Abstract
An obligate intermediate during microRNA (miRNA) biogenesis is an ∼22-nucleotide RNA duplex, from which the mature miRNA is preferentially incorporated into a silencing complex. Its partner miRNA* species is generally regarded as a passenger RNA, whose regulatory capacity has not been systematically examined in vertebrates. Our bioinformatic analyses demonstrate that a substantial fraction of miRNA* species are stringently conserved over vertebrate evolution, collectively exhibit greatest conservation in their seed regions, and define complementary motifs whose conservation across vertebrate 3′-UTR evolution is statistically significant. Functional tests of 22 miRNA expression constructs revealed that a majority could repress both miRNA and miRNA* perfect match reporters, and the ratio of miRNA:miRNA* sensor repression was correlated with the endogenous ratio of miRNA:miRNA* reads. Analysis of microarray data provided transcriptome-wide evidence for the regulation of seed-matched targets for both mature and star strand species of several miRNAs relevant to oncogenesis, including mir-17, mir-34a, and mir-19. Finally, 3′-UTR sensor assays and mutagenesis tests confirmed direct repression of five miR-19* targets via star seed sites. Overall, our data demonstrate that miRNA* species have demonstrable impact on vertebrate regulatory networks and should be taken into account in studies of miRNA functions and their contribution to disease states.
Keywords: 3′ UTR, microRNA*, post-transcriptional repression
INTRODUCTION
microRNAs (miRNAs) are an abundant class of ∼22 nucleotide RNAs, derived from endogenous transcripts bearing short inverted repeats, that perform critical roles as post-transcriptional repressors in diverse higher eukaryotes (Flynt and Lai 2008; Bartel 2009). An obligate step in miRNA biogenesis is the cleavage of pre-miRNA hairpins by Dicer RNAse III enzymes to yield ∼22-nt duplexes. By convention, the mature miRNA is defined as the duplex strand that accumulates to a higher steady-state level than its partner strand, which is termed the miRNA* species. The higher steady-state level of miRNAs relative to miRNA* species is presumed to reflect their preferred incorporation into Argonaute (Ago) complexes, and thus into post-transcriptional regulatory networks.
A key indicator of the endogenous function of miRNAs is the conservation of 7-nt sequences, usually within 3′ untranslated regions (3′ UTRs), that exhibit Watson-Crick pairing to positions 2–8 of the mature miRNA (the “seed” region) (Lai 2002; Lewis et al. 2003; Brennecke et al. 2005; Krek et al. 2005). Complementarity to positions 2–7 of the miRNA also yields signal for evolutionary constraint, and the activity of both types of seed matches improves if there is an adenosine across from the first miRNA position and/or the site lies in a structurally open context (Bartel 2009). Application of these criteria to multispecies genome alignments yields evidence that a majority of mammalian transcripts bear conserved sites for one or more miRNAs (Friedman et al. 2009). The extent of the miRNA target network is broader yet, when considering the possibility for unrecognized miRNA genes and functional sites that are poorly conserved, lack seed matches, or that occur outside of 3′ UTRs (Bartel 2009; Brodersen and Voinnet 2009).
On the other hand, the potential regulatory activity of miRNA* species has received comparatively limited attention. Nevertheless, it was recognized early on that both strands of some artificial siRNA duplexes could direct target cleavage, in both Drosophila (Nykanen et al. 2001) and mammalian systems (Elbashir et al. 2001a,b). Indeed, a major improvement in siRNA specificity came with the implementation of rules, derived in part from analysis of miRNA/miRNA* strand selection, that force the asymmetric incorporation of siRNA strands into Ago complexes (Khvorova et al. 2003; Schwarz et al. 2003). However, as many miRNA* species accumulate to substantial levels in vivo, endogenous miRNA genes do not universally exclude miRNA* species from functional complexes (Ruby et al. 2007; Azuma-Mukai et al. 2008; Czech et al. 2008; Ender et al. 2008; Okamura et al. 2008; Goff et al. 2009; Chiang et al. 2010).
Studies in Drosophila revealed that a substantial fraction of miRNA genes are highly conserved along both miRNA and miRNA* sequences and that seed matches of both miRNA and miRNA* species exhibit preferential conservation in 3′ UTRs (Okamura et al. 2008). Although mature strand miRNAs far outnumber miRNA* species in the Drosophila miRNA effector AGO1 (Czech et al. 2008) and miRNA* species are also strongly loaded into the siRNA effector AGO2 (Czech et al. 2009; Okamura et al. 2009; Ghildiyal et al. 2010), miRNA* strands exert detectable impact on miRNA-type target networks in this species. Similar functionality of vertebrate miRNA* species has been proposed (Ro et al. 2007; Chiang et al. 2010; Schulte et al. 2010), although not systematically tested. We provide here broad evidence from comparative genomics and experimental assays demonstrating that many vertebrate miRNA* species exhibit hallmarks of incorporation into endogenous regulatory networks. Importantly, we identified clear signatures of miRNA* target regulation in transcriptome-wide studies and confirmed direct repression of 3′-UTR targets of miRNA* species. Although the regulatory reach of miRNA* species is less than that of miRNAs, their impact is demonstrable and therefore relevant to vertebrate gene evolution, miRNA gain-of-function experiments, and miRNA biology in normal and disease states.
RESULTS
Experimental assessment of mature miRNA* species
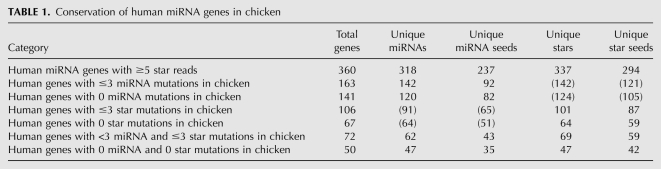
Until recently, many vertebrate miRNA* species evaded direct experimental detection due to their low expression. This has changed with the availability of large data sets of small RNA sequences from next-generation methods. Mapping of tens of millions of reads from several published studies (see Materials and Methods) to miRBase annotations (http://www.mirbase.org/) yielded a set of ∼2 million mature strand reads and ∼78,500 star strand reads from human, and ∼4 million mature strand reads and ∼160,000 star strand reads from mouse (Supplemental Table 1), or ∼4% star reads in both species. From these, we defined 360 genes for which the miRNA* species was deemed confidently assessed, based on a dominant 5′ end being represented by at least 5 reads (Supplemental Data Set 1). Because there are many duplicated miRNA loci, which more typically yield identical mature miRNAs than identical star species, these “well-annotated” miRNA genes actually comprise 318 distinct mature miRNAs and 337 distinct star species (Table 1). These data identified a number of unannotated star species and revealed some discrepancies with miRBase annotations (Supplemental Data Set 2).
TABLE 1.
Conservation of human miRNA genes in chicken
Among well-conserved miRNA genes, mature strands exhibited preference for 5′ U, and to a lesser extent 5′ A (Supplemental Fig. 1). This is consistent with the structural preference of human AGO2 to bind 5′ U and A with much greater affinity than other nucleotides (Frank et al. 2010) and the fact that strand selection is influenced by duplex thermodynamic asymmetry, so that the predominance of weak base pairs at the 5′ ends of miRNAs is coupled to their preferred selection as guide strands (Khvorova et al. 2003; Schwarz et al. 2003). Their partner miRNA* strands instead exhibited preference for 5′ A and 5′ C. The 5′-nt biases of miRNA and star strands were slightly more pronounced when analyzing the set of miRNA genes that are highly conserved among vertebrates (Supplemental Fig. 1), suggesting that these represent preferred characteristics of miRNA genes that have been retained during vertebrate evolution. Notably, both miRNA and miRNA* strand populations strongly disfavored 5′ G, suggesting that miRNA* strands are under selection to avoid a nucleotide feature that is strongly avoided by recognized miRNA regulatory strands.
