent evidence links Cbf5 to tRNA biogenesis, likely through its stand-alone functionality. In yeast, a mutation in Cbf5 caused mislocalization of precursor tRNA, which suppresses tRNA gene-mediated transcriptional silencing (Kendall et al. 2000). In Euglena gracilis, Cbf5 was copurified with the selenocysteine tRNA (Russell et al. 2004). These activities reflect the findings in archaea where Cbf5 can modify tRNA substrate in vitro and raise the possibility of a dual functional role of Cbf5. By using small RNA hairpin substrates with or without ACA nucleotides, we now show that the Cbf5-Nop10-Gar1 complex is unable to function as a stand-alone pseudouridine synthase on hairpin substrates containing ACA trinucleotides in stems. The crystal structure of the Cbf5-Nop10-Gar1 in complex with an ACA-containing substrate provides the structural basis for its discrimination of substrates studied here. The unique PUA domain of Cbf5 is specialized to interact with ACA trinucleotide independent of the main body of the H/ACA RNA, which disrupts the required RNA structure for catalysis. Clearly, ACA trinucleotide located outside the stem of the hairpin should not disrupt the RNA structure and may in fact facilitate the Cbf5-substrate interaction. The fact that the 3′ terminal CCA trinucleotide of tRNA was found to significantly enhance the stand-alone activity of Cbf5 (Muller et al. 2007) provides some support to this proposal given the close resemblance of CCA to ACA. It appears that the stand-alone activity of Cbf5 is a reminder activity before it was specialized to bind H/ACA RNA. This is consistent with the fact that archaea contain another tRNA U55 pseudouridine synthase, pfuPus10, whose activity has been demonstrated in vitro (Roovers et al. 2006) and that eukaryotic Cbf5 has no demonstrated stand-alone activity.
MATERIALS AND METHODS
Protein expression, purification, and crystallization
Escherichia coli BL21 cells containing the plasmid encoding E. coli pseudouridine synthase TruB with a polyhistidine tag (a kind gift from Dr. Brian G. Miller at Florida State University) were grown and harvested by standard methods. TruB was purified by a method consisting of Ni-NTA, ion exchange, and size exclusion chromatography similar to those reported previously (Hoang and Ferre-D'Amare 2001). Purified TruB in a storage buffer containing 20 mM HEPES-KOH pH 7.5, 150 mM KCl, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 1 mM 2-mercaptoethanol, and 5% glycerol was concentrated to 100 mg/mL and stored at −80°C. Pyrococcus furiosus Cbf5-Nop10-Gar1 complex was co-expressed and purified as previously described (Rashid et al. 2006). The TSL 22-mer RNA (GGCGACGGUUCGAAUCCGUGGC), the TSL-ACA 22-mer RNA (GGUGUCGGUUCGAAUCCGACAC), the TSL 17-mer RNA (CGGUGUUCGAUCCACCG), and the TSL-ACA 17-mer RNA (CUGUGUUCGAUCCACAG) substrates were ordered from Integrated DNA Technology (IDT) and dissolved in filtered water. For crystallization, the TSL-ACA 17mer RNA oligomer at 1 mM was first heated at 90°C for 5 min in a buffer containing 10 mM HEPES pH 7.5 and 1 mM EDTA, and cooled down on the ice. The protein complex was added at a 1:1.8 molar ratio to RNA to a final concentration of 12 mg/mL. The RNA-protein complex was then incubated at room temperature for 1 h before crystallization trials. Crystals were obtained after 2 d from a well solution containing 120 mM magnesium acetate tetrahydrate, 50 mM MES PH 5.6, 20% 2-Methyl-2,4-pentanediol at 30°C by the hanging-drop vapor diffusion method. The crystals from the original mother liquor could be directly frozen in a liquid nitrogen stream for diffraction studies. A single wavelength diffraction data set was collected at the southeast regional collaborative access team (SER-CAT) beamline 22ID and was processed by the program suite HKL2000 (Otwinowski and Minor 1997).
