Abstract
Toll-like receptor (TLR) activation is important in immune responses and in differentiation of hematopoietic stem cells. We detected mRNA expression of TLRs 1, 2, 3, 5, and 6, but not TLRs 4, 7, 8, and 9 in murine (m)ESC line E14, and noted high cell surface protein expression of TLR2, but not TLR4, for mESC lines R1, CGR8, and E14. ESC lines were cultured in the presence of leukemia inhibitory factor (LIF). Pam3Cys enhanced proliferation and survival of the 3 ESC lines. In contrast, lipopolysaccharide (LPS) decreased proliferation and survival. Pam3Cys and LPS effects on proliferation and survival were blocked by antibody to TLR2, suggesting that effects of both Pam3Cys and LPS on these mESC lines were likely mediated through TLR2. E14 ESC line expressed MyD88. Pam3Cys stimulation of E14 ESCs was associated with induced NF-κB translocation, enhanced phosphorylation of IKK-α/β, and enhanced mRNA, but not protein, expression of tumor necrosis factor-α, interferon-γ, and IL-6. TLR2 activation by Pam3Cys or inhibition by LPS was not associated with changes in morphology or expression of alkaline phosphatase, Oct4, SSEA1, KLF4, or Sox2, markers of undifferentiated mESCs. Our studies identify TLR2 as present and functional in E14, R1, and CGR8 mESC lines.
Introduction
Mouse (m) embryonic stem cell (ESC) lines are derived from the inner cell mass of a blastocyst. Because ESCs have the capacity to differentiate into cells of 3 germ layers, they have potential for regenerative medicine [1,2]. In the presence of leukemia inhibitory factor (LIF), mESCs are maintained in an immature undifferentiated state [1–3]. Better understanding of factors that modulate/regulate ESC function may help in future efforts toward realizing the utility of ESCs for regenerative medicine. We recently demonstrated that mESC lines growing in the presence of LIF produce a number of biologically active cytokines and chemokines that are active on hematopoietic progenitor and other cell types [4]. Moreover, the mESCs have receptors for and produce and respond to some of these factors, such as stromal cell derived factor-1 (SDF-1/CXCL12), the ligand for CXCR4 [5].
Toll-like receptors (TLRs) are important for innate immune system recognition of pathogen-associated molecular patterns (PAMPs). This initiates a primary response toward fighting pathogens, and in recruitment of adaptive immune responses [6–16]. Active TLRs are expressed on mesenchymal stromal/stem cells [17], and on immature subsets of hematopoietic stem cells and progenitor cells [18]. We hypothesized that mESC lines would express TLRs and that some were functional. In this present report, we demonstrate that murine (m)ESCs express certain TLRs, and demonstrate that on the E14 mESC line, TLR2 serves as a functional receptor on LIF-maintained immature cells, which can be activated by Pam3Cys, a TLR2 ligand, to enhance proliferation, survival, NF-κB translocation, phosphorylation of IKK-α/β, and mRNA expression for selected cytokines, without inducing differentiation. Although we did not detect mRNA or cell surface TLR4, lipopolysaccharide (LPS) had suppressive activity on proliferation and induced apoptosis of mESCs.
Materials and Methods
Cell culture
Wild-type ESC lines E14, R1, and CGR8 were cultured on gelatinized plates in Dulbecco's modified Eagle's medium (DMEM) with 15% ESC qualified fetal bovine serum (Gibco-BRL, Grand Island, NY), 5.5 × 10−2 mM β-mercaptoethanol (Gibco-BRL), and 103 U/mL of LIF (Chemicon, Temecula, CA). Raw 264.7, a mouse macrophage cell line, was purchased from ATCC (Manassas, VA) and cultured in DMEM (Gibco).
Primers
RT-PCR primers were designed and optimized as previously reported [19]. Primers were purchased from Invitrogen (Carlsbad, CA).
RNA Extraction
The 5 × 105 E14 mESCs were seeded in 60-mm culture dishes and grown to confluency. Total cellular RNA was extracted using the Qiagen RNeasy Kit™ according to manufacturer's instructions (Qiagen Inc., Valencia, CA). RNA was stored in RNAse-free water at −80°C.
DNase Treatment
RNA samples were DNase-treated using Qiagen DNase free™ according to manufacturer's instructions (Qiagen Inc., Valencia, CA).
Reverse transcriptase-polymerase chain reaction
Expression of TLRs 1–9 and GAPDH was measured using a semiquantitative RT-PCR one-step AccessQuick™ RT-PCR system (Promega, Madison, WI). The oligonucleotide primers used for TLRs 1–9 and GAPDH have been reported [19]. Total RNA was isolated from the E14 mESC line and Raw 264.7 cells using RNeasy minicolumns (Qiagen, Valencia, CA). All RNA samples were treated with RNase-free DNase I (Qiagen) to remove genomic–DNA contamination and were quantified by spectrophotometric analysis. RNA integrity was confirmed by agarose gel electrophoresis. Using 1 μg of total RNA as the template for each reaction, RT-PCR was accomplished by using a polymerase kit (Access RT-PCR; Promega, Madison, WI). Cycling conditions were as follows: 1 min and 30 s of initial denaturation at 95°C, followed by 8 cycles of 30 s at 95°C, 15 s at 60°C, and 30 s at 72°C. After the initial 8 cycles, the 30-s 72°C extension cycle was increased 3 s per cycle for 25 cycles. During the 40th cycle, the 72°C extension was 3 min to complete the RT-PCR. Reactions were also amplified in the absence of reverse transcriptase as negative controls. PCR products were electrophoresed on 1.5% agarose gels. Each DNA band was visualized by staining with ethidium bromide. Experiments were done in triplicate.