Sequences of small RNA that immunoprecipitate (IP) with mammalian Ago proteins have been reported, including hAgo2- and hAgo3-IP data from Jurkat cells (Azuma-Mukai et al. 2008) and hAgo1- and hAgo2-IP data from HEK293 cells (Ender et al. 2008). These data sets showed that some miRNA* species are present in Ago complexes at substantial levels (Supplemental Data Set 4), including miR-142-5p(*), miR-17*, let-7d*, and mir-93* in hAgo2-Jurkat, and mir-142-5p, mir-629*, mir-93*, let-7d*, and mir-130b* in hAgo3-Jurkat. Indeed, this profiling yielded more let-7d* than let-7d in hAgo3, more miR-629* in hAgo3 than all but six other mature miRNAs, and more miR-142-5p(*) in hAgo2 than all but seven other mature miRNAs. In total, the miRNA mappings from the combined Ago-IP data contained 3.3% star reads, comparable to what was observed from mappings across all mammalian total RNA data sets. These data provide evidence that miRNA* species contribute substantially to the diversity of abundant small RNAs resident in endogenous human Ago protein complexes.
Characteristics of miRNA gene evolution in vertebrates
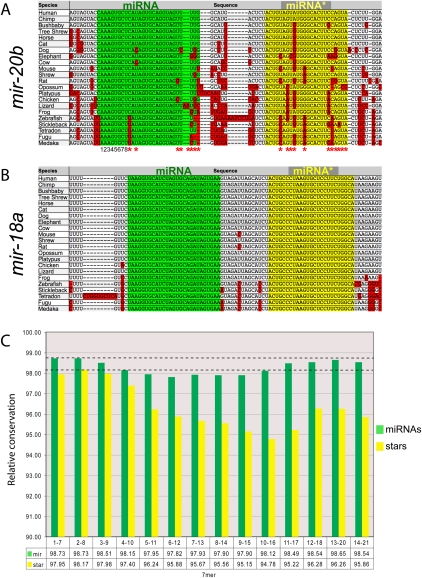
We visualized these dominant cloned miRNA and miRNA* species with respect to 28-way alignments of vertebrate genomes available from the UC Santa Cruz Genome Browser (http://genome.ucsc.edu/). We implemented color-coded schemes for conserved and diverged nucleotides along various regions of the primary miRNA hairpin, which made evolutionary trends in the miRNA and miRNA* species evident upon cursory visual inspection (Fig. 1).
FIGURE 1.
Evolutionary profiles of well-conserved vertebrate miRNA genes. (A,B) Multispecies alignments of two miRNA genes that are conserved from humans to fish. (Green) Mature strands; (yellow) star strands; (red) nucleotides diverged with respect to human. (A) mir-20b is highly conserved; its mature product has sustained a few positions of divergence, but none involve its seed (nucleotides 1–8). On the other hand, miR-20b* has accumulated many more positions of divergence, including in seed nucleotides in its fish orthologs. (B) mir-18a is perfectly conserved along both miRNA and star arms among all vertebrates, from human to fish. Such extreme constraint is suggestive of conserved regulatory activities of both small RNAs produced by mir-18a. (C) Sequence divergence in 7-nt windows across 106 miRNA genes whose star arms sustained ≤3 diverged positions between human and chicken. We note three observations: (1) miRNA strands (green) are better conserved than star strands (yellow); (2) the ends of both miRNA and star strands are better conserved than their central regions; and (3) the 2–8 seed windows exhibit highest conservation along miRNA and star sequences (dotted reference line); the mature 1–7 and 2–8 windows had similar scores.
As expected, the majority of human miRNA sequences with reasonably abundant reads were highly conserved across many vertebrate genomes (Supplemental Data Set 3). In cases in which the mature miRNA diverged, changes in the seed region (2–8) were preferentially avoided (e.g., mir-20b) (Fig. 1A). Also as expected, miRNA* species were overall less conserved than their partner strands, as illustrated by mir-20b. In the case of miR-20b*, its multiple divergence events were consistent with the notion that its predominant role is to maintain overall hairpin structure. However, a substantial fraction of human miRNA* species were, indeed, highly conserved, if not invariant, across broad swaths of vertebrate evolution. For example, both miRNA and miRNA* species of mir-18a were perfectly conserved from all mammals down to all five of the sequenced fish genomes (Fig. 1B). Such constraint cannot easily be explained by a merely structural requirement for miR-18a*.
This level of star-sequence constraint was not peculiar to mir-18a. We assessed miRNA conservation between mammalian genomes and an avian (chicken) genome, a “distant” outgroup with great utility for identifying miRNA binding sites that are conserved across vertebrates (Krek et al. 2005; Lewis et al. 2005). We defined 163 miRNA loci that are well-conserved between mammals and chicken, by the criterion of three or fewer mutations across the mature strand sequence, and 106 miRNA loci with a similar level of conservation across their star strands (Table 1). Of these, 141 mature strand miRNAs and 67 star strand species had no mutations between mammals and chicken, and the intersection of these gene sets comprised 50 loci with no nucleotide changes from mammals to chicken in either miRNA or star. Therefore, a majority of miRNA genes that are preserved between human and chicken maintain stringent constraints on their star species, and scores of star species are invariant across this evolutionary distance.
We quantified the conservation of consecutive 7-nt windows across cohorts of miRNA and miRNA* sequences (Fig. 1C), using a branch length score metric that takes into account the relative divergence of a given species from human (Miller et al. 2007). Analysis of vertebrate miRNA genes whose star species were well-conserved between mammals and chicken (three or fewer positions of divergence) revealed four general trends. First, miRNA strands were more highly conserved than miRNA* strands, consistent with the notion that a single strand is selected as the predominant trans-regulatory species from a given small RNA duplex. Second, both ends of miRNA/miRNA* duplexes were preferentially conserved relative to nucleotides at the center of the duplex, consistent with the necessity to maintain precision in Drosha and Dicer cleavage sites and/or the relative sorting of miRNA/miRNA* species by virtue of thermodynamic duplex asymmetry (Khvorova et al. 2003; Schwarz et al. 2003). Third, the middle regions of miRNA strands were generally more conserved than with miRNA* strands; this region was recently reported to mediate a subclass of regulatory interactions via “centered sites” (Shin et al. 2010). Fourth, we observed that the most highly conserved 7-mer along both miRNA strands and star strands was the 2–8 window.
These features parallel those previously observed across the evolution of Drosophila miRNA genes (Okamura et al. 2008). In particular, the increased constraint in the canonical seed window of the collected star species argues against a purely structural role of mammalian miRNA* strands, since selective pressure to maintain its pairing to the miRNA seed might otherwise have been expected to yield preferred constraint in the miRNA* 3′ region. Instead, the similar and marked seed constraints of mature miRNA sequences (Lewis et al. 2003; Okamura et al. 2008) and star sequences (Fig. 1C) are suggestive of endogenous trans-regulatory roles for both RNAs in the Dicer-cleaved duplex.
Detection of preferred miRNA* seed conservation among vertebrate 3′ UTRs
Although there is not a perfect correlation, the influence of miRNA species on 3′-UTR evolution is correlated with their expression: miRNAs that are more highly expressed generally have more conserved targets than do miRNAs that accumulate to lower levels (Lewis et al. 2003; Ruby et al. 2007). Therefore, in spite of their characteristic evolutionary constraints, the relatively low levels of miRNA* species raised the question of whether they were abundant enough to exert measurable impact on transcript evolution. On the other hand, seemingly “lowly expressed” miRNAs can serve critical endogenous functions (Johnston and Hobert 2003); thus, low gross accumulation does not necessarily imply lack of endogenous usage.
We therefore tested the potential impact of miRNA* species on vertebrate 3′-UTR evolution by analyzing the conservation of miRNA and miRNA* seed matches across various cohorts of vertebrate genomes. We assessed the relative conservation of the approximately 16,000 7-mers across alignments of mammalian and avian orthologous 3′ UTRs (Chen and Rajewsky 2006). We selected control seeds to compare against each genuine miRNA or miRNA* seed and were careful to screen out all seeds with 7-mer and 6-mer + t1a complementarity to miRNA or miRNA* seeds from appearing in control cohorts (see Materials and Methods).
