Abstract
Cytokinins are plant hormones involved in regulation of diverse developmental and physiological processes in plants whose molecular mechanisms of action are being intensely researched. However, most rapid responses to cytokinin signals at the proteomic and phosphoproteomic levels are unknown. Early cytokinin responses were investigated through proteome-wide expression profiling based on image and mass spectrometric analysis of two-dimensionally separated proteins and phosphoproteins. The effects of 15 min treatments of 7-day-old Arabidopsis thaliana seedlings with four main cytokinins representing hydroxyisopentenyl, isopentenyl, aromatic, and urea-derived type cytokinins were compared to help elucidate their common and specific function(s) in regulating plant development. In proteome and phosphoproteome maps, significant differences were reproducibly observed for 53 and 31 protein spots, respectively. In these spots, 96 proteins were identified by matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry (MALDI-TOF/TOF MS), providing a snapshot of early links in cytokinin-regulated signalling circuits and cellular processes, including light signalling and photosynthesis, nitrogen metabolism, the CLAVATA pathway, and protein and gene expression regulation, in accordance with previously described cytokinin functions. Furthermore, they indicate novel links between temperature and cytokinin signalling, and an involvement of calcium ions in cytokinin signalling. Most of the differentially regulated proteins and phosphoproteins are located in chloroplasts, suggesting an as yet uncharacterized direct signalling chain responsible for cytokinin action in chloroplasts. Finally, first insights into the degree of specificity of cytokinin receptors on phosphoproteomic effects were obtained from analyses of cytokinin action in a set of cytokinin receptor double mutants.
Keywords: Arabidopsis thaliana, cytokinin, phosphoproteome, proteome
Introduction
Cytokinins were first identified by their ability to promote division in cultured plant cells (Miller et al., 1955). They have since been shown to play roles in diverse aspects of plant growth and development including cell division, shoot initiation, apical meristem function, and vascular formation (Mok and Mok, 2001).
Naturally occurring cytokinins are adenine derivatives substituted at the N6 position with an isoprenoid or aromatic side chain. Isoprenoid cytokinins are the most abundant cytokinins, while aromatic cytokinins, including N6-benzyladenine (BA), are minor components of the cytokinin pool (Strnad, 1997). Isoprenoid cytokinins are either of the isopentenyl (iP) type, with an isopentenyl N6 side chain, or of the zeatin (Z) type, with a hydroxylated isopentenyl N6 side chain in either trans (t-Z) or cis (c-Z) configuration. Reduction of the double bond in the side chain results in dihydrozeatin (DZ) (Brzobohatý et al., 1994; Mok and Mok, 2001). In addition, structurally unrelated (synthetic) phenylurea-type cytokinins, for example thidiazuron (TDZ), show high activity in most cytokinin bioassays (Mok and Mok, 2001).
The diversity of compounds exerting cytokinin activity might underlie the molecular fine tuning of their numerous functions. Indeed, differences in biological activities of specific cytokinins have long been recognized, for instance in growth and morphogenic responses (e.g. Mok et al., 1978; Sujatha and Reddy, 1998; Lexa et al., 2003), but molecular mechanisms underlying between-cytokinin differences in activity are just emerging.
Between-cytokinin differences in activity can be at least partially explained by differences in the receptors that perceive them and trigger biological responses. Cytokinin perception and signalling apparently evolved from bacterial two-component phosphorelays (Ferreira and Kieber, 2005). Binding of cytokinins to the Arabidopsis sensor hybrid histidine kinases AHK2, AHK3, and AHK4/CRE1/WOL1 initiates a phosphorelay in which Arabidopsis histidine-containing phosphotransfer proteins (AHPs) are phosphorylated then translocated into the nucleus, where they transfer the phosphate to Arabidopsis type-B response regulators (ARRs) (Kakimoto, 2003; Rashotte et al., 2003; Kiba et al., 2005; Choi and Hwang, 2007). The latter play roles in mediating transcriptional responses to cytokinin, including rapid induction of another class of response regulators, type-A ARRs (Rashotte et al., 2003), which act as negative regulators of the primary signal transduction pathway (Argueso et al., 2009). The first evidence for differential ligand specificity of cytokinin receptors has been obtained from their characterization in bacterial expression systems (Spíchal et al., 2004; Yonekura-Sakakibara et al., 2004; Romanov et al., 2006).
Global genome expression profiling of cytokinin action in Arabidopsis has yielded a genome-wide view of changes in abundance of cytokinin-responsive transcripts that might be relevant for the many biological processes governed by cytokinins (Hoth et al., 2003; Rashotte et al., 2003; Brenner et al., 2005). However, since changes in transcript abundance are not necessarily linearly related to changes in levels and/or activities of corresponding proteins, proteome profiling can provide valuable complementary information regarding molecular mechanisms linking cytokinin signals and their diverse effects in plants. In addition to protein abundance, post-translational modifications (PTMs) of proteins are crucial determinants of protein activity and subcellular location. Phosphorylation is a key PTM; at least 5% of the Arabidopsis thaliana genome is involved in regulating protein phosphorylation (Laugesen et al., 2004), indicating that nearly all aspects of cell function may involve reversible phosphorylation.
A set of proteins involved in cytokinin-induced photomorphogenesis has been identified by proteomic analysis (Lochmanová et al., 2008). In addition, rapid alterations of the phosphoproteome following cytokinin treatment have been examined in the moss Physcomitrella patens (Heintz et al., 2006), although comprehensive interpretation of the data was hindered by gaps in knowledge of its genome sequence. Nevertheless, our understanding of early cytokinin-responsive proteins and protein PTMs is still rudimentary. Hence, further analysis of proteome and phosphoproteome alterations caused by cytokinins before proteins encoded by the immediate cytokinin response genes (Brenner et al., 2006) accumulate significantly is needed to elucidate aspects of cytokinin signalling and action networks that cannot be deduced solely from transcriptome profiling. Therefore, proteomic analysis was applied to identify early cytokinin response proteins and phosphoproteins in Arabidopsis seedlings treated with four main cytokinins—t-zeatin (t-Z), isopentenyladenine (i-P), 6-benzylaminopurine (BA), and thidiazuron (TDZ). Detection of proteins involved in processes known to be regulated by cytokinins validated the experimental approach, and unexpected cytokinin targets were identified. Contributions of specific cytokinin receptors to the phosphoproteome alterations were assessed by examining effects of the cytokinins in ahk2ahk3, ahk2cre1, and ahk3cre1 mutants.
