Abstract
We describe the localization of the recently identified glucose transporter GLUTx1 and the regulation of GLUTx1 in the hippocampus of diabetic and control rats. GLUTx1 mRNA and protein exhibit a unique distribution when compared with other glucose transporter isoforms expressed in the rat hippocampus. In particular, GLUTx1 mRNA was detected in hippocampal pyramidal neurons and granule neurons of the dentate gyrus as well as in nonprincipal neurons. With immunohistochemistry, GLUTx1 protein expression is limited to neuronal cell bodies and the most proximal dendrites, unlike GLUT3 expression that is observed throughout the neuropil. Immunoblot analysis of hippocampal membrane fractions revealed that GLUTx1 protein expression is primarily localized to the intracellular compartment and exhibits limited association with the plasma membrane. In streptozotocin diabetic rats compared with vehicle-treated controls, quantitative autoradiography showed increased GLUTx1 mRNA levels in pyramidal neurons and granule neurons; up-regulation of GLUTx1 mRNA also was found in nonprincipal cells, as shown by single-cell emulsion autoradiography. In contrast, diabetic and control rats expressed similar levels of hippocampal GLUTx1 protein. These results indicate that GLUTx1 mRNA and protein have a unique expression pattern in rat hippocampus and suggest that streptozotocin diabetes increases steady-state mRNA levels in the absence of concomitant increases in GLUTx1 protein expression.
The family of facilitative glucose transporter (GLUT) proteins is responsible for the entry of glucose into cells throughout the periphery and the brain (1, 2). In the central nervous system (CNS), glucose transporters play an essential role in neuronal homeostasis, because glucose represents the primary energy source for the brain (3, 4). Metabolic disorders that disrupt glucose delivery or utilization in the CNS may adversely affect neuronal activity, particularly cognitive function. For example, glucose utilization is reduced in Alzheimer's disease (AD) (5), and the cognitive impairments associated with AD can be ameliorated to some extent by glucose administration (6) and insulin therapy (7). Similarly, diabetes mellitus (type 1 or insulin-dependent diabetes) is associated with cognitive impairments in humans and in animal models of hyperglycemia (8). Reductions in glucose transport in the CNS may be particularly endangering to the hippocampus, a region that has been identified as an important integration center in the development of learning and memory (9). In view of the special metabolic requirements of the hippocampus, it is not unexpected that hippocampal neurons exhibit robust expression of the neuron-specific glucose transporter GLUT3 (10) as well as the insulin-sensitive glucose transporter GLUT4 (11).
Recent cloning of a novel mammalian glucose transporter was stimulated by the unexpected phenotypes of GLUT2 and GLUT4 knockout mice (12, 13) and by the ability to search databases for sequence similarities with GLUTs 1–5 (14). GLUTx1, also referred to as GLUT8, exhibits between 29 and 32% identity with rat GLUTs 1–5, and transport activity of GLUTx1 expressed in Xenopus oocytes was inhibited by cytochalasin B (14, 15). Subsequent studies suggested that expression or translocation of GLUTx1 to the plasma membrane may be hormonally regulated (14–16). Northern blot analysis demonstrated that GLUTx1 mRNA is expressed most abundantly in testis, as well as several brain regions, including the hippocampus (14). In an attempt to determine whether this novel glucose transporter assists GLUT3 and GLUT4 in meeting the metabolic demands of the hippocampus, we have examined the localization of GLUTx1 mRNA and protein in the hippocampus by in situ hybridization histochemistry and immunohistochemistry, respectively. In addition, we also have examined the regulation of GLUTx1 mRNA and protein in the hippocampus of streptozotocin (STZ) diabetic rats.
Materials and Methods
Animal Protocols.
Diabetes was produced in adult male Sprague–Dawley rats by i.p. injections of STZ (35 mg/kg; Sigma) dissolved in citrate buffer (0.1 M, pH 4.2) administered on 3 consecutive days; control animals were injected with vehicle only. The development of hyperglycemia was confirmed by measuring fasting glucose concentrations, using the glucose oxidase method [Glucose (trinder) kit; Sigma]. STZ-treated rats exhibited plasma glucose levels 3–4 times above control rats (control = 110.48 mg/dl, STZ = 353.05 mg/dl; P ≤ 0.0001). On the seventh day of STZ-induced hyperglycemia, rats were killed, blood was collected, and the brains were removed quickly. For in situ hybridization histochemistry, the brains from control and diabetic rats (n = 8) were frozen on dry ice and stored at −70°C until use. For immunoblot analysis, vehicle-treated rats and diabetic rats (n = 8) were decapitated, brains were removed quickly, and the hippocampus was microdissected, frozen on dry ice, and stored at −70°C until use.
