Abstract
In Caenorhabditis elegans, the transcripts of many genes are trans-spliced to an SL1 spliced leader, a process that removes the RNA extending from the transcription start site to the trans-splice site, thereby making it difficult to determine the position of the promoter. Here we use RT-PCR to identify promoters of trans-spliced genes. Many genes in C. elegans are organized in operons where genes are closely clustered, typically separated by only ∼100 nucleotides, and transcribed by an upstream promoter. The transcripts of downstream genes are trans-spliced to an SL2 spliced leader. The polycistronic precursor RNA is processed into individual transcripts by 3′ end formation and trans-splicing. Although the SL2 spliced leader does not appear to be used for other gene arrangements, there is a relatively small number of genes whose transcripts are processed by SL2 but are not close to another gene in the same orientation. Although these genes do not appear to be members of classical C. elegans operons, we investigated whether these might represent unusual operons with long spacing or a different, nonoperon mechanism for specifying SL2 trans-splicing. We show transcription of the entire region between the SL2 trans-spliced gene and the next upstream gene, sometimes several kilobases distant, suggesting that these represent exceptional operons. We also report a second type of atypical “alternative” operon, in which 3′ end formation and trans-splicing by SL2 occur within an intron. In this case, the processing sometimes results in a single transcript, and sometimes in two separate mRNAs.
Keywords: C. elegans, operons, transcription start site, trans-splicing, 3′end formation, Ur element
INTRODUCTION
The transcripts of around 70% of the genes in Caenorhabditis elegans are trans-spliced (Blumenthal 2005). Trans-splicing is a process closely related to cis-splicing of introns (Hannon et al. 1991). A 5′ fragment of a newly transcribed pre-mRNA is removed and replaced with a capped spliced leader sequence, donated by a specialized snRNP, the spliced leader (SL) snRNP (Krause and Hirsh 1987; Bektesh and Hirsh 1988; Liou and Blumenthal 1990). Trans-splicing occurs at an unpaired 3′ splice site upstream of the coding sequence of a pre-mRNA (Conrad et al. 1991, 1993a,b). During trans-splicing, the RNA upstream of the trans-splice site, known as an outron, becomes attached to the SL snRNP to form a branched molecule, analogous to the intron lariat formed during cis-splicing (Bektesh and Hirsh 1988). The branched RNA is presumably quickly debranched and degraded (Conrad et al. 1991). Since trans-splicing is a very efficient process, it has proven to be difficult to identify transcriptional start sites upstream of genes whose RNA is trans-spliced, which, in C. elegans, is most genes. However, recent whole-genome ChIP-Chip and ChIP-seq experiments have identified sites of RNA polymerase II poised for transcription, and sites of accumulation of specialized or modified histones specific for promoter regions (Whittle et al. 2008; Baugh et al. 2009). Because chromatin IP experiments are a relatively blunt tool, these experiments provide only a rough idea of where the promoter is. In this manuscript, we show that we can use high density RT-PCR to locate the 5′ ends of outrons, which mark transcription start sites. Using this tool, we can calculate a relatively precise location of promoters in a trans-splicing organism.
Trans-splicing has allowed the evolution of operons: assemblages of two to eight identically oriented genes, clustered tightly along the chromosome (Spieth et al. 1993). Typically, genes within an operon are separated by only about 100 base pairs (bp) (Blumenthal et al. 2002). All genes in an operon are transcribed as a single polycistronic RNA from an upstream promoter. The polycistronic RNA is then processed into individual mRNAs by 3′ end formation of the upstream gene accompanied by trans-splicing of the downstream gene.
Transcripts from nonoperon genes or from genes located in the most upstream position in an operon are generally trans-spliced with a spliced leader donated from the SL1 snRNP (Bruzik et al. 1988; Van Doren and Hirsh 1988). Transcripts from genes located in downstream positions in an operon are trans-spliced to a spliced leader donated by the SL2 snRNP (Huang and Hirsh 1989; Spieth et al. 1993). This process is necessary for polycistronic pre-mRNA processing and for the addition of caps to transcripts from downstream genes. It is also mechanistically coupled to 3′ end formation of the upstream RNA (Kuersten et al. 1997). An upstream polyadenylation signal (AAUAAA) appears to play a role in SL2-specific trans-splicing (Liu et al. 2001). The RNA between the cleavage site of the upstream gene and the trans-splice site of the downstream gene is known as the intercistronic region (ICR). It also contains several features that facilitate specific downstream RNA SL2 trans-splicing. In a typical 110-nt ICR, ∼50 nucleotides (nt) downstream from the 3′ cleavage site is a sequence known as the Ur element (Huang et al. 2001; Liu et al. 2003). Originally characterized as a ∼20-nt U-rich sequence, this element has been recently shown to be composed of two distinct components: a stem–loop of variable sequence followed closely by one or more copies of the consensus sequence UAYYUU (Lasda et al. 2010). Both of these features are necessary for SL2 trans-splicing in vitro. It has been shown that the SL2 snRNP directly or indirectly associates with the Ur element, which activates it and places it in proximity for trans-splicing at the downstream 3′ splice site.
Genes in operons have been identified by the fact that they are very close together, with ICRs of ∼100 nt, and are SL2 trans-spliced. However, there are occasional examples of genes that do not appear to be in canonical operons but whose transcripts are processed nonetheless by SL2 trans-splicing. For example, the gene nduf-5, which encodes a subunit of the mitochondrial NADH dehydrogenase complex, is overwhelmingly (86%) SL2 trans-spliced (Hillier et al. 2009; Allen et al. 2011), but this gene lies 2.6 kilobases (kb) downstream from the nearest upstream gene in the same orientation. Likewise, the gene K01C8.6 encodes a mitochondrial ribosomal protein. Deep sequencing indicates that it is also mainly (89%) SL2 trans-spliced, but it is located 2.3 kb downstream from the nearest identically oriented gene. Thus, the RNAs from these genes are processed as if they are downstream in operons, but the genes do not appear to be in operons. It has been previously proposed that RNA from such genes may be processed through an alternative operon-independent SL2 trans-splicing mechanism (Blumenthal et al. 2002).
We have used RT-PCR to search for transcriptional start sites upstream of these genes. We show that they are, in fact, in unusual operons. For nduf-5 and K01C8.6, even though the distance to the nearest upstream genes is much greater than the 110 bp typically observed, our analysis shows that RNA is transcribed along the entire region between these genes and the genes upstream. This phenomenon is not seen in similarly oriented nonoperon genes. Importantly, in constructs in which ∼600 nt of K01C8.6 or nduf-5 upstream sequence dictate the trans-splicing of a GFP or GFP-fusion transcript, the resulting RNA is overwhelmingly SL1 trans-spliced, indicating that the SL2 trans-splicing seen in these genes' natural context is due to their position downstream in operons.
