Abstract
Organisms with the same genome can inherit information in addition to that encoded in the DNA sequence — this is known as epigenetic inheritance. Epigenetic inheritance is responsible for many of the phenotypic differences between different cell types in multicellular organisms. Work by many investigators over the past decades has suggested that a great deal of epigenetic information might be carried in the pattern of post-translational modifications of the histone proteins, although this is not as well established as many believe. For example, it is unclear whether and how the histones, which are displaced from the chromosome during passage of the replication fork and are often exchanged from the DNA template at other times, carry information from one cellular generation to the next. Here, we briefly review the evidence that some chromatin states are indeed heritable, and then focus on the mechanistic challenges that remain in order to understand how this inheritance can be achieved.
Introduction
Epigenetic inheritance
Because many definitions of the word ‘epigenetic’ can now be found in the literature, it is important to start this review by pointing out that we use the term in the Holliday [1] sense — traits that are mitotically heritable without a change in DNA sequence are called epigenetically heritable. Epigenetic inheritance is increasingly appreciated as a major contributor to processes from development to metabolism to oncogenesis. Whereas inheritance of DNA sequence is conceptually straightforward thanks to the elegance of the complementary base-pairing between paired DNA strands, the mechanistic basis underlying most epigenetic inheritance systems is less clear. Multiple epigenetic information carriers have been proposed including transcription factors, prions, cytosine methylation patterns, small RNAs, and chromatin structure [2–4]. Although some proposed information carriers, such as DNA methylation, are well established as heritable marks [1,5,6], the idea that post-translational histone modifications are the mechanistic basis for inheritance is more controversial. Here, we will explore the current information regarding the mechanism by which chromatin states might be inherited.
Chromatin as a carrier of epigenetic information
It is commonly believed that the packaging of eukaryotic genomes into chromatin provides a carrier of mitotically heritable information. This concept is not as well supported as many believe, as covered in detail in an excellent review from Ptashne [7••]. We will not reiterate those arguments, but agree with the central point that chromatin per se is seldom heritable. Instead we briefly review the evidence for cases in which chromatin might be the epigenetic information carrier. In general, it is worth noting that most epigenetic inheritance paradigms that implicate chromatin tend to involve heritable repression, rather than heritable activity, of downstream genes.
Two broad lines of evidence suggest that chromatin carries epigenetic information. First, stable epigenetically heritable cell types in multicellular organisms are frequently correlated with alternative chromatin states. For example, the β-globin gene is maintained in open chromatin in definitive erythroid cells, in closed chromatin in liver cells, and when each cell divides this packaging state is maintained; the converse is true for the albumin gene. These chromatin states are therefore epigenetically inherited (as are aspects of cell morphology and other defining features of cell type), but this does NOT necessarily mean that chromatin itself carries heritable information during cell division. The second major line of evidence for chromatin as an epigenetic information carrier is genetic — in Drosophila, Trithorax, and Polycomb class mutants that fail to epigenetically maintain cell identity encode proteins involved in histone modifying complexes [8,9]. Chromatin regulators are also genetically implicated in epigenetic inheritance systems in unicellular organisms — variegated repression of subtelomeric genes requires histone deacetylases in Saccharomyces cerevisiae and P. falciparum [10,11], and subtelomeric loci in the epigenetic OFF state are packaged into a distinctive chromatin structure that differs significantly from the packaging state of the epigenetic ON state [12,13].
These types of evidence have led to the widespread idea that chromatin itself carries information during genomic replication. However, a situation in which a DNA-linked mark like cytosine methylation carried information at replication, but required downstream histone deacetylation as an effector function, would produce both types of observations described above. For example, yeast that have recently experienced galactose activate GAL genes more rapidly in response to galactose than do naïve yeast [14•,15•]. This heritable ‘memory’ genetically requires several chromatin regulators, but elegant heterokaryon analysis reveals that memory is carried in the cytoplasm [16••]. Thus, in this system chromatin modifiers are genetically required for an epigenetic state (for unclear reasons), and chromatin structure differs in naïve and memory-induced cells, yet the information is transferable via the cytoplasm.