In Situ Hybridization Histochemistry.
In situ hybridization studies were performed as described (17) by using sense and antisense probes generated from rat GLUTx1 cDNA corresponding to bp 1,300–2,087. Antisense riboprobes were labeled with [35S]UTP by using T7 RNA polymerase; sense probes were labeled with [35S]UTP by using T3 RNA polymerase. Slides were exposed to Kodak X-Omat film for autoradiography and subsequently dipped in Kodak NTB-2 emulsion for single-cell emulsion autoradiographic analysis. All autoradiographic films and emulsion slides were developed and processed for analysis at identical time points. Autoradiographic film optical density analysis and quantification of reduced silver grains were performed as described (18). The values obtained represent the average of measurements taken from 2–3 sections per slide and two slides per animal (control and STZ, n = 8). Statistical analysis was performed by using an unpaired t test; significance was accepted at P ≤ 0.05.
GLUTx1 Antibody Generation.
Polyclonal antisera to the carboxyl-terminal 11 aa [LEQITAHFEGR] of the GLUTx1 protein were generated by using a previously described technique (19). The synthetic GLUTx1 peptide (Research Genetics, Huntsville, AL) was linked to keyhole limpet hemacyanin (Pierce) and was administered to New Zealand rabbits by s.c. injection.
Immunohistochemistry.
Immunohistochemical analysis was performed as described (20). GLUTx1 primary antisera were used at a concentration of 1:1,000. For competition studies, immunizing peptide was added to the immune serum at increasing concentrations (0.5–2.4 mg/ml) and incubated for 1 h at 4°C. The competed serum then was used at a dilution of 1:1,000.
Immunoblot Analysis.
Hippocampal plasma membrane and intracellular membrane-containing fractions were separated by sucrose density centrifugation as described (21). Proteins (50 μg) were separated by 10% SDS/PAGE, transferred to nitrocellulose (NC) membranes and blocked in TBS plus 5% nonfat dry milk for 60 min. NC membranes then were incubated with GLUTx1 primary antisera (1:1,000 in TBS/5% nonfat dry milk) for 90 min at room temperature (RT) with gentle shaking. NC membranes were washed with TBS plus 0.05% Tween 20 (TBST) three times for 5 min with gentle shaking and then subjected to a high-stringency wash (200 mM urea/100 mM glycine/1% Triton X-100, pH = 7.5) for 5 min, followed by a final 5-min wash of TBST. NC membranes then were incubated with peroxidase-labeled anti-rabbit secondary antibodies (1:20,000; Vector Laboratories) for 60 min at RT with gentle shaking. NC membranes were washed as described above and developed by using enhanced chemiluminescence reagents (Amersham Pharmacia) as described by the manufacturer. Optical density analysis of GLUTx1 bands was performed as described (21); statistical comparisons were based on at least four separate immunoblot assays. Statistical analysis was performed by using an unpaired t test; significance was accepted at P ≤ 0.05.
Results
Localization of GLUTx1 mRNA in Rat Hippocampus.