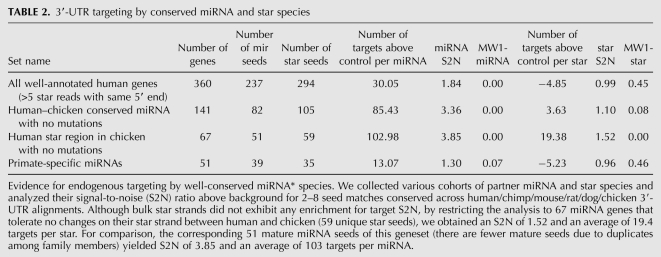
We started by analyzing a set of human miRNA genes with well-annotated star species (as described earlier, with a predominant star 5′ end represented by at least five reads), encompassing 237 distinct miRNA seeds and 294 miRNA* seeds. This set exhibited 1.84-fold signal-to-noise (S2N) in enrichment for conserved matches for miRNA seeds, but no enrichment (0.99 S2N) for star seeds (Table 2); only unique seeds were analyzed in this and all subsequent tests. This gene set includes a number of miRNA species that are not conserved in chicken, and certainly many more star species that are relatively poorly conserved even among vertebrates. That we observed substantial S2N for mature strand targeting, despite inclusion of irrelevant miRNAs, reflects the strong signals achieved by seed matches to well-conserved miRNAs. Reciprocally, the fact of no enrichment above background for star targets across this aggregate set provides confidence that we selected appropriate control 7-mers and indicates that conserved star targets, if they exist, cannot overcome the large background of irrelevant seeds analyzed in this set. We performed additional analysis of a set of 51 primate-specific miRNA genes for evidence of target sites conserved to chicken. As expected, these yielded almost no S2N for targets across the 39 distinct miRNA seeds or the 35 distinct star seeds, again indicating that our control sets were selected appropriately.
TABLE 2.
3′-UTR targeting by conserved miRNA and star species
A more relevant assessment in screening for target sites that are conserved human–chicken is to restrict the analysis to those miRNA genes that are conserved in chicken. Using this set of 141 miRNA genes, comprising 82 distinct mature miRNA strand seeds and 105 star strand seeds, we observed a strong increase in miRNA targeting, as expected (Table 2). This set identified conserved seed matches at a S2N of 3.36, with an average of 85 targets/miRNA seed above background, similar to previous assessments of vertebrate seed matches conserved from human to chicken (Lewis et al. 2005). The corresponding set of star species yielded a slight increase in star S2N to 1.10, with an average of 3.6 targets/star above background, although this was not significant by the one-sided Mann-Whitney test (Table 2).
As this set contains a number of miRNA genes whose star strand has diverged substantially in chicken, we further restricted the analysis to the 67 miRNA genes whose star strand was unchanged between human and chicken (comprising 51 unique mature miRNA seeds and 59 unique star seeds). This group achieved 3.85 S2N for mature strand seeds and 1.52 S2N for star seeds, with an average of 103 targets/miRNA and 19.4 targets/star (Table 2). The level of star targeting for these gene set was now highly significant by the one-sided Mann-Whitney test comparing the distribution of S2N of star seeds to control seeds. We conclude that miRNA* species exhibit modest overall signal for 3′-UTR targeting relative to miRNA species, but that miRNA* targeting is considerable among the scores of miRNA* seeds that have been highly conserved during vertebrate evolution.
miRNA* species can repress targets via perfect sites and seed matches
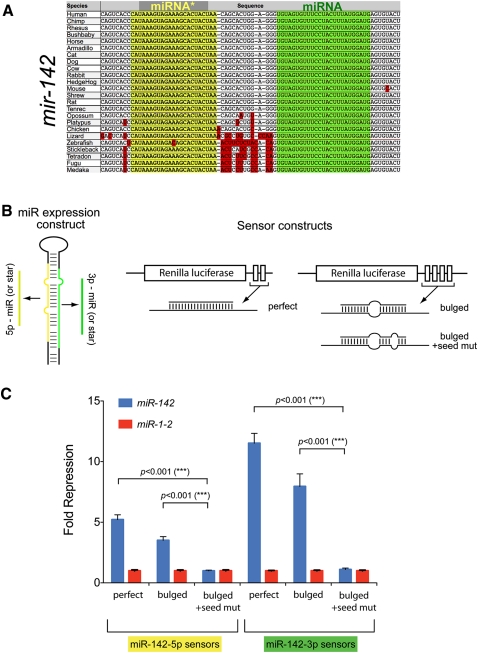
We selected mir-142 for initial functional tests, since it exhibited several traits that epitomized dual miRNA/miRNA* regulatory function. The ratio of mir-142 miR:star reads is low in both human and mouse data sets, both miR-142-5p and miR-142-3p are highly represented among endogenous RNAs associated with independent hAgo2-IP and hAgo3-IP experiments (Azuma-Mukai et al. 2008), and both mature sequences are perfectly conserved among vertebrates ranging from humans to fish (Fig. 2A). Such properties strongly suggested that both small RNA products have been selected for endogenous usage as regulatory species.
FIGURE 2.
Validation of the regulatory activity of mature strand and star strand targets of mir-142. (A) mir-142 has been exceptionally conserved along both its miRNA (green) and star (yellow) strands; positions of divergence (red) reside in the pre-miRNA flanks or in the terminal loop. (B) Schematics of artificial sensors for miR-142-5p or miR-142-3p, containing either two perfect target sites, four bulged sites, or four bulged sites with seed mismatches. (C) Target repression by both miR-142-5p and miR-142-3p is seed-dependent. Sensors were assayed and normalized as described in Figure 4. Introduction of the mir-142 expression plasmid yields robust repression of both perfect and bulged sensors, but mutation of seed nucleotides abolishes target repression.
We tested siRNA-type Renilla luciferase reporters containing two perfect sites for either miRNA or miRNA* species of mir-142, as well as miRNA-type sensors containing four target sites with central bulges (Fig. 2B). Renilla reporter activities were normalized to internal firefly luciferase reporters to control for transfection efficiency and then normalized with respect to a non-cognate mir-1-2 expression construct to control for potential nonspecific effects of miRNA overexpression. These tests revealed that mir-142 repressed perfect miR-142-5p and miR-142-3p sensors approximately fivefold to 11-fold, respectively (Fig. 2C). We also observed threefold to eightfold repression of their bulged sensors, a lower amount that is consistent with the more efficient silencing mediated by target cleavage. To demonstrate that the repression of the bulged sensors was due to typical miRNA-mediated seed matching, we made another pair of bulged constructs carrying three point mutations in the seed-matching region (Fig. 2B). Now, mir-142 was unable to repress either target construct (Fig. 2C), indicating that the repression of the bulged sensors reported on the activity of both miRNA and miRNA* species of mir-142 on conventional seed-driven regulatory interactions.
Generalization of the regulatory capacity of miRNA* species
We next sought to test how broadly the regulatory capacity of miRNA* species applied. Having validated the repressive activity of miR-142* using both perfect and seed-matched sensors, we performed subsequent experiments using perfect sensors as these provided greater sensitivity. We used mir-1-2 as a non-cognate control for these tests; mir-1-2 itself was compared against mir-199a-2 and an empty miRNA expression vector as non-cognate comparisons, and it was seen to be similarly highly active by both controls. In these tests of perfect sensors, we considered normalized sensor repression >2 standard deviations above the average non-cognate sensor baseline to represent confident repression.