Beyond intellectual interest, the actual information carrier in epigenetic inheritance systems is of practical importance — there is widespread clinical interest in reversing pathological epigenetic states such as inappropriate silencing of tumor suppressors [17]. The appeal of such a therapeutic modality, beyond having yet another ‘druggable’ molecular target, is that in principle blocking a pathological epigenetic state need only be a transient chemotherapeutic regimen, thus allaying some of the drawbacks of long-term chemotherapy.
All this said, there are several scenarios where it seems chromatin has not been excluded as the actual epigenetic information carrier. First, in the case of S. pombe mating-type silencing, substantial evidence argues that chromatin state per se is heritable [18,19]. Most importantly, epigenetic transcriptional states at the S. pombe mating-type locus are heritable in cis during both mitosis and meiosis [20]. Histone modification-bound proteins such as heterochromatin protein 1 (HP1) homolog Swi6 are the likely information carriers (although absolutely ruling out an underlying DNA modification is still difficult).
The most stringent way to rule out an unidentified information carrier that re-establishes chromatin de novo every S phase is to recapitulate chromatin replication in vitro with purified components. Polycomb group proteins are involved in the maintenance of repression during development in many organisms [9]. In living cells, artificial targeting of the methyltransferase-containing PRC2 complex to a reporter construct has been used to explore inheritance of silencing [21]. Here, prior expression of GAL4-EED resulted in stable H3K27me3 and PRC1 binding and repression of the reporter gene, suggesting that, once recruited, the PRC1 complex was being retained at the reporter via a local positive feedback. Importantly, Francis et al. have recently shown using an in vitro SV40/human cell extract replication system that the Polycomb PRC1 complex can remain associated with DNA as it undergoes coordinated DNA replication and chromatin assembly [22••], even in the presence of replicating competitor templates. This key result proves that Polycomb association can be inherited during in vitro replication, thereby disfavoring a role for some cryptic underlying carrier.
Chromatin dynamics at the replication fork
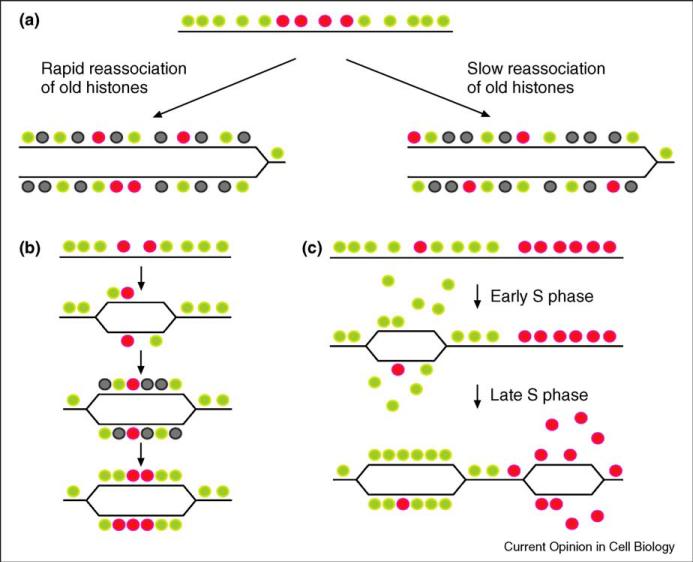
Inheritance of chromatin states is subject to several influences that do not affect standard genetic inheritance. First, during genomic replication the passage of the replication fork disrupts histone–DNA contacts, and old histones must reassociate with daughter chromosomes at a location close to their original location on the mother chromosome (Figure 1a). Otherwise, locus-specific epi-genetic information would be randomly shuffled every generation. Second, old histones only account for one half of the histones on each new genome, and the remaining histones are newly synthesized during each S phase. This implies some information passage from old to new histones (Figure 1b,c), as otherwise old chromatin states would rapidly be diluted by new histones.
Figure 1.
Processes likely to tune the precision of inheritance of chromatin states. For all figures, green and red circles represent histones in different modification states, while gray represents free nucleosomes drawn from the nucleoplasmic pool. (a) Dispersal of old nucleosomes determines the precision of chromatin inheritance. Examples show two distinct dispersal patterns, using red histones to indicate a potentially heritable modification. If histones displaced by the replication machinery rapidly reassociate with the chromosome following DNA synthesis, then short domains of a given histone modification state will be heritable (left), while slow reassociation will result in spreading and dilution of the old histones in a particular state (right). (b, c) Two models for feedback from old nucleosomes to new. In (b), new nucleosomes are modified by histone modifying enzymes recruited locally by old nucleosomes. In (c), green represents acetylated nucleosomes and red represents poorly acetylated nucleosomes. Regions of the genome associated with highly acetylated nucleosomes are replicated early in S phase, concordant with the acetylation of newly synthesized histones. Late in S phase, hypoacetylated chromatin is replicated, and new histones recruited to those regions are deacetylated. These models may be distinguished based on how small domains of histone modification behave during S phase.