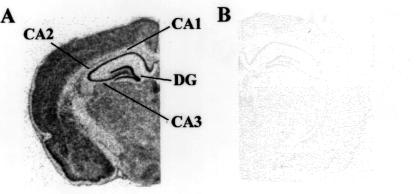
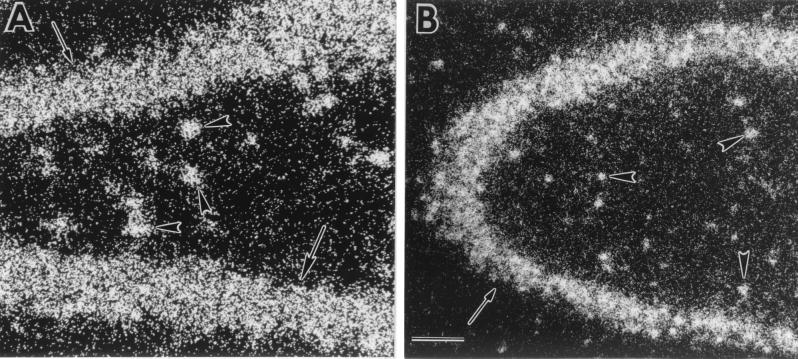
Autoradiographic film analysis revealed that GLUTx1 mRNA exhibits a similar pattern of distribution in the rat hippocampus when compared with the neuron-specific glucose transporter GLUT3 (17). GLUTx1 mRNA expression is observed in CA1, CA2, and CA3 pyramidal neurons of the hippocampus and the granule neurons of the dentate gyrus (Fig. 1A). The specificity of the antisense-labeled probe was demonstrated with use of 35S-labeled sense probe, which did not detect GLUTx1 mRNA (Fig. 1B). GLUTx1 mRNA was also detected in other neuronal populations, including the cerebral cortex (Fig. 1A) and the hypothalamus (data not shown). Single-cell emulsion autoradiographic analysis revealed that GLUTx1 mRNA expression also was detected in nonprincipal cells of the rat hippocampus. GLUTx1 mRNA was detected as reduced silver grains deposited over hippocampal nonprincipal cells in the rat hilus (Fig. 2A), as well as in stratum radiatum and stratum lucidum of CA1 of Ammon's Horn and in nonprincipal cells in CA3 (Fig. 2B). In contrast, as described in our previous studies (17), GLUT3 mRNA is observed exclusively in the principal neurons of the rat hippocampus.
Figure 1.
Autoradiographic analysis of GLUTx1 mRNA expression in rat brain. (A) GLUTx1 expression in rat brain as detected by use of 35S-labeled antisense riboprobe. GLUTx1 mRNA is detected in the pyramidal neurons of CA1, CA2, and CA3 of Ammon's Horn, as well as in granule neurons of the dentate gyrus (DG). (B) Adjacent section incubated with 35S-labeled sense riboprobe fails to detect GLUTx1 mRNA in rat brain.
Figure 2.
Representative dark-field photomicrographs of emulsion autoradiographic analysis for GLUTx1 mRNA expression in the rat hippocampus. (A) Reduced silver grains for GLUTx1 were detected over granule neurons of the dentate gyrus (arrows) and nonprincipal cells in the hilus (arrowheads). (B) Reduced silver grains of GLUTx1 mRNA over CA3 pyramidal neurons (arrows) and nonprincipal cells (arrowheads). (Bar = 100 μ.)
Regulation of GLUTx1 mRNA Expression in the Hippocampus of Diabetic Rats.
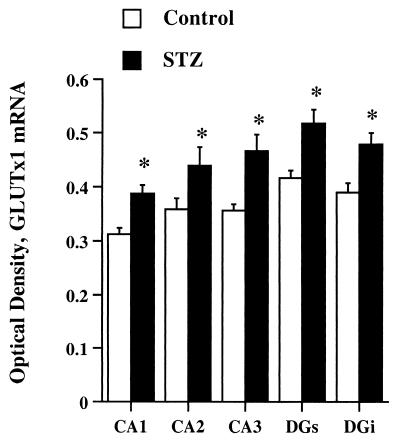
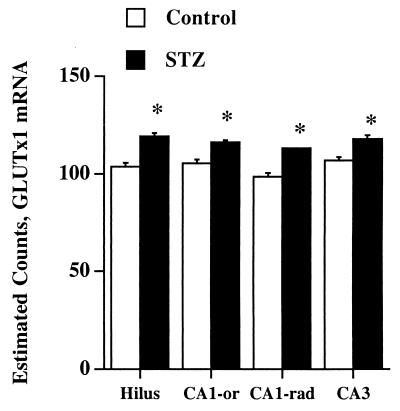
The regulation of GLUTx1 mRNA expression was examined by in situ hybridization histochemistry in the hippocampus of STZ diabetic rats. Autoradiographic film image analysis revealed that GLUTx1 mRNA levels were increased in CA1, CA2, and CA3 pyramidal neurons in diabetic rats compared with vehicle-treated controls (Fig. 3). Similarly, GLUTx1 mRNA expression was increased in granule neurons of the superior and inferior blades of the dentate gyrus (DGs and DGi, respectively; Fig. 3). GLUTx1 mRNA levels also were increased in the cerebral cortex of diabetic rats compared with vehicle-treated controls (data not shown). Single-cell emulsion autoradiographic analysis was performed to determine the regulation of GLUTx1 mRNA expression in hippocampal nonprincipal cells of diabetic rats. As shown in Fig. 4, quantification of reduced silver grains determined that GLUTx1 mRNA levels were increased in hippocampal nonprincipal cells in the hilus, CA1 oriens, CA1 radiatum, and CA3 of diabetic rats compared with control rats. Collectively, these results demonstrate that GLUTx1 mRNA levels are increased in principal neurons, as well as nonprincipal cells in the hippocampus of STZ-treated rats, illustrating that GLUTx1 mRNA levels are up-regulated in response to hyperglycemia and/or insulinopenia as in type 1 diabetes.
