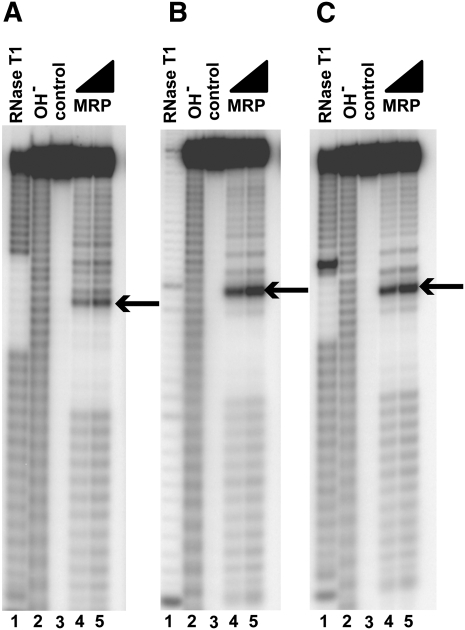
FIGURE 3.
RNase MRP cleavage of partially randomized sequences. Random sequences were used to eliminate the potential influence of the upstream and downstream sequences or secondary structures on the results. (A) 5′-N19-UUUU-H*H-AUC-N13 -3′. (B) 5′-N19-UUUU-H*H-CUC-N13 -3′. (C) 5′-N19-UUUU-H*H-CGC-N13-3′. In all panels, N is a random nucleotide (G, A, U, C); H is a random mix of A, U, C; asterisk (*) designates the cleavage site (indicated by an arrow in the figure). (Lane 1) Digest with RNase T1 (a marker); (lane 2) alkaline hydrolysis (a marker); (lane 3) untreated RNA substrate; (lanes 4,5) substrates digested with RNase MRP. Substrates were 5′-end-labeled with 32P and separated on a 6% denaturing polyacrylamide gel.