To make an analogy between chromatin replication and DNA replication, the first problem, that of histone dissociation, is akin to having DNA nucleotides diffuse relative to one another after the two strands are separated. The second issue, that of communication between old and new nucleosomes, is analogous to asking what the equivalent of base pairing is when nucleosomes are the ‘bases.’ Going further in this vein, one might ask what the ‘mutation’ rate is for inherited chromatin states, which might well translate into ON → OFF phenotypic epimutation rates. We discuss these issues in greater detail below.
What happens to nucleosomes during genomic replication?
After DNA replication, newly synthesized DNA must be packaged into chromatin [23]. However, in addition to providing the template for assembly of new nucleosomes, the passage of the replication fork also disrupts previously existing nucleosomes, dissociating H3/H4 tetramers from H2A/H2B dimers [24], and disrupting histone–DNA contacts [25]. Nucleosomes undergo some level of disassembly as the replication fork passes, but the exact results in terms of histone composition and position are yet undefined. Of course, if nucleosomes completely dissociated and became freely soluble then epigenetic inheritance of chromatin would become conceptually difficult. Evidence that nucleosomes do not completely dissociate comes from in vitro studies using the SV40 replication system — here, two competition studies using unlabeled DNA indicated that nucleosomes were not transferred to competitor DNA [26,27], although transfer of histones in trans was later observed at an 5–10-fold excess of competitor [25].
Following passage of the replication fork, parental H3/H4 molecules reassociate with one of the two daughter strands in a seemingly random fashion [28–31], accounting for half of the required nucleosome density over the two new genomes. In vivo, psoralen crosslinking studies show that nucleosomes associate with daughter chromosomes rapidly after the passage of the replication fork, with nucleosome-depleted DNA confined to ~1 kb surrounding the replication fork [31–33]. Translational positions of nucleosomes are established rapidly after passage of the replication fork at the yeast rDNA locus [33], although whether rapid positioning occurs globally or only at strong nucleosome positioning sequences [34] is unknown. Interestingly, in the absence of new histones, old nucleosomes appear to associate with daughter chromosomes in stretches, suggesting cooperative binding of nucleosomes to DNA [31,35]. Despite the rapid reassociation of nucleosomes with daughter genomes, it is unknown whether old nucleosomes normally reassociate close to their original positions on the mother chromosome. If nucleosomes do remain associated with their previous location, then how histones are kept from diffusing away is unclear (Figure 1a) — candidate factors include the histone chaperone CAF-1 (see below), or the Mcm proteins at the replication fork [36].
An important question regarding recruitment-based models is the relevant unit of chromatin inheritance. H2A/H2B are replaced during G1 much more extensively than H3/H4 [24,37–39], and since H3/H4-modifying enzymes are implicated in most putative chromatin inheritance paradigms, we simply consider H3/H4 here — do H3/H4 tetramers stay intact during genomic replication, or do H3/H4 dimers split apart? The vast majority of studies argue against tetramer splitting (reviewed in [40•]). However, this debate has recently been reopened by the observation that nascent H3/H4 complexes are dimers [41], in part because the histone chaperone Asf1 prevents H3/H4 tetramer formation [42,43]. A major open question is whether nucleosome diversity is generated based on dimeric intermediates. Do newly synthesized H3/H4 dimers ever mix with old ones? Crosslinking studies [29,30] suggest that this is not a common pathway, but could those bulk studies have missed mixing at critical loci? And if new dimers are only combined with new, how is mixing prevented?