Figure 3.
Autoradiographic image analysis of hippocampal GLUTx1 mRNA expression in STZ rats compared with vehicle-treated control rats. Statistical analysis revealed that GLUTx1 mRNA levels were significantly increased in diabetic rats in CA1 pyramidal neurons, CA2 pyramidal neurons, CA3 pyramidal neurons, granule neurons of the superior blade of the dentate gyrus (DGs), and granule neurons of the inferior blade of the dentate gyrus (DGi) compared with vehicle-treated controls (* = P ≤ 0.05).
Figure 4.
Statistical analysis of emulsion autoradiography for GLUTx1 mRNA expression in nonprincipal cells of the rat hippocampus. Diabetic rats exhibited significant increases in GLUTx1 mRNA expression compared with vehicle-treated controls in nonprincipal cells of the hilus, CA1-radiatum, CA1-oriens, and CA3 (* = P ≤ 0.05).
Immunohistochemical Localization of GLUTx1 in Rat Hippocampus.
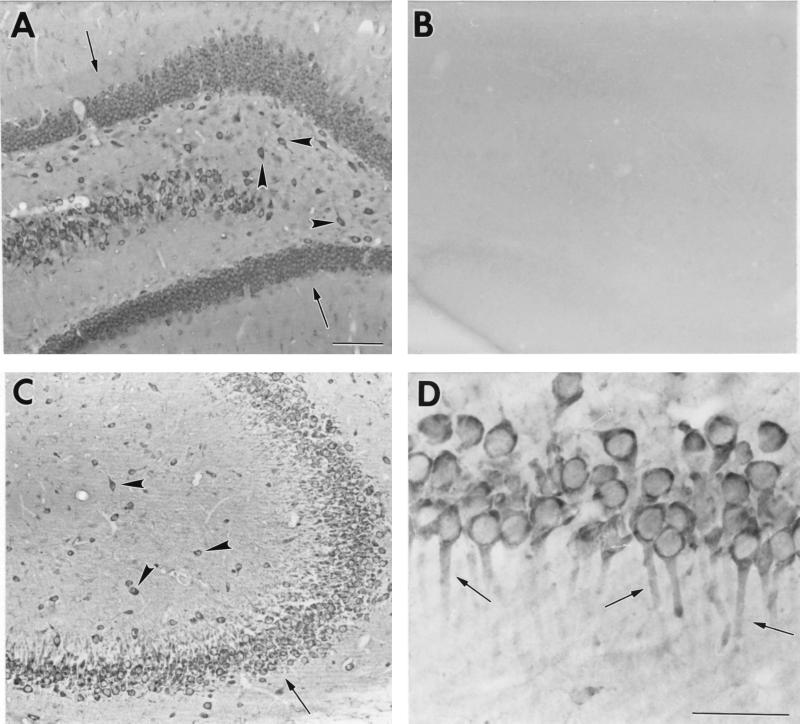
GLUTx1-specific antisera were used in immunohistochemical analysis to localize GLUTx1 protein in the rat hippocampus. GLUTx1 immunoreactivity (IR) exhibited a similar distribution as GLUTx1 mRNA, in that GLUTx1 IR was detected in principal neurons as well as nonprincipal cells of the rat hippocampus. Specifically, as shown in Fig. 5A, GLUTx1 IR was observed in the granule neurons of the dentate gyrus (Fig. 5, arrows) and nonprincipal cells of the rat hilus (Fig. 5, arrowheads); GLUTx1 primary antisera preabsorbed with immunogen did not detect GLUTx1 protein in the rat hilus (Fig. 5B). GLUTx1 IR was detected in CA3 pyramidal neurons and nonprincipal cells of CA3 (Fig. 5C). Higher magnification revealed that GLUTx1 IR is predominantly cytoplasmic and associated specifically with neuronal cell bodies and the most proximal dendritic branches in CA1 pyramidal neurons (Fig. 5D, arrows).
Figure 5.
