Abstract
Multi-drug resistant (MDR), pathogenic Gram-negative bacteria pose a serious health threat, and novel antibiotic targets must be identified to combat MDR infections. One promising target is the zinc-dependent metalloamidase UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase (LpxC), which catalyzes the committed step of lipid A (endotoxin) biosynthesis. LpxC is an essential, single copy gene that is conserved in virtually all Gram-negative bacteria. LpxC structures, revealed by NMR and X-ray crystallography, demonstrate that LpxC adopts a novel ‘β-α-α-β sandwich’ fold and encapsulates the acyl chain of the substrate with a unique hydrophobic passage. Kinetic analysis revealed that LpxC functions by a general acid-base mechanism, with a glutamate serving as the general base.
Many potent LpxC inhibitors have been identified, and most contain a hydroxamate group targeting the catalytic zinc ion. Although early LpxC-inhibitors were either narrow-spectrum antibiotics or broad-spectrum in vitro LpxC inhibitors with limited antibiotic properties, the recently discovered compound CHIR-090 is a powerful antibiotic that controls the growth of E. coli and P. aeruginosa, with an efficacy rivaling that of the FDA-approved antibiotic ciprofloxacin. CHIR-090 inhibits a wide range of LpxC enzymes with sub-nanomolar affinity in vitro, and is a two-step, slow, tight-binding inhibitor of A. aeolicus and E. coli LpxC. The success of CHIR-090 suggests that potent LpxC-targeting antibiotics may be developed to control a broad range of Gram-negative bacteria.
Keywords: LpxC, CHIR-090, antibiotic, lipid A, lipopolysaccharide, endotoxin, catalysis, mechanism
Introduction
Pathogenic bacteria pose a persistent health threat, and untreated infections are a major cause of mortality and morbidity throughout the world. One example is Salmonella enterica serotype typhi, which causes 16 million new typhoid fever cases and 600,000 fatalities worldwide each year, with higher per capita death rates in developing countries [1]. Proper medical care and distribution of antibiotics are likely to reduce mortality. However, in regions where antibiotics are available, multi-drug resistant (MDR) pathogens are emerging as serious health threats, including Pseudomonas aeruginosa and Acinetobacter baumannii [2]. To combat MDR and otherwise recalcitrant bacteria, novel antibiotics that inhibit previously unexploited targets must be identified [3]. The unique and essential zinc-dependent metalloamidase UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase (LpxC), which catalyzes the committed step of lipid A biosynthesis, has emerged as an attractive antibiotic target.
Lipid A is a glucosamine-based phospholipid that comprises the outer monolayer of the outer membrane in Gram-negative bacteria. It is the membrane anchor of lipopolysaccharide (LPS) and is essential to protect Gram-negative bacteria against external agents such as antibiotics and detergents [4]. With few exceptions, bacteria lacking lipid A are not viable, whereas mutants with reduced lipid A biosynthetic capacity grow slowly and are hypersensitive to a wide range of antibiotics [5]. In addition to its role as an integral membrane component, lipid A is a powerful endotoxin, capable of eliciting life-threatening septic shock that is lethal in roughly 50% of the cases [6, 7]. Indeed, septicemia is the tenth leading cause of fatality and is responsible for 1.4% of all deaths (>30,000) in the United States alone each year [8]. Therefore, an effective therapeutic inhibiting lipid A biosynthesis promises to cure Gram-negative bacterial infections and sensitize infectious bacteria to other antibiotics. As an added benefit, therapeutics targeting endotoxin production may reduce the complications of septic shock during treatment.
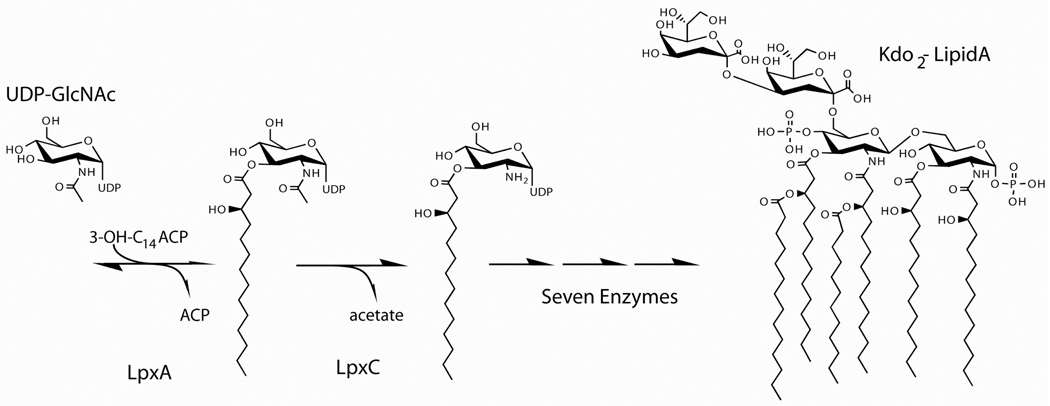
Lipid A is synthesized in the cytosol on the inner surface of the inner membrane by nine unique enzymes (Figure 1) [4]. Although the biosynthesis begins with the acylation of UDP-GlcNAc catalyzed by LpxA, this reaction is thermodynamically unfavorable [9]. Thus, the first committed step of lipid A biosynthesis is the deacetylation reaction catalyzed by LpxC.