Materials and methods
Plant material, growth conditions, and cytokinin treatment
Seeds of A. thaliana ecotype Columbia (Col-0), and ahk2ahk3, ahk2cre1, and ahk3cre1 double mutants (provided by Professor Thomas Schmülling, Free University of Berlin) were surface-sterilized and sown on Uhelon 120T (Silk & Progress, Czech Republic) mesh placed on 1% (w/v) agar containing Murashige and Skoog (MS) medium (pH 5.7) supplemented with 5×10−4% (v/v) dimethylsulphoxide (DMSO), stratified at 4 °C for 3 d, and cultivated at 21 °C/19 °C day/night temperatures, with a 16 h photoperiod (90 μmol m−2 s−1 light intensity) for 7 d. On the seventh day (after the first 2 h of the day period), the Uhelon mesh with the seedlings was transferred onto liquid MS medium supplemented with (i) 5×10−4% (v/v) DMSO (mock buffer); (ii) 5 μM individual cytokinins (BA, TDZ, iP, and t-Z; Duchefa) in DMSO (final concentration, as for the mock); (iii) 30 μM D600 and 60 μM LaCl3 (Sigma); or (iv) 30 μM D600, 60 μM LaCl3 (Sigma), and 5 μM t-Z, and incubated for 15 min. The concentrations of the calcium signalling inhibitors (D600 and LaCl3) followed Saunders and Hepler (1983) who observed disruption of cytokinin-induced bud formation in the moss Funaria in response to them. Seedlings were rapidly harvested, dried, then frozen and ground in liquid nitrogen.
Protein extraction
Total protein was extracted from frozen seedlings (250–300 mg) by acetone/trichloroacetic acid (TCA) extraction (Damerval et al., 1986). Dried protein was solubilized for 2 h at 30 °C in SOL buffer: 7 M urea, 2 M thiourea, 2% (w/v) CHAPS, 90 mM dithiothreitol (DTT). Insoluble matter was removed by centrifugation (15 000 g for 10 min) and the protein concentration was determined (Bradford, 1976) (Sigma-Aldrich, http://www.sigmaaldrich.com/) after diluting 1 μl of the total protein extract in 1 ml of reaction mix to prevent the SOL buffer interfering with the Bradford assay. Solubilized protein was then diluted 1:1 with rehydration solution [SOL supplemented with 1% (v/v) ampholytes pH 3–10, 0.2% (w/v) bromophenol blue] and loaded onto IPG strips (Bio-Rad, http://www.bio-rad.com/).
For phosphoproteome analysis, an isolation procedure was established using a PhosphoProtein Purification Kit (Qiagen, http://www.qiagen.com/). Briefly, 350–400 mg of seedlings ground in liquid nitrogen were extracted with 4 ml of lysis buffer supplemented with protease inhibitors and benzonase (Qiagen kit). Each sample was then diluted to 25 ml with lysis buffer, applied to an affinity column and processed according to the supplier's manual (Qiagen). Protein concentration was determined by the Bradford assay. Desalted phosphoproteins in TRIS-HCl buffer (pH 7.0) were diluted with rehydration solution:SOL (1:1) and loaded onto IPG strips.
2D gel electrophoresis
Proteins were separated essentially as previously described (Lochmanová et al., 2008). Briefly, portions containing 500 μg of protein or 150 μg of phosphoprotein were applied to 18 cm and 7 cm IPG strips, respectively, with a linear pH gradient (4–7), the strips were rehydrated for 16 h at room temperature in buffer containing the extracts, then the proteins were isoelectrically focused at 22 °C in six steps in a PROTEAN IEF Cell unit (Bio-Rad): 150 V (30 min), 300 V (60 min), 600 V (60 min), 1500 V (180 min), 3500 V (300 min), and 10 000 V to 80 000 Vh for long strips; 150 V (20 min), 300 V (20 min), 600 V (20 min), 1500 V (20 min), 3000 V (20 min), and 4000 V up to 12 000 Vh for short strips. The strips were then treated with buffers containing DTT and iodoacetamide (Sigma-Aldrich) to reduce and alkylate the proteins, which were then separated by 11% polyacrylamide SDS–PAGE with the following settings: 50 V (120 min) followed by 100 V (16 h) for large gels (proteome analysis), and 100 V (10 min) followed by 150 V (50 min) for small gels (phosphoproteome analysis), using a PROTEAN Plus Dodeca Cell, and a Mini-PROTEAN 3 Dodeca Cell (Bio-Rad), respectively.
Protein staining and image analysis
Gels were stained with colloidal Bio-Safe Coomassie G-250 (Bio-Rad) and scanned with a Bio-Rad GS-800 Calibrated Densitometer (400 dpi and 700 dpi for large and small gels, respectively). Acquired images were analysed using Decodon Delta 2D software (http://www.decodon.com). Three, six, three, and four biological replicates were used in the 2-DE total proteome comparisons of the wild type, phosphoproteome comparisons of the wild type, phosphoproteome comparisons of the ahk double mutant, and phosphoproteome comparisons of wild-type samples in the presence of calcium signalling inhibitors, respectively. Cytokinin responses of proteins corresponding to detected spots were deemed significant if there was a cytokinin/mock, BA/TDZ, BA/iP, or BA/t-Z spot volume ratio of ±1.4 or more (for at least one variant), with t-test values ≥95% and similar profiles in (i) ≥2 biological replicates for total protein comparisons (with three parallel SDS–PAGE analyses for each treatment, i.e. 15 parallel SDS–PAGE analyses for each biological replicate); (ii) ≥3 biological replicates for phosphoproteome comparisons (with two parallel SDS–PAGE analyses per treatment, i.e. 10 parallel SDS–PAGE analyses for each biological replicate); (iii) three biological replicates for phosphoproteome comparisons in the ahk double mutants (with two parallel SDS–PAGE analyses per treatment, i.e. 8 parallel SDS–PAGE analyses for each biological replicate); or (iv) ≥2 biological replicates for phosphoproteome comparisons in the wild type in the presence of calcium signalling inhibitors. Only spots with significant and reproducible changes were considered for mass spectroscopic identification. The experimental design is outlined schematically in Supplementary Fig. S1 available at JXB onluine.