Representative bright-field photomicrographs of GLUTx1 immunoreactivity in the rat hippocampus. (A) GLUTx1 immunoreactivity in granule neurons of the dentate gyrus (arrows) and nonprincipal cells of the hilus (arrowheads). (B) Preabsorption of GLUTx1 antisera eliminates GLUTx1 immunoreactivity in the hilus. (C) Low-power magnification of GLUTx1 immunoreactivity in pyramidal neurons (arrows) and nonprincipal cells (arrowheads) in CA3 of Ammon's Horn. (D) Higher-power magnification of GLUTx1 immunolabeling showing cytoplasmic labeling and GLUTx1 immunoreactivity in the proximal dendrites of CA1 pyramidal neurons (arrows). (Bars = 100 μ.)
Regulation of GLUTx1 Protein Expression in the Hippocampus of Diabetic Rats.
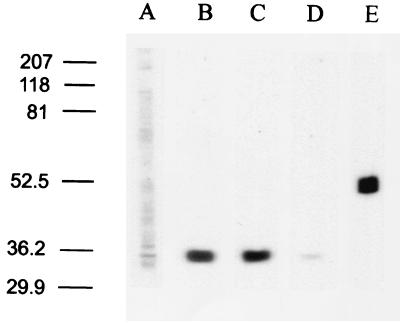
To determine whether GLUTx1 protein expression was regulated in type 1 diabetes, we measured GLUTx1 immunoreactive protein levels in hippocampal membrane fractions isolated from diabetic rats and control rats. Immunoblot analysis of hippocampal plasma membrane fractions and mitochondrial fractions prepared from control rats did not exhibit appreciable levels of GLUTx1 (Fig. 6A; data not shown). GLUTx1 expression similarly was absent in plasma membrane fractions and mitochondrial fractions isolated from STZ-treated rats. In hippocampal cytoplasmic fractions, GLUTx1 IR was detected as a single band at approximately 35 kDa in vehicle-treated control rats (Fig. 6B) and STZ diabetic rats (Fig. 6C). Preabsorption of GLUTx1 antisera with immunizing peptide eliminated this 35-kDa band (Fig. 6D). Successful isolation of the intracellular fraction was verified by immunoblot analysis by using antisera raised against proteins found exclusively in the cytoplasm, including glutamic acid decarboxylase, calretinin (data not shown), and β-tubulin (Fig. 6E). Autoradiographic image analysis of the 35-kDa protein in the cytoplasmic fractions revealed that GLUTx1 expression was nearly identical in STZ diabetic rats and vehicle-treated controls (STZ = 97.8% of control; t6 = 0.554, P = 0.599).
Figure 6.
Immunoblot analysis of membrane fractions isolated from rat hippocampus using GLUTx1 antisera. Lane A, GLUTx1 primary antisera (1:1,000) weakly immunodetect a 35-kDa protein in hippocampal plasma membrane fractions; lanes B and C, GLUTx1 antisera immunodetect a single protein at approximately 35 kDa in hippocampal intracellular membrane-containing fractions isolated from control rats (lane B) and STZ diabetic rats (lane C); lane D, preabsorption of GLUTx1 antisera with blocking peptide eliminates the detection of this 35-kDa protein in control cytoplasmic fractions; lane E, β-tubulin mAb (Sigma) immunodetects a single protein at approximately 52 kDa in cytoplasmic fractions, confirming the successful isolation of hippocampal cytoplasmic fractions from hippocampal plasma membrane fractions. (All lanes equal 50 μg of protein; lanes are representative of at least four separate immunoblot experiments.)
Discussion
The current study provides a description of the localization of GLUTx1 mRNA and protein in the rat brain. Although our study focused upon the hippocampus, GLUTx1 mRNA and protein displayed a ubiquitous distribution in neuronal cell bodies throughout the brain, including the cortex and the hypothalamus. In the hippocampus, GLUTx1 mRNA and protein are expressed in both principal and nonprincipal neurons. In addition, we found that steady-state levels of GLUTx1 mRNA are increased in the hippocampus of STZ rats whereas GLUTx1 protein levels are unchanged. We previously reported similar findings for GLUT3, in that GLUT3 mRNA levels were increased in the hippocampus of STZ diabetic rats, without a concomitant increase in GLUT3 protein levels (17). The absence of detectable changes in GLUTx1 protein in the presence of increased steady-state levels of GLUTx1 mRNA does not eliminate the possibility that GLUTx1 protein is modulated in STZ-treated rats. Protein recycling or protein stability may be modulated by hyperglycemia or insulinopenia, resulting in the maintenance of basal expression of GLUTx1 protein. Previous studies have reported that glucose transporter degradation may be greatly affected by a variety of metabolic challenges, including STZ-induced diabetes (22). Similarly, increased steady-state levels of GLUTx1 mRNA in the hippocampus of STZ rats may result from changes in stability or half-life as well as increased transcription of GLUTx1.