IL-6, TNF-α, and IFN-γ mRNA and protein expression
Total RNA was isolated by TRIZOL preparation followed by phenol chloroform/isoamyl alcohol extraction and ethanol precipitation. Changes in IL-6, TNF-α, and IFN-γ RNA levels in mESCs were analyzed by quantitative real-time PCR. Relative changes in IL-6, TNF-α, and IFN-γ were determined using the 2−ΔΔCt method. Data are expressed as fold change. To test our qRT-PCR primers, we analyzed cytokine expression in mouse macrophage cells stimulated with TLRs 2 and 4 ligands. Accuracy of these primers was validated when we observed large levels of cytokine expression in mouse macrophage cDNA, which has been well characterized.
In order to measure cytokine expression by enzyme-linked immunosorbent assay (ELISA), 5 × 105 ESCs were seeded in 24-well plates. Twenty-four hours later, the media was replaced with DMEM with or without the ligands for TLR2 or TLR4, respectively. IL-6, TNF-α, and IFN-β (Ready-SET-Go! ELISA kit; eBioscience San Diego, CA) were determined by ELISA according to manufacturer's instruction. Standard curves were established using mouse recombinant IL-6, TNF-α, and IFN-β, respectively. The assay detection limit was 4 pg/mL.
Primary antibodies and TLR ligands
Primary antibodies were: isotype control PE rat IgG2a (eBioscience 17-4331), isotype control APC rat IgG2b (eBioscience 12-4321), TLR2/CD282 anti-mouse clone 6C2 (eBioscience 17-9021), and TLR4/MD2 anti-mouse (eBioscience 12-9924), p-IKK-α/β (Cell Signaling, Denver, MA; 2697S), myeloid-derived factor 88 (MyD88) (Abcam, Cambridge, MA; ab 2068), and total IKK-α/β (Santa Cruz, Santa Monica, CA; sc7607), anti-NF-κB p65 (Upstate Cell Signaling Solutions, Temecula, CA; 0701049995), PARP (Cell Signaling; 9542), and ERK1/2 (Cell Signaling; 9102). TLR ligands included: TLR2 agonist, Pam3Cys, and TLR4 agonist, bacterial LPS from Salmonella typhosa, obtained from Sigma-Aldrich, St. Louis, MO.
Flow cytometric analysis for TLR2 and TLR4
An aliquot of 1 × 106 cells was washed in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) (PBS–1% BSA) 3 times. Next, 100 μL of staining buffer (PBS containing 1% BSA and 0.5% ethylenediaminetetraacetic acid [EDTA]) was added to the cell pellet along with 5 μL of either TLR4-PE or TLR2-APC antibody for 1 h in the dark at 4°C. Cells were washed 3 times with wash buffer and 300 μL of wash buffer was added to cells, and cells were analyzed by flow cytometry. IgG2a was used as an isotype control for TLR4-PE and IgG2b was used as an isotype control for TLR2-APC.
Flow cytometric analysis for Oct4, SSEA1, Sox2, and KLF4
Wild-type ESC lines E14, R1, and CGR8 were cultured with and without TLRs 2 and 4 agonists in the presence of LIF. Cells were collected after days 1, 2, 3, 4, and 5 of proliferation assay and after 30 min, 1 h, and 4 h of TLR agonist treatment. An aliquot of 1 × 106 cells was washed in PBS containing 1% BSA (PBS–1% BSA) and incubated with anti-mouse CD16/CD32 receptor monoclonal antibody at 1 μg/100 μL (Pharmagen, San Diego, CA) to block nonspecific binding of immunoglobulins to mouse Fc-III/II receptors and cells used for SSEA1 antibody staining. Cells analyzed for SSEA1 were incubated with a 1:20 dilution of monoclonal anti-SSEA1 (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at 4°C. Cells were then washed and incubated with a 1:100 dilution of FITC:goat anti-mouse IgM antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and analyzed. Remaining cells were fixed with Cytoperm/Cytofix (BD Biosciences, San Jose, CA) and stained with a 1:100 dilution rabbit–mouse Oct3/4 polyclonal antibody (Chemicon, Temecula, CA) and KLF4 and Sox2 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at 4°C in the dark. Cells were washed 3 times with 1 mL of 1× Perm/Wash Buffer, followed by staining with 1:100 dilution of FITC:goat anti-mouse IgG antibody (Santa Cruz Biotechnology). Finally, cells were washed 3 times with 1× Perm/Wash Buffer and resuspended in 300 μL of 1× Perm/Wash Buffer for FACScan analysis (Becton Dickinson, Sunnyvale, CA).
Evaluation of cell proliferation
To determine proliferation rate, cells were detached using Trypsin–EDTA (Gibco, Grand Island, NY). mESCs were plated 100,000 cells per plate in a 60-cm2 gelatin-treated dish, and varying concentrations of test material were added the day after plating. After 24, 48, 72, and 96 h, cells were trypsinized and stained with trypan blue (Cellgro, CA). Total viable cell numbers were counted by hemocytometer.