Information transfer from old histones to new histones
After replication, old and new nucleosomes will often be neighbors [30]. Multiple models exist for the transfer of chromatin information from old histones to newly synthesized histones, of which we consider two. First, parental nucleosomes (carrying a given modification) could recruit chromatin modulators to adjacent newly synthesized nucleosomes, recreating even very small (~2–4 nucleosome) domains of histone modification [40•,44,45]. Second, the ‘timing’ model (Figure 1c) rests on the observation that hyperacetylated chromatin is replicated early in S phase, while hypoacetylated chromatin is replicated late in S phase (see [46] for review). If hyperacetylated histones are loaded onto DNA replicated early in S phase and hypoacetylated histones are loaded late in S phase, then broad domains of acetylation could be replicated [47].
The idea that positive feedback in chromatin inheritance is provided by modifying enzymes that are recruited by the very modifications they create is a very popular one. In S. cerevisiae, subterlomeric silencing requires the Sir complex, which deacetylates H4K16 [45,48,49], and preferentially binds to deacetyl H4K16 [44,50–52]. In S. pombe, silencing of the heterochromatic loci requires the H3K9 methylase Clr4, and the HP1 homolog Swi6, which binds to H3K9me3 [18,19]. In flies and mammals, Polycomb group proteins play major roles in memory of repressive states, and mammalian PRC2 contains both a H3K27 methylase (Ezh2) and a H3K27me3-binding subunit (Eed) [53]. Consistent with the recruitment model, most well-characterized loci subject to epigenetic inheritance are associated with relatively long domains of histone modifications (see [54] for review). Inheritance of domains of many modified nucleosomes may provide cells with an error correction mechanism to decrease epimutation rates.
Despite the clear appeal of recruitment models, they are difficult to reconcile with some observations. For instance, the Eed component of the PRC2 H3K27 methylase complex binds not only to H3K27me3, but also to other ‘repressive’ histone marks such as H3K9me3 [53]. Thus, in the simplest model of recruitment, one would expect ‘crosstalk’ to result in cooccurrence of K9 and K27 methylation, yet this is not observed in mapping studies. Furthermore, not every positive feedback loop that propagates chromatin marks laterally results in heritable chromatin states. For instance, a K4 methylase complex subunit, WDR5, directly associates with H3K4me3, and is required for global H3K4me3 [55]. Despite this positive feedback, typical H3K4me3 domains are quite short in vivo,and H3K4me3 patterns are generally quite plastic, correlating with the presence of RNA polymerase [54]. Therefore, the combination of modifying enzymes and modification-binding proteins is a common paradigm for building chromatin structures, but not unique to truly inherited modules.
PCNA-linked interactions at replication forks may foster epigenetic inheritance
Assembly of nucleosomes onto newly replicated DNA is carried out by a number of proteins, including the chromatin assembly complex CAF-1 which is recruited to the replication fork by the sliding clamp protein PCNA [56]. CAF-1 is required for fully efficient epigenetic silencing at the yeast subtelomeric and mating loci [57–59] while mutations in PCNA that diminish its interaction with CAF-1 also reduce heterochromatic repression [60]. It is not certain whether the primary role of CAF-1 is in the replication of silent chromatin, in the stable maintenance of silenced chromatin structures during cell cycle progression, or both. Evidence exists for both roles: in budding yeast, CAF-1 is required for stable silencing of the mating loci even in G1-arrested cells, demonstrating that its contribution to heterochromatin stability is not limited to S phase [61].
One hypothesis for CAF-1s role in epigenetic stability during S phase may be that this chaperone prevents excessive spreading of old histones from scrambling locus-specific epigenetic information (Figure 1a). An alternative hypothesis is that CAF-1 is required for feedback from old to new histones, and CAF-1 has specific protein interactions consistent with this idea. For example, during S phase, human SetDB1 associates with CAF-1 and mono-methylates H3–K9 before histone deposition [62•]. After deposition, the monomethyl K9 is likely di-methylated and tri-methylated by Suv39 enzymes, creating binding sites for HP1. Notably, CAF-1 itself can recruit HP1 to pericentric heterochromatin [63,64], supporting the notion that CAF-1 contributes to a positive feedback network that favors maintenance of heterochromatic structures during replication. In another case of PCNA recruitment of positive feedback networks, the DNA methyltransferase DNMT1 is recruited to replication forks by binding to PCNA [65]. DNMT1 also interacts with G9a [66], a major H3K9-monomethyltransferase. This interaction results in monomethylation of H3K9 at sites of replication, with further methylation by Suv39H1 providing binding sites for HP1.