The intracellular localization of GLUTx1 raises the important question of whether GLUTx1 translocates to the plasma membrane and, if so, what stimulus leads to this translocation. The initial description of GLUTx1 characterized a dileucine motif at the amino terminus that may serve as an internalization signal that limits GLUTx1 translocation to the plasma membrane (14). Using mouse blastocysts and confocal microscopy, GLUTx1 was shown to translocate from a cytoplasmic location to the vicinity of the plasma membrane in a manner analogous to the translocation of GLUT4 (16). Alternatively, GLUTx1 may remain sequestered in the cytoplasmic compartment to assist in the maintenance of glucose concentrations of intracellular organelles. Immunohistochemical and immunoblot analysis performed in the current study strengthens the hypothesis that GLUTx1 is retained in an intracellular compartment, consistent with previous reports (14–16). The experimental approaches used in our study cannot eliminate the possibility that GLUTx1 is associated with the plasma membrane; nonetheless, our results suggest that GLUTx1 is localized to the cytoplasmic compartment and may be associated with intracellular organelles. In support of this hypothesis, electron microscopic analysis of hippocampal neurons has revealed that in addition to being localized to the plasma membrane, GLUT3 and GLUT4 are associated with intracellular organelles, such as mitochondria (GLUT3), vesicles (GLUT3 and GLUT4), and the rough endoplasmic reticulum (GLUT4) (23, 24).
As described above, the localization of GLUT3 and GLUT4 has been well established in the rat hippocampus by immunohistochemical techniques at the light and electron microscopic levels. GLUT3 has been localized to pre- and postsynaptic nerve terminals and small neuronal processes, but is relatively absent from neuronal cell bodies and apical dendrites (10, 24, 25). Like GLUTx1, GLUT4 is associated with neuronal cell bodies in hippocampal pyramidal neurons (23, 26); GLUT4 is also localized to a select population of parvalbumin-positive interneurons, but GLUT4 immunoreactivity was not detected in other hippocampal nonprincipal cell types (26). The absence of a ubiquitous distribution of glucose transporters in nonprincipal cells is somewhat paradoxical because the metabolic demands of neuronal cell bodies are often greater than the surrounding neuropil (27). Our immunohistochemical studies demonstrate that GLUTx1 is a neuronal cell body-specific glucose transporter and suggests that GLUTx1 may support the glucose requirements of hippocampal neuronal cell bodies.
In the hippocampus, GABAergic interneurons serve as the major postsynaptic targets of the mossy fiber pathway, which projects from the dentate gyrus to the apical dendrites of CA3 pyramidal neurons (28). Previous studies from our laboratory have demonstrated that stress or exposure to stress levels of glucocorticoids leads to the reversible dendritic remodeling of the apical dendrites of CA3 pyramidal neurons (29, 30). Administration of GABA receptor agonists inhibits stress-induced dendritic remodeling, suggesting that hippocampal nonprincipal cells play an important role in stress-induced morphological changes (31). GLUTx1 expression may be increased in hippocampal interneurons and permit enhanced glucose utilization during periods of heightened neuronal activity involved in dendritic remodeling. In this regard, diabetic rats exhibit morphological changes in CA3 pyramidal neurons similar to euglycemic rats subjected to stress levels of glucocorticoids (32). Because interneurons represent a subtype of nonprincipal neurons, future studies will be required to more accurately define the phenotype of GLUTx1-positive nonprincipal cells in the rat hippocampus.