TLR-dependent proliferation in vitro
One hundred thousand mouse embryonic stem cells were used for proliferation assay. Fifty micrograms per milliliter of the T2.5 antibody (a purified blocking antibody against mouse TLR2 (CD282); Biolegend, San Diego, CA; Catalog # 121802) were applied for 1 h prior to challenge with control medium, 10 μg/mL of Pam3Cys, or 10 μg/mL of LPS from S. typhosa (Sigma, St. Louis, MO). After 24, 48, and 72 h culture of cells with the TLR ligands, mESCs were counted each day to determine total cell numbers.
Survival/Apoptosis assays for ESCs
ESC growth depends on serum. After withdrawal of serum from plates, 95% of ESCs die within 96 h [4,5]. Reagents were added at the beginning of the experiments, and ESC cultures were initiated without serum in 1% methylcellulose-based DMEM with 5.5 × 10−2 mM 2-ME and 103 U/mL LIF (Chemicon, Temecula, CA) at 2,000 cells/mL. Serum was added at 24, 48, or 96 h to each group and colonies scored 7 days later. After 7 days, ESCs were collected and the undifferentiated status of the cells checked by staining of the cells with anti-mouse Oct4, Sox2, KLF, and SSEA1 antibody.
To analyze mESCs undergoing apoptosis, cell cultures were subjected to serum withdrawal in the presence of LIF. Reagents were added at the beginning of cultures as followed: control medium, TLR2 ligand, Pam3Cys (10 μg/mL), TLR4 ligand, LPS (10 μg/mL), and SDF-1/CXL12 (200 ng/mL). Cells were collected at days 1, 2, 3, and 4 after serum withdrawal and were stained with Annexin V (BD Biosciences, San Jose, CA). After withdrawal of serum for 4 days, mESCs were stained with undifferentiated markers Oct4, KLF4, SSEA1, and Sox2, respectively, to determine if the cells remained undifferentiated, at least as assessed by these markers.
Western blot analysis
Cells were plated in 60-mm culture dishes and grown to 80%–90% confluency at 37°C before being treated. mESCs were washed once with PBS, and protein was isolated using NucBuster Protein Extraction Kit (Novagen, Madison, WI) or Mammalian protein extraction reagent (Pierce, Rockford, IL) containing 1% protease inhibitor (Calbiochem; LaJolla, CA) and 1% phosphatase inhibitor cocktail set II (Calbiochem; 524625). Lysates were clarified by centrifugation at 14,000 rpm at 4°C for 10 min. Protein concentrations were measured by DC Protein Assay Kit (BioRad Hercules, CA). Lysates (20 μg) were separated on a 4%–12% SDS–polyacrylamide gel and transferred to PVDF membranes (Amersham Bioscience Piscataway, NJ). Membranes were blocked with 5% nonfat dried milk in 1× Tris-buffered saline (TBS) containing 0.2% Tween-20 (TBST) at room temperature for 1 h, and blots were incubated overnight with primary antibodies. Blots were then washed in TBST and incubated with species-specific IgG conjugated to horseradish peroxidase (1:2,000; Cell Signaling Beverly, MA; cs 7074) for 1 h at room temperature. Antigen–antibody complexes were visualized after exposure to X-ray film by enhanced chemiluminescence for 5 min at room temperature (GE Healthcare, Piscataway, NJ).
Statistical analysis
Significant differences were determined by t-test comparisons for at least 3 experiments each, with triplicates performed for each experiment, unless otherwise noted.
Results
Expression of TLRs in mESC lines
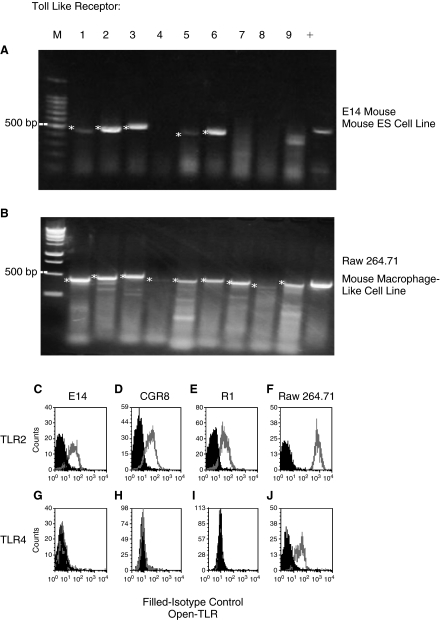
Examination of the E14 mESC line by RT-PCR demonstrated mRNA expression of TLRs 1, 2, 3, 5, and 6, but not TLRs 4, 7, 8, and 9 (Fig. 1A). As a positive control, we evaluated the mouse macrophage-like cell line, Raw 264.71 for expression of TLR mRNA. As shown in Figure 1B, Raw 264.71 cells expressed mRNAs for TLRs 1–9. Based on these results, cell surface expression of TLR2 protein and as a control, TLR4 protein were assessed in 3 different mESC lines, E14, CGR8, and R1 using specific TLR2 or TLR4 antibodies and flow cytometry. Protein expression of TLR2 (Fig. 1C–1E), was detected and the percentage of TLR2-expressing cells averaged 64 ± 8, 68 ± 7, and 48 ± 24 (mean ± 1 SD for 3 separate experiments each) for the E14, CGR8, and R1 mESC lines. In contrast, we did not detect expression of TLR4 protein (Fig. 1G–1I). We detected surface protein expression of both TLRs 2 and 4 on Raw 264.71 (Fig. 1F, 1J). Because TLR2 was expressed on these mESC lines, we mainly focused our efforts to determine whether the TLR2 ligand, Pam3Cys, had functional effects on proliferation and survival of the E14 mESC line. We also evaluated possible effects of LPS.