The gene F58G1.2 encodes a zinc-finger protein of unknown function. It is 6.2 kb downstream from the nearest similarly oriented gene. Although the 5′ end of the RNA transcribed from this gene is SL1 trans-spliced, as would be expected, the RNA is also sometimes SL2 trans-spliced at the 3′ end of the first intron, as if this point marked the beginning of an RNA sequence corresponding to a downstream gene in an operon. This gene arrangement turns out to be what we are calling an “alternative operon.” We show that 3′ end formation and polyadenylation do sometimes occur after the first exon in this RNA. When this happens, downstream RNA is processed by SL2 trans-splicing and this presumably results in production of a separate protein. We identify several additional examples of alternative operons. Thus, using RT-PCR to map transcriptional start sites, as well as modEncode data, including histone variant Htz peaks and RNA polymerase II (pol II)-ChIP data, 3′ end formation sites, and RNA deep sequencing reads, our results expand our understanding of how genes are organized throughout the C. elegans genome.
MATERIALS AND METHODS
Worm strains and maintenance
Worms were maintained and grown as described (Brenner 1974; Sulston and Brenner 1974). Transgenic worms carrying extra chromosomal arrays were produced as described (Mello et al. 1991; Spieth et al. 1993), except DNA was injected at a concentration of 50 ng/μL. Stable transformant lines were obtained and verified by PCR, using the gene-specific forward primers 5′ CTTTACTTTTTGAAATTCGTTTGTCTGAAATTC 3′ (K01C8.6) or 5′ CCGAAAAATCGATATTTTGCTTCTCAACTC 3′ (nduf-5) and the GFP sequence-specific reverse primers 5′ CATGGAACAGGTAGTTTTCCAGTAGTGC 3′ (for RT) and 5′ ATTTGTGCCCATTAACATCACC 3′ (for PCR). The strain BL8102 carries the K01C8.6∷GFP array, and the strains BL8103 and BL8104 carry the nduf-5>∷GFP array.
Plasmid construction
The K01C8.6- and nduf-5>∷GFP fusion constructs were built from the sur-5>∷GFP reporter plasmid, pTG96 (Gu et al. 1998). pTG96 was digested with SphI and SacI to remove the sur-5 promoter fragment. To construct the nduf-5>∷GFP vector, N2 DNA was amplified using the forward primer 5′ GCGCGCGCATGCGGGTAATTCTGAAATTATTACGGGAACAC 3′ and the reverse primer 5′ GCGCGCGAGCTCCTGGAATTTTGGAATTTTATTATGAGTTGAGAAGC 3′ (in which the underlined nucleotides are SphI and SacI restrictions sites included in the primer sequence). The resulting 602-bp product was digested with SphI and SacI and ligated into the plasmid backbone upstream of the GFP coding sequence. To construct K01C8.6>∷GFP, N2 DNA was amplified with the forward primer 5′ GCGCGCGCATGCGACCATTCGGCGTAACTGCGC 3′ and the reverse primer 5′ GCGCGCGAGCTCTTGAAATGTCGTGGAGTTGGACGTG 3′. The 675-bp product was digested with SphI and SacI and ligated into the plasmid backbone to produce an in-frame fusion. All constructs were verified by sequencing.
RNA preparation and RT-PCR analysis
N2 and mutant RNA was isolated and RT-PCR performed as previously described (Lasda et al. 2010), using gene-specific primers (Supplemental Table S1). PCR products were analyzed by agarose gel electrophoresis.
RESULTS
Localization of transcription start sites of trans-spliced genes
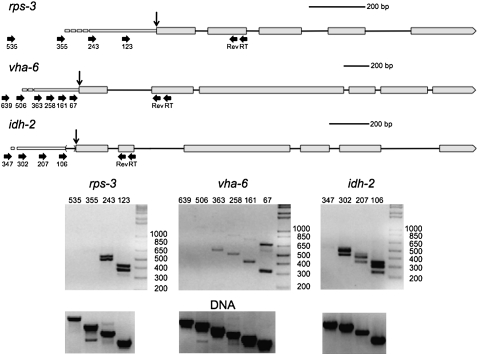
Because trans-splicing is an efficient process, the unspliced precursor is difficult to capture. Therefore, the transcriptional start site at the 5′ end of the outron of trans-spliced genes is usually unknown. To determine whether RT-PCR could amplify the small amount of unprocessed RNA from trans-spliced transcripts, we examined RNA from three highly expressed genes: rps-3 (a small ribosomal subunit protein), vha-6 (a subunit of a vacuolar ATPase), and idh-2 (isocitrate dehydrogenase) (Fig. 1). In the idh-2 transcript a 5′ splice site upstream of the 3′ trans-splice site results in occasional excision of an intron in the 5′ UTR and production of a non-trans-spliced transcript whose transcriptional start site can be identified in ESTs. Thus, idh-2 serves as a positive control, since the location of the promoter is known.
FIGURE 1.
Localization of transcriptional start sites of trans-spliced genes by RT-PCR. Schematic representation of the rps-3, vha-6, and idh-2 pre-mRNAs, including their proposed outrons. In all figures, exons are indicated by boxes, and introns by lines. Trans-splice sites are indicated by vertical arrows. The forward and reverse primer sites are indicated by horizontal arrows. Scale bars are shown for each diagram. Primers for reverse transcription (RT) and reverse primers for PCR (Rev) are indicated under the second exons. Forward primers are named according to their location (in bp) upstream of the trans-splice sites. Since the exact transcriptional start site is known only to fall somewhere between the most 5′ primer that gives a product and the next upstream one that does not, the 5′ ends of the outrons are depicted as dashes. Gels (below) show RT-PCR products using the primers shown above. Some of the RNA has been cis-spliced as well as trans-spliced, so there are two bands in each lane: one including an intron and one without. The bands resulting from intron-containing RNA are visible but faint with primers 363, 258, and 161 on the vha-6 gel. RNA from the idh-2 gene produces extra bands in each lane, since the trans-splice site sometimes serves as the 3′ splice site for the removal of a small intron upstream of the first coding exon. The lowest band is at the position expected for removal of both introns; the largest band is at the position expected for removal of neither intron; and the middle band is at the position expected for removal of either intron. The forward primers used in the reactions are shown along the tops of the gels. Gels of the products of PCRs from C. elegans (N2) genomic DNA (lower gels) verify the specificity of the primer pairs used in the analysis.