Structure determination
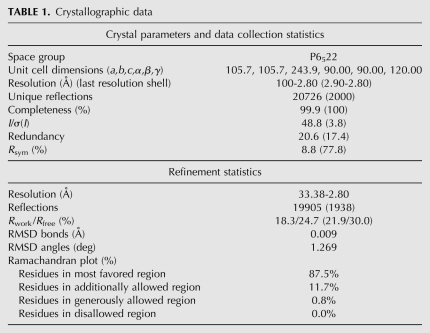
The structure was solved by the molecular replacement method using the previously published coordinates of Cbf5-Nop10-Gar1 complex as the search probe (PDBid: 2EY4) (Rashid et al. 2006). The substrate RNA was not added at initial stages of refinement and model building in order to avoid model bias. At later stages of refinement, a computed 3Fo-2Fc density map phased with protein coordinates allowed the RNA to be built using COOT (Emsley and Cowtan 2004) and O (Jones et al. 1991) programs. The structure was refined using the program PHENIX (Adams et al. 2002) and validated using PROCHECK (Laskowski et al. 1993). All figures were prepared using PyMOL (Delano, 2002). Data collection and refinement statistics are included in Table 1. Final coordinates and structure factors have been deposited in the Protein Data Bank (http://www.rcsb.org) (PDBid 3MQK).
TABLE 1.
Crystallographic data
Pseudouridylation activity assay
The RNA substrates were similarly annealed as described above. The annealed substrate at 100 μM was mixed with TruB or the Cbf5-Nop10-Gar1 complex at 3 μM final concentration and the reaction mix was incubated at 37°C (TruB) or 70°C (Cbf5 complex) for 2 h in a reaction buffer containing 10 mM HEPES PH 7.5, 100 mM NaCl, and 1 mM dithiothreitol. The RNA was then purified by phenol extraction and ethanol precipitation as previously described (Baker et al. 2005) and digested by nuclease P1 (United States Biological) and alkaline phosphatase (Roche Applied Science). The digested nucleosides were loaded onto a Microsorb-MV C18 column (Varian, 100-5 250 × 4.6 mm) connected to a UV detector at 254 nm. To separate the nucleosides, the column was equilibrated by buffer A (20 mM ammonium acetate, PH 6.0) and the nucleosides were eluted by buffer B (40% acetonitrile in water) with a gradient of 0 min, 0% B; 30 min, 50% B; 35 min, 90% B; 40 min, 0% B; 50 min, 0% B at a 1.8ml/min flow rate. An in-line electrospray ionization-time of flight (ESI-TOF) mass spectrometer (JMS-T100LC “AccuTOF,” JEOL Ltd.) was used to analyze the eluted nucleosides.
ACKNOWLEDGMENTS
This work was supported in part by a National Institutes of Health grant R01 GM66958-01 (H.L.). J. Zhou is a predoctoral fellow of the American Heart Association, Florida/Puerto Rico Affiliate (0815267E). B. Liang is a predoctoral fellow of the American Heart Association, Florida/Puerto Rico Affiliate (0415179B). X-ray diffraction data were collected from the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2415811.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC 2002. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Baker DL, Youssef OA, Chastkofsky MI, Dy DA, Terns RM, Terns MP 2005. RNA-guided RNA modification: Functional organization of the archaeal H/ACA RNP. Genes Dev 19: 1238–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin AG, Smith L, Fournier MJ 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834 [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Henry Y, G'Elugne JP, Caizergues-Ferrer M, Kiss T 1997. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J 16: 4770–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier B, Muller S, Branlant C 2005. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res 33: 3133–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Blasco MA, Greider CW 2000. Secondary structure of vertebrate telomerase RNA. Cell 100: 503–514 [DOI] [PubMed] [Google Scholar]
- Delano WL 2002. The PyMOL Molecular Graphics System. http://www.pymol.org/
- Dragon F, Pogacic V, Filipowicz W 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol Cell Biol 20: 3037–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Li L, Lu J, Wang W, Ye K 2009. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell 34: 427–439 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fu D, Collins K 2003. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol Cell 11: 1361–1372 [DOI] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809 [DOI] [PubMed] [Google Scholar]
- Girard JP, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B 1992. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J 11: 673–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton CS, Greco TM, Vizthum CA, Ginter JM, Johnston MV, Mueller EG 2006. Mechanistic investigations of the pseudouridine synthase RluA using RNA containing 5-fluorouridine. Biochemistry 45: 12029–12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne JP, Caizergues-Ferrer M 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J 17: 7078–7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang C, Ferre-D'Amare AR 2001. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: Nucleotide flipping by an RNA-modifying enzyme. Cell 107: 929–939 [DOI] [PubMed] [Google Scholar]