Timing model
A completely different idea for positive feedback from old to new histone states is a model based on replication timing. Here, the basic observation is that highly acetylated chromatin replicates early in S phase, and poorly acetylated chromatin replicates late in S phase. Elegant microinjection studies in tissue culture cells showed that reporter plasmids injected early during S phase were assembled into acetylated chromatin and were competent for transcription in the next cell cycle, whereas reporters injected late in S phase were assembled in repressive chromatin and were not transcribed [47,67]. Thus, regions designated for early replication timing could self-propagate if there was a limited pool of highly acetylated newly synthesized histones available each S phase. An implication of this model is that the unit of chromatin inheritance would be domains whose length would be set by the genomic distance replicated during each window of availability of a given histone modification.
Computational modeling of epigenetic inheritance of chromatin state
Computational modeling has proven a valuable tool for understanding key aspects of many signaling cascades, and for making predictions about key reactions in the pathway. Recently, systematic in silico modeling of chromatin inheritance has been carried out using the S. pombe mating locus as a specific model [68••]. In this model, a region of 60 nucleosomes was modeled, with parameters such as feedback strength, cooperativity, distance over which feedback acts, and number of modifying cycles per cell cycle. At around the same time, a model treated Sir complex propagation in S. cerevisiae [69••], focusing on the requirements for bistability of heterochromatin domains. A subsequent more general chromatin inheritance model [70••] came to the conclusion that systems with multiple marks are naturally subject to nonlinearities, as was the case in the S. pombe model.
Together, these models made a variety of interesting predictions. First, all models agreed in a requirement for cooperativity in producing a bistable system. This is consistent with a general requirement for ‘ultrasensitivity’ in systems that transform graded stimuli into bistable all or none outputs [71–73]. The molecular basis for cooperativity likely differs between different candidate chromatin inheritance systems. For example, in the case of S. pombe silencing, cooperativity was inadvertently built in to the original model from Dodd et al. by including the competing acetylation and methylation of H3 lysine 9. In contrast, in S. cerevisiae there is no known competing modification on H4 lysine 16, although several features of Sir complex biochemistry provide candidates for the required cooperativity, such as the Sir2 deacetylation byproduct O-acetyl ADP-ribose [49,74], which enhances the affinity of Sir complex for chromatin [51,75]. Alternatively, competing modifications such as H3K4me3 or H3K79me3 also provide a ‘three state’ solution (79+/Sir–, 79–/Sir–, 79–/Sir+) that eliminates the need for an explicit cooperativity term.
Another interesting prediction of the first model was that bistability was very difficult to achieve when feedback only occurred between adjacent nucleosomes — when nucleosomes only influenced their direct neighbors, resulting chromatin domains behaved as ‘patches’ of modified nucleosomes of varying size that grew and contracted as a random walk. The idea that lateral spreading of heterochromatin complexes might involve short loops or other interactions beyond adjacent nucleosomes has some support (reviewed in [76]). Additional predictions regarding the relationship between heterochromatin domain length, silencing factor concentration, and silencing stability differ depending on whether silencing is constrained between boundaries as at mating loci, or spreading into chromosomes as from telomeres [68••,69••].
Perspective
Recent advances have illuminated much about the behavior of chromatin during replication, and have identified numerous histone modification systems that contain positive feedback loops of modifying enzymes linked to modification-binding modules. What remains to be determined is whether any of these feedback systems actually result in heritable epigenetic states (and why others do not), and the replication dynamics of chromatin that allow some old nucleosomes to influence the state of newly synthesized nucleosomes. Finally, a great deal of evidence not surveyed here points toward abundant crosstalk between epigenetic modalities such as cytosine methylation, small RNAs, and chromatin, and future work will be needed to determine the contribution of chromatin-mediated feedback loops in epigenetic stability and plasticity.
Acknowledgements
This work was supported by NIH grants to OJR and PDK.