We used sets of gene-specific primers: two nested reverse primers in the second exon of each of the three genes and several forward primers at increasing distances upstream of the trans-splice site (Fig. 1, thick black arrows in the gene diagrams) Following reverse transcription with the most 3′ primer, we performed PCRs on the cDNA using each of these upstream forward primers and the nested 3′ primer. Importantly, for each gene, primers closer to the trans-splice site always gave products of the expected size, but at some point as more upstream primers were used, no products were found (even though these same primers always gave products with the genomic DNA controls shown in the lower panels). This indicates that this procedure is using the unprocessed precursor for trans-splicing as the template. RT- reactions were run in parallel with all RT-PCRs to ensure that all products were RNA dependent (data not shown.) We calculate the approximate outron length as falling within the interval between the most 5′ primer producing a PCR product and the next upstream primer that did not give a product (because it lies upstream of the transcriptional start site.) This procedure should be able to identify the most distal transcription start site, but if there are multiple start sites, the proximal ones would not be distinguishable. By this method, we were able to calculate that the outron of rps-3 extends into the interval between 243 and 355 nt upstream of the trans-splice site (Fig. 1, cf. gene diagrams and gels). The outron of vha-6 extends into the interval from 363 to 506 nt upstream, and the outron of idh-2 extends into the interval between 302 and 347 nt upstream of the trans-splice site. This result corresponds nicely with idh-2's known transcriptional start site of 304 nt upstream of the trans-splice site (from published ESTs, WormBase, WS215). The very faint band with the 347 oligonucleotide may indicate that there is a small amount of transcription coming from somewhere further upstream as well. The pairs of bands in some lanes result from RNA that has undergone cis-splicing of the first introns (lower band) and RNA that has not yet been spliced (upper band).
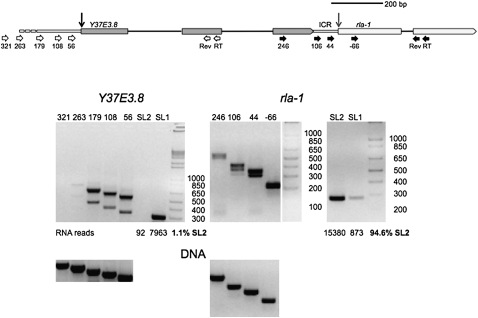
Analysis of a known operon
To determine whether RNA from genes in operons could be differentiated from that produced from genes with outrons by this technique, we examined RNA from the well-characterized rla-1 operon (Fig. 2). In this case, unprocessed RNA transcribed from the promoter upstream of this operon should extend through both Y37E3.8 and rla-1. RT-PCR using nested reverse primers in the second exon of rla-1 and forward primers located upstream in the ICR between the genes and in the final exon of Y37E3.8 shows that RNA does indeed extend from the upstream gene into the downstream gene. Moreover, the mature mRNA from rla-1 is almost exclusively SL2 trans-spliced, as would be expected from a downstream gene in an operon. In contrast, RT-PCR analysis of the RNA upstream of Y37E3.8, the first gene in the operon, indicates the presence of a typical outron, extending from somewhere in the region of 263–321 nt upstream of the trans-splice site. The mature RNA from this gene is SL1 trans-spliced, as expected.
FIGURE 2.
RT-PCR analysis of transcripts from an operon. Diagram of the Y37E3.8 operon. Y37E3.8 is the upstream gene and rla-1 is the downstream gene. The genes are separated by a 110-bp intercistronic region (ICR), depicted as a narrow bar between them. The Y37E3.8 RNA trans-splice site is indicated by a black vertical arrow and the rla-1 RNA trans-splice site is indicated by a gray vertical arrow. Primers are measured from these trans-splice sites and are indicated by white horizontal arrows for Y37E3.8 and black horizontal arrows for rla-1. Gels of RT-PCR products show that, as expected, Y37E3.8 RNA is trans-spliced to SL1 whereas rla-1 RNA is trans-spliced primarily to SL2. The analysis of the transcriptional start site is as shown in Figure 1. The start site for Y37E3.8 transcription can be estimated, while rla-1 transcription proceeds from the upstream gene, as would be expected for a downstream gene in an operon. Some RNA has been cis-spliced, so two bands are present. RNA reads are taken from Illumina sequencing of polyadenylated RNA, by Hillier et al. (2009), and the percentage of SL2 trans-spliced RNA is calculated from this data.
Analysis of SL2 trans-spliced genes not downstream in typical operons
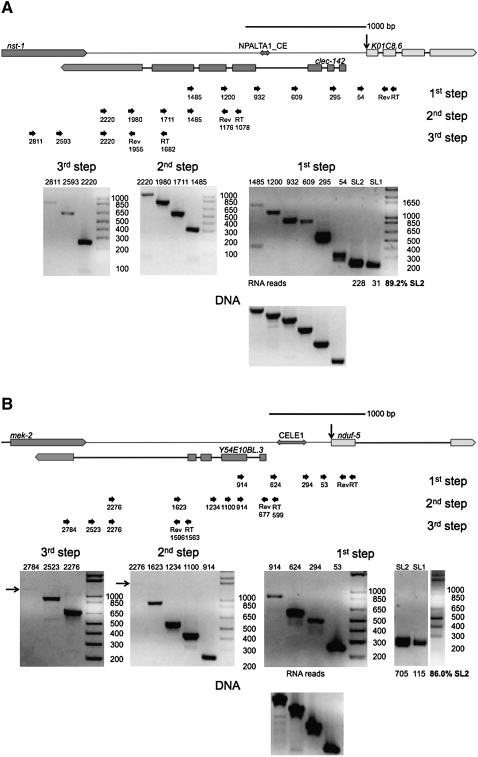
SL2 trans-splicing at outron trans-splice sites is extremely rare (<1%) (Allen et al. 2011). Since K01C8.6, nduf-5, and F58G1.2 RNA are all frequently SL2 trans-spliced, but none of them appear to be downstream in operons, we hypothesized that either they are downstream in unusual operons; or they are not in operons but are trans-spliced by SL2 through an operon-independent mechanism. To distinguish between these two possibilities, we analyzed their unprocessed RNAs to determine whether transcriptional start sites could be identified upstream of the genes. First, we examined the RNA from K01C8.6 by RT-PCR (Fig. 3A). Analysis of this gene shows that its mRNA is mostly SL2 trans-spliced, as expected based on the deep sequencing (Fig. 3A, cf. lanes SL1 and SL2; Allen et al. 2011). Using reverse primers in the second exon, we detected continuous RNA from 1485 nt upstream of the trans-splice site (first step gel). The products generated by forward primers 932, 1200, and 1485 are shorter than expected, based on the known sequence and the sizes observed with genomic DNA as a template (lower panel). These PCR products encompass the site of a NPALTA1_CE transposon. It has previously been shown that some C. elegans transposons can be imprecisely excised from the RNA by an unknown process that is clearly not spliceosomal splicing since the ends of the excised RNA do not match canonical splice sites (Rushforth et al. 1993; Rushforth and Anderson 1996). We have sequenced the PCR products from this region and determined that a similar event appears to be occurring to this RNA (data not shown). However, these bands could also be due to reverse transcriptase or PCR skipping over the inverted sequences.
FIGURE 3.