FIG. 1.
Toll-like receptor (TLR) mRNA (A, B) and protein (C–E and G–I) expression in mouse embryonic stem cell (mESC) line E14 and macrophage cell line, 264.71 (F, J). For A and B: * denotes positive bands; –, RT control samples without reverse transcriptase were negative control for all primers (data not shown); +, GAPDH expression was used as a positive control. Results are for 1 of at least 3 reproducible experiments each.
Effects of Pam3Cys, a ligand for TLR2, on mouse E14 ESC line proliferation
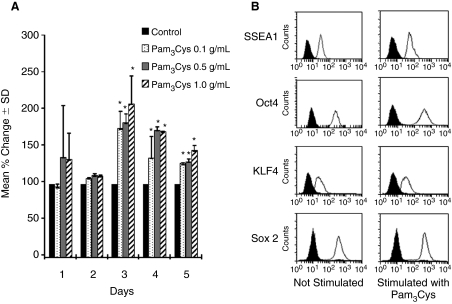
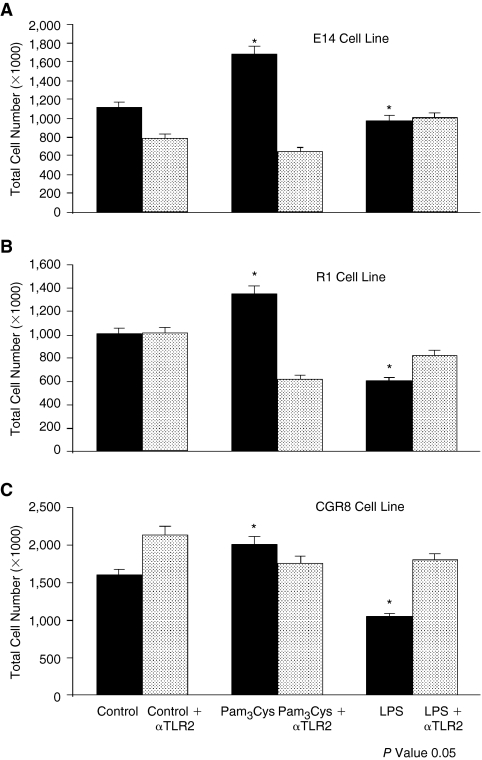
To determine whether TLR2 ligand influenced proliferation of mESC, mESCs were stimulated with 0.1, 0.5, and 1.0 μg/mL Pam3Cys, in the presence of LIF. Cells stimulated with Pam3Cys manifested a significant enhancement in total cell numbers starting at day 3 compared with those treated with control medium (Fig. 2A). Blocking/neutralizing antibodies against TLR2 (T2.5) counteracted the enhanced proliferation induced by Pam3Cys for all 3 cell lines (Fig. 3A–3C), demonstrating that the Pam3Cys effect was likely mediated through TLR2. Although we did not detect mRNA or protein expression of TLR4, we found that LPS had significant suppressive effects on numbers of E14, R1, and CGR8 mESC lines, an effect also counterbalanced or blocked by TLR2 antibody, suggesting that the LPS-suppressive effects may also be mediated through TLR2. We compared levels of SSEA1, Oct4, KLF4, and Sox2 mESC markers with and without TLR2 ligand stimulation for 3 days in order to determine if the Pam3Cys influenced protein expression of these immature ESC markers (Fig. 2B). The 1.0 μg/mL Pam3Cys showed no significant effect on expression of mESC markers SSEA1, Oct4, KLF4, and Sox2 as compared with mESCs cultured in the presence of control medium. Alkaline phosphatase was expressed and this expression was not changed by Pam3Cys (data not shown). There was also no change in expression of SSEA1, Oct4, KLF4, Sox2, or alkaline phosphatase staining in response to LPS (data not shown). Thus, treatment of cells with Pam3Cys or LPS, while having effects on proliferation, apparently did not influence differentiation of the cells in the presence of LIF at least at the level of expression of these immature cell markers.
FIG. 2.
Influence of Pam3Cys on proliferation of mouse embryonic stem cell (mESC) line E14. (A) Mean % control ± standard deviation (SD) of cell number compared with control for each day. Percent changes are based on control numbers (×103) in 3 separate experiments (3 replicates per experiment) of cells at Day 1 of: 60, 132, 203; at Day 2 of: 340, 510, 680; at Day 3 of: 1,132, 1,200, 1,064; at Day 4 of: 4,715, 1,885, 3,300; and at Day 5 of: 5,803, 8,960, 2,645. (B) Expression of stem cell markers by flow cytometry. Black is isotype control without and with stimulation of cells with Pam3Cys. This is representative of 3 experiments.
FIG. 3.