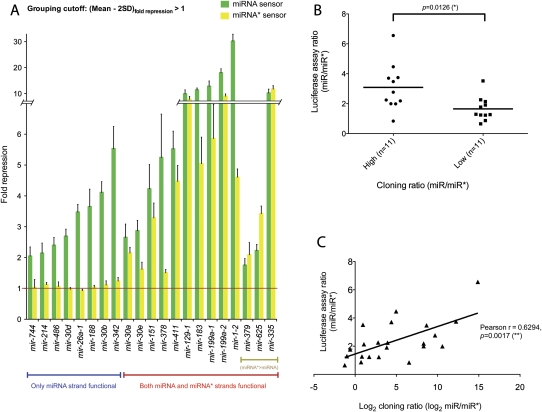
In total, we tested partner miRNA and miRNA* sensors for 22 different miRNA genes (Figs. 2, 3). For all mature strand miRNA sensors, we observed greater than twofold (and up to 30-fold) repression, consistent with the expectation that all bona fide miRNA genes should produce a functional regulatory RNA. However, we also observed that 14 star strand sensors were confidently repressed upon introduction of a cognate miRNA expression construct, although some by less than twofold (Fig. 3A). Therefore, a majority of miRNA genes tested generated functional regulatory RNAs from both left and right hairpin arms.
FIGURE 3.
General tests of the dual regulatory capacity of miRNA genes. (A) Renilla luciferase sensors containing four antisense matches to either the mature miRNA or star species of a given miRNA gene were tested for their response to a cognate pri-miRNA expression plasmid in HeLa cells. Sensor values were normalized to an internal firefly luciferase transfection control and then represented as the fold repression relative to the sensor level in the presence of a non-cognate miRNA expression plasmid (usually mir-1-2; the control for mir-1-2 was mir-199a-2). We deemed a miRNA expression plasmid to be “dual function” if the mean repression value was at least two standard deviations above 1. Values are derived from two independent sets of quadruplicate transfection experiments performed on different batches of cells; standard deviations are shown. Some miRNA constructs were only capable of repressing the mature strand sensor, but a majority of constructs could repress both mature strand and star sensors. Three genes for which the inferred miRNA* species (based on meta-analysis of published library data) yielded slightly higher repression than their partner miRNA species are segregated to the right. (B) Correlation of ectopic miRNA sensor tests and endogenous cloning ratios. We collected small RNA reads from the 22 loci tested in Figure 2 and this figure and compared their miRNA:miRNA* cloning ratios with their miRNA:miRNA* sensor repression ratios. A rank analysis was performed to group genes with higher cloning ratio (i.e., more asymmetric accumulation of the duplex strands) or lower cloning ratio (i.e., more balanced accumulation of the two strands). The correlation with higher versus lower sensor repression ratios was statistically significant. (C) A linear regression was performed between the miRNA:miRNA* sensor repression ratio and the log2(miRNA:miRNA*) cloning ratio. The correlation was significant according to both Pearson's tests.
We recognize that these sensor studies relied on overexpressed miRNAs, whose sorting might not be identical to their endogenous counterparts. The fact that eight miRNAs were functional only on their mature miRNA strand suggested that inappropriate loading of star strands was not a pervasive issue, since this analysis clearly identified many functional asymmetric miRNA genes. To probe this further, we compared the relative levels of repression mediated by partner miRNA and miRNA* species with their endogenous ratios of miRNA:miRNA* accumulation summed across a diverse set of published libraries (see Materials and Methods).
A caveat to this analysis is that the reproducibility of most miRNA cloning data reported in the literature is unknown. For example, there could be variability among data sets due to technical differences in cloning or sequencing protocols or biological variability between different tissue samples assayed. Nevertheless, even with such caveats, we observed statistically significant correlations. Using group rankings to segment miRNA genes into pools with higher or lower miRNA:miRNA* ratios, we observed a good correlation (P-value <0.0126) with their ratio of miRNA:miRNA* sensor activity (Fig. 3B). In particular, many genes whose endogenous miRNA:miRNA* cloning ratios were most biased (e.g., mir-30b with more than 7000 mature reads and three star reads, or mir-26a-1 with more than 50,000 mature reads and no star reads) failed to repress their star sensors in the ectopic test.
We also performed a linear regression analysis of the miRNA:miRNA* cloning ratio and sensor ratio (Fig. 3C). We analyzed the log2 values of cloning ratios, since the ratio of their cloned reads varied over 3–4 orders of magnitude, while the ratio of fold repression between miRNA and miRNA* varied less than sevenfold. This analysis yielded a Pearson's correlation of r = 0.629, which was again significant (p = 0.0017). We conclude from the sensor studies that a majority of tested miRNA genes have the capacity to repress targets complementary to both miRNA and miRNA* species, and that the functional sorting of small RNAs produced from these expression constructs was reasonably correlated with their endogenous accumulation.
Transcriptome-wide evidence for repression by miRNA* species
We wished to extend our experimental results to the genome-wide level. It has commonly been observed that following up-regulation of an individual miRNA, the transcriptome exhibits preferential down-regulation of mRNAs bearing cognate seed matches (Lim et al. 2005). This strategy for functional miRNA analysis is convenient in that does not depend on endogenous miRNA levels and has achieved widespread usage to identify potential target genes, or even merely to gain evidence for miRNA activity (e.g., in delivery trials). Recently, careful analysis of such data has also revealed the de-repression of endogenous miRNA targets (Khan et al. 2009), indicating that microarray profiling provides a sensitive platform to detect gene expression changes beyond simple targeting by mature miRNAs.
Only a few published studies reported the enforced expression of miRNA* sequences. One such study by the Linsley group reported microarray expression profiles of HCT116 and DLD1 cells following transfection with either mature miRNA or star strand of mir-17 using duplex mimics (Linsley et al. 2007). In their experiments, the partner strands were reciprocally altered so that either miR-17-5p (mature) or miR-17-3p (star) was specifically delivered to active regulatory complexes. In principle, this should permit the discrimination of miRNA and star targets of mir-17, although only miR-17-5p targets were analyzed in this study. In particular, they reported that mir-17-5p represses many targets that are shared by its family members miR-20a and miR-106b (Linsley et al. 2007), providing support for the notion that the seed region is a major determinant of target selectivity (Lai 2002; Lewis et al. 2003; Stark et al. 2003).
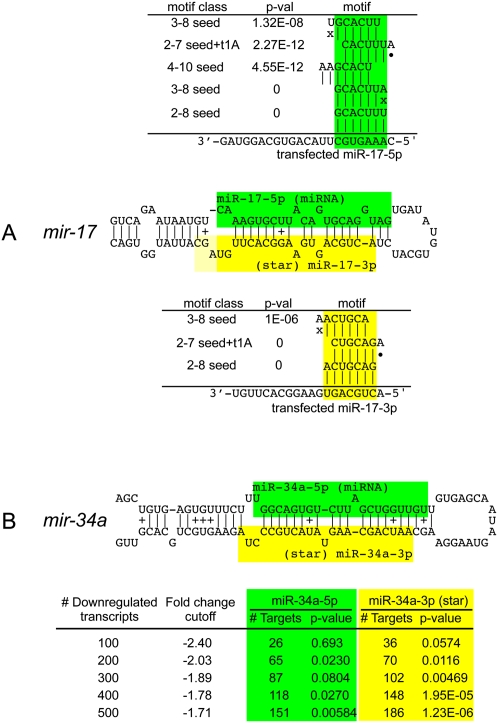
We applied miREDUCE (Sood et al. 2006) to the miR-17-5p data set and confirmed the result that the sequence motif that was best-correlated with down-regulated transcripts (and reciprocally absent from up-regulated transcripts) was the miR-17-5p seed (GCACUUU) (Fig. 4A). Several related motifs (including 2–7 seeds with t1A anchors, other offset 6-mer or 7-mer motifs, and some G:U seeds) were also highly correlated with target down-regulation (see also Supplemental Table 2). On the other hand, using the miR-17-3p (star) data set, we observed very strong enrichment for repression of star seed target genes. Indeed, the miR-17-3p (star) seed was the top correlated motif, with again several related motifs being very highly correlated (Fig. 4A).
FIGURE 4.