Detection of RNA along the entire region upstream of SL2 trans-spliced genes. (A) The genomic arrangement of K01C8.6 is depicted above. This gene is ∼2.1 kb downstream from nst-1. The gene clec-142 lies on the opposite strand between these two genes. The transposon NPALTA1_CE is indicated as a double arrow upstream of K01C8.6. Gels of overlapping RT-PCRs indicate that RNA is transcribed along the entire region between K01C8.6 and the nearest upstream gene, nst-1. The analysis was done in three steps to examine the entire region between the genes. The first step gel and the RNA deep sequencing data show that K01C8.6 RNA is preferentially SL2 trans-spliced, as would be expected if this gene were downstream in an operon. Products extending from the 1485, 1200, and 932 primers are shorter than expected. This is presumably due to splicing of the transposon NPALTA1_CE from the RNA, as has been previously described with other transposons (Rushforth et al. 1993; Rushforth and Anderson 1996) or due to a reverse transcription or PCR artifact at the inverted repeats. Because all reverse transcription was performed with strand-specific primers and the reverse transcriptase heat inactivated prior to PCR, clec-142 mRNA cannot be detected in these assays. (B) The genomic arrangement of nduf-5 is depicted above. This gene is ∼2.6 kb downstream from mek-2. The gene Y54E10BL.3 lies on the opposite strand between these two genes. The transposon CELE1 is indicated as a double arrow upstream of nduf-5. Gels of overlapping RT-PCRs indicate that RNA is transcribed along the entire region between nduf-5 and the nearest upstream gene, mek-2. The analysis was done in three steps to examine the entire region between the genes. Arrows mark the site of faint products on the gels. The first step gel and the RNA deep sequencing data show that nduf-5 RNA is preferentially SL2 trans-spliced, as would be expected if this gene were downstream in an operon. Products extending from the 624 and 914 primers are shorter than expected, presumably due to splicing of the CELE1 transposon from the RNA or an artifact of reverse transcription or PCR at the inverted repeats. Because all reverse transcription was performed with strand-specific primers and reverse transcriptase heat-inactivated prior to PCR, Y54E10BL.3 mRNA cannot be detected in these assays.
Because these products were nearing the size limit of unprocessed products detectable in RT-PCRs primed from the introns of highly expressed RNA (fat-1, Y69A2AR.18; data not shown), we examined RNA upstream of this gene in steps. We constructed a second set of nested reverse primers at 1078 and 1176 nt upstream of the trans-splice site. From them, we detected RNA up to −2220 (Fig. 3A, second step gel.) Likewise, from a third set of reverse primers at −1682 and −1955, we detected RNA up to −2811 (Fig. 3A, third step gel.) The longest two products shown in this gel (2593 and 2811) lie within the nst-1 gene, indicating that unprocessed RNA extends from this gene downstream to K01C8.6, as would be expected in an operon.
We performed a similar analysis on transcripts of nduf-5 (Fig. 3B). Since the single intron in this gene is slightly over 1 kb in length, we designed an initial set of reverse primers in the first exon, then proceeded upstream in steps, as described above. The mRNA from this gene is also preferentially SL2 trans-spliced (Fig. 3B, cf. lanes SL1 and SL2). We detected RNA 914 nt upstream of the trans-splice site (Fig. 3B, first step gel). The products generated by the 624 and 914 forward primers are shorter than expected, presumably due to the presence of a CELE1 transposon in this region, which is cut from the RNA, as determined by sequencing of the shortened PCR product RNA or is missing from the PCR product due to a reverse transcriptase or PCR artifact occurring at the inverted repeats. We constructed a second set of reverse primers 599 and 677 nt upstream, and, from these, were able to detect RNA up to −2276 (Fig. 3B, second step gel). Finally, from another set of reverse primers at −1563 and −1596, we detected RNA up to −2784, within the mek-2 gene (Fig. 3B, third step gel), indicating that the entire region between mek-2 and nduf-5 is transcribed, consistent with the hypothesis that they form an operon. However, we show below that this spacing, with a gene in between on the opposite strand, is not a sufficient condition for SL2 trans-splicing or for diagnosis of an operon.
The context of the trans-splice site determines its specificity
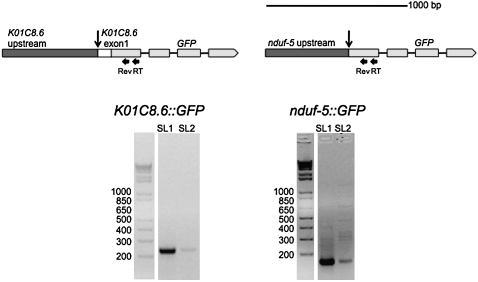
As an independent test of the idea that the regions upstream of the trans-splice sites of K01C8.6 and nduf-5 could be responsible for their receiving SL2 instead of SL1, we created constructs to examine their transcriptional processing outside of their normal genomic context (Fig. 4). We assumed that if these genes were like SL1 trans-spliced genes except with a signal for SL2, they would have a nearby promoter and an outron. We fused the 561 nt upstream of the trans-splice site and the first exon of K01C8.6 in frame to GFP, and we fused the 578 nt upstream of the trans-splice site of nduf-5 to GFP. We isolated RNA from transgenic strains carrying these constructs in extrachromosomal tandem arrays, and analyzed this RNA by RT-PCR. Although we expected this amount of upstream sequence to be enough to contain any elements necessary to direct SL2 trans-splicing, analysis of both constructs revealed little expression, based on our inability to see any GFP fluorescence. However, RT-PCR analysis indicated that there is in fact a low level of transcription. However, this transcription was coming from an unknown transcription start site outside the insert, based on RT-PCR analysis like that shown in Figure 3 (data not shown). Interestingly, this RNA is almost exclusively SL1 trans-spliced instead of the SL2 trans-splicing that occurs when these genes are in their natural contexts (Fig. 4, K01C8.6>∷GFP and nduf-5>∷GFP gels.) It is not clear why there is no detectable GFP, but we assume it is due to production of such a low level of mRNA from these transgenes. This experiment shows that the sequences upstream of these genes are not sufficient to specify the SL2 trans-splicing associated with the RNA from these two genes in their normal context. This observation also supports the idea that these are downstream genes in unusual, long spaced, operons.
FIGURE 4.
Expression of K01C8.6 and nduf-5 GFP fusion constructs in transgenic worms. Constructs in which the region upstream of nduf-5 or K01C8.6 was fused to the GFP gene are depicted above. For K01C8.6, 561 nt upstream of the trans-splice site along with the first exon of the gene were fused in frame to GFP. For nduf-5, the 578 nt upstream of the trans-splice site of the gene were fused to GFP. RNA was isolated from transgenic strains expressing these constructs and analyzed by RT-PCR. The mRNA is almost exclusively SL1 trans-spliced, indicating that the endogenous chromosomal context of nduf-5 and K01C8.6 is important in directing SL2 trans-splicing.