Footnotes
This review comes from a themed issue on Nucleus and gene expression Edited by Ana Pombo and Dave Gilbert
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Holliday R. The inheritance of epigenetic defects. Science. 1987;238:163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- 2.Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128:655–668. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 3.Jablonka E, Lamb MJ. Epigenetic Inheritance and Evolution: The Lamarckian Dimension. Oxford University Press; Oxford; New York: 1995. [Google Scholar]
- 4.Jablonka E, Lamb MJ. Evolution in Four Dimensions: Genetic, Epigenetic, Behavioral, and Symbolic Variation in the History of Life. MIT Press; Cambridge, Mass: 2005. [Google Scholar]
- 5.Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- 6.Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, Whitelaw E. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7••.Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–R236. doi: 10.1016/j.cub.2007.02.030. [An excellent review from Mark Ptashne exploring the evidence for chromatin as an epigenetic information carrier. Most cases of proposed chromatin inheritance are revealed to be explainable by alternative hypotheses.] [DOI] [PubMed] [Google Scholar]
- 8.Cavalli G, Paro R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999;286:955–958. doi: 10.1126/science.286.5441.955. [DOI] [PubMed] [Google Scholar]
- 9.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 10.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 11.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 12.de Bruin D, Kantrow SM, Liberatore RA, Zakian VA. Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol Cell Biol. 2000;20:7991–8000. doi: 10.1128/mcb.20.21.7991-8000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venditti S, Di Stefano G, D'Eletto M, Di Mauro E. Genetic remodeling and transcriptional remodeling of subtelomeric heterochromatin are different. Biochemistry. 2002;41:4901–4910. doi: 10.1021/bi016052y. [DOI] [PubMed] [Google Scholar]
- 14•.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [This paper as well as Refs. [15•, 16••] showed that memory of galactose exposure is epigenetically heritable in yeast. In both cases, chromatin regulators were genetically required for this memory.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Kundu S, Horn PJ, Peterson CL. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [This paper as well as Refs. [14•, 16••] showed that memory of galactose exposure is epigenetically heritable in yeast. In both cases, chromatin regulators were genetically required for this memory.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Zacharioudakis I, Gligoris T, Tzamarias D. A yeast catabolic enzyme controls transcriptional memory. Curr Biol. 2007;17:2041–2046. doi: 10.1016/j.cub.2007.10.044. [This paper showed that galactose memory was carried in the cytoplasm. Along with Refs. [14•, 15•], this shows that even when chromatin is heavily implicated in a memory system, it may not be the actual carrier of information during S phase.] [DOI] [PubMed] [Google Scholar]
- 17.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 18.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 19.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 20.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 21.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 22••.Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [In this paper, Francis and colleagues investigated the replication of Polycomb-bound templates in an in vitro replication system. Polycomb was maintained at replicating SV40 templates even in the presence of excess competitor DNA, demonstrating that chromatin-bound complexes can be passed to daughter chromosomes in cis despite the disruption caused by replication fork passage.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim UJ, Han M, Kayne P, Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 1988;7:2211–2219. doi: 10.1002/j.1460-2075.1988.tb03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson V. In vivo studies on the dynamics of histone–DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990;29:719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- 25.Gruss C, Wu J, Koller T, Sogo JM. Disruption of the nucleosomes at the replication fork. EMBO J. 1993;12:4533–4545. doi: 10.1002/j.1460-2075.1993.tb06142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randall SK, Kelly TJ. The fate of parental nucleosomes during SV40 DNA replication. J Biol Chem. 1992;267:14259–14265. [PubMed] [Google Scholar]
- 27.Krude T, Knippers R. Transfer of nucleosomes from parental to replicated chromatin. Mol Cell Biol. 1991;11:6257–6267. doi: 10.1128/mcb.11.12.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cusick ME, DePamphilis ML, Wassarman PM. Dispersive segregation of nucleosomes during replication of simian virus 40 chromosomes. J Mol Biol. 1984;178:249–271. doi: 10.1016/0022-2836(84)90143-8. [DOI] [PubMed] [Google Scholar]
- 29.Jackson V. Deposition of newly synthesized histones: new histones H2A and H2B do not deposit in the same nucleosome with new histones H3 and H4. Biochemistry. 1987;26:2315–2325. doi: 10.1021/bi00382a037. [DOI] [PubMed] [Google Scholar]
- 30.Jackson V. Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry. 1988;27:2109–2120. doi: 10.1021/bi00406a044. [DOI] [PubMed] [Google Scholar]