Protein identification
Proteins were identified as previously described (Hradilová et al., 2010) with minor modifications. Briefly, selected protein spots were digested with trypsin. The dried tryptic peptides were each dissolved in 10 μl of 0.1% trifluoroacetic acid and purified using ZipTip C18 tips. The eluate was mixed with 1 vol. of 10 mg ml−1 α-cyano-4-hydroxycinnamic acid (CHCA) in 50% (v/v) acetonitrile and 0.1% trifluoroacetic acid for spotting onto sample plates, and dried for matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry (MALDI-TOF/TOF MS) analysis. To demonstrate phosphorylation of selected peptides, phosphopeptides were first enriched from tryptic peptides dissolved in 10% acetonitrile and 0.1% acetic acid using IMAC tips (Millipore) containing iron ions. After loading, the tips were washed with 10% acetonitrile and 0.1% acetic acid, then rinsed with water. Phosphopeptides were eluted by 0.3 N ammonium hydroxide and measured using 15 mg ml−1 2,5-dihydroxybenzoic acid in 50% (v/v) acetonitrile and 6% phosphoric acid solution as a matrix.
Alkaline phosphatase treatment was used to confirm the phosphorylation of the phosphopeptides according to Larsen et al. (2001). Briefly, IMAC-purified phosphopeptides were incubated with 0.05 U μl−1 alkaline phosphatase in 50 mM NH4HCO3, pH 7.8 at 37 °C for 30 min then acidified with 2.5 μl of 5% trifluoroacetic acid. Phosphopeptides were identified by single or multiple 80 Da (HPO3) losses in MALDI-TOF-MS following alkaline phosphatase treatment, for mono- and multiphosphorylated peptides, respectively.
MALDI-TOF/TOF measurements were performed with an Applied Biosystems 4700 Proteomic Analyzer (Applied Biosystems, http://www.appliedbiosystems.com/) equipped with an Nd:YAG laser (355 nm) operated with 3–7 ns pulses and 200 Hz firing rate in positive reflectron mode for both MS and MS/MS analyses. The accelerating voltage in the ion source for MS and MS/MS analyses was set at 20 kV and 8 kV, respectively. Acquired sequences were searched against the NCBInr sequence database (version 09/2009) using Mascot (http://www.matrixscience.com/), and peaks generated from the acquired mass spectra by the Peak-to-Mascot function incorporated in the software. In the MS analyses, peaks in the 900–4000 m/z range with signal to noise (S/N) ratios >4 were sought. In the MS/MS analyses, peaks with S/N ratios >4 in the m/z range from 68 m/z up to 20 m/z units lower than their precursors’ m/z values were used. The resulting peak lists contained information from both MS and MS/MS runs concerning fragmentation patterns of selected precursors. Parameters for both MS and MS/MS data searches in Mascot were: taxonomy, Arabidopsis thaliana; enzyme, trypsin; allowed missed cleavages, 1 [except for the peptide VGKDSKDKELKEAFK of endoplasmin homologue (SHD), where allowed missed cleavages were set to 4]; fixed modification, carbamidomethyl (C); variable modifications, none or Phospho (ST) and Phospho (Y) (for searching phosphopeptides); peptide tolerance, 0.5 Da; MS/MS tolerance, 0.5 Da; peptide charge, (+1); instrument, MALDI-TOF/TOF. Protein matches in MS/MS identification were considered valid if there was at least one peptide with a Mascot score corresponding to identity or extensive homology (P <0.05). Protein scores were derived from ion scores as a non-probabilistic basis for ranking protein hits by Mascot. Similar parameters were set for peptide mass fingerprint analysis—only protein matches with Mascot scores indicating extensive homology were accepted.
Gene ontology
Gene ontology was evaluated by BiNGO 2.3 in Cytoscape 2.6.2, with data from the NCBI (http://www.ncbi.nlm.nih.gov) and TAIR (http://www.arabidopsis.org) databases.
Results
Identification of early cytokinin response proteins
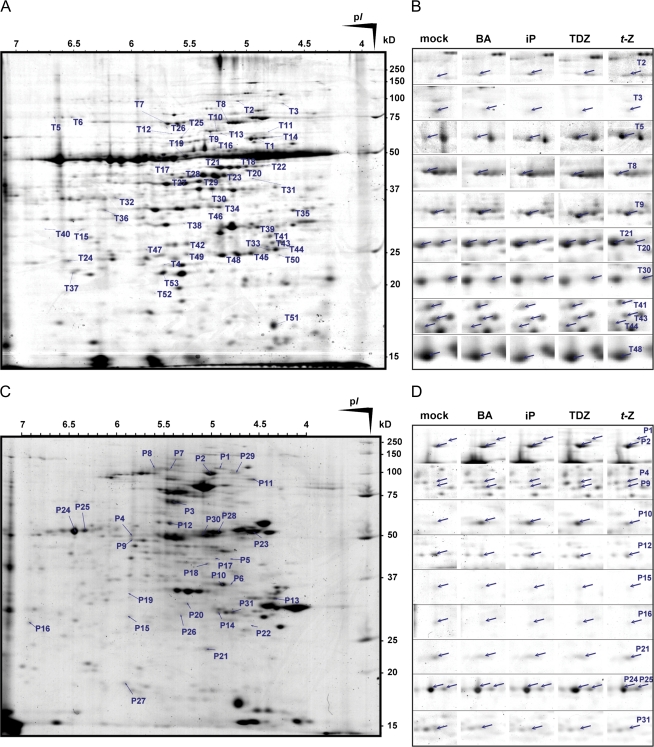
To identify early cytokinin response proteins, 7-day-old Arabidopsis seedlings were treated (separately) with four main cytokinins (BA, iP, TDZ, and t-Z) at a concentration of 5 μM for 15 min. Total proteins were then extracted and subjected to 2-DE (Fig. 1A, B). Image analysis of the resulting proteome maps revealed >850 reproducibly resolved spots in gels over pI and molecular mass ranges of 4–7 and 10–120 kDa, respectively, then proteome patterns of seedlings treated with the individual cytokinins were compared separately with the proteome patterns of seedlings treated with mock buffer. Significant differences (P <0.05) in all biological replicates were found for 160 resolved spots, but only 53 spots were reproducibly significant in two or more independent experiments and were then subjected to protein identification. Altogether, 67 proteins were identified in the 53 spots, including 10 protein mixtures and a non-dissociated heterodimer consisting of small and large Rubisco subunits (T13), by MALDI-TOF/TOF MS analysis followed by Mascot database searches of the full NCBI Arabidopsis protein database (Table 1; Supplementary Table S1 at JXB online). The ratio of numbers of up-regulated to down-regulated proteins was ∼1:2. Identified protein spots are marked in protein maps shown in Fig. 1A, and corresponding partial amino acid sequences are listed in Supplementary Table S3. Protein identifications and relative fold changes based on mean percentage volumes of each of these spots are presented in Table 1. The apparent strength of effects of the cytokinins on expression of the early cytokinin response proteins decreased in the order BA=TDZ>t-Z=iP.