The genomic context is not sufficient to specify SL2 trans-splicing
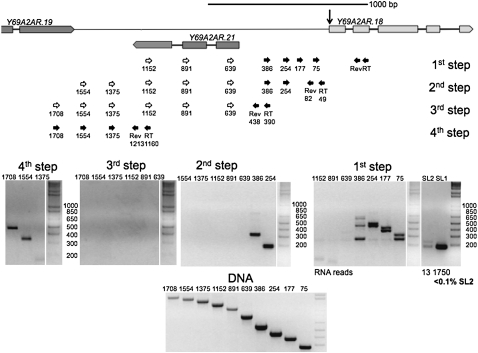
The genomic contexts surrounding K01C8.6 and nduf-5 are very similar. Each gene is ∼2.5 kb downstream from the nearest same-strand gene. Furthermore, genes on the opposite DNA strand can be found within this region in both instances. To determine if there is something special about this orientation that either caused SL2 trans-splicing of the RNA produced from downstream genes and/or produced RNA throughout the region, we examined a third gene, Y69A2AR.18, which shares a similar genomic orientation but is normally trans-spliced to SL1 (Fig. 5). We designed a set of reverse primers in its second exon and proceeded upstream, as above. As expected, the mRNA produced is preferentially SL1 trans-spliced (Fig. 5, cf. lanes SL1 and SL2). Furthermore, RNA can be detected only up to 386 nt upstream of the trans-splice site, as is typical of an outron. Construction of a second set of reverse primers at 49 and 82 nt upstream of the trans-splice site produced similar results (Fig. 5, second step gel.) We also designed a third set of reverse primers at 390 and 438 nt upstream of the trans-splice site. PCRs with upstream forward primers failed to generate a product, as would be expected if these primers were upstream of the Y69A2AR.18 transcriptional start site, even though the primers produced product when supplied with a DNA template (Fig. 5, cf. third step gel and DNA comparison gel.) A fourth set of reverse primers at 1160 and 1213 nt upstream of the trans-splice site did produce products with upstream forward primers. These products most likely represent unprocessed RNA transcribed from the upstream gene, since it has been shown that RNA polymerase II transcription can continue for over a kilobase downstream from the polyadenylation signal before termination in C. elegans (Haenni et al. 2009). Thus, genomic arrangement cannot account for processing of K01C8.6 and nduf-5 RNA, since Y69A2AR.18 is in a very similar context but is neither co-transcribed with an upstream gene nor trans-spliced to SL2.
FIGURE 5.
Analysis of the region upstream of Y69A2AR.18. The genomic arrangement of Y69A2AR.18 is similar to that of K01C8.6 and nduf-5. It is located ∼1.6 kb downstream from Y69A2AR.19, and the gene Y69A2AR.21 lies between these genes on the opposite DNA strand. Gels of overlapping RT-PCRs indicate that RNA is not transcribed in the region between Y69A2AR.18 and the nearest upstream gene, Y69A2AR.19. Instead, RNA from this gene has a defined outron. The first step gel and RNA deep sequencing data show that Y69A2AR.18 mRNA is preferentially SL1 trans-spliced, as would be expected in a nonoperon gene. The RNA identified in the fourth step RT-PCR is consistent with a termination product of the upstream gene, as has been previously observed (Haenni et al. 2009).
A gene that is SL2 trans-spliced at the 3′ end of the first intron: An “alternative” operon
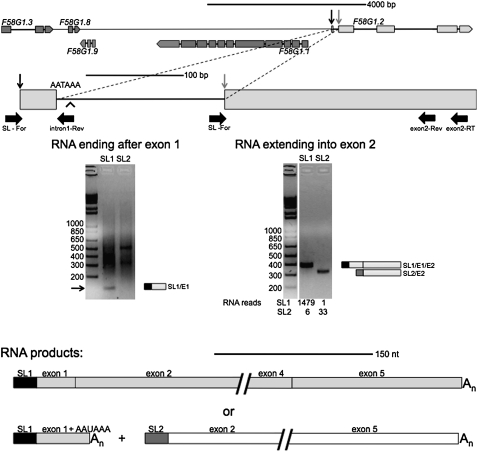
The gene F58G1.2 is a member of another uncommon class of genes. It is apparently not downstream in an operon, but it sometimes undergoes SL2 trans-splicing at the cis-splice site at the 3′ end of an intron. We have shown recently that although there is often a very low level of trans-splicing at the 3′ ends of long introns, this trans-splicing is SL1 trans-splicing (Allen et al. 2011). Therefore, in an unusual situation like this one, where a relatively high level of SL2 trans-splicing is occurring, a different phenomenon is likely. F58G1.2 has an apparent 3′ end formation signal, AATAAA, near the 5′ end of the same intron. We hypothesized that 3′ end formation accompanied by SL2 trans-splicing 120 nt downstream sometimes occur in this intron, thereby creating an operon of two separate genes out of what is otherwise a single gene. This would represent a case of alternative splicing/3′ end formation accompanied by trans-splicing, which has not previously been described in C. elegans. We therefore examined the RNA products of this gene by RT-PCR. We first reverse-transcribed the RNA using an oligo dT primer, and then used a reverse primer just downstream from the first exon and encompassing the polyadenylation site in conjunction with either an SL1- or SL2-specific primer (Fig. 6, exon 1 gel). A 130-bp SL1 trans-spliced product indicates that, at times, cleavage and polyadenylation occur near the 3′ end of the first exon. A recent analysis of the C. elegans 3′UTRome (Mangone et al. 2010) has also demonstrated use of a 3′ end formation site in this location (Fig. 6, arrowhead). This RNA could conceivably encode a 22-amino acid protein, but we believe it is unlikely to do so because this short open reading frame is not conserved in closely related species of Caenorhabditis (unpublished observation).
FIGURE 6.