Transcriptome-wide evidence for target repression directed by partner miRNA and star species. (A) Target repression by mature (green) and star (yellow) products of mir-17. Linsley et al. (2007) reported microarray profiles for cells transfected with mimics for either miR-17-5p (mature) or miR-17-3p (star), but only analyzed the mature strand response. Using miREDUCE, we observe that the motifs that are most highly correlated with down-regulated transcripts in these data sets are the miR-17-5p seed (2–8 or 3–8) and the miR-17-3p seed (2–8 or 2–7seed + t1A), respectively, all with P-values = 0. Other highly statistically enriched motifs in down-regulated transcripts include various seed variants (see also Supplemental Table 2). (B) Mendell and colleagues reported microarray profiles following infection with a retroviral mir-34a construct (Chang et al. 2007) and reported that the miR-34a seed was enriched among the 100 most down-regulated transcripts. miREDUCE on this transcript set yielded statistically significant enrichment of miR-34a 2-7 seed (green) among the top 200 down-regulated transcripts, but this enrichment increased when larger sets of down-regulated transcripts were analyzed. These same transcript subsets yielded even stronger enrichment for miR-34a* seeds (yellow) across all comparable cohorts of down-regulated genes, peaking at 1.23E-06 in the top 500 most down-regulated transcripts.
In principle, the transfection experiments might be biased by the designed asymmetry of mimic duplexes. We were therefore interested to validate these results with data from pri-miRNA expression experiments. The Mendell group performed microarray analysis following infection of HCT116 cells with a control retrovirus or a mir-34a expression construct (Chang et al. 2007). Their experimental conditions yielded only threefold to fourfold up-regulation of miR-34a, which might reflect an expression change that is more physiological than obtained with miRNA transfection. Still, it was noted that many more transcripts changed under these conditions than in typical miRNA transfection experiments (Lim et al. 2005; Linsley et al. 2007), presumably indicating that these data reflected a substantial indirect changes in gene expression (Chang et al. 2007). Consistent with this suggestion, we ran miREDUCE over a range of parameters (see Materials and Methods) and discovered several 3′-UTR motifs that were significantly correlated with the gene expression changes but none that matched miRNA seeds (Supplemental Table 2).
Despite the presence of indirect effects, Mendell and colleagues reported that when they focused on the most down-regulated transcripts, they found a modest enrichment of miR-34a 2–7 seed matches (Chang et al. 2007). Analysis of the top 200 down-regulated transcripts confirmed this trend (Fig. 4B). Interestingly, we observed even stronger enrichment for the miR-34a seed among larger cohorts of down-regulated transcripts, peaking at p = 0.00584 across the top 500. Perhaps unexpectedly, analysis of the same cohorts of down-regulated transcripts revealed even stronger enrichment for miR-34a* 2–7 seed matches. This preferred enrichment for miR-34a* seeds over mature miR-34a seeds was stronger across each set of down-regulated transcripts analyzed, peaking at p = 1.23E-06 among the top 500 down-regulated transcripts (Fig. 4B). We conclude that retroviral expression of pri-mir-34a results in repression of both miR-34a and miR-34a* targets.
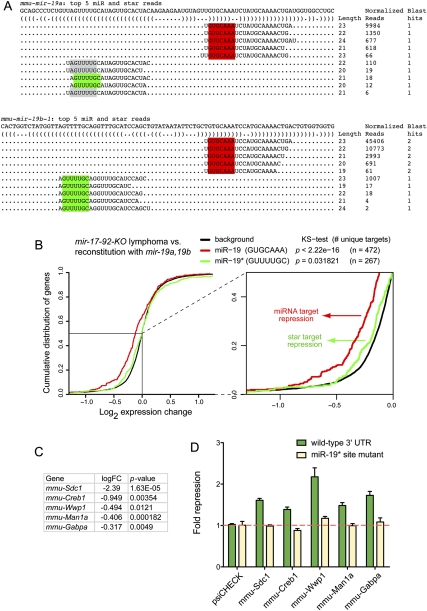
In a final set of analyses, we examined microarray data using Myc-induced lymphoma cells deleted for the mir-17→92 cluster, or reconstituted with retrovirus expressing the two mir-19 genes in this cluster, mir-19a and mir-19b-1 (i.e., mir-19a/b). We were particularly interested in mir-19a/b as it was recently shown to be the critical oncogenic component of this miRNA cluster (Mu et al. 2009; Olive et al. 2009; Mavrakis et al. 2010). The mature products of the mir-19 genes predominate over their star species by 60- to 75-fold and share a common seed (GTGCAAA), suggesting that this sequence should exert the dominant signature on the transcriptome for mir-19-derived small RNAs. Their star regions are distinct: miR-19a* has two major 5′ isomiRs, one of which is shared with miR-19b-1* (GUUUUGC). Since mir-19b-1* is expressed at least eightfold higher than miR-19a*, this suggests that the alternate miR-19a* seed (AGUUUUG) may contribute correspondingly less to endogenous gene regulation (Fig. 5A; Supplemental Fig. 2).
FIGURE 5.
Evidence for targeting by endogenous miR-19 and miR-19*. (A) Dominant read counts of mmu-mir-19a and mmu-mir-19b-1 analyzed from six data sets reported in GSE11724 (Marson et al. 2008; see also Supplemental Fig. 2). Both genes generate a mature species with a shared seed (red box); miR-19a* generates two 5′ isomiRs, one of which is shared with miR-19b-1* (green box); note that the GUUUUGC miR-19* seed is by far the dominant star seed generated by the mir-19 genes. (B) Cumulative distribution function of gene expression in lymphoma cell lines deleted for the mir-17→92 cluster compared to those re-expressing mir-19a/b under retroviral control. Transcripts whose 3′ UTRs contained exclusively miR-19 (GUGCAAA) or miR-19* (GUUUUGC) seed sites are segregated for analysis; transcripts containing both types of seed matches were discarded to avoid ambiguity in assigning the targeting species. Transcripts with miR-19 seed sites (red) or miR-19* seed sites (green) were down-regulated in the presence of mir-19 relative to background gene expression of all other transcripts (black); this trend was more substantial for the mature strand miR-19 but still statistically significant for star strand targets. The region boxed in the main graph is expanded to illustrate this trend more clearly. (C) Fold change of transcript down-regulation for selected targets bearing conserved miR-19* seed matches. Following multiple hypothesis correction by the false discovery rate (FDR), log fold changes with adjusted P-values (FDR <%5) were considered significant; all of these gene expression changes were highly significant. (D) Confirmation of direct repression by miR-19*. 3′-UTR target sensors and matched mutants bearing specific point changes within the miR-19* seed matches were tested for their response to transfection of mir-19a/b in HeLa cells; sensor activities were normalized to their level in the presence of functional mir-1-2 construct. All five were repressed in a manner that was completely dependent on the integrity of the miR-19* seed matches (cf. wild-type sensors in green with mutant sensors in tan).
We analyzed the cumulative distribution of differential gene expression between the two cell lines with or without mir-19a/b and segregated those putative targets containing exclusively perfect miR-19 seed matches or miR-19b-1* (GUUUUGC) seed matches; transcripts containing both types of sites were excluded to avoid confounding attribution of miRNA regulation. As reported earlier using all miR-19 targets (Mu et al. 2009), the distribution of transcripts with exclusive miR-19 seed matches was strongly down-regulated in the presence of mir-19a/b. In addition, we observed a modest but statistically significant trend (assessed by Kolmogorov-Smirnov test) for down-regulation of exclusive miR-19* targets as well (Fig. 5B). Finally, similar tests with matches to the miR-19a* isomiR seed (AGUUUUG) targets failed to reveal a significant shift (data not shown), consistent with its very low endogenous accumulation. We conclude that physiological expression of mir-19a/b results in transcriptome-wide down-regulation of both miR-19 targets, as expected, but also targets of the dominant miR-19* species as well.
To validate the regulation of miR-19* targets directly, we analyzed sensors of candidate target genes. We generated a set of Renilla reporters fused to the 3′ UTRs of five different transcripts whose levels were decreased in the presence of mir-19a/b, and that bore conserved miR-19* seed matches (Fig. 5C; Supplemental Fig. 3). These were co-transfected with mir-19a/b into HeLa cells and normalized against mir-1-2 as a non-cognate control. Their response was modest, between ∼1.5-fold and twofold, but detectable in all cases (Fig. 5D). To verify that such repression was indeed directly mediated by miR-19*, we mutated the target sites in these sensors. This abolished their response to mir-19 in all cases (Fig. 5D), demonstrating direct repression via star target sites in these 3′ UTRs.