- Hoang C, Chen J, Vizthum CA, Kandel JM, Hamilton CS, Mueller EG, Ferre-D'Amare AR 2006. Crystal structure of pseudouridine synthase RluA: Indirect sequence readout through protein-induced RNA structure. Mol Cell 24: 535–545 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kendall A, Hull MW, Bertrand E, Good PD, Singer RH, Engelke DR 2000. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc Natl Acad Sci 97: 13108–13113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148 [DOI] [PubMed] [Google Scholar]
- Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D 1998. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev 12: 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM 1993. Procheck: A program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Li L, Ye K 2006. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443: 302–307 [DOI] [PubMed] [Google Scholar]
- Liang B, Xue S, Terns RM, Terns MP, Li H 2007. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat Struct Mol Biol. 14: 1189–1195 [DOI] [PubMed] [Google Scholar]
- Liang B, Kahen EJ, Calvin K, Zhou J, Blanco M, Li H 2008. Long-distance placement of substrate RNA by H/ACA proteins. RNA 14: 2086–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Zhou J, Kahen E, Terns RM, Terns MP, Li H 2009. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat Struct Mol Biol 16: 740–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manival X, Charron C, Fourmann JB, Godard F, Charpentier B, Branlant C 2006. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5-aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Res 34: 826–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ 1995. The small nucleolar RNAs. Annu Rev Biochem 64: 897–934 [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Collins K 2000. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol Cell 6: 361–371 [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 19: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Fourmann JB, Loegler C, Charpentier B, Branlant C 2007. Identification of determinants in the protein partners aCBF5 and aNOP10 necessary for the tRNA:Psi55-synthase and RNA-guided RNA:Psi-synthase activities. Nucleic Acids Res 35: 5610–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Leclerc F, Behm-Ansmant I, Fourmann JB, Charpentier B, Branlant C 2008. Combined in silico and experimental identification of the Pyrococcus abyssi H/ACA sRNAs and their target sites in ribosomal RNAs. Nucleic Acids Res 36: 2459–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM 2003. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc Natl Acad Sci 100: 12648–12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phannachet K, Huang RH 2004. Conformational change of pseudouridine 55 synthase upon its association with RNA substrate. Nucleic Acids Res 32: 1422–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phannachet K, Elias Y, Huang RH 2005. Dissecting the roles of a strictly conserved tyrosine in substrate recognition and catalysis by pseudouridine 55 synthase. Biochemistry 44: 15488–15494 [DOI] [PubMed] [Google Scholar]
- Rashid R, Liang B, Baker DL, Youssef OA, He Y, Phipps K, Terns RM, Terns MP, Li H 2006. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol Cell 21: 249–260 [DOI] [PubMed] [Google Scholar]
- Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L 2006. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res 34: 4293–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res 31: 869–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AG, Schnare MN, Gray MW 2004. Pseudouridine-guide RNAs and other Cbf5p-associated RNAs in Euglena gracilis. RNA 10: 1034–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedaliere CJ, Mueller EG 2004. Not all pseudouridine synthases are potently inhibited by RNA containing 5-fluorouridine. RNA 10: 192–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci 99: 7536–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I 2001. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413: 432–435 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Gray MW 2000. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res 28: 2342–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Luhrmann R 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4: 1549–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]