Analysis of RNA from F58G1.2, an alternative operon. The genomic orientation of F58G1.2. This gene lies 6.3 kb downstream from the nearest upstream gene oriented in the same direction (upper diagram). The SL1 trans-splice site is indicated by a black vertical arrow and the SL2 trans-splice site is indicated by a gray vertical arrow in both the upper and lower diagrams. RNA transcribed from this gene is SL1 trans-spliced upstream of the sequence annotated as the first exon, as indicated by the RT-PCRs shown in the gel on the left and the SL1 lane of the gel on the right. It is sometimes SL2 trans-spliced at the second exon, as shown by the SL2 lane of the gel on the right, as well as the 33 SL2 reads reported in Hillier et al. (2009). The alternative polyadenylation site (AATAAA) is near the 5′ end of the first intron, and the 3′ end formation site identified in the C. elegans 3′ UTRome (Mangone et al. 2010) is indicated by an arrowhead. RNA was reverse transcribed with an oligo dT primer, and the cDNA amplified with an F58G1.2 intron 1-specific reverse primer and either an SL1- or SL2-specific forward primer (left gel). The arrow indicates a band of ∼130 bp in the SL1 lane only, the size expected for trans-splicing at the 5′ end of the first exon. The smear above the band presumably comes from amplification of multiple RNAs by oligo dT and SL primers. The gel on the right shows RNA reverse transcribed with an F58G1.2-specific primer (at the 3′ end of exon 2), and the cDNA amplified with an F58G1.2 exon 2-specific primer and either an SL1- or SL2-specific forward primer. The SL1 trans-spliced product is ∼362 bp, the size expected for a product where trans-splicing occurred upstream of exon 1 and the intron was cis-spliced. The SL2 trans-spliced product is ∼297 bp, the size expected for a product where trans-splicing occurred at exon 2. The possible mRNA products are shown below: A single SL1 trans-spliced product transcribed from the entire gene (gray), or a short SL1 trans-spliced product containing sequence from mainly the first exon of the gene (gray) and a long SL2 trans-spliced product transcribed from the downstream exons (white).
RT-PCR with a gene-specific reverse primer set nested in the second exon along with an SL1-specific forward primer revealed that SL1 trans-spliced product can also extend through this exon (Fig. 6, exon 2 gel). In this case, the 362-bp product indicates that intron 1 is spliced from the RNA and the entire gene is transcribed and processed as a single gene. In contrast, PCR with an SL2-specific forward primer produces a 297-bp product, whose size indicates that trans-splicing has occurred at the beginning of what is annotated as the second exon. RNA processing in this manner is consistent with this gene being downstream in an operon when 3′ end formation occurs in the first intron of the gene. The SL2 trans-spliced RNA also contains an open reading frame, but we have not examined its ultimate fate. This analysis indicates that this gene can be processed as either a single gene by SL1 trans-splicing upstream of the first exon and canonical intron splicing, or it can be processed as an operon, in which the first exon and a bit of the intron following is processed as the upstream gene and the second exon onward is processed as a downstream gene. We are calling this alternative RNA processing situation an “alternative operon.”
DISCUSSION
Mapping promoters of trans-spliced genes
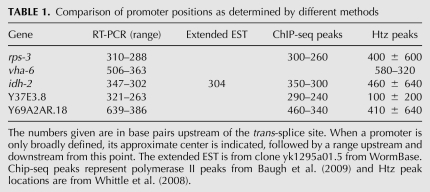
About 60%–70% of C. elegans genes are trans-spliced (Allen et al. 2011). It has been difficult to determine where transcription of these genes begins, because the promoter is upstream of the 5′ end of the outron, rather than the 5′ end of the first exon. Trans-splicing is a relatively efficient process (in most cases), so an unprocessed precursor has been found only rarely. Here we have shown that such precursors can be detected by RT-PCR, and that the 5′ end of the precursor can be at least roughly localized. Furthermore, this is evidence that the 5′ RNA ends really do identify promoter locations. A recently published paper examining growth arrest in starved C. elegans L1 larvae employed RNA pol II ChIP, followed by deep sequencing, to map the location of RNA polymerase II across the genome (Baugh et al. 2009). The investigators observed significant starvation-induced accumulations of polymerase just upstream of many genes—typically those thought to have some function in growth. They concluded that, in such cases, these peaks marked polymerase sequestered at the promoters of the genes. Such peaks exist upstream of rps-3, idh-2, Y37E3.8, and Y69A2AR.18. The centers of all of these peaks correspond well with the promoter positions we mapped by RT-PCR (Table 1). Furthermore, ChIP arrays, using antibodies recognizing the histone variant Htz, known to mark active promoters, have provided evidence for the existence of promoters upstream of many genes (Whittle et al. 2008; ModEncode data in WormBase). Aligning these peaks to the genes studied in this report identifies peaks upstream of rps-3, vha-6, idh-2, Y37E3.8, and Y69A2AR.18 that also correspond at least roughly with the promoter positions mapped by RT-PCR (Table 1). Finally, several ESTs for idh-2 extending upstream of the trans-splice site are also consistent with the 5′ end of the RT-PCR products. Thus, we have confidence that this technique can be used to accurately map the 5′ ends of outrons to reasonably precise positions depending on how many separate upstream PCR primers are employed.
TABLE 1.
Comparison of promoter positions as determined by different methods
The varieties of operon in C. elegans
Since the discovery of operons in C. elegans, they have been characterized primarily by four features: (1) the existence of a single upstream promoter; (2) the close proximity of genes; (3) SL2 trans-splicing of the downstream genes; and (4) the presence of a Ur element within the ICR that is required for SL2 trans-splicing (Spieth et al. 1993; Huang et al. 2001; Lasda et al. 2010). Indeed, these features characterize almost all operons. While some genes are in hybrid operons containing internal promoters, most downstream operon genes lack an adjacent promoter (Huang et al. 2007; Allen et al. 2011). However, as the C. elegans genome has been thoroughly annotated, a few intriguing exceptions to this pattern have emerged.
After the initial characterization of operons in C. elegans, a second type of operon, known as the SL1-type operon, was discovered (Williams et al. 1999). In these operons, genes are not separated by an ICR; instead the trans-splice site of the downstream gene immediately follows the polyadenylation site of the upstream gene, such that the poly(A) tail is added to the last base in the trans-splice site, as if the free 3′ end created by trans-splicing is the substrate for polyadenylation. Furthermore, RNA transcribed from the downstream gene is trans-spliced to SL1. Since this RNA contains no ICR, no Ur element exists to specify SL2 trans-splicing to the downstream RNA. These operons are characterized by an exceptionally long polypyrimidine tract that was shown to be required for SL1 trans-splicing at the downstream RNA.
Here we show that other operon arrangements also exist in the C. elegans genome, although they appear to be quite rare. Sequences containing a spliced leader were aligned and identified (Allen et al. 2011), and the total number of SL1 and SL2 trans-spliced transcripts for each gene was tabulated (SL1 and SL2 data in Figs. 1–3 and 5). In this analysis, virtually all SL2 trans-splicing was found to occur in conventional operons, with a gene in the same orientation just upstream. However, analysis of these data also revealed some exceptions to this rule. Genes whose RNA received an SL2 spliced leader, even though there were no genes in the same orientation closely spaced upstream, were identified and a few of these were chosen for further study. Although they are very rare compared to canonical operons (as SL1-type operons are) they suggest that there are numerous ways to process polycistronic pre-mRNAs and that C. elegans utilizes many of these ways. Specifically, we have identified two new operon arrangements: operons with atypically long ICRs, and alternative operons.