Fig. 1.
Effects of cytokinin treatment on the proteome and phosphoproteome of Arabidopsis seedlings. (A) Average two-dimensional gel electrophoresis proteome map of 7-day-old Arabidopsis seedlings treated with cytokinin/mock buffer for 15 min. Differentially regulated protein spots are indicated. See Table 1, and Supplementary Table S1 at JXB online, for detailed information on the corresponding identified proteins. Proteins (500 μg) were separated in the first and second dimensions by IPG (18 cm strips, pH 4–7) followed by 11% SDS–PAGE then visualized by Bio-Safe Coomassie G250 staining. Isoelectric points (pI) and migrating positions of molecular mass (kDa) markers are marked. (B) Examples of spots corresponding to the differentially regulated proteins in Arabidopsis seedlings treated with 5 μM cytokinin (BA, iP, TDZ, or t-Z) or mock buffer for 15 min. For details see Materials and methods. (C) Average 2-DE phosphoproteome map of 7-day-old Arabidopsis seedlings treated with cytokinin/mock buffer for 15 min. Differentially regulated protein spots are indicated. See Table 2, and Supplementary Table S2, for detailed information on the corresponding identified proteins. Phosphoprotein fractions were obtained using a PhosphoProtein Purification Kit. Phosphoproteins (150 μg) were separated in the first and second dimensions by IPG (7 cm strips, pH 4–7) followed by 11% SDS–PAGE then visualized by Bio-Safe Coomassie G250 staining. Isoelectric points (pI) and relative migrating positions of molecular mass (kDa) markers are marked. (D) Examples of spots corresponding to the differentially regulated phosphoproteins in Arabidopsis seedlings treated with 5 μM cytokinin (BA, iP, TDZ, or t-Z) or mock buffer for 15 min. For details see Materials and methods. (This figure is available in colour at JXB online.)
Table 1.
Early cytokinin response proteins of Arabidopsis
| Spot/protein no. | AGI code | Protein name | MALDI-MS (protein score/%cov/pep#) | Relative fold change |
|||
| BA | iP | TDZ | t-Z | ||||
| T1 | At2g28000 | Rubisco large subunit-binding protein subunit α, chloroplastic | 666/32/12 | –1.5±0.23 | –1.5±0.23 | –1.4±0.21 | –1.4±0.21 |
| T2 | At4g24190 | Endoplasmin homologue (SHD) | 27/1/1 | 1.5±0.10 | 1.8±0.20 | 1.6±0.32 | 1.4±0.28 |
| T3 | At5g56000 | Heat shock protein 81-4 | 153/8/5 | –2.0±0.20 | –1.5±0.23 | –2.0±0.30 | –1.3±0.20 |
| T4 | At1g19570 | Glutathione S-transferase DHAR1, mitochondrial | 35/17/2 | –1.4±0.21 | –1.3±0.20 | –1.3±0.20 | –1.5±0.23 |
| T5 | At5g17920 | Cobalamin-independent methionine synthase | 49/6/3 PMF: 127/23/11 | 1.4±0.14 | 1.4±0.26 | 1.6±0.33 | 1.7±0.30 |
| T6 | At5g17920 | Cobalamin-independent methionine synthase | 55/7/3 PMF: 104/21/9 | 1.3±0.26 | 1.4±0.28 | 1.5±0.30 | 1.4±0.21 |
| T7 | At3g60750 | Putative transketolase | 236/17/4 | –1.5±0.10 | –1.5±0.30 | –1.6±0.30 | –1.4±0.20 |
| T8 | At5g02500 | Heat shock cognate 70 kDa protein 1 | 104/10/3 | –1.3±0.20 | –1.4±0.21 | –1.5±0.23 | –1.3±0.20 |
| T9 | At2g30950 | Cell division protease FtsH homologue 2, chloroplastic | 98/8/3 | 1.5±0.30 | 1.4±0.28 | 1.3±0.26 | 1.4±0.21 |
| T10 | At2g30950 | Cell division protease FtsH homologue 2, chloroplastic | 101/9/3 | 1.4±0.10 | 1.7±0.12 | 1.5±0.30 | 1.8±0.10 |
| T11 | At5g60640 | Protein disulphide isomerase-like protein | 96/25/8 | –2.5±0.38 | –2.0±0.30 | –1.6±0.24 | –2.0±0.30 |
| T12 | AtCg00120 | ATP synthase subunit α, chloroplastic | 114/26/9 | 1.3±0.26 | 1.4±0.28 | 1.4±0.21 | 1.4±0.21 |
| T13 | At5g38420 | Rubisco small chain 2β, chloroplastic | 45/8/1 | –1.4±0.21 | –1.3±0.20 | –1.3±0.20 | –1.4±0.21 |
| AtCg00490 | Rubisco large chain | PMF: 159/34/14 | |||||
| T14 | At1g21750 | Probable protein disulphide-isomerase 1 | 41/6/2 | –1.7±0.23 | –1.4±0.21 | –1.6±0.30 | –1.4±0.15 |
| T15 | At1g20020 | Ferredoxin-NADP reductase, leaf 2, chloroplastic | 383/29/7 | 1.6±0.32 | 1.3±0.30 | 1.0±0.30 | 1.0±0.32 |
| T16 | AtCg00120 | ATP synthase subunit α, chloroplastic | 238/20/6 | 1.5±0.30 | 1.2±0.24 | 1.2±0.22 | 1.4±0.28 |
| T17 | At5g08690 | ATP synthase subunit β-2, mitochondrial | 84/12/4 | –1.3±0.20 | –1.2±0.20 | –1.2±0.18 | –1.5±0.23 |
| T18 | AtCg00490 | Rubisco large chain | 269/22/7 | 2.0±0.40 | 1.5±0.30 | 2.0±0.40 | 1.5±0.30 |
| T19 | AtCg00480 | ATP synthase subunit β, chloroplastic | 128/10/4 | –1.3±0.24 | –1.4±0.30 | –1.5±0.23 | –1.4±0.21 |
| T20 | At2g39730 | Rubisco activase, chloroplastic | 714/39/10 | 1.4±0.28 | 1.5±0.34 | 1.5±0.30 | 1.4±0.25 |
| T21 | At2g39730 | Rubisco activase, chloroplastic | 465/17/5 | 1.4±0.24 | 1.5±0.30 | 1.3±0.26 | 1.6±0.25 |