In addition to morphological changes, GLUTx1 expression may be intimately involved in a variety of hippocampal functions, including glucoregulation and cognitive function. For example, microinjection of glucose into the hippocampus increases acetylcholine release, an effect that is potentiated during task learning in rats, suggesting that glucose can amplify the activity of the hippocampal cholinergic system (33). Conversely, reductions in hippocampal cholinergic activity are associated with cognitive impairments in STZ rats (34). Such results suggest that a dynamic interplay exists between hippocampal glucoregulation, activation of the cholinergic system, and cognitive function. Central to hippocampal glucoregulation and, ultimately, cognitive performance is the regulation and function of hippocampal glucose transporters. Our studies have shown that GLUT3 and GLUTx1 mRNA expression is up-regulated in the hippocampus of STZ rats, possibly to compensate for decreases in glucose transport in diabetes. However, our other studies also have shown that GLUT3 is a target of diabetes-induced increases in oxidative stress in the hippocampus (21), suggesting that compensatory increases in glucose transporter expression may not alleviate the metabolic endangerment produced by hyperglycemia. The isolation of a glucose transporter that is expressed in neuronal cell bodies provides another mechanism through which the metabolic demands of the brain may be fulfilled. Accordingly, future studies will be necessary to determine the functional role of GLUTx1 in regard to how this glucose transporter may contribute to the glucoregulatory and cognitive functions of the hippocampus.
Acknowledgments
This work was supported by Juvenile Diabetes Foundational International Grant 10-2000-134 (L.P.R.), Consejo Nacional de Investigaciones Cientificas y Técnicas de Argentina (C.A.G. and G.G.P.), an Irma T. Hirschl career scientist award (M.J.C.), and National Institutes of Health Grants MH59251 (S.E.A.), DK47425 and HL58119 (M.J.C.), and MH42156 (B.S.M.).
Abbreviations
- GLUT
glucose transporter
- AD
Alzheimer's disease
- STZ
streptozotocin
- IR
immunoreactivity
References
- 1.Vannucci S J, Maher F, Simpson I A. Glia. 1997;21:2–21. doi: 10.1002/(sici)1098-1136(199709)21:1<2::aid-glia2>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 2.Maher F, Vannucci S J, Simpson I A. FASEB J. 1994;8:1003–1011. doi: 10.1096/fasebj.8.13.7926364. [DOI] [PubMed] [Google Scholar]
- 3.Lund-Anderen H. Physiol Rev. 1979;59:305–310. doi: 10.1152/physrev.1979.59.2.305. [DOI] [PubMed] [Google Scholar]
- 4.Pardridge W M. Physiol Rev. 1983;63:1481–1535. doi: 10.1152/physrev.1983.63.4.1481. [DOI] [PubMed] [Google Scholar]
- 5.Piert M, Koeppe R A, Giordani B, Berent S, Kuhl D E. J Nucl Med. 1996;37:201–208. [PubMed] [Google Scholar]
- 6.Craft S, Dagogo-Jack S E, Wiethop B V, Murphy C, Nevins R T, Fleischman S, Rice V, Newcomer J W, Cryer P E. Behav Neurosci. 1993;6:926–940. doi: 10.1037//0735-7044.107.6.926. [DOI] [PubMed] [Google Scholar]
- 7.Craft S, Newcomer J, Kanne S, Dagogo-Jack S E, Cryer P, Sheline Y, Luby J, Dagogo-Jack A, Anderson A. Neurobiol Aging. 1996;17:123–130. doi: 10.1016/0197-4580(95)02002-0. [DOI] [PubMed] [Google Scholar]
- 8.Gispen W H, Biessels G-J. Trends Neurosci. 2000;23:542–549. doi: 10.1016/s0166-2236(00)01656-8. [DOI] [PubMed] [Google Scholar]
- 9.Bliss T V P, Collingridge G L. Nature (London) 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 10.McCall A L, Van Bueren A M, Moholt-Siebert M, Cherry N J, Woodward W R. Brain Res. 1994;659:292–297. doi: 10.1016/0006-8993(94)90896-6. [DOI] [PubMed] [Google Scholar]
- 11.LeLoup C, Arluison M, Kassis N, Lepetit N, Cartier N, Ferré P, Pénicaud L. Mol Brain Res. 1996;38:45–53. doi: 10.1016/0169-328x(95)00306-d. [DOI] [PubMed] [Google Scholar]