Operons with long spacing
We identified several examples of genes with SL2 trans-splicing but with no nearby upstream gene, and we chose two of these genes for closer examination. We hypothesized that these could represent operons with long spacing or result from a nonoperon mechanism of specifying SL2. In the latter situation, we would have expected transcripts from these genes to begin with outrons, and these outrons might have contained some sort of signal for SL2 trans-splicing. However, our investigations showed that the genes apparently did not have an outron; instead, transcription occurred throughout the region from an upstream gene to the trans-splice site of the downstream gene, a distance of 2–2.5 kb. Furthermore, when 500–600 bp of the upstream regions of these two genes were fused to GFP and expressed from transgenic arrays, they were found to be expressed at a very low level from an unknown site outside of the insert and to be trans-spliced to SL1, not SL2 as they are in their natural context. This suggests they lack an outron signal capable of specifying SL2. Thus, all of the experimental data are consistent with the idea that these are actually newly discovered operons, operons with extraordinarily long ICRs.
In addition to this data, there are other lines of evidence that indicate that these represent true operons. The published pol II ChIP-seq data shows no evidence of polymerase peaks in the ∼2.5 kb regions between K01C8.6 and its upstream gene, nst-1, or between nduf-5 and its upstream gene, mek-2 (Baugh et al. 2009). However, a polymerase peak is clearly present upstream of mek-2, where the operon promoter ought to be. Furthermore, the Lieb lab data showing Htz occupancy provide little support for the existence of promoters between the genes (Whittle et al. 2008). Although a peak exists upstream of K01C8.6, this peak may mark the promoter used to drive clec-142, which is located upstream on the opposite strand. There is no Htz peak upstream of nduf-5, but a peak can be seen upstream of mek-2, which should be the operon promoter. Finally, the ModEncode data, based on immunoprecipitation experiments with antibodies to histones that mark promoter locations also indicate a lack of promoters between these genes.
In operons the expression of downstream genes is controlled by a single upstream promoter. Transcription from this promoter results in SL2 trans-splicing of the downstream polycistronic pre-mRNA. Although the limits of RT-PCR prevented a definitive conclusion that a single RNA spans the region between K01C8.6 and nduf-5 and their upstream genes, the fact that RNA is transcribed throughout this entire region, while being absent in a similarly oriented nonoperon gene, suggests continuous transcription. Undoubtedly, as more information about the C. elegans genome is obtained, additional unusual operons will be discovered. We considered the possibility that the long spacing might be due to a relatively recent change in the C. elegans genome, but quite similar spacing exists between the orthologous genes in Caenorhabditis briggsae, a nematode about 100 million years distant from C. elegans (data not shown). In both of the cases we analyzed, the long ICR contains transposons. In 23/25 previously annotated operons in which the downstream gene is highly (>75%) SL2 trans-spliced and is >1.5 kb downstream from the upstream gene, the ICRs also contain transposons and/or other repetitive DNA (MA Allen, pers. comm.). We hypothesize that these repetitive elements somehow make these regions permissive for long ICRs. Although operons with long spacing are rare, knowledge of their existence broadens the range of gene arrangements that may potentially be operons. At this time, it seems safe to conclude that if a C. elegans gene is trans-spliced by SL2 to any significant degree, it should be annotated as part of a possible operon with the nearest gene upstream in the same orientation.
Requirements for SL2 trans-splicing
Although the mechanism controlling SL2 specificity is not completely understood, it is clear that the operons with long spacing have many of the features found necessary for transcription and processing in operons. Importantly, the intercistronic regions appear to contain Ur elements located within the ∼100 bp immediately preceding the trans-splice site. The Ur element consists of a short stem/loop of variable sequence, followed immediately by the consensus sequence UAYYUU. This element interacts with the SL2 snRNP (the source of the SL2 spliced leader), possibly by base pairing with its 5′ splice site, to direct downstream SL2-specific trans-splicing (Lasda et al. 2010). The possible long ICR upstream of K01C8.6 has the sequence GUGAAUAAUCCCAAAAAAUUAAUUUAUCUUUACUUU 44 nt upstream of the trans-splice site, in which the underlined nucleotides have the potential to form a stem with a single adenosine bulge, and the bold-faced nucleotides match the UAYYUU consensus. Furthermore, 46 nt upstream of this apparently perfect match to the Ur element consensus is a perfect match to the AAUAAA consensus polyadenylation site. Additionally, the ICR of nduf-5 contains the sequence UUUUCUGAUUAUCUCUGAAAAAAAAACCGAAAAAUCGAUAUUUU, which is also a consensus Ur element 58 nt upstream of the trans-splice site. An AAAAAA, which has a one-base mismatch to the perfect polyadenylation site, precedes this perfect Ur element motif by 41 nt. This AAAAAA sequence is sometimes used to specify 3′ end formation in C. elegans (Blumenthal and Steward 1997). The existence and position of these sequence elements may allow the RNA processing machinery bound to the polymerase to process this downstream gene as if it were in a typical short ICR.
Although it is unknown how transcription termination is prevented along this ∼2.5-kb ICR, it is possible that the complex secondary structure of the RNA, in part mediated by the extended stem/loops formed by numerous inverted repeats at the end of the transposons and other repetitive elements found throughout this region, may inhibit termination efficiently enough to allow trans-splicing and subsequent stabilization of the RNA from the downstream gene. It is important to emphasize that although the polyadenylation signals appear to be present ∼100 bp upstream of the trans-splice sites in both of these apparent operons, we found no evidence for 3′ end formation actually occurring at these sites. This suggests that if the signal for SL2 specificity involves 3′ end formation machinery, it is the presence of the machinery at that location rather than the 3′ end formation itself that is responsible.
Alternative operons
Trans-splicing sometimes occurs at a low level at intron 3′ splice sites (Allen et al. 2011). This splicing is almost always SL1 trans-splicing, and it usually occurs at long first introns. It is presumed that this splicing results from the splicing machinery misinterpreting a long intron as an outron. However, we were intrigued by a few examples of SL2 trans-splicing at intron splice sites, and we have analyzed here one of these genes, hypothesizing that these also represent noncanonical, or hidden, operons. Here we showed that one such case appears to be an alternative operon. The first intron is sometimes spliced out, but at other times, 3′ end formation occurs just downstream from the intron 5′ splice site. This appears to be accompanied by SL2 trans-splicing at the intron 3′ splice site only 114 bp further downstream.