| T22 | At3g54050 | Fructose-1,6-bisphosphatase, chloroplastic | 100/12/4 | –1.3±0.15 | –1.2±0.18 | –1.5±0.08 | –1.5±0.10 |
| T23 | At5g35630 | Glutamine synthetase, chloroplastic/mitochondrial | PMF: 72/29/5 | –1.3±0.20 | –1.2±0.18 | –1.4±0.21 | –1.2±0.18 |
| T24 | At4g02520 | Glutathione S-transferase PM24 | 230/45/8 | 1.7±0.20 | 1.9±0.10 | 1.5±0.30 | 1.3±0.26 |
| At5g61410 | Ribulose-5-phosphate-3-epimerase | 302/19/3 | |||||
| T25 | At1g09780 | 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase 1 | 435/25/9 | 1.4±0.28 | –1.2±0.13 | –1.4±0.20 | –1.3±0.20 |
| T26 | At5g26000 | Myrosinase | PMF: 61/13/5 | –1.5±0.24 | –1.6±0.24 | –1.5±0.23 | –1.3±0.12 |
| T27 | At3g18780 | Actin-2 | 147/23/5 | –1.6±0.24 | –1.5±0.14 | –1.5±0.23 | –1.4±0.22 |
| At1g49240 | Actin-8 | 147/23/5 | |||||
| At5g35630 | Glutamine synthetase, chloroplastic/mitochondrial | 133/15/3 | |||||
| T28 | At4g20360 | Elongation factor Tu, chloroplastic | 66/7/2 | –1.5±0.23 | –1.3±0.20 | –1.8±0.26 | –1.5±0.10 |
| T29 | At2g39730 | Rubisco activase, chloroplastic | 396/22/5 | 1.4±0.20 | 1.4±0.15 | 1.4±0.30 | 1.4±0.21 |
| T30 | At1g32060 | Phosphoribulokinase, chloroplastic | 287/26/7 | –1.6±0.15 | –1.4±0.22 | –1.5±0.10 | –1.5±0.20 |
| T31 | At3g12780 | Phosphoglycerate kinase | 254/17/5 | –1.5±0.20 | –1.5±0.10 | –1.5±0.10 | –1.6±0.20 |
| T32 | At3g52930 | Fructose-bisphosphate aldolase | 318/23/6 | 1.6±0.32 | 1.7±0.21 | 2.1±0.42 | 2.2±0.30 |
| T33 | At2g43910 | Thiol methyltransferase, putative | 64/13/2 | –1.8±0.27 | –1.6±0.24 | –1.1±0.22 | –1.2±0.30 |
| T34 | At3g09200 | 60S Acidic ribosomal protein P0-2 | 59/14/2 | –1.4±0.21 | –1.7±0.22 | –2.1±0.31 | –2.0±0.30 |
| T35 | At1g30230 | Elongation factor 1- δ 1 | 897/46/8 | –1.5±0.20 | –1.4±0.23 | –1.5±0.15 | –1.4±0.21 |
| T36 | At2g05990 | Enoyl-[acyl-carrier-protein] reductase | 206/23/6 | 1.7±0.15 | 1.7±0.34 | 1.6±0.32 | 1.7±0.02 |
| T37 | At3g10920 | Superoxide dismutase [Mn], mitochondrial | 462/38/7 | –1.2±0.19 | –1.4±0.08 | –1.4±0.21 | –1.3±0.20 |
| At2g47730 | Glutathione S-transferase 6, chloroplastic | 147/35/4 | |||||
| T38 | At3g16420 | PBP1 | 35/6/1 | –1.4±0.21 | –1.2±0.18 | –1.4±0.21 | –1.3±0.20 |
| T39 | At3g53460 | 29 kDa ribonucleoprotein, chloroplastic | 71/4/1 | –1.4±0.22 | –1.4±0.21 | –1.6±0.24 | –1.4±0.21 |
| T40 | At4g28520 | Cruciferin 3 | 475/41/10 | –2.4±0.36 | 2.7±0.54 | 1.1±0.30 | –1.3±0.20 |
| T41 | At5g38480 | 14-3-3-like protein GF14 ψ | 403/39/6 | –1.5±0.23 | –1.3±0.20 | –1.6±0.25 | –1.2±0.18 |
| At1g22300 | 14-3-3-like protein GF14 ϵ | 326/26/5 | |||||
| T42 | At5g14740 | β-Carbonic anhydrase 2 | 333/35/5 | 1.6±0.20 | 1.6±0.10 | 1.7±0.34 | 1.9±0.38 |
| T43 | At2g37220 | Putative ribonucleoprotein, chloroplastic | 440/24/6 | –1.4±0.21 | –1.2±0.11 | –1.1±0.17 | –1.2±0.18 |
| At5g50250 | Putative 31 kDa ribonucleoprotein, chloroplastic | 85/7/2 | |||||
| T44 | At5g10450 | 14-3-3-like protein GF14 λ | 39/9/1 | 1.9±0.35 | 1.5±0.30 | 1.4±0.24 | 1.4±0.28 |
| T45 | At2g34430 | Photosystem II type I chlorophyll a/b-binding protein (LHB1B1) | 229/31/3 | –1.6±0.24 | –1.7±0.26 | –1.1±0.20 | –1.5±0.23 |
| At2g34420 | Photosystem II type I chlorophyll a/b-binding protein (LHB1B2) | 229/31/3 | |||||
| T46 | At2g21330 | Fructose-bisphosphate aldolase (FBA1) | 105/12/3 | –1.2±0.23 | –1.3±0.20 | –1.5±0.23 | –1.6±0.24 |
| At4g38970 | Fructose-bisphosphate aldolase (FBA2) | 76/19/4 | |||||
| T47 | At1g54870 | Glucose and ribitol dehydrogenase homologue 1 | PMF: 61/31/5 | –1.5±0.23 | –1.5±0.23 | –1.5±0.23 | –1.4±0.21 |
| T48 | At1g29910, At1g29920 | Chlorophyll a-b-binding protein 165/180, chloroplastic (CAB2/3) | 419/35/4 | –1.5±0.23 | –1.5±0.30 | –1.4±0.21 | –1.3±0.20 |
| At1g29910, At1g29930 | Chlorophyll a-b-binding protein 2, chloroplastic (CAB1) | ||||||
| T49 | At3g55440 | Triosephosphate isomerase, cytosolic | 122/23/3 | –1.4±0.22 | –1.5±0.23 | –2±0.30 | –1.4±0.15 |
| T50 | At3g14290 | Proteasome subunit α type-5-B | 121/27/4 | –1.5±0.23 | –1.7±0.30 | –1.2±0.20 | –1.5±0.23 |
| At1g53850 | Proteasome subunit α type-5-A | 88/21/3 | |||||
| T51 | At3g27830 | 50S Ribosomal protein L12-1, chloroplastic | 313/36/3 | –1.6±0.18 | –1.1±0.17 | –1.3±0.20 | –1.3±0.20 |
| At3g27850 | 50S Ribosomal protein L12-3, chloroplastic | ||||||
| T52 | At4g38680 | Glycine-rich protein 2/cold shock domain protein 2 | 74/21/2 | –1.6±0.10 | –1.5±0.23 | –1.6±0.24 | –1.3±0.20 |
| T53 | At1g61520 | LHCA3 (PSI type III chlorophyll a/b-binding protein); chlorophyll binding | 207/13/3 | –1.3±0.20 | –1.4±0.20 | –1.3±0.32 | –1.7±0.26 |