Antibody to toll-like receptor (TLR)2 blocks proliferation-inducing effects of Pam3Cys and suppressive effects of lipopolysaccharide (LPS) (Day 3) on: (A) E14, (B) R1, and (C) CGR8 mouse embryonic stem cells (mESC) lines. Results are shown for 3 experiments for A and 2 experiments each for B and C. mESCs were treated with neutralizing antibody to TLR2 for 1 h prior to stimulation with 10 μg/mL of Pam3Cys or LPS.
Influence of Pam3Cys on NF-κB nuclear translocation, downstream signaling, and cytokine production
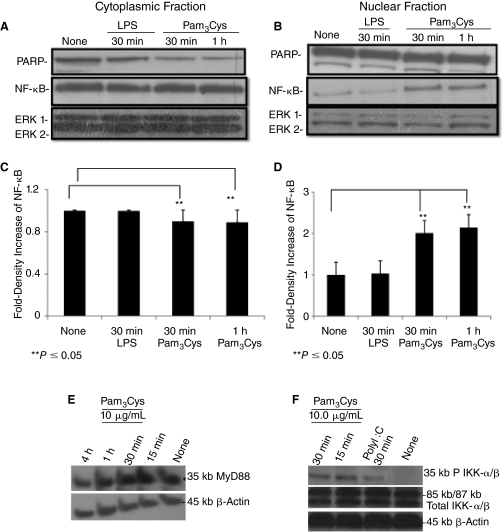
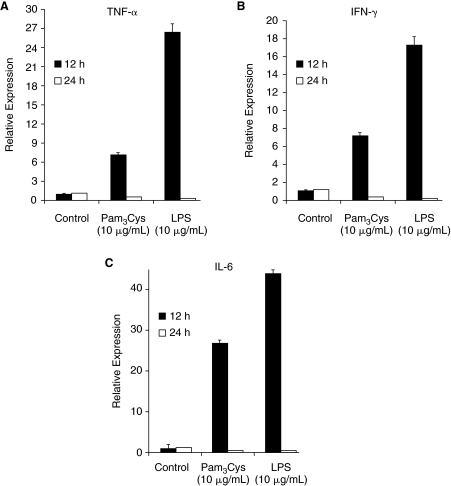
TLR ligand stimulation is known to induce translocation of NF-κB from the cytosol to the nucleus in a number of different cell types [17,20], resulting in NF-κB-dependent gene expression. We treated mESCs with Pam3Cys for 0, 30, and 60 min. As shown in Figure 4A–4D, Pam3Cys, but not LPS, induced translocation of NF-κB to the nucleus within 30 min. This indicates the functional status of TLR2 on mESCs at an intracellular level, perhaps by way of MyD88, which is expressed in these cells (Fig. 4E). Pam3Cys enhanced/induced IKK-α/β phosphorylation (Fig. 4F). We also tested poly I:C an activator of the internal TLR3. Interestingly, poly I:C also enhanced/induced phosphorylation of IKK-α/β (Fig. 4F). Expression of total ERK1/2 was not influenced by either Pam3Cys or LPS (Fig. 4A, 4B). We evaluated downstream effects of NF-κB signaling by assessing production of proinflammatory cytokines at an mRNA level by qRT-PCR and at a protein level by ELISA analysis in E14 mESCs. As shown in Figure 5A–5C, there is a significant quantitative increase in TNF-α, IFN-γ, and IL-6 mRNA expression when mESCs were treated for 12 h with Pam3Cys. The mRNA expression of TNF-α, IFN-γ, and IL-6 was no longer detectable by 24 h after cell exposure to Pam3Cys. However, we did not detect protein release into conditioned medium of either TNF-α, IFN-γ, or IL-6 18 h after Pam3Cys simulation (data not shown). LPS significantly, and to a greater extent than Pam3Cys, enhanced mRNA expression of TNF-α, IFN-γ, and IL-6 (Fig. 5), without enhancing release of the proteins for these cytokines (data not shown).
FIG. 4.
Pam3Cys induces NF-κB nuclear translocation and phosphorylation of IKK-α/β in mouse embryonic stem cell (mESC) line E14. Once cells reached confluence, 10 μg/mL Pam3Cys or 1 μg/mL of lipopolysaccharide (LPS) was added. mESCs were harvested 30 min or 1 h later. Cytoplasmic (A and C) and nuclear (B and D) fractions were quantified, run on SDS-PAGE gel and blotted with anti-NF-κB p65, PARP, or total ERK1/2 antibodies. Autoradiographs were quantified by densitometry (C and D). Expression of PARP was used as a measure of cytoplasmic contamination of nuclear material. (E) Expression of MyD88. (F) IKK-α/β phosphorylation at 15 and 30 min with Pam3Cys and at 30 min with poly I:C. Anti-total IKK-α/β is shown. Anti-β-actin was used as a loading control. Results are representative of at least 3 experiments each.
FIG. 5.
Toll-like receptor (TLR) ligands enhance mRNA levels of cytokines (A–C). qRT-PCR analysis of mRNA levels was performed after stimulation with Pam3Cys and lipopolysaccharide (LPS) (10 μg/mL each). Anti-β-actin antibody was used as a loading control. P ≤ 0.05 for Toll-like receptor (TLR) ligand stimulation at 12 h. Results shown are the average of 3 experiments.