Figure 1. The lipid A biosynthesis pathway.
The biosynthesis of Kdo2-Lipid A in E. coli requires nine enzymes, beginning with the LpxA-catalyzed acylation of UDP-GlcNAc. Because this reaction is thermodynamically unfavorable, the second reaction catalyzed by LpxC (deacetylation) is the committed step of lipid A biosynthesis.
While each of the first six enzymes in this pathway is a potential antibiotic target, LpxC is particularly attractive due to its regulatory role in lipid A biosynthesis [10, 11]. Either increasing or decreasing LpxC activity is lethal to E. coli [11–14]. Additionally, LpxC is highly conserved among Gram-negative bacteria, but shares no sequence or structural homology with any mammalian proteins. This uniqueness should permit the development of a highly specific inhibitor, with limited off-target affinity and toxicity. In this review, we will describe the structure, enzymology and inhibition of LpxC, with an emphasis on the development of potent LpxC-specific antibiotics.
Discovery of LpxC as a zinc metalloamidase
The lpxC locus was originally identified in a penicillin-sensitive strain from a screen of chemically mutagenized penicillin-resistant E. coli [5, 15]. The mutation, named envA for envelope mutant A, exhibited slow, filamentous growth with cell division stalling during separation. It was noted that this strain was hypersensitive to many antibiotics. Later, envA harboring strains were shown to have reduced LpxC activity (5% of wild type) and slightly reduced LPS content (~70% of wild type) [16].
Our current knowledge of the LpxC mechanism and structure is primarily derived from studies using LpxC proteins from E. coli (EcLpxC) and the hyperthermophillic bacterium A. aeolicus (AaLpxC). The discovery of a class of EcLpxC inhibitors containing a zinc-chelating hydroxamate moiety was the first indication that LpxC is a zinc-dependent enzyme [17]. LpxC activity was inhibited by dipicolinic acid and EDTA [18]. Zinc, cobalt, nickel or manganese substitution restored activity, but plasma emission spectroscopy indicated that only zinc was present in purified samples. Similar to other zinc amidases, excess zinc was inhibitory. Genetic analysis of EcLpxC and AaLpxC identified two likely zinc ligands (H79 and H238, by EcLpxC numbering; H74 and H226 of AaLpxC), and two possibilities for a third zinc ligand (H265 or D246; H253 or D234 of AaLpxC) [note: D242 instead of D246 or H265 was later shown to be the true zinc ligand (D230 of AaLpxC)] [19]. Extended X-ray absorption fine structure (EXAFS) spectroscopic analysis using LpxC suggested that zinc is coordinated by two oxygen and two nitrogen atoms [20]. Because zinc-coordinated water was thought to be necessary for catalysis, the remaining three zinc ligands of LpxC were presumably H79, H238 and D242 (H74, H226 and D230 of AaLpxC). This specific coordination pattern represented a novel zinc-binding motif.
LpxC adopts a novel structural fold
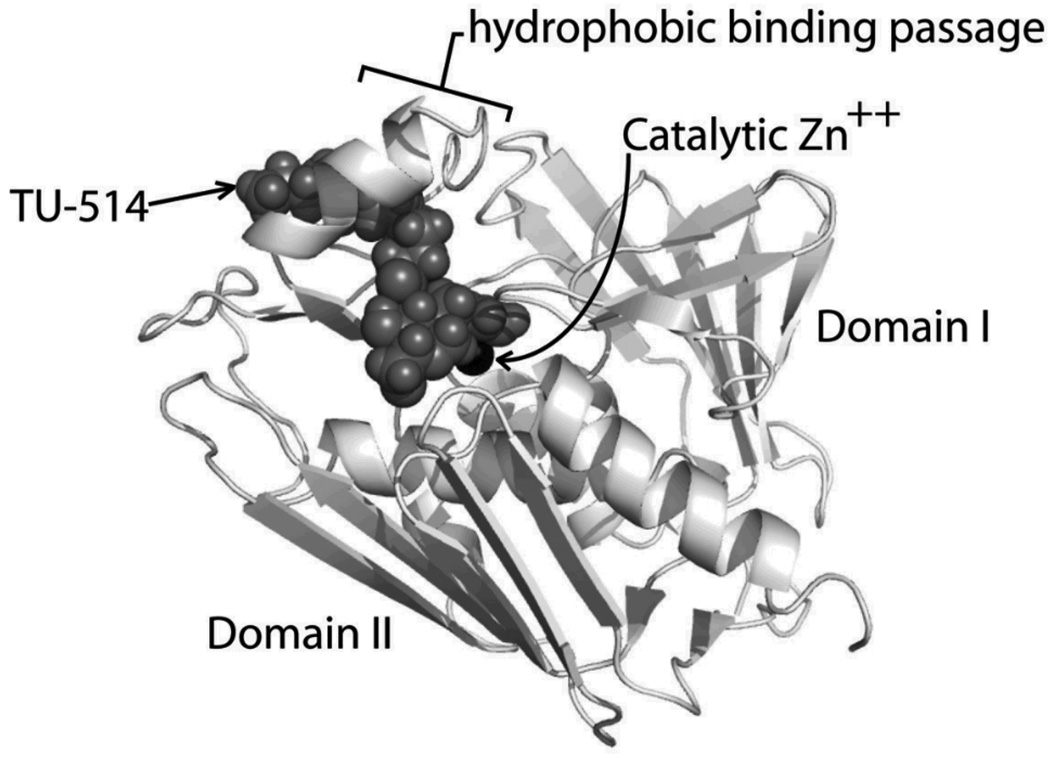
The studies of LpxC catalysis have been greatly facilitated by the availability of high-resolution structural information [21–24]. The structure of LpxC is characterized by a novel “β-α-α-β sandwich” fold where four mostly internal alpha helices are sandwiched between two beta sheets (Figure 2) [25, 26]. Two domains of the molecule have the same fold, each containing one five-stranded β-sheet and two α-helices. The β-sheet of Domain I is severely distorted while the sheet of Domain II is relatively flat. Each domain contains a unique insert, with the Domain I insert forming a small antiparallel β-sheet and the Domain II insert forms a hydrophobic binding passage that encapsulates the acyl chain of a substrate analog (TU-514, highlighted in Figure 2) [24]. It was proposed that this unusual substrate recognition mechanism explains the 20,000-fold greater affinity of LpxC for the substrate (UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc) over a compound lacking only the acyl chain (UDP-GlcNAc) [18].