Altogether, these analyses of three different cancer-relevant miRNA genes demonstrate that star species can mediate target modulation that is measurable across the transcriptome and could be validated using conventional sensor assays usually used to test “mature” miRNA target sites. These results were robust in both miRNA “mimic” transfection experiments, as well as in more physiological expression strategies using retrovirally delivered pri-miRNAs. These data indicate that the regulatory activity of miRNA* species should be considered to reveal the full picture of miRNA-mediated gene regulation.
DISCUSSION
miRNA* activity contributes to the vertebrate miRNA target network
In this study, we showed that well-conserved vertebrate miRNA* species follow the same principles as their partner miRNA species in that seed nucleotides 2–8 exhibit preferred conservation along the small RNA, and 3′-UTR seed matches exhibit excess conservation over control heptamers. We directly confirmed the ability of a majority of miRNA genes to repress designed sensors for both miRNA and miRNA* species, consistent with the recent observation of diverse miRNA* species in mammalian Ago-IPs. Indeed, we easily observed the repressive consequence of miRNA* activity in multiple transcriptome-wide expression profiles as well as individual 3′-UTR sensors demonstrating star-seed-dependent regulation. Although miRNA* species contribute less to gene regulatory networks than their partner miRNA strands, as indicated by their smaller signal-to-noise ratios in target predictions and their smaller transcriptome signatures, our data indicate that their regulatory influence is demonstrable by the major computational and experimental strategies used to assess the function of their partner mature miRNA strands.
Our systematic studies in vertebrates parallel the results from our Drosophila studies (Okamura et al. 2008) and greatly extend recent reports indicating functional targets of specific mammalian miRNA* species, such as miR-9a* (Packer et al. 2008; Yoo et al. 2009), miR-199* (Kim et al. 2008; Lee et al. 2009; Lin et al. 2009), miR-30a* (Ro et al. 2007), and miR-18a* (Tsang and Kwok 2009). Altogether, these data indicate that the regulatory impact of miRNA* species on mammalian genomes is much more substantial than is currently envisaged. We do not advocate discarding miRNA/miRNA* terminology, since this nomenclature provides important information regarding strand preference of miRNA biogenesis, which is almost always asymmetric. On the other hand, it is abundantly clear that miRNA* species cannot be ignored as regulatory molecules and support the notion of a broader usage of “5p-3p” nomenclature that acknowledges fluidity rather than sole use of “miRNA:star” nomenclature.
Why hasn't an appreciable extent of vertebrate miRNA* activity been detected by earlier genome-wide studies? Target repression is strongly influenced by the cellular concentration of the cognate regulatory RNA. Therefore, if endogenous strand selection pathways strongly prefer the miRNA over the miRNA*, then direct regulatory changes will mostly reflect the regulation of mature strand targets. However, the fact of low miRNA* accumulation does not necessarily imply that they are irrelevant to gene regulation, any more than genuine miRNAs with low and/or regionally specific expression should be inferred to be “less functional” than highly and/or broadly expressed miRNAs. Indeed, high-throughput profiling of Ago binding sites revealed that significant enrichment for miRNA seed matches in Ago mRNA-CLIP tags was observed for only a few tens of the most highly expressed miRNAs, out of hundreds of miRNAs detected in the same complexes (Chi et al. 2009; Hafner et al. 2010).
On the other hand, miRNA* function was readily observed in gain-of-function tests using miRNA expression constructs, including settings with only a few fold up-regulation of the miRNAs in question. In the literature, a preferred method of elevating miRNA activity has not been through primary miRNA transcription, but instead by transfection or injection of miRNA/miRNA* duplexes (Lim et al. 2005; Giraldez et al. 2006; Wang and Wang 2006; Linsley et al. 2007; Selbach et al. 2008). As it happens, such duplexes were almost always designed to force the incorporation of miRNA strands, by mutating miRNA* strands so as to prevent their loading into Ago complexes. Therefore, most of the existing tests in the literature were poorly suited to report on miRNA* targeting in vertebrates. Indeed, commercially available libraries of miRNA “mimics” designed to up-regulate miRNA activity (e.g., Ambion “Pre-miRs,” QIAGEN “miScript miRNA Mimics,” and Dharmacon “miRIDIAN mimics”) are mutated in their star strands, ostensibly to promote “correct” strand incorporation by RISC. Such mutations alter endogenous miRNA duplex strand selection and prevent repression of endogenous star strand targets. As these “mimic” reagents are currently in predominant use in the literature, the effects of star strand target regulation may continue to be hidden from studies of mammalian miRNA function.
Consequences of miRNA* activity during disease
The mir-34 family is a functional component of the p53 target network (Chang et al. 2007; He et al. 2007; Raver-Shapira et al. 2007; Tarasov et al. 2007), and there is great interest in using miR-34 as a therapeutic for p53-compromised tumors. Although “mimic” studies focus on directing mature miR-34 into regulatory complexes, our studies indicate that expression of pri-mir-34 is associated with direct repression of both miR-34 and miR-34* targets. We hypothesize that a fuller rescue of mir-34 loss-of-function might be provided by endogenous duplexes, instead of the star-mutated versions typically used as mimics.
Reciprocally, miRNA* activity may prove even more broadly relevant to gain-of-function settings. The ectopic expression of many miRNAs induces developmental defects; for example, enforced expression of mir-142 in hematopoietic progenitors results in abnormally high commitment to T-lymphoid lineages (Chen et al. 2004). Given our data on the balanced output of functional miRNAs from both arms of mir-142, the potential functional contribution of both miR-142-5p and miR-142-3p deserves study. It is worth noting that alternative Drosha processing has diversified the functional output of mir-142 even further (Wu et al. 2009).
Many cancers are due to genomic amplification or gain-of-function. For example, elevated levels of the “oncomir-1 cluster,” comprising the mir-17, mir-18, mir-19, mir-20, mir-92 genes, underlies various hematopoietic and solid cancers (Ota et al. 2004; He et al. 2005; O'Donnell et al. 2005). Five of the six genes in this cluster have highly conserved miRNA* species that exhibit seed constraint (Supplemental Data Set 2), and multiple members appear to have compelling star functions. miR-18a* was reported to regulate K-ras (Tsang and Kwok 2009), and endogenous miR-17* is abundant in murine Argonaute complexes (Azuma-Mukai et al. 2008). Transgenic activation of mir-17 affects tissue growth, and in such animals, miR-17* is differentially and more highly up-regulated in certain tissues than mature miR-17 (Shan et al. 2009). Our transcriptome analysis indicates broad regulatory effects or miR-17* (Fig. 4). Finally, and perhaps most significantly, mir-19 is the critical oncogenic component of this cluster (Mu et al. 2009; Olive et al. 2009; Mavrakis et al. 2010). We provided evidence for transcriptome-wide response of miR-19* targets from microarray profiling, and a cohort of miR-19* targets was shown to be directly repressed by mir-19a/b via conserved miR-19* star sites in conventional sensor assays (Fig. 5).
We hypothesize that settings involving deregulated miRNA loci, such as the mir-17→92 cluster, likely involve the inappropriate repression of miRNA* targets. More generally, our computational and experimental studies support the notion that endogenous miRNA* species have measurable impact on mammalian gene expression and evolution. We conclude that evaluation of the regulatory activities of miRNA star species is necessary for a full understanding of the miRNA regulatory network.