Influence of Pam3Cys on survival of mESC colony-forming cells (CFCs)
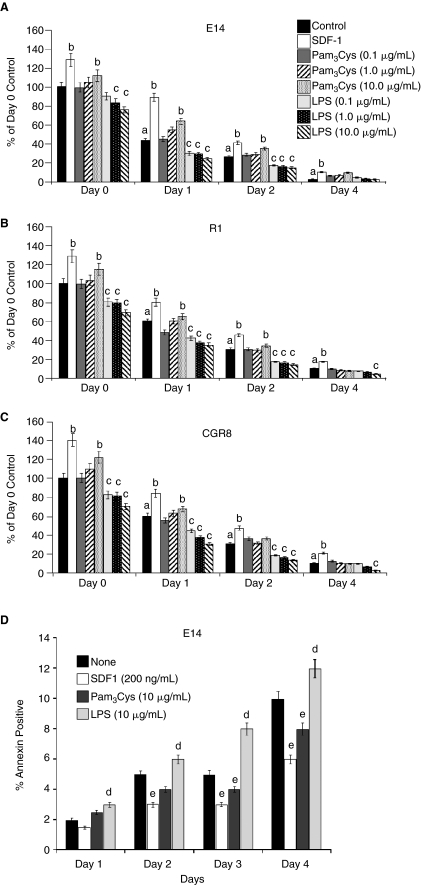
In order to determine if Pam3Cys or LPS had an effect on proliferation and survival of mESC line E14, R1, and CGR8 CFCs, we plated mESCs in semi-solid culture medium in the presence of LIF, and either SDF-1/CXCL12 or varying concentrations of Pam3Cys or LPS (Fig. 6A–6C). We used SDF-1/CXCL12 as a positive control, as we had previously reported that SDF-1/CXCL12 enhanced proliferation and survival of mESCs [5]. Survival of mESC was evaluated after delayed addition of serum [4,5]. Serum was added to cells at either time 0, or the addition of serum was delayed until days 1, 2, or 3 after the cells were placed in the incubator. As shown in Figure 6A–6C, SDF-1/CXCL12 enhanced numbers of mESC line colonies when serum was present at day 0, and also enhanced the survival of the mESC line CFC, similar to our previous report [5]. Pam3Cys, at a concentration of 10.0, but not 0.1 or 1.0 μg/mL, modestly but significantly enhanced numbers of mESC line CFC when serum was added at day 0, and LPS at concentrations as low as 0.1 to 1.0 μg/mL decreased mESC line CFC. Pam3Cys (10 μg/mL) enhanced and LPS at 0.1 to 10.0 μg/mL decreased survival of mESC line CFC after delayed addition of serum. Pam3Cys at 10 μg/mL decreased and LPS at 10 μg/mL increased apoptosis of cells in which serum addition was delayed until Days 2 to 4 (Fig. 6D). Thus, Pam3Cys enhanced proliferation and survival, and decreased apoptosis of mESC line E14 CFC, although effects of Pam3Cys were not as potent as that of SDF-1/CXCL12, and LPS had the opposite effect.
FIG. 6.
Influence of Pam3Cys and lipopolysaccharide (LPS) on proliferation (serum added on day 0) and on survival of mouse embryonic stem cell (mESC) colony-forming cells in the presence of leukemia inhibitory factor (LIF), but after cells were subjected to delayed addition of serum. (A) E14, (B) R1, and (C) CGR8 mESCs were cultured without serum, and serum was added on either day 0, 1, 2, or 4 after the start of culture. Colonies formed by mESCs were counted 7 days after addition of serum. (D) Apoptosis assay of E14 mESCs as assessed by percent Annexin V-positive cells. For A–C: a, significant decrease from day 0 control; b, significant increase from control of that day; c, significant decrease from control of that day. For D: d, significant increase for that day; e, significant decrease for that day. P ≤ 0.05 is considered significant for A–D. Results shown are for 3 experiments each for A and D, and for 2 experiments each for B and C, with 3 replicate plates scored for each experiment.
Discussion
TLRs, critical components of the innate and adaptive immune responses [6–16], are expressed on mesenchymal stromal/stem cells [17], hematopoietic stem cells, progenitor cells [18], and other cell types [20]. Very recently, others have shown that the D3 mESC line expresses selected TLRs [21]. While it is not entirely clear why mESC lines express TLRs, mESC lines have been shown to express certain cytokine receptors, as well as produce cytokines that are active on mESC lines, as well as on hematopoietic progenitor cells [4,5]. Since our original studies on mESC lines [4,5], we have been interested in a potential role for TLRs and their ligands on mESC line function. Our study addresses mRNA expression of TLRs in the E14, CGR8, and R1 mESCs, protein expression of TLRs 2 and 4, and the functional activities of Pam3Cys, a ligand for TLR2, and LPS on the E14 mESC line.