Spot no., spot number (as given in Fig. 1A); AGI code, accession number in the TAIR database; Protein name, entry name in the NCBI database; %cov, percentage of protein coverage; pep#, number of peptides; PMF, peptide mass fingerprint; Relative fold change, fold change relative to the mock control (calculated by DECODON DELTA 2D software) ±SE. Full information on the proteins including their classification, peptide sequences and peak list is given in Supplementary Tables S1 and S3 at JXB online.
Previously, cytokinin early response transcripts were identified following 15 min treatment of 7-day-old Arabidopsis seedlings with 5 μM BA. Here it was confirmed that levels of type-A ARR genes (ARR3 and ARR5) increased following BA treatment in the experimental set-up employed using quantitative RT-PCR (P. Souček, unpublished data) as outlined in Souček et al. (2007).
Identification of early cytokinin response phosphoproteins
A fraction of phosphoproteins phosphorylated at serine and threonine residues was isolated from seedlings treated with cytokinins using a Quiagen Phosphoprotein enrichment kit with an optimized procedure, as outlined above. In addition, cytokinin receptor double mutants (ahk2ahk3, ahk2cre1, and ahk3cre1) treated with 5 μM t-Z for 15 min were analysed to assess how much the individual cytokinin receptors contribute to phosphoproteome regulation. Phosphoprotein fractions were subjected to 2-DE, and image analysis was used to reveal phosphoproteins differentially regulated by cytokinins (Fig. 1C, D), essentially as described above for early cytokinin response proteins. Of 450 reproducibly resolved spots in phosphoproteome maps from wild-type samples, significant differences (P <0.05) in all 10 independent experiments (including the four pilot experiments using only one cytokinin each) were found for 90 resolved spots (for a schematic representation of the experimental design see Supplementary Fig. S1 at JXB online). Reproducible significant changes in at least three biological replicates were found for 31 spots. Subsequently, >90% of them were reproducibly resolved in phosphoproteome maps displaying phosphoproteins from each of the three cytokinin receptor double mutants (Table 3).
Table 3.
Regulation of the early cytokinin response phosphoproteins by t-Z in the cytokinin receptor double mutants ahk2cre1, ahk3cre1, and ahk2ahk3
| Spot/protein no. | AGI code | Protein name | Relative fold change |
|||
| ahk2ahk3 | ahk2cre1 | ahk3cre1 | Wild type | |||
| Significant response apparently mediated by a single cytokinin receptor | ||||||
| P1 | At1g22530 | Patellin-2 (PATL-2) | –1.1±0.13 | 1.5±0.25 | 1.1±0.04 | 1.9±0.06 |
| P2 | At4g24190 | Endoplasmin homologue (SHD) | 1.2±0.30 | 1.4±0.10 | 1.0±0.30 | 1.7±0.20 |
| P3 | At5g56030 | Heat shock protein 81-2 | 1.5±0.08 | 1.0±0.05 | 1.3±0.32 | 2.0±0.15 |
| P4 | At5g11170 | DEAD-box ATP-dependent RNA helicase 15 | 1.4±0.06 | 1.0±0.03 | 1.0±0.18 | 1.5±0.15 |
| P7 | X | X | 1.0±0.26 | 1.5±0.07 | 1.0±0.26 | 2.5±0.13 |
| P9 | At1g09640 | Probable elongation factor 1-γ 1 | 1.5±0.05 | 1.0±0.12 | 1.0±0.15 | 1.5±0.05 |
| P11 | At5g60640 | Protein disulphide isomerase-like protein | 1.0±0.28 | –1.4±0.10 | –1.1±0.15 | –1.8±0.09 |
| P12 | AtCg00120 | ATP synthase subunit α, chloroplastic | –1.5±0.19 | –1.3±0.09 | –1.2±0.11 | –1.5±0.06 |
| P14 | At2g39990 | eIF2 (eukaryotic translation initiation factor) | 1.0±0.25 | –1.1±0.17 | –1.5±0.30 | –2.0±0.12 |
| P18 | At5g56030 | Heat shock protein 81-2/3/4 | 1.2±0.35 | –1.4±0.01 | –1.0±0.15 | –1.5±0.05 |
| P21 | At3g51880 | HMGB1 | –1.5±0.15 | –1.3±0.01 | 1.0±0.20 | –1.6±0.14 |
| P27 | At1g26630 | eIF5A-2 (eukaryotic translation initiation factor) | 1.5±0.08 | –1.1±0.14 | –1.2±0.29 | 1.5±0.05 |
| P28 | At1g20010 | Tubulin β-5 chain | 1.7±0.23 | 1.0±0.26 | 1.0±0.05 | 1.6±0.07 |
| P29 | X | X | 1.0±0.08 | 1.6±0.24 | 1.2±0.11 | 1.7±0.05 |
| P31 | At3g09200 | 60S Acidic ribosomal protein P0-2 | –1.1±0.11 | –1.4±0.02 | –1.2±0.16 | –1.6±0.10 |
| Significant response apparently mediated by two cytokinin receptors | ||||||
| P13 | AtCg00490 | Rubisco large chain | 1.6±0.21 | –1.6±0.57 | +/– | –2.0±0.23 |
| At1g67090 | Rubisco small chain 1A, chloroplastic | |||||
| P15 | At5g14740 | β-Carbonic anhydrase 2 | 1.0±0.18 | –1.4±0.14 | –1.5±0.34 | –1.4±0.10 |
| P19 | At3g16420 | PBP1 | –1.2±0.16 | –1.5±0.21 | –1.4±0.13 | –1.3±0.20 |
| P24 | AtCg00490 | Rubisco large subunit | +/– | –1.5±0.26 | 2.0±0.14 | 1.3±0.15 |
| P25 | AtCg00490 | Rubisco large subunit | 1.4±0.12 | 1.4±0.21 | 1.0±0.17 | 1.3±0.15 |