- 12.Katz E B, Stenbit A E, Hatton K, Depinho R, Charron M J. Nature (London) 1995;377:151–155. doi: 10.1038/377151a0. [DOI] [PubMed] [Google Scholar]
- 13.Guillam M T, Hummler E, Schaerer E, Yeh J I, Birnbaum M J, Beermann F, Schmidt A, Deriaz N, Thorens B. Nat Genet. 1997;17:327–330. doi: 10.1038/ng1197-327. [DOI] [PubMed] [Google Scholar]
- 14.Ibberson M, Uldry M, Thorens B. J Biol Chem. 2000;275:4607–4612. doi: 10.1074/jbc.275.7.4607. [DOI] [PubMed] [Google Scholar]
- 15.Doege H, Schürmann A, Bahrenberg G, Brauers A, Joost H G. J Biol Chem. 2000;275:16275–16280. doi: 10.1074/jbc.275.21.16275. [DOI] [PubMed] [Google Scholar]
- 16.Carayannopoulos M O, Chi M M-Y, Cui Y, Pingsterhaus J M, McKnight R A, Mueckler M, Devaskar S U, Moley K H. Proc Natl Acad Sci USA. 2000;97:7313–7318. doi: 10.1073/pnas.97.13.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reagan L P, Magariños A M, Lucas L R, Van Bueren A, McCall A L, McEwen B S. Am J Physiol. 1999;276:E879–E886. doi: 10.1152/ajpendo.1999.276.5.E879. [DOI] [PubMed] [Google Scholar]
- 18.Reagan L P, McKittrick C R, McEwen B S. Neuroscience. 1999;91:211–219. doi: 10.1016/s0306-4522(98)00615-0. [DOI] [PubMed] [Google Scholar]
- 19.Baldini G, Hohman R, Charron M J, Lodish H F. J Biol Chem. 1991;266:4037–4040. [PubMed] [Google Scholar]
- 20.Alves S E, Weiland N, Hayashi S, McEwen B S. J Comp Neurol. 1998;391:322–334. [PubMed] [Google Scholar]
- 21.Reagan L P, Magariños A M, Yee D K, Szweda L I, Van Bueren A, McCall A L, McEwen B S. Brain Res. 2000;862:292–300. doi: 10.1016/s0006-8993(00)02212-5. [DOI] [PubMed] [Google Scholar]
- 22.Kim S-S, Bae J W, Jung C Y. Am J Physiol. 1994;267:E132–E139. doi: 10.1152/ajpendo.1994.267.1.E132. [DOI] [PubMed] [Google Scholar]
- 23.Messari S E, LeLoup C, Quignon M, Brisorgueil M-J, Penicaud L, Arluison M. J Comp Neurol. 1998;399:492–512. doi: 10.1002/(sici)1096-9861(19981005)399:4<492::aid-cne4>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 24.Leino R L, Gerhart D Z, Van Bueren A M, McCall A L, Drewes L R. J Neurosci Res. 1997;49:617–626. doi: 10.1002/(SICI)1097-4547(19970901)49:5<617::AID-JNR12>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 25.Gerhart D Z, Leino R L, Borson N D, Taylor W E, Gronlund K M, McCall A L, Drewes L R. Neuroscience. 1995;66:237–246. doi: 10.1016/0306-4522(94)00544-f. [DOI] [PubMed] [Google Scholar]
- 26.Aplet J, Melhorn G, Schliebs R. J Neurosci Res. 1999;57:693–705. [PubMed] [Google Scholar]
- 27.Duelli R, Maurer M H, Staudt R, Heiland S, Duembgen L, Kuschinsky W. Brain Res. 2000;858:338–347. doi: 10.1016/s0006-8993(00)01942-9. [DOI] [PubMed] [Google Scholar]
- 28.Acsády L, Kimondi A, Sík A, Freund T, Buzsáki G. J Neurosci. 1998;18:3386–3403. doi: 10.1523/JNEUROSCI.18-09-03386.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woolley C S, Gould E, McEwen B S. Brain Res. 1990;531:225–231. doi: 10.1016/0006-8993(90)90778-a. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe Y, Gould E, McEwen B S. Brain Res. 1992;588:341–345. doi: 10.1016/0006-8993(92)91597-8. [DOI] [PubMed] [Google Scholar]
- 31.Magariños A M, Deslandes A, McEwen B S. Eur J Pharmacol. 1999;371:113–122. doi: 10.1016/s0014-2999(99)00163-6. [DOI] [PubMed] [Google Scholar]
- 32.Magariños A M, McEwen B S. Proc Natl Acad Sci USA. 2000;97:11056–11061. doi: 10.1073/pnas.97.20.11056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragozzino M E, Pal S N, Unick K, Stefani M R, Gold P E. J Neurosci. 1998;18:1595–1601. doi: 10.1523/JNEUROSCI.18-04-01595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blokland A, Jolles J. Pharmacol Biochem Behav. 1993;44:491–494. doi: 10.1016/0091-3057(93)90497-h. [DOI] [PubMed] [Google Scholar]