Using semiquantitative RT-PCR, we found that the E14 mESC line growing in the presence of LIF expresses mRNA for TLRs 1, 2, 3, 5, and 6, but not TLRs 4, 7, 8, and 9. This is a slightly different display of TLRs than that demonstrated for the D3 mESC line by others [21], where RT-PCR analysis showed mRNA for TLRs 2–6, but not for TLR1 or TLRs 7–9. This distinguishes different expression of mRNA for TLRs 1 and 4 between the D3 and our E14, R1, and CGR8 cell lines. Consistent with lack of mRNA expression of TLR4, we did not detect cell surface protein expression for TLR4 on either the E14, CGR8, and R1 mESC lines. Others did find TLR4 expression on the D3 mESC line [21]. These differences in TLR mRNA and protein expression may be due to the cell lines themselves, or perhaps to subtle differences in how the cell lines were maintained and grown. In this context, it has been demonstrated that expression of TLR4 varies in the D3 mESC line and is regulated by epigenetic modifications [22], and thus there can be differences in expression of TLR4 even between different D3 mESC lines.
Most importantly, we have defined an active role for TLR2 and its ligand, Pam3Cys, on the E14, R1, and CGR8 mESC lines. Pam3Cys enhanced proliferation and cell survival, and decreased apoptosis in LIF-cultured cells, without apparent changes in the immature phenotype of the cells as assessed by cell morphology, and expression of SSEA1, Oct4, KLF4, Sox2, and alkaline phosphatase. These stimulating effects were completely blocked by antibody to TLR2, suggesting that TLR2 mediated the Pam3Cys effects. Some of these enhancing effects may be, at least in part, mediated by translocation of NF-κB [23,24], and events downstream of NF-κB, including phosphorylation of IKK-α/β, as well as induced/enhanced expression of mRNA for TNF-α, IFN-γ, and IL-6, as assessed by real-time/quantitative PCR. These cytokines have many functional activities, including effects on the hematopoietic system [25], although we did not detect, within the limits of our ELISA assay, released TNF-α, IFN-γ, or IL-6 protein into mESC line containing culture medium in the presence or absence of Pam3Cys. If these 3 cytokines are having effects on proliferation and/or survival on the E14, R1, and CGR8 mESC lines, it would likely be through an autocrine-type interaction within the mESCs, unless cytokine levels below that which we can detect are active, or working in synergy with each other and/or other released cytokines [5].
In contrast to the D3 mESC line in which a large percentage of the cells expressed TLR4, and LPS modestly enhanced proliferation of these cells as detected by bromodeoxyuridine (BrdU) incorporation [21], our mESC lines that showed neither protein expression of TLR4 on the cell surface nor TLR mRNA responded to LPS with decreased cell proliferation, survival, and enhanced apoptosis. LPS while a ligand for TLR4, has also been reported to act through other TLRs [26–28]. That the suppressive effects of LPS on proliferation and survival of the 3 mESC lines were blocked by antibodies to TLR2 suggests that LPS effects on these cells may also be mediated through TLR2. In contrast to Pam3Cys effects, the LPS effects did not reflect nuclear translocation of NF-κB. Our results with poly I:C (Fig. 4F), an activator of TLR3, suggest that TLR3 is also functionally active in the E14 mESC line. Poly I:C enhanced/induced phosphorylation of IKK-α/β.
Murine ESC lines have been and continue to be useful models to study stem cell function and responsiveness to cytokines/ligands. Although the biological significance of functional TLRs in mESC lines is not yet known, further investigation of these cells should shed new information on the self-renewing, pluripotent state of ESCs that may translate into useful information for other stem cell types, and their modulation.
Acknowledgments
These studies were supported by the following US Public Health Service grants from the NIH to HEB: R01 HL56416 and R01 HL67384. TT was originally supported by NIH R25 GM067592 (“Bridges to the Doctorate” minority student program) and is currently supported by NIH R25 GM079657 (“Indiana University Initiative for Maximizing Student Diversity”) to HEB.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Doetschman TC. Eistetter H. Katz M. Schmidt W. Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 2.Odorico JS. Kaufman DS. Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 3.Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995;7:862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 4.Guo Y. Graham-Evans B. Broxmeyer HE. Murine embryonic stem cells secrete cytokines/growth modulators that enhance cell survival/anti-apoptosis and stimulate colony formation of murine hematopoietic progenitor cells. Stem Cells. 2006;24:850–856. doi: 10.1634/stemcells.2005-0457. [DOI] [PubMed] [Google Scholar]
- 5.Guo Y. Hangoc G. Bian H. Pelus LM. Broxmeyer HE. SDF-1/CXCL12 enhances survival and chemotaxis of murine embryonic stem cells and production of primitive and definitive hematopoietic progenitor cells. Stem Cells. 2005;23:1324–1332. doi: 10.1634/stemcells.2005-0085. [DOI] [PubMed] [Google Scholar]