Figure 2. The structure of LpxC.
The three-dimensional structure of LpxC is characterized by a unique “β-α-α-β sandwich” fold. This structure was solved with one bound molecule of the substrate analog, TU-514, that mimics the hexose ring and acyl chain of the substrate. The α-helix of the hydrophobic binding passage encapsulates the acyl chain of TU-514 (pdb-1XXE; this figure was generated using Pymol).
Supporting previous biochemical evidence, the NMR and crystal structures demonstrated that the three non-water zinc ligands were two histidines and one aspartate. This motif is characterized by the sequence HKΦΦD, where Φ indicates a hydrophobic residue necessary for proper helix packing [26]. This motif is distinct, and differs from previously described zinc metalloamidases that coordinate zinc with HEXXH, HXXE or HXXEH. The conserved lysine residue has been implicated in UDP binding and will be discussed later.
Interestingly, AaLpxC crystal structures containing TU-514 or the C16 fatty acid palmitate reveal the possibility of unusual zinc coordination geometry for LpxC [21]. While three crystal structures (zinc-inhibited, imidazole- and cacodylate-bound) and EXAFS evidence were consistent with tetracoordinate zinc, two complexes (TU-514- and palmitate-bound) are suggestive of pentacoordinate zinc. Interestingly, an AaLpxC crystal structure containing imidazole shows that 50% of the AaLpxC molecules contained imidazole and tetracoordinate zinc, and 50% contained two bound water molecules and pentacoordinate zinc instead. It is possible that LpxC zinc coordination switches from tetrahedral geometry in the unliganded enzyme to pentacoordinate (square pyramidal) in the TU-514 bound form. Alternatively, the unliganded enzyme may contain a pentacoordinate zinc atom, with two zinc-bound water molecules and three amino acid side chains. Certainly, careful studies will be necessary to resolve the geometry of zinc coordination by LpxC without substrate, during catalysis, and when bound by a hydroxamate-containing inhibitor. Structural biology using LpxC from different species will determine whether this zinc coordination inconsistency is unique to AaLpxC, or a true characteristic of the LpxC catalytic mechanism and structure.
In addition to the zinc coordination motif and the acyl-binding loop, two regions of the active site were identified that may function in substrate binding and catalysis. A basic patch, characterized by K143, K239, K262 and the ionized form of H265 (AaLpxC R137, K227, R250, H253), is adjacent to the catalytic zinc and opposite the hydrophobic passage described in the previous paragraph. This region is well suited to interact with the negative charges on the diphosphate moiety of the substrate. Additionally, a hydrophobic patch formed by F161, F192, and F194 (F155, F180 and F182 of AaLpxC) resides between the basic patch and the hydrophobic passage. In addition to the passage and the basic patch, the hydrophobic patch will be an important region of the protein to target when designing novel LpxC inhibitors.
Very recently, AaLpxC crystal structures containing UDP revealed the nucleotide-binding site [27, 28]. The HKΦΦD lysine 239 (K227 of AaLpxC) stabilized the negatively charged diphosphate, and the uracil and α-phosphate moieties interacted with other conserved “basic patch” residues described earlier [27]. However, biochemical evidence suggests that nucleotide binding by itself is weak, with an equilibrium dissociation constant in the millimolar range. Supporting this idea, UDP-GlcNAc (the LpxA substrate that lacks the acyl chain) is a poor LpxC substrate, with a KM >20,000-fold greater than the native substrate. Furthermore, NMR titration of AaLpxC with millimolar UDP and UMP failed to find any perturbed resonances [24]. Therefore, in order to bind substrate with high specificity, the majority of the binding energy must come from the interaction of the acyl chain and protein [18]. However, there has been no report of the UDP moiety binding energy contribution to KM; specifically interesting is a KM comparison of acyl-GlcNAc to UDP-acyl-GlcNAc. Another interesting comparison would be a measurement of the affinity of UDP for diverse LpxC proteins, as it is possible that other LpxC orthologs bind UDP more tightly than AaLpxC.