MATERIALS AND METHODS
Annotating mature products from small RNA data libraries
Small RNA reads were collected from several previously published studies of human (Landgraf et al. 2007; Azuma-Mukai et al. 2008; Ender et al. 2008; Friedlander et al. 2008; Morin et al. 2008), mouse (Calabrese et al. 2007; Babiarz et al. 2008; Baek et al. 2008; Marson et al. 2008; Stark et al. 2008; Tam et al. 2008; Watanabe et al. 2008), dog (Friedlander et al. 2008), and chicken (Glazov et al. 2008; Rathjen et al. 2009). Small RNA reads were clipped if necessary and mapped to known miRNA precursors (miRBase) using custom python scripts. The mapping process requires a 100% match. A mature product or star was annotated as confident if the most abundant read comprised at least five reads with the same 5′ end. Hairpins were folded using RNAfold from the Vienna Package (Hofacker 2003).
Evolutionary analysis across miRNA gene windows
We retrieved 28-way multiZ alignments for each miRNA precursor from the UCSC Genome Browser (http://genome.ucsc.edu/). From these alignments we also extracted human, chimp, mouse, dog, and chicken data; made manual adjustments to optimize alignments; and color-coded the output (mature miRNAs in green, miRNA* species in yellow, positions of divergence in red). Complete alignment data can be found in Supplemental Data Set 2. In addition, the 28-way and 6-way alignments are also accessible at http://michaeldphillips.com/mam_mirstar/, using the user login: referee and password: s14ha3i.
The conservation of each base was calculated by pairwise comparison of each of chimp, mouse, dog, and chicken with Homo sapiens hg18 as the reference. Matches were scored a 1, while mismatched or gapped nucleotides were scored as a 0. To capture the significance of evolutionary distance across the species, we weighted this score using distances from the standard 28-way vertebrate phylogeny (Miller et al. 2007). For each 7-nt window across the miRNA-miRNA* sequence, we summed the seven base scores and rescaled them from 0 to 100, with 100 representing full conservation across all five species.
We noted that many cloned chicken miRNAs are missing from the UCSC multiZ alignments, probably because many conserved non-coding RNAs are located in non-syntenic regions of the chicken and human genomes (International Chicken Genome Sequencing Consortium 2004). We therefore supplemented the human–chicken highly conserved miRNA geneset with additional chicken miRNAs that were missing from UCSC alignments, but recovered from deep sequencing of chicken libraries (Glazov et al. 2008; Rathjen et al. 2009). This read evidence is summarized in Supplemental Data Set 5.
Analysis of conserved target properties
Although many strategies exist to analyze miRNA targeting computationally, the major signal in comparative genomic scans is mostly accounted for conserved 7-mer matching to miRNA seeds (positions 2–8) (Bartel 2009). We therefore focused on this class of potential conserved miRNA binding site.
To generate sets of control seeds, we tallied the occurrence of all (47 = 16,384) possible heptamers across annotated hg18 3′ UTRs (Chen and Rajewsky 2006). We generated controls for each miRNA or miRNA* 2–8 heptamer by randomly selecting 50 control heptamers whose frequency in hg18 3′ UTRs was within 10% of the original heptamer (Krek et al. 2005). The control seed sets were filtered so as to remove any 7-mers that matched known miRNA or miRNA* seeds, or polyadenylation sites (AAUAAA and AUUAAA).
We also explored methods for generating control sets using permuted seeds with matching of dinucleotide frequencies, as earlier performed in vertebrate target analyses (Lewis et al. 2003) and in our studies of Drosophila targets (Okamura et al. 2008). We observed overall similar results with this method but found that many seeds did not generate sizable control seeds that satisfied the tolerance for frequency in the transcriptome. We therefore preferred the former strategy, which has been used in several target studies in various animal clades (Grun et al. 2005; Krek et al. 2005; Lall et al. 2006).
To identify instances of conserved 7-mers (i.e., “targets”), we updated our earlier alignments of human/chimp/mouse/rat/dog/chicken 3′ UTRs using previously described methods (Krek et al. 2005; Chen and Rajewsky 2006). miRNA and star seeds were analyzed non-redundantly (i.e., duplicate seeds in family members were not tested). The ratio of target number between a genuine miRNA or miRNA* seed and its control cohort of 50 seeds was taken as its signal-to-noise ratio (S2N). The average S2N across various groupings of partner miRNA and miRNA* seeds was then calculated.
Generation of miRNA expression constructs
Approximately 500-nt pri-miRNA fragments were amplified from HeLa cell genomic DNA by polymerase chain reaction and cloned downstream from GFP in pcDNA6.2/N-EmGFP-TOPO (Invitrogen). Expression of miRNAs is under the control of the CMV promoter. The primer sequences of miRNAs are listed in Supplemental Table 3 in the “miRNA expression primers” worksheet.
Generation of luciferase sensor constructs
For miRNA perfect sensors, oligonucleotide pairs were designed to contain two antisense copies of an miRNA or miRNA* species, and 5′-NotI- and 3′-XhoI-compatible ends. These target sequences were cloned into the NotI–XhoI sites downstream from the Renilla luciferase coding region in a modified psiCHECK2 vector (Okamura et al. 2007), which has an internal firefly luciferase gene for normalization (Promega). The sensor sequences for the different miRNAs are listed in Supplemental Table 3 in the “miRNA sensor primers” worksheet.
For the mir-142 sensor set, oligonucleotide pairs were designed to contain a single bulged or mutant miR-142 target site flanked by SalI and XhoI restriction sites at the 5′ and 3′ ends, respectively. To anneal complementary oligonucleotide_A and _B, 50 μL of 100 μM A and B was mixed and placed for 5 min at 95°C, for 20 min at 65°C, and for 20 min at room temperature. 5′ phosphates were generated by incubating the resulting double-stranded DNA products with T4 PNK in T4 ligase buffer for 30 min at 37°C. Oligomerization was performed by incubating the prepared duplexes with T4 DNA ligase in the presence of SalI and XhoI, which proofread the ligation reaction for correct orientation. Four-copy-site multimers were purified from low-melting-point agarose gels and cloned into the modified psiCHECK2 vector. The artificial bulged and mutant sensor sequences of miR-142 are listed in Supplemental Table 3 in the “mir-142 sensor primers” worksheet.
For 3′-UTR sensors of miR-19* targets, we chose a set of genes that were down-regulated in the microarray analysis of mir-17-92 knockout lymphoma cell lines before and after reconstitution with mir-19a/b retrovirus (Mu et al. 2009), and that contained conserved miR-19a/b* target seeds. We cloned ∼250-bp 3′-UTR fragments containing the miR-19a/b* sites downstream from Renilla luciferase in psiCHECK2. To generate mutant 3′-UTR sensors, separate PCR reactions were carried out first with wild-type forward and mutant reversed primers, and mutant forward and wild-type reversed primers, respectively. The resulting two PCR fragments, which have several overlapping nucleotides flanking the mutant sites, were gel-purified and used as template in the presence of wild-type forward and reversed primers for the second PCR reaction. The PCR products were gel-purified and cloned as before.
HeLa cell transfection and luciferase sensor assay
HeLa cells were seeded into 96-well plates at the density of 1 × 104 per well 24 h before transfection. Cells were transfected in quadruplicate with 40 ng of cognate or non-cognate miRNA plasmids and 10 ng of psiCHECK2 sensor plasmids using FuGENE6 Transfection Reagent (Roche Applied Science). Twenty-four hours after transfection, cells were lysed with 75 μL of Passive Lysis 5× Buffer (Promega). Seventy-microliter cell lysates were subjected to the Dual-Glo Luciferase Assay System (Promega), which was analyzed on a Veritas plate luminometer (Turner Biosystems). Fold repression was normalized to parallel sensor assays transfected with non-cognate miRNA. GFP-mir-1-2 was used as a control for most miRNA constructs; empty vector or GFP-mir-199a-2 was used for GFP-mir-1-2 experiments. Data from two independent batches of quadruplicated experiments were pooled, and the mean and standard deviation of each miRNA sensor assay were calculated.
Correlation analysis of miRNA/star accumulation and sensor regulation
XY correlation
We plotted log2 of the [miR/miR*] ratio of cloning counts from human libraries ([miR/miR*]human cloning counts) against the [miR/miR*] ratio of fold repression ([miR/miR*]fold repression) derived from a luciferase sensor assay. The data were subjected to linear regression analysis and correlation using Pearson's tests.