| P26 | At1g76180 | Dehydrin ERD14 | 1.5±0.15 | –1.4±0.32 | –1.3±0.27 | –1.3±0.16 |
| P30 | At5g44340 | Tubulin β-4 chain | 1.4±0.20 | 1.4±0.07 | 1.3±0.11 | 1.4±0.15 |
| Non-significant response to t-Z in wild type and/or mutants | ||||||
| P10 | At1g76180 | Dehydrin ERD14 | +/– | –1.3±0.33 | +/– | 1.3±0.07 |
| P16 | At5g43830 | GATase like protein | 1.1±0.20 | 1.0±0.20 | –1.1±0.22 | –1.5±0.10 |
| P17 | At5g56030 | Heat shock protein 81-2 | 1.0±0.16 | 1.0±0.18 | 1.0±0.05 | 1.0±0.30 |
| P20 | At5g42790 | Proteasome subunit a type-1-A | 1.2±0.08 | 1.0±0.01 | 1.2±0.14 | –1.5±0.08 |
| P22 | At3g09200 | 60S Acidic ribosomal protein P0-2 | 1.0±0.16 | 1.0±0.30 | 1.0±0.19 | –1.2±0.20 |
| P23 | At1g20440 | Dehydrin COR47 | 1.0±0.02 | –1.2±0.34 | 1.0±0.22 | 1.1±0.18 |
| At4g26110 | NAP1 | |||||
Spot no., spot number (as given in Fig. 1C); AGI code, accession number in the TAIR database; Protein name, entry name according to the NCBI database; Relative fold change, fold change relative to the mock control (calculated by DECODON DELTA 2D software) ±SE; +/–, inconsistent regulation in three biological replicas (down-, up-, and non-regulated in the individual biological replicas). Full information on the phosphoproteins including their classification, peptide sequences, and peak list is given in Supplementary Tables S2 and S4 at JXB online.
In total, 29 proteins were identified in these spots, including two protein mixtures, by MALDI-TOF/TOF MS followed by Mascot database searches of the full NCBI protein database (Table 2; Supplementary Table S2 at JXB online). Phosphorylation has been previously reported for 22 of these proteins (Table 2; PhosPhAt 3.0, http://phosphat.mpimp-golm.mpg.de). Here, phosphorylation was confirmed for 60S acidic ribosomal protein P0-2 (P22) and endoplasmin homologue (SHD; P2) by comparing MS spectra of their IMAC-purified peptides VEEKEESDEEDYGGDFGLFDEE and VGKDSKDKELKEAFK, respectively, before and after alkaline phosphatase treatment (Supplementary Fig. S2). Serine was previously shown to be a phosphorylation site on the peptide of 60S acidic ribosomal protein P0-2 (Laugesen et al., 2006), but phosphorylation of endoplasmin homologue (SHD) has not been previously reported. In addition, 29 of the 31 spots were stained by the phosphoprotein-specific stain Phos-tag™ (Supplementary Fig. S3). The ratio of numbers of up-regulated to down-regulated phosphoproteins was ∼1:1. Alterations in levels of individual phosphoproteins may result from phosphorylation/dephosphorylation events and/or modulation of turnover rates of the phosphoproteins. Identified protein spots are marked in protein maps shown in Fig. 1C, and examples of spots containing phosphoproteins differentially regulated by the individual cytokinins in Fig. 1D. The corresponding partial amino acid sequences are listed in Supplementary Table S4. Protein identifications and relative fold changes based on mean percentage volumes of these spots are presented in Tables 2 and 3 for phosphoproteins of the wild type and cytokinin receptor double mutants, respectively. Apparent strength of effects of the cytokinins on expression of the early cytokinin response phosphoproteins decreased in the order BA>TDZ>t-Z=iP. Responses specific for a single receptor were found in 15 spots, while seven spots were regulated by two individual receptors (Table 3). The remaining spots were either non-significantly or inconsistently (down-, up-, and non-regulated in the individual biological replicas) regulated. Interestingly, regulation was apparent in the mutants for four spots (P19, P24, P25, and P26) that remained below cut-off limits in wild-type seedlings. The opposite regulation was found for spots P13, P26 (ahk2ahk3 and ahk2cre1), and P24 (ahk2cre1 and ahk3cre1), suggesting receptor interactions in response regulation. Further, loss of consistent regulation in the double mutants for two spots regulated by t-Z in the wild type (P16, P20) was observed, implying that simultaneous activity of at least two receptors may be needed for correct regulation of the corresponding proteins. In addition, responses of a fraction of the 15 spots primarily regulated by a single receptor to cytokinin treatment were lower in all three double mutants than in wild-type plants, suggesting they may require simultaneous activity of one or more other receptor(s) for a full response. The highest numbers of regulated spots were found in the ahk2cre1 (14) mutant, followed by ahk2ahk3 (11) and ahk3cre1 (four).
Table 2.