LpxC exhibits dynamic behavior in solution and the acyl chain-binding passage is likely disordered in the unliganded enzyme. Coggins and coworkers reported that portions of unliganded AaLpxC (containing only zinc) were unstructured and incorporating TU-514 ordered the complex [26]. Consistent with this report, X-ray diffraction data from AaLpxC crystals contain electron density consistent with myristate occupying the acyl chain-binding passage, even though no ligand was added to the protein solution [25]. It is probable that ordering of the substrate-binding loop was essential for crystallization. Consideration of the dynamic behavior of LpxC will be imperative to understand the structure / function relationship of LpxC catalysis. Future LpxC structural investigations should reveal the unliganded LpxC conformation (bound only to zinc), and may identify dynamic regions of the molecule. The conformation and dynamics of these integral motifs in solution may be studied using solution NMR techniques.
Enzymology of LpxC
The three-dimensional structural information of AaLpxC spurred investigation of the catalytic mechanism. The LpxC pH rate-dependency shows a bell-shaped curve similar to other metalloamidases, indicative of one catalytic base with a pK1 fitted to 6.1 and one catalytic acid with a pK2 fitted to 8.0 [22]. Based on genetic studies, two groups published mechanisms that are similar in nature, but disagree over the function of a conserved histidine and threonine in the active site [21–23]. Both models propose that LpxC functions by a general acid-base mechanism (Figure 3), with glutamate 78 (E73 of AaLpxC) as the catalytic general base. McClerren and coworkers reported that the E73A mutant of AaLpxC lacked the acidic limb of the pH-rate profile. However, the active group responsible for the basic limb of the pH profile has not been definitively identified.
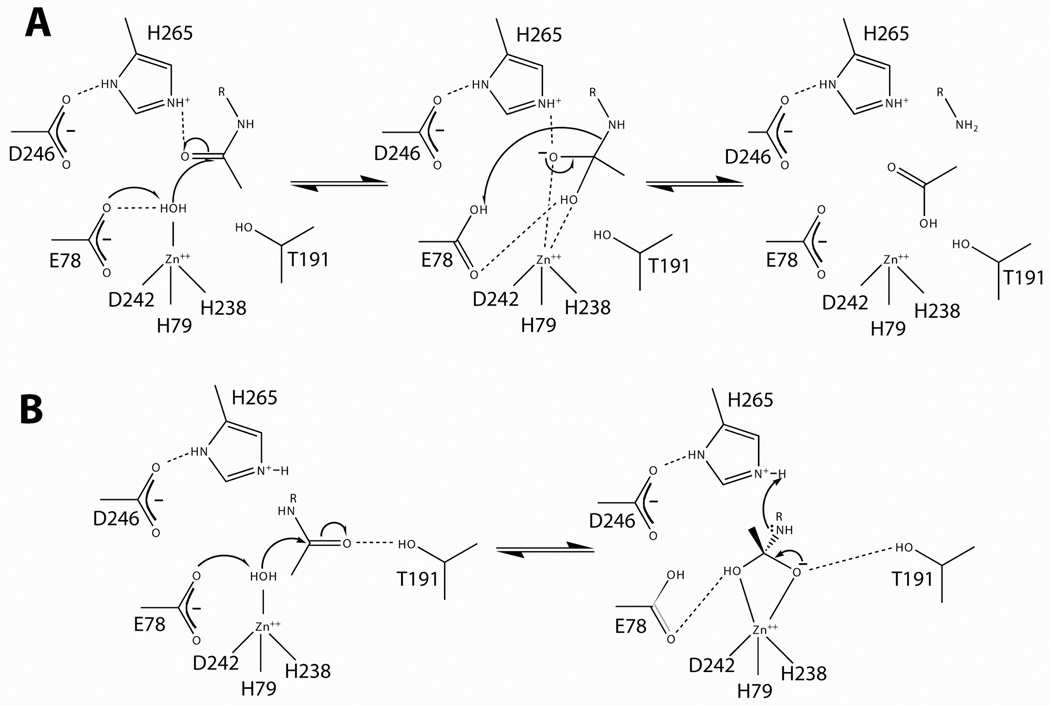
Figure 3. Proposed LpxC mechanisms.
Panel A. The LpxC catalytic mechanism proposed by McClerren and coworkers (2005). This proposal suggests that E78 abstracts a proton from zinc-bound water, thereby activating the water for attack on the carbonyl carbon. The proposal presented here suggests that H265 stabilizes the oxyanion intermediate, and E78 later donates a proton to the terminal amine. Panel B. An alternate hypothesis by Hernick and coworkers (2005) suggests that T191 stabilizes the oxyanion intermediate and H265 donates a proton to the liberated amine.
By the current mechanism, E78 abstracts a proton from zinc-bound water, thereby activating the water molecule for nucleophilic attack on the scissile amide linkage of the substrate (Figure 3A). This attack results in the formation of an oxyanion bound to a tetrahedral carbon. The terminal amine of the glucosamine moiety next abstracts the proton from E78, resulting in the collapse of the tetrahedral carbon intermediate, releasing acetate from UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc. Finally, both products are released and the protein catalyst is regenerated [22]. The mechanism of McClerren and coworkers suggests that H265 stabilizes the oxyanion intermediate, but neither donates nor abstracts a proton during the catalytic cycle. An NMR titration of H265 (H253 in AaLpxC) demonstrated a pKa of at least 8.5 for this residue [24]. These data suggest that H265 has an elevated pKa, consistent with its proposed role in stabilizing the oxyanion intermediate.