A polymerase peak and an Htz peak are both present upstream of the first exon of F58G1.2, but not within the first intron, consistent with the conclusion that transcription starts only upstream of the first exon. Furthermore, the 3′UTR transcriptome data (Mangone et al. 2010) show 3′ end formation occurring 14 nt downstream from the AAUAAA near the 5′ splice site of the intron. Finally, a Ur element can also be found within the first intron. The proposed ICR contains the sequence GGUUCCAAGAAACCUAGUUAUUAUCACUAUUUGAUAUUUAUUUC 41 nt upstream of the 3′ splice. Although there is no perfect UAYYUU consensus, several sites (bold face) with only a single mismatch (in lowercase) are present immediately downstream from the stem/loop. The AAUAAA polyadenylation signal precedes this motif by 43 nt. Thus, all of the characteristics of an operon are present to cause this intron to act alternatively as an operon ICR. Interestingly, we have identified other alternative operons, each of which has been previously annotated as a single gene, but each of which also produces an additional set of transcripts with the upstream RNA polyadenylated within an intron and the downstream RNA trans-spliced at the intron 3′ splice site. None of these three, hdac-6, sma-9, and smg-6, involve the first intron; they are all cases of an intron somewhere within the gene being alternatively spliced and polyadenylated. In the case of sma-9 it has recently been shown that this alternative processing is functionally significant (Yin et al. 2010). This rare sort of operon has been independently discovered by the Bartel lab as well (Jan et al. 2010).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We are grateful to members of the Blumenthal lab for helpful suggestions and for a critical reading of the manuscript. This research was supported by research grant GM42432 from the National Institute of General Medical Sciences.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2447111.
REFERENCES
- Allen MA, Hillier LW, Waterston RH, Blumenthal T 2011. A global analysis of C. elegans trans-splicing. Genome Res (in press). doi: 10.1101/gr.113811.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Demodena J, Sternberg PW 2009. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 324: 92–94 [DOI] [PubMed] [Google Scholar]
- Bektesh SL, Hirsh DI 1988. C. elegans mRNAs acquire a spliced leader through a trans-splicing mechanism. Nucleic Acids Res 16: 5692 doi: 10.1093/nar/16.12.5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T 2005. Trans-splicing and operons. In WormBook (ed. The C. elegans Research Community), Wormbook; doi: 10.1895/wormbook.1.5.1. http://www.wormbook.org [Google Scholar]
- Blumenthal T, Steward K 1997. RNA processing and gene structure. In C. elegansII (ed. Riddle DL), pp. 117–145 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, Thierry-Mieg J, Thierry-Mieg D, Chiu WL, Duke K, Kiraly M, et al. 2002. A global analysis of Caenorhabditis elegans operons. Nature 417: 851–854 [DOI] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzik JP, Van Doren K, Hirsh D, Steitz JA 1988. Trans splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature 335: 559–562 [DOI] [PubMed] [Google Scholar]
- Conrad R, Thomas J, Spieth J, Blumenthal T 1991. Insertion of part of an intron into the 5′ untranslated region of a Caenorhabditis elegans gene converts it into a trans-spliced gene. Mol Cell Biol 11: 1921–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Liou RF, Blumenthal T 1993a. Conversion of a trans-spliced C. elegans gene into a conventional gene by introduction of a splice donor site. EMBO J 12: 1249–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Liou RF, Blumenthal T 1993b. Functional analysis of a C. elegans trans-splice acceptor. Nucleic Acids Res 21: 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Orita S, Han M 1998. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol Cell Biol 18: 4556–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni S, Sharpe HE, Gravato Nobre M, Zechner K, Browne C, Hodgkin J, Furger A 2009. Regulation of transcription termination in the nematode Caenorhabditis elegans. Nucleic Acids Res 37: 6723–6736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Maroney PA, Nilsen TW 1991. U small nuclear ribonucleoprotein requirements for nematode cis- and trans-splicing in vitro. J Biol Chem 266: 22792–22795 [PubMed] [Google Scholar]
- Hillier LW, Reinke V, Green P, Hirst M, Marra MA, Waterston RH 2009. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res 19: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Hirsh D 1989. A second trans-spliced RNA leader sequence in the nematode Caenorhabditis elegans. Proc Natl Acad Sci 86: 8640–8644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Kuersten S, Deshpande AM, Spieth J, MacMorris M, Blumenthal T 2001. Intercistronic region required for polycistronic pre-mRNA processing in Caenorhabditis elegans. Mol Cell Biol 21: 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Pleasance ED, Maydan JS, Hunt-Newbury R, O'Neil NJ, Mah A, Baillie DL, Marra MA, Moerman DG, Jones SJ 2007. Identification and analysis of internal promoters in Caenorhabditis elegans operons. Genome Res 17: 1478–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP 2010. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature doi: 10.1038/nature09616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Hirsh D 1987. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell 49: 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S, Lea K, MacMorris M, Spieth J, Blumenthal T 1997. Relationship between 3′ end formation and SL2-specific trans-splicing in polycistronic Caenorhabditis elegans pre-mRNA processing. RNA 3: 269–278 [PMC free article] [PubMed] [Google Scholar]
- Lasda EL, Allen MA, Blumenthal T 2010. Polycistronic pre-mRNA processing in vitro: snRNP and pre-mRNA role reversal in trans-splicing. Genes Dev 24: 1645–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou RF, Blumenthal T 1990. Trans-spliced Caenorhabditis elegans mRNAs retain trimethylguanosine caps. Mol Cell Biol 10: 1764–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Huang T, MacMorris M, Blumenthal T 2001. Interplay between AAUAAA and the trans-splice site in processing of a Caenorhabditis elegans operon pre-mRNA. RNA 7: 176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kuersten S, Huang T, Larsen A, MacMorris M, Blumenthal T 2003. An uncapped RNA suggests a model for Caenorhabditis elegans polycistronic pre-mRNA processing. RNA 9: 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V 1991. Efficient gene transfer in C.elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J 10: 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushforth AM, Anderson P 1996. Splicing removes the Caenorhabditis elegans transposon Tc1 from most mutant pre-mRNAs. Mol Cell Biol 16: 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushforth AM, Saari B, Anderson P 1993. Site-selected insertion of the transposon Tc1 into a Caenorhabditis elegans myosin light chain gene. Mol Cell Biol 13: 902–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T 1993. Operons in C. elegans: Polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell 73: 521–532 [DOI] [PubMed] [Google Scholar]
- Sulston JE, Brenner S 1974. The DNA of Caenorhabditis elegans. Genetics 77: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren K, Hirsh D 1988. Trans-spliced leader RNA exists as small nuclear ribonucleoprotein particles in Caenorhabditis elegans. Nature 335: 556–559 [DOI] [PubMed] [Google Scholar]
- Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD 2008. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet 4: e1000187 doi: 10.1371/journal.pgen.1000187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Xu L, Blumenthal T 1999. SL1 trans splicing and 3′-end formation in a novel class of Caenorhabditis elegans operon. Mol Cell Biol 19: 376–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Yu L, Savage-Dunn C 2010. Alternative trans-splicing of Caenorhabditis elegans sma-9/schnurri generates a short transcript that provides tissue-specific function in BMP signaling. BMC Mol Biol 11: 46 doi: 10.1186/1471-2199-11-46 [DOI] [PMC free article] [PubMed] [Google Scholar]