Grouping analysis
Twenty-two tested miRNAs were ranked according to the [miR/miR*]human cloning counts from high to low. “Group High” includes the first 11 miRNAs, and “Group Low” includes the last 11 miRNAs. [miR/miR*]fold repression of miRNAs that grouped together were plotted on the same column. We used the two-sided Mann-Whitney test to perform statistical tests on Group High and Group Low. All statistics and charts were generated using GraphPad Prism.
miREDUCE analysis
Microarray data for miR-17 (Series GSE14831) and miR-34a (Series GSE7754) experiments were downloaded from GEO. Affymetrix or Rosetta probe names were translated to Ensembl transcript IDs, and P-values were computed in R using a χ2 test. We ran miREDUCE (http://www.mdc-berlin.de/en/research/research_teams/systems_biology_of_gene_regulatory_elements/projects/mireduce/index.html) and set the motif length parameter to either 6/6 (only 6-mer motifs), 6/7 (either 6-mer or 7-mer motifs) or 7/7 (only 7-mer motifs), consistent with current models of miRNA target recognition. As described earlier (Sood et al. 2006), miREDUCE fits the expression data to the motif counts of all genes detected on the array, including those that are not significantly down-regulated and those that are up-regulated. This yields increased statistical power over simply searching for enriched motifs in the down-regulated geneset, since it leverages information regarding the depletion of motifs in up-regulated genes, as well as minor changes in gene expression that fail to meet standard cutoffs for down-regulation.
In the case of the mir-34a microarray data, miREDUCE (Sood et al. 2006) failed to retrieve known miRNA seeds when run on the full set of expression data, consistent with previous analysis (Chang et al. 2007) and probably reflecting a great deal of indirect changes in gene expression between the queried cell states. We therefore performed the more restricted analysis by searching for motifs that were enriched among the top down-regulated genes. We tested a range of cutoffs from 100 to 500, with optimal results obtained with the top 500 down-regulated genes. It was not possible to go above this range because of the limited number of significantly down-regulated genes.
CDF analysis
We used gene expression data from experiments in which the expression of the mir-17→92 cluster and mir-19a/b was reconstituted in Myc-driven B-cell lymphoma cell lines bearing a conditional knockout allele of the mir-17→92 cluster (Mu et al. 2009). In each case, the cumulative distribution of the log expression changes for the predicted miRNA and miRNA* targets was compared to that of a background gene set that had no predicted targets. Targets were predicted and scored using miRanda-mirSVR method (Betel et al. 2010) and restricted for perfect seed complementarity. Empirical cumulative distributions were computed using the R ecdf function, and P-values were computed by the Kolmogorov-Smirnov non-parametric test. All predicted target sets were mutually exclusive such that genes with target sites of multiple miRNAs were excluded from the comparison. This helped to ensure that the observed log expression change of a particular predicted target cohort was attributable to specific miRNA species under examination, as opposed to any other miRNA species in the comparison sets.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Peter Linsley and Irena Ivanovska for assistance with microarray data, and Michael Chen for mapping some small RNA libraries. K.C.C. thanks N. Rajewsky and M. Siegal for support. A.C.S. and M.D.P. were funded in part by a David and Lucile Packard Foundation Fellowship; A.V. was funded by the Kimmel Cancer Research Foundation, Geoffrey Beene Cancer Foundation, and NCI R01-10355609; K.C.C. was funded by NIH R00HG004515-02; E.C.L. was funded by the Alfred Bressler Scholars Fund, the Burroughs Wellcome Foundation, the Starr Cancer Consortium (I3-A139), and NIH R01-GM083300.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2537911.
REFERENCES
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC 2008. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci 105: 7964–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 11: R90 doi: 10.1186/gb-2010-11-8-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM 2005. Principles of microRNA–target recognition. PLoS Biol 3: e85 doi: 10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O 2009. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol 10: 141–148 [DOI] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA 2007. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci 104: 18097–18102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26: 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N 2006. Deep conservation of microRNA–target relationships and 3′UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb Symp Quant Biol 71: 149–156 [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86 [DOI] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB 2009. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel J, Sachidanandam R, et al. 2008. An endogenous siRNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ 2009. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell 36: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T 2001a. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T 2001b. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 20: 6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G 2008. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet 9: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B 2010. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465: 818–822 [DOI] [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26: 407–415 [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD 2010. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 16: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML 2008. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res 18: 957–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Davila J, Swerdel MR, Moore JC, Cohen RI, Wu H, Sun YE, Hart RP 2009. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS ONE 4: e7192 doi: 10.1371/journal.pone.0007192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D, Wang Y-L, Langenberger D, Gunsalus KC, Rajewsky N 2005. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comp Biol 1: 0051–0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. 2005. A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. 2007. A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL 2003. Vienna RNA secondary structure server. Nucleic Acids Res 31: 3429–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716 [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845–849 [DOI] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS 2009. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 27: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, Choung S, Kim YJ, Choi YC 2008. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2). J Biol Chem 283: 18158–18166 [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, Macmenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. 2005. Combinatorial microRNA target predictions. Nat Genet 37: 495–500 [DOI] [PubMed] [Google Scholar]
- Lai EC 2002. microRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 30: 363–364 [DOI] [PubMed] [Google Scholar]
- Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, et al. 2006. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol 16: 460–471 [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Shatseva T, Jeyapalan Z, Du WW, Deng Z, Yang BB 2009. A 3′-untranslated region (3′UTR) induces organ adhesion by regulating miR-199a* functions. PLoS ONE 4: e4527 doi: 10.1371/journal.pone.0004527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Lin EA, Kong L, Bai XH, Luan Y, Liu CJ 2009. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem 284: 11326–11335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, et al. 2007. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol 27: 2240–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, et al. 2010. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol 12: 372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W, Rosenbloom K, Hardison RC, Hou M, Taylor J, Raney B, Burhans R, King DC, Baertsch R, Blankenberg D, et al. 2007. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res 17: 1797–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A 2009. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev 23: 2806–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309–321 [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC 2008. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol 15: 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Liu N, Lai EC 2009. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 36: 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L 2009. miR-19 is a key oncogenic component of mir-17-92. Genes Dev 23: 2839–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M 2004. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res 64: 3087–3095 [DOI] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL 2008. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci 28: 14341–14346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen T, Pais H, Sweetman D, Moulton V, Munsterberg A, Dalmay T 2009. High throughput sequencing of microRNAs in chicken somites. FEBS Lett 583: 1422–1426 [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M 2007. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26: 731–743 [DOI] [PubMed] [Google Scholar]
- Ro S, Park C, Young D, Sanders KM, Yan W 2007. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res 35: 5944–5953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte JH, Marschall T, Martin M, Rosenstiel P, Mestdagh P, Schlierf S, Thor T, Vandesompele J, Eggert A, Schreiber S, et al. 2010. Deep sequencing reveals differential expression of microRNAs in favorable versus unfavorable neuroblastoma. Nucleic Acids Res 38: 5919–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee SP, Siragam V, et al. 2009. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol 11: 1031–1038 [DOI] [PubMed] [Google Scholar]
- Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP 2010. Expanding the microRNA targeting code: Functional sites with centered pairing. Mol Cell 38: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N 2006. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci 103: 2746–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM 2003. Identification of Drosophila microRNA targets. PLoS Biol 1: e60 doi: 10.1371/journal.pbio.0000060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, Bartel DP, Cohen SM, Kellis M 2008. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev 22: 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H 2007. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 6: 1586–1593 [DOI] [PubMed] [Google Scholar]
- Tsang WP, Kwok TT 2009. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis 30: 953–959 [DOI] [PubMed] [Google Scholar]
- Wang X, Wang X 2006. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res 34: 1646–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Wu H, Ye C, Ramirez D, Manjunath N 2009. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS ONE 4: e7566 doi: 10.1371/journal.pone.0007566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR 2009. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460: 642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]