Consistent with this hypothesis, the AaLpxC H265A mutant protein had ~1000-fold reduced activity when compared to the wild-type enzyme. However, pH-rate profiles of H265A singly-mutated EcLpxC and AaLpxC proteins did not result in the loss of a basic limb, suggesting that H265 does not contribute to pK2 [22].
The proposed mechanism of Hernick and coworkers deviates by suggesting that H265 (H253 of AaLpxC) donates a proton to the amine of the leaving group, rather than E78 in the mechanism described above (Figure 3B) [23]. Additionally, this hypothesis suggests that T191 (T179 of AaLpxC), rather than H265, stabilizes the oxyanion intermediate through a hydrogen bond. The effect of mutating T191 has not been reported, and the catalytic role of this residue and H265 remains to be investigated.
Zinc-bound water ionization may explain the observed pK2, where zinc-bound water deprotonation leaves a catalytically inactive hydroxide ion. Consistent with this suggestion, adding an inhibitor that inactivates zinc-hydroxide dependent enzymes, NaF, failed to inhibit LpxC activity [22]. Zinc-bound water would be the active species necessary for both proposed mechanisms presented above, where E78 abstracts a proton from zinc-bound water to trigger the reaction.
Inhibition of lipid A biosynthesis through LpxC
Because LpxC catalyzes the committed reaction in the biosynthesis of an essential molecule, lipid A, it is a prime target for the development of novel antibiotics. All of the LpxC inhibitors described are competitive with the substrate, though there is no reason to exclude the future identification of an uncompetitive or noncompetitive inhibitor. Not all inhibitors, however, are effective antibiotics. In this section we will discuss the most important advances in the development of a potent LpxC inhibitor.
L-161,240
In the late 1980s, investigators at Merck Research Laboratories discovered the first group of potent lipid A biosynthesis inhibitors when screening E. coli cells for compounds that inhibited 14C-galactose uptake [17]. One compound (L-573,656) that inhibited LPS accumulation was a hydroxamic acid attached to a 2-phenyloxazaline ring. L-573,656 was assayed against all nine enzymes of lipid A biosynthesis and shown to specifically inhibit LpxC activity. Analogs of L-573,656 were synthesized and the most potent compound, L-161,240, was found to be a competitive inhibitor with a dissociation constant (Ki) of 50 nM for EcLpxC (Figure 4). This optimized compound was as effective as ampicillin and more effective than rifampicin or erythromycin in inhibiting E. coli growth, and killed 99.9% of the cells within four hours. Additionally, L-161–240 effectively protected mice against septicemia when challenged with a lethal dose of E. coli. Unfortunately, it was ineffective against Pseudomonas aeruginosa or Serratia marcescens. Using the L-161,240 insensitive LpxC activity from P. aeruginosa expressed from a low copy plasmid, Mdluli and coworkers demonstrated that LpxC is the primary target of L-161,240 in E. coli cells [29].
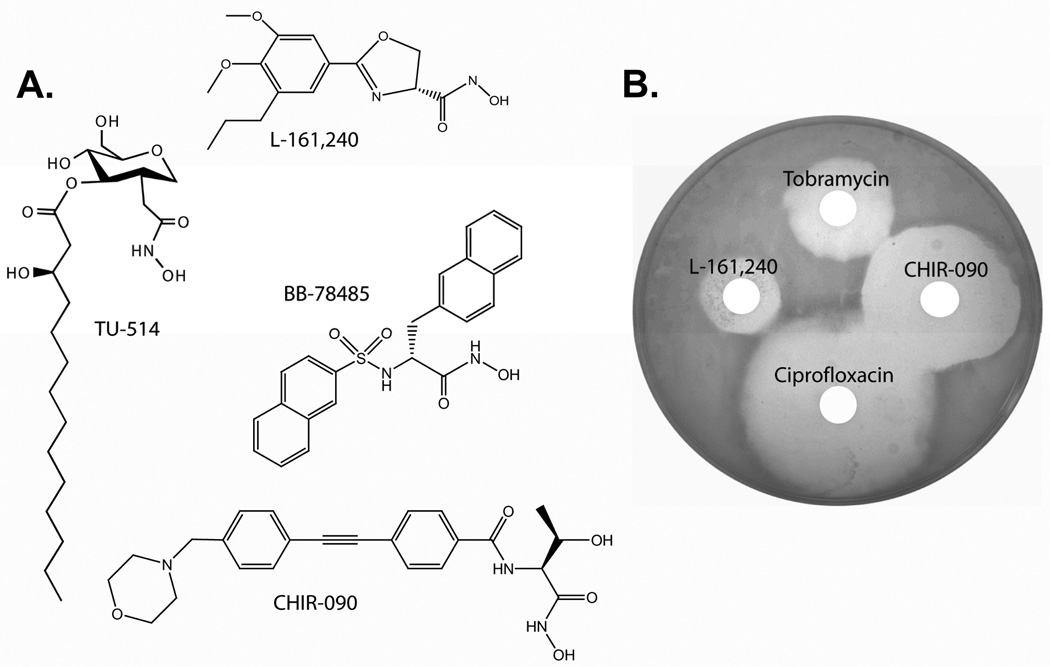
Figure 4. The chemical structures and antibiotic properties of LpxC inhibitors.