- 6.Heil F. Hemmi H. Hochrein H. Ampenberger F. Kirschning C. Akira S. Lipford G. Wagner H. Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 7.Hoebe K. Du X. Georgel P. Janssen E. Tabeta K. Kim SO. Goode J. Lin P. Mann N. Mudd S. Crozat K. Sovath S. Han J. Beutler B. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 8.Honda K. Murao N. Ibuki T. Kamiya HO. Takano Y. The role of spinal muscarinic acetylcholine receptors in clonidine-induced anti-nociceptive effects in rats. Biol Pharm Bull. 2003;26:1178–1180. doi: 10.1248/bpb.26.1178. [DOI] [PubMed] [Google Scholar]
- 9.Krieg AM. Efler SM. Wittpoth M. Al Adhami MJ. Davis HL. Induction of systemic TH1-like innate immunity in normal volunteers following subcutaneous but not intravenous administration of CPG 7909, a synthetic B-class CpG oligodeoxynucleotide TLR9 agonist. J Immunother. 2004;27:460–471. doi: 10.1097/00002371-200411000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Lebon A. Adler P. Bernhard C. Boris AV. Pimenov AV. Maljuk A. Lin CT. Ulrich C. Keimer B. Magnetism, charge order, and giant magnetoresistance in SrFeO(3-delta) single crystals. Phys Rev Lett. 2004;92:037202. doi: 10.1103/PhysRevLett.92.037202. [DOI] [PubMed] [Google Scholar]
- 11.Lund J. Sato A. Akira S. Medzhitov R. Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meier A. Kirschning CJ. Nikolaus T. Wagner H. Heesemann J. Ebel F. Toll-like receptor (TLR) 2 and TLR4 are essential for Aspergillus-induced activation of murine macrophages. Cell Microbiol. 2003;5:561–570. doi: 10.1046/j.1462-5822.2003.00301.x. [DOI] [PubMed] [Google Scholar]
- 13.Diebold SS. Kaisho T. Hemmi H. Akira S. Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi O. Hoshino K. Kawai T. Sanjo H. Takada H. Ogawa T. Takeda K. Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 15.Underhill DM. Ozinsky A. Smith KD. Aderem A. Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc Natl Acad Sci USA. 1999;96:14459–14463. doi: 10.1073/pnas.96.25.14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Underhill DM. Ozinsky A. Hajjar AM. Stevens A. Wilson CB. Bassetti M. Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 17.Pevsner-Fischer M. Morad V. Cohen-Sfady M. Rousso-Noori L. Zanin-Zhorov A. Cohen S. Cohen IR. Zipori D. Toll-like receptors and their ligands control mesenchymal stem cell functions. Blood. 2007;109:1422–1432. doi: 10.1182/blood-2006-06-028704. [DOI] [PubMed] [Google Scholar]
- 18.Nagai Y. Garrett KP. Ohta S. Bahrun U. Kouro T. Akira S. Takatsu K. Kincade PW. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derbigny WA. Hong SC. Kerr MS. Temkit M. Johnson RM. Chlamydia muridarum infection elicits a beta interferon response in murine oviduct epithelial cells dependent on interferon regulatory factor 3 and TRIF. Infect Immun. 2007;75:1280–1290. doi: 10.1128/IAI.01525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolls A. Shechter R. London A. Ziv Y. Ronen A. Levy R. Schwartz M. Toll-like receptors modulate adult hippocampal neurogenesis. Nat Cell Biol. 2007;9:1081–1088. doi: 10.1038/ncb1629. [DOI] [PubMed] [Google Scholar]
- 21.Lee SH. Hong B. Sharabi A. Huang XF. Chen SY. Embryonic stem cells and mammary luminal progenitors directly sense and respond to microbial products. Stem Cells. 2009;27:1604–1615. doi: 10.1002/stem.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zampetaki A. Xiao Q. Zeng L. Hu Y. Xu Q. TLR4 expression in mouse embryonic stem cells and in stem cell-derived vascular cells is regulated by epigenetic modifications. Biochem Biophys Res Commun. 2006;347:89–99. doi: 10.1016/j.bbrc.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 23.Delhalle S. Blasius R. Dicato M. Diederich M. A beginner's guide to NF-kappaB signaling pathways. Ann N Y Acad Sci. 2004;1030:1–13. doi: 10.1196/annals.1329.002. [DOI] [PubMed] [Google Scholar]
- 24.Liang Y. Zhou Y. Shen P. NF-kappaB and its regulation on the immune system. Cell Mol Immunol. 2004;1:343–350. [PubMed] [Google Scholar]
- 25.Shaheen M. Broxmeyer HE. The humoral regulation of hematopoiesis. In: Hoffman R, editor; Benz EJ Jr, editor; Shattil SJ, editor; Furie B, editor; Silberstein LE, editor; McGlave P, editor; Heslop H, editor; Anastasi J, editor. Hematology: Basic Principles and Practice. 5th. Elsevier Churchill Livingston; Philadelphia, PA: 2009. pp. 253–275. Part III, Chapter 24. [Google Scholar]
- 26.Dziarski R. Wang Q. Miyake K. Kirschning CJ. Gupta D. MD-2 enables Toll-like receptor 2 (TLR2)-mediated responses to lipopolysaccharide and enhances TLR2-mediated responses to Gram-positive and Gram-negative bacteria and their cell wall components. J Immunol. 2001;166:1938–1944. doi: 10.4049/jimmunol.166.3.1938. [DOI] [PubMed] [Google Scholar]
- 27.Erridge C. Pridmore A. Eley A. Stewart J. Poxton IR. Lipopolysaccharides of Bacteroides fragilis, Chlamydia trachomatis and Pseudomonas aeruginosa signal via toll-like receptor 2. J Med Microbiol. 2004;53(Pt 8):735–740. doi: 10.1099/jmm.0.45598-0. [DOI] [PubMed] [Google Scholar]
- 28.Alhawi M. Stewart J. Erridge C. Patrick S. Poxton IR. Bacteroides fragilis signals through Toll-like receptor (TLR) 2 and not through TLR4. J Med Microbiol. 2009;58(Pt 8):1015–1022. doi: 10.1099/jmm.0.009936-0. [DOI] [PubMed] [Google Scholar]