- 31.Sogo JM, Stahl H, Koller T, Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- 32.Gasser R, Koller T, Sogo JM. The stability of nucleosomes at the replication fork. J Mol Biol. 1996;258:224–239. doi: 10.1006/jmbi.1996.0245. [DOI] [PubMed] [Google Scholar]
- 33.Lucchini R, Wellinger RE, Sogo JM. Nucleosome positioning at the replication fork. EMBO J. 2001;20:7294–7302. doi: 10.1093/emboj/20.24.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shrader TE, Crothers DM. Effects of DNA sequence and histone–histone interactions on nucleosome placement. J Mol Biol. 1990;216:69–84. doi: 10.1016/S0022-2836(05)80061-0. [DOI] [PubMed] [Google Scholar]
- 35.Cremisi C, Chestier A, Yaniv M. Assembly of SV40 and polyoma minichromosomes during replication. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):409–416. doi: 10.1101/sqb.1978.042.01.043. [DOI] [PubMed] [Google Scholar]
- 36.Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 37.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 38.Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [An excellent review on the fate of H3/H4 tetramers during replication.] [DOI] [PubMed] [Google Scholar]
- 41.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 42.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antczak AJ, Tsubota T, Kaufman PD, Berger JM. Structure of the yeast histone H3–ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics. BMC Struct Biol. 2006;6:26. doi: 10.1186/1472-6807-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carmen AA, Milne L, Grunstein M. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J Biol Chem. 2002;277:4778–4781. doi: 10.1074/jbc.M110532200. [DOI] [PubMed] [Google Scholar]
- 45.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 46.McNairn AJ, Gilbert DM. Epigenomic replication: linking epigenetics to DNA replication. Bioessays. 2003;25:647–656. doi: 10.1002/bies.10305. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H. Establishment of transcriptional competence in early and late S phase. Nature. 2002;420:198–202. doi: 10.1038/nature01150. [DOI] [PubMed] [Google Scholar]
- 48.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 49.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson A, Li G, Sikorski TW, Buratowski S, Woodcock CL, Moazed D. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol Cell. 2009;35:769–781. doi: 10.1016/j.molcel.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell. 2009;33:323–334. doi: 10.1016/j.molcel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Sinha M, Watanabe S, Johnson A, Moazed D, Peterson CL. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell. 2009;138:1109–1121. doi: 10.1016/j.cell.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 56.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 57.Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 58.Monson EK, de Bruin D, Zakian VA. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci U S A. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Enomoto S, McCune-Zierath PD, Gerami-Nejad M, Sanders MA, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 61.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62•.Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, Imhof A, Nakatani Y, Dent SY, Almouzni G. The HP1alpha–CAF1–SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10:769–775. doi: 10.1038/embor.2009.90. [Outlines a two-step pathway in which the S phase complex of CAF-1/SetDB1 provides H3K9-monomethylated histones during heterochromatin replication, followed by trimethylation by the Suv39 enzyme. The CAF-1 interaction with the H3K9me3-binding protein HP1 makes this a positive feedback loop.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 64.Quivy JP, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni G. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 2004;23:3516–3526. doi: 10.1038/sj.emboj.7600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA–(cytosine-5) methyltransferase–PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 66.Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lande-Diner L, Zhang J, Cedar H. Shifts in replication timing actively affect histone acetylation during nucleosome reassembly. Mol Cell. 2009;34:767–774. doi: 10.1016/j.molcel.2009.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68••.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [See annotation to [70••].] [DOI] [PubMed] [Google Scholar]
- 69••.Sedighi M, Sengupta AM. Epigenetic chromatin silencing: bistability and front propagation. Phys Biol. 2007;4:246–255. doi: 10.1088/1478-3975/4/4/002. [See annotation to Ref. [70••].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70••.David-Rus D, Mukhopadhyay S, Lebowitz JL, Sengupta AM. Inheritance of epigenetic chromatin silencing. J Theor Biol. 2009;258:112–120. doi: 10.1016/j.jtbi.2008.12.021. [A series of computational models that explore key contributors to epi-genetic heritability of chromatin states. Key predictions are that cooperativity is necessary for the bistability of chromatin domains, and that chromatin modifying enzymes must affect nonadjacent nucleosomes in order for bistable domains to exist.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferrell JE., Jr Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 72.Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 73.Huang CY, Ferrell JE., Jr Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1996;93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci U S A. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 76.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]