Panel A. LpxC inhibitors are structurally diverse, though all shown here contain a hydroxamate moiety. The E. coli LpxC Ki for each compound is shown. Each compound, except TU-514, is an effective antibiotic. Panel B. A disc-diffusion assay with 10 µg of each compound applied to a E. coli W3110 bacterial lawn demonstrates that L-161,240 and CHIR-090 are effective antibiotics. Drug-resistant colonies are visible in the zone of L-161,240 growth control distal to ciprofloxacin, suggesting that resistance to this compound is readily selected. There are no resistant colonies visible in the zone of growth control for any other compound tested here.
One concern with a hydroxamate-containing compound such as L-161,240 was that in the patient the hydroxamate could be reduced to hydroxylamine, a toxic breakdown product. As a result, phenyl-isoxazaline compounds containing thiols, phosphinates and heterocyclic amines were synthesized and assayed against LpxC [30]. Replacing the hydroxamate (CONHOH) with a phosphonate (PO3H2) improved the inhibition of the phenyl-isoxazaline scaffold but did not inhibit E. coli growth in a bacterial disc test.
Though a hydroxamate may give rise to a toxin, the potential benefit of a hydroxamate-based antibiotic may greatly outweigh a brief exposure to low levels of hydroxylamine, particularly because modern high-potency antibiotics are prescribed for acute infections and rarely chronically administered.
An additional concern with L-161,240 and its derivatives was that these compounds were narrow spectrum antibiotics, and did not control P. aeruginosa or Serratia marcescens. To address this, analogs of L-161,240 were synthesized and assayed against P. aeruginosa LpxC (PaLpxC) [31]. These compounds were varied by substitutions on the phenyl ring distal to the hydroxamate. A few of these compounds demonstrated moderate improvements in potency, but the in vivo activity of these compounds was not established. The most potent hits from this study had IC50 values near 100 nM, and contained groups at the 3 and 4 position of the phenyl ring and maintained the oxygen lone electron pair in the oxazoline ring.
TU-514
In an effort to develop broad-spectrum LpxC inhibitors, Jackman and coworkers designed a series of substrate analogs that mimic the hexose and acyl chain moieties, replacing the acetate with a hydroxamate [32]. Of this series, TU-514 inhibited the broadest range of LpxC activities and was one of the most potent inhibitors in the series, with a Ki of 1 nM at pH 7.4 and 650 nM at pH 5.5 for AaLpxC and 650 nM at pH 5.5 and 7.4 for EcLpxC [32, 33]. The structure of the AaLpxC/TU-514 complex provided insight into how the substrate was bound in the active site. While TU-514 inhibited most LpxC enzymes with similar potency, including PaLpxC, it had no apparent antibiotic activity. The long hydrophobic acyl chain may have prevented TU-514 from penetrating the membrane [32].
BB-78484 and BB-78485
A leap forward in the search for LpxC inhibitors came in 2002, when two novel hydroxamate-based, branched sulfonamide compounds were reported [34]. These compounds, BB-78484 and BB-78485 were unique in that they contained two hydrophobic moieties around a hydroxamate core (Figure 4). The dissociation constants of BB-78484 and BB-78485 were estimated to be 50 nM and 20 nM for EcLpxC, respectively, suggesting that BB-78485 was the most potent LpxC inhibitor at that time [35]. BB-78484 is a bactericidal inhibitor of E. coli growth with a minimal inhibitory concentration (MIC) of 2 µg / mL. Unlike the phenyl-isoxazaline compounds, BB-78484 and BB-78485 inhibited the growth of a broad range of Gram-negative bacteria, including Serratia, Burkholderia and Klebsiella. However, neither compound inhibited the growth of P. aeruginosa.
Colonies resistant to BB-78484 or L-161,240 are visible on plates in a disc-diffusion assay, suggesting that resistant colonies are present at a low frequency (1 in 107–109 bacteria). Some of these colonies found on a plate containing 16 µg / mL (8x MIC) of the bactericidal compound BB-78484 were isolated and the source of the resistance was investigated. Two open reading frames encoding LpxC and FabZ were probed for changes in the amino acid-coding DNA sequence. FabZ catalyzes the dehydration of R-3-hydroxymyristoyl-ACP and represents a branch point of acyl-ACP going to phospholipid or lipid A biosynthesis. In short, increased flux through FabZ favors phospholipid biosynthesis, whereas reduction of FabZ activity diverts more acyl-ACP to LPS biosynthesis. Most of the resistant colonies contained mutations that altered the amino acid sequence of FabZ, suggesting that reducing this activity perturbed the lipid A / phospholipids balance, overcoming the bactericidal effect of BB-78484-reduced LpxC activity. Interestingly, one colony that contained a wild-type copy of FabZ and a mutant LpxC protein (I38T) was isolated. The role of this LpxC mutation remains to be investigated.
CHIR-090
The novel N-aroyl-l-threonine hydroxamate CHIR-090 was disclosed by Andersen and coworkers in 2004 and characterized in 2005 by McClerren and coworkers as a potent inhibitor of AaLpxC activity [33, 36]. As the most effective LpxC-targeting antibiotic, CHIR-090 controlled the growth of both E. coli and P. aeruginosa with potency comparable to tobramycin and ciprofloxacin in a bacterial disc diffusion assay. Unlike any of the conventional LpxC inhibitors previously described, where the on- and off-rates of inhibitor binding and dissociation were essentially instantaneous, CHIR-090 is a slow, tight-binding inhibitor of AaLpxC. Additionally, CHIR-090 is a two-step, slow, tight-binding inhibitor with a Ki describing the reversible initial interaction of 1.0 nM followed by an apparently irreversible enzyme/inhibitor isomerization. This isomerization has a half-life of 56 seconds and was non-covalent according ESI-MS analysis. It was later shown that CHIR-090 inhibited EcLpxC in a similar but fully reversible manner, with a Ki of 5 nM and a Ki* describing the dissociation constant after isomerization of 0.5 nM (A. Barb, P. Zhou and C.R.H. Raetz, unpublished).
CHIR-090 also strongly inhibits P. aeruginosa, Neisseria meningitidis and Helicobacter pylori LpxC activity, but is a less effective inhibitor of Rhizhobium leguminosarum with a Ki of 0.34 µM (A. Barb, P. Zhou and C.R.H. Raetz, unpublished). CHIR-090 does not control the growth of R. leguminosarum or related bacteria in the Rhizobeaceae family. Using the CHIR-090-insensitive LpxC gene from R. leguminosarum, CHIR-090 resistance was engineered into E. coli W3110. An E. coli colony containing only the RlLpxC ORF replacing the wild type LpxC ORF on the bacterial chromosome grew on LB medium containing 10 µg / mL CHIR-090 (40x the E. coli MIC). This result suggests that LpxC is the primary target of CHIR-090 in vivo.
Future LpxC Studies
Treatment of bacterial infections that are recalcitrant to contemporary antibiotics will require drugs that exploit novel antibiotic targets [3, 37]. Currently, no antibiotics targeting LpxC or lipid A biosynthesis are clinically available. With the need for novel antibiotics steadily growing, the structure-based studies of LpxC / inhibitor complexes will assist in developing clinically useful compounds. Currently, the NMR and crystal structures of AaLpxC provide the basic fold of LpxC, but they do not account for minor structural deviations that are present in other LpxC orthologs and may be critical when designing novel inhibitors. Currently there is no precise structural model of LpxC from a pathogenic organism. Additionally, a structural characterization of the potent inhibitors (CHIR-090, BB-78485 and L-161,240) will be invaluable to define the molecular basis of the LpxC / inhibitor interaction and designing novel and potentially chimeric inhibitors. A structural description of both inhibition complexes (the rapidly forming EI complex and the slowly forming EI* complex) will be critical to reveal the molecular details of the CHIR-090 / LpxC interactions. A complete structural, kinetic and thermodynamic picture of enzyme:inhibitor complex isomerization may permit the logical design of a slow, tight-binding LpxC inhibitor as a potent antibiotic for treating Gram-negative bacterial infections.
Acknowledgements
We thank Dr. C.R.H. Raetz for a critical reading of the manuscript. This work is supported by the National Institutes of Health (grant AI055588 to P.Z.).
References
- 1.Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. N. Engl. J. Med. 2002;347:1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 2.Paterson DL. Am. J. Infect. Control. 2006;34:S20–S28. doi: 10.1016/j.ajic.2006.05.238. [DOI] [PubMed] [Google Scholar]
- 3.Projan SJ, Youngman PJ. Curr. Opin. Microbiol. 2002;5:463–465. doi: 10.1016/s1369-5274(02)00364-8. [DOI] [PubMed] [Google Scholar]
- 4.Raetz CRH. Escherichia coli and Salmonella : cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1035–1063. [Google Scholar]
- 5.Normark S, Boman HG, Matsson E. J. Bacteriol. 1969;97:1334–1342. doi: 10.1128/jb.97.3.1334-1342.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller SI, Ernst RK, Bader MW. Nat. Rev. Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 7.Aderem A, Ulevitch RJ. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 8.Anderson RN, Smith BL. National Vital Statistics Reports. 2005;53:1–90. [PubMed] [Google Scholar]
- 9.Anderson MS, Bull HG, Galloway SM, Kelly TM, Mohan S, Radika K, Raetz CRH. J. Biol. Chem. 1993;268:19858–19865. [PubMed] [Google Scholar]
- 10.Luo M, Lin H, Fischbach M, Liu D, Walsh C, Groves J. ACS Chemical Biology. 2006;1:29–32. doi: 10.1021/cb0500034. [DOI] [PubMed] [Google Scholar]
- 11.Sorensen PG, Lutkenhaus J, Young K, Eveland SS, Anderson MS, Raetz CRH. J. Biol. Chem. 1996;271:25898–25905. doi: 10.1074/jbc.271.42.25898. [DOI] [PubMed] [Google Scholar]
- 12.Ogura T, Inoue K, Tatsuta T, Suzaki T, Karata K, Young K, Su LH, Fierke CA, Jackman JE, Raetz CRH, Coleman J, Tomoyasu T, Matsuzawa H. Mol. Microbiol. 1999;31:833–844. doi: 10.1046/j.1365-2958.1999.01221.x. [DOI] [PubMed] [Google Scholar]
- 13.Fuhrer F, Langklotz S, Narberhaus F. Mol. Microbiol. 2006;59:1025–1036. doi: 10.1111/j.1365-2958.2005.04994.x. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan NF, Donachie WD. J. Bacteriol. 1984;160:724–732. doi: 10.1128/jb.160.2.724-732.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young K, Silver LL, Bramhill D, Cameron P, Eveland SS, Raetz CRH, Hyland SA, Anderson MS. J. Biol. Chem. 1995;270:30384–30391. doi: 10.1074/jbc.270.51.30384. [DOI] [PubMed] [Google Scholar]
- 16.Grundstrom T, Normark S, Magnusson KE. J. Bacteriol. 1980;144:884–890. doi: 10.1128/jb.144.3.884-890.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onishi HR, Pelak BA, Gerckens LS, Silver LL, Kahan FM, Chen MH, Patchett AA, Galloway SM, Hyland SA, Anderson MS, Raetz CRH. Science. 1996;274:980–982. doi: 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- 18.Jackman JE, Raetz CRH, Fierke CA. Biochemistry. 1999;38:1902–1911. doi: 10.1021/bi982339s. [DOI] [PubMed] [Google Scholar]
- 19.Jackman JE, Raetz CRH, Fierke CA. Biochemistry. 2001;40:514–523. doi: 10.1021/bi001872g. [DOI] [PubMed] [Google Scholar]
- 20.McClure CP, Rusche KM, Peariso K, Jackman JE, Fierke CA, Penner-Hahn JE. J. Inorg. Biochem. 2003;94:78–85. doi: 10.1016/s0162-0134(02)00611-6. [DOI] [PubMed] [Google Scholar]
- 21.Gennadios HA, Whittington DA, Li X, Fierke CA, Christianson DW. Biochemistry. 2006;45:7940–7948. doi: 10.1021/bi060823m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClerren AL, Zhou P, Guan Z, Raetz CRH, Rudolph J. Biochemistry. 2005;44:1106–1113. doi: 10.1021/bi048001h. [DOI] [PubMed] [Google Scholar]
- 23.Hernick M, Gennadios HA, Whittington DA, Rusche KM, Christianson DW, Fierke CA. J. Biol. Chem. 2005;280:16969–16978. doi: 10.1074/jbc.M413560200. [DOI] [PubMed] [Google Scholar]
- 24.Coggins BE, McClerren AL, Jiang L, Li X, Rudolph J, Hindsgaul O, Raetz CRH, Zhou P. Biochemistry. 2005;44:1114–1126. doi: 10.1021/bi047820z. [DOI] [PubMed] [Google Scholar]
- 25.Whittington DA, Rusche KM, Shin H, Fierke CA, Christianson DW. Proc. Natl. Acad. Sci. USA. 2003;100:8146–8150. doi: 10.1073/pnas.1432990100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coggins BE, Li X, McClerren AL, Hindsgaul O, Raetz CRH, Zhou P. Nat. Struct. Biol. 2003;10:645–651. doi: 10.1038/nsb948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gennadios HA, Christianson DW. Biochemistry. 2006 doi: 10.1021/bi0619021. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buetow L, Dawson A, Hunter WN. Acta Cryst. 2006;F62 doi: 10.1107/S1744309106041893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mdluli KE, Witte PR, Kline T, Barb AW, Erwin AL, Mansfield BE, McClerren AL, Pirrung MC, Tumey LN, Warrener P, Raetz CRH, Stover CK. Antimicrob. Agents. Chemother. 2006;50:2178–2184. doi: 10.1128/AAC.00140-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pirrung MC, Tumey LN, Raetz CRH, Jackman JE, Snehalatha K, McClerren AL, Fierke CA, Gantt SL, Rusche KM. J. Med. Chem. 2002;45:4359–4370. doi: 10.1021/jm020183v. [DOI] [PubMed] [Google Scholar]
- 31.Kline T, Andersen NH, Harwood EA, Bowman J, Malanda A, Endsley S, Erwin AL, Doyle M, Fong S, Harris AL, Mendelsohn B, Mdluli K, Raetz CRH, Stover CK, Witte PR, Yabannavar A, Zhu S. J. Med. Chem. 2002;45:3112–3129. doi: 10.1021/jm010579r. [DOI] [PubMed] [Google Scholar]
- 32.Jackman JE, Fierke CA, Tumey LN, Pirrung M, Uchiyama T, Tahir SH, Hindsgaul O, Raetz CRH. J. Biol. Chem. 2000;275:11002–11009. doi: 10.1074/jbc.275.15.11002. [DOI] [PubMed] [Google Scholar]
- 33.McClerren AL, Endsley S, Bowman JL, Andersen NH, Guan Z, Rudolph J, Raetz CRH. Biochemistry. 2005;44:16574–16583. doi: 10.1021/bi0518186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clements JM, Coignard F, Johnson I, Chandler S, Palan S, Waller A, Wijkmans J, Hunter MG. Antimicrob Agents Chemother. 2002;46:1793–1799. doi: 10.1128/AAC.46.6.1793-1799.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Y, Prusoff WH. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 36.Andersen NH, Bowman J, Erwin AL, Harwood EA, Kline T, Mdluli K, Pfister KB, Shawar R, Wagman A, Yabannavar A. World Intellectual Property Organization WO 2004/062601 A2. Emeryville CA: Chiron; 2004. p. 324. [Google Scholar]
- 37.Projan SJ. Curr. Opin. Pharmacol. 2002;2:513–522. doi: 10.1016/s1471-4892(02)00197-2. [DOI] [PubMed] [Google Scholar]