Early cytokinin response phosphoproteins of Arabidopsis
| Spot/protein no. | AGI code | Protein name | PhosPhAt database | MALDI-MS (protein score/%cov/pep#) | Relative fold change |
|||
| BA | iP | TDZ | t–Z | |||||
| P1 | At1g22530 | Patellin-2 (PATL-2) | + | 42/1/3 | 2.0±0.05 | 2.1±0.10 | 1.8±0.08 | 1.9±0.06 |
| P2 | At4g24190 | Endoplasmin homologue (SHD) | 76/10/7 | 1.9±0.07 | 1.4±0.11 | 1.7±0.09 | 1.7±0.20 | |
| P3 | At5g56030 | Heat shock protein 81-2 | + | 227/10/5 | 2.0±0.15 | 1.7±0.06 | 1.6±0.05 | 2.0±0.15 |
| P4 | At5g11170 | DEAD-box ATP-dependent RNA helicase 15 | 33/4/2 | 1.6±0.18 | 1.7±0.05 | 1.8±0.06 | 1.5±0.15 | |
| P5 | At5g22650 | Histone deacetylase HDT2 | + | 86/11/3 | 1.6±0.13 | 2.0±0.31 | 2.3±0.28 | 1.6±0.18 |
| P6 | X | X | ? | X | 2.3±0.15 | 2.5±0.34 | 2.5±0.26 | 2.0±0.21 |
| P7 | X | X | ? | X | 1.7±0.15 | 2.0±0.32 | 1.7±0.21 | 2.5±0.13 |
| P8 | X | X | ? | X | 2.2±0.10 | 1.7±0.09 | 1.6±0.05 | 1.4±0.08 |
| P9 | At1g09640 | Probable elongation factor 1-γ 1 | 84/10/3 | 2.0±0.08 | 1.6±0.06 | 1.5±0.07 | 1.5±0.05 | |
| P10 | At1g76180 | Dehydrin ERD14 | + | 50/20/2 | 1.7±0.04 | 2.1±0.13 | 2.3±0.15 | 1.3±0.07 |
| P11 | At5g60640 | Protein disulphide isomerase-like protein | 333/22/9 | –1.5±0.05 | –1.4±0.07 | –1.6±0.09 | –1.8±0.09 | |
| P12 | AtCg00120 | ATP synthase subunit α, chloroplastic | + | 701/25/9 | –1.3±0.06 | –1.8±0.11 | –1.5±0.10 | –1.5±0.06 |
| P13 | AtCg00490 | Rubisco large chain | + | 80/12/5 | –1.5±0.09 | –1.7±0.15 | –1.9±0.12 | –2.0±0.23 |
| At1g67090 | Rubisco small chain 1A, chloroplastic | + | 78/19/3 | |||||
| P14 | At2g39990 | eIF2 (eukaryotic translation initiation factor) | 139/17/3 | –1.6±0.05 | –1.5±0.17 | –1.2±0.25 | –2.0±0.12 | |
| P15 | At5g14740 | β-Carbonic anhydrase 2 | + | 383/25/4 | –1.5±0.08 | –1.5±0.09 | –1.4±0.08 | –1.4±0.10 |
| P16 | At5g43830 | GATase-like protein | + | 119/9/2 | –1.4±0.04 | –2.0±0.06 | –1.5±0.05 | –1.5±0.10 |
| P17 | At5g56030 | Heat shock protein 81-2 | + | 574/19/11 | –2.4±0.20 | –2.5±0.33 | –2.6±0.24 | 1.0±0.30 |
| P18 | At5g56030 | Heat shock protein 81-2/3/4 | + | 63/8/4 | –1.7±0.08 | –1.6±0.06 | –2.0±0.30 | –1.5±0.05 |
| P19 | At3g16420 | PBP1 | + | 70/12/2 | –2.5±0.35 | –1.8±0.18 | –1.8±0.25 | –1.3±0.20 |
| P20 | At5g42790 | Proteasome subunit a type-1-A | 94/27/4 | –1.9±0.23 | –2.0±0.15 | –1.5±0.08 | –1.5±0.08 | |
| P21 | At3g51880 | HMGB1 | + | 48/7/2 | –1.6±0.10 | –1.4±0.12 | –1.3±0.07 | –1.6±0.14 |
| P22 | At3g09200 | 60S Acidic ribosomal protein P0-2 | + | 124/18/3 | –2.5±0.22 | –2.0±0.32 | –2.0±0.21 | –1.2±0.20 |
| P23 | At1g20440 | Dehydrin COR47 | + | PMF: 106/43/9 | 1.7±0.08 | 1.6±0.15 | 1.3±0.15 | 1.1±0.18 |
| At4g26110 | NAP1 | + | PMF: 61/23/7 | |||||
| P24 | AtCg00490 | Rubisco large subunit | + | 291/22/7 | 1.6±0.10 | 1.6±0.08 | 1.4±0.18 | 1.3±0.15 |
| P25 | AtCg00490 | Rubisco large subunit | + | 317/16/7 | –1.6±0.07 | –1.6±0.05 | –1.2±0.40 | 1.3±0.15 |
| P26 | At1g76180 | Dehydrin ERD14 | + | 87/25/3 | –1.8±0.05 | –1.6±0.08 | –1.6±0.05 | –1.3±0.16 |
| P27 | At1g26630 | eIF5A-2 (eukaryotic translation initiation factor) | + | 95/28/3 | 1.7±0.08 | 1.4±0.14 | 1.3±0.10 | 1.5±0.05 |
| P28 | At1g20010 | Tubulin β-5 chain | 223/14/5 | 1.6±0.11 | 1.5±0.29 | 1.8±0.07 | 1.6±0.07 | |
| P29 | X | X | ? | X | 1.3±0.23 | 1.5±0.07 | 1.7±0.05 | 1.7±0.05 |
| P30 | At5g44340 | Tubulin β-4 chain | + | 488/23/11 | 1.4±0.15 | 1.4±0.05 | 1.5±0.04 | 1.4±0.15 |
| P31 | At3g09200 | 60S Acidic ribosomal protein P0-2 | + | 309/16/3 | –1.3±0.20 | –1.4±0.08 | –1.5±0.10 | –1.6±0.10 |
Spot no., spot number (as given in Fig. 1C); AGI code, accession number in the TAIR atabase; PhosPhAt, the Arabidopsis Protein Phosphorylation Site Database (Heazlewood et al., 2008); Protein name, entry name in the NCBI database; %cov, percentage of protein coverage; pep#, number of peptides; PMF, peptide mass fingerprint; Relative fold change, fold change relative to the mock control (calculated by DECODON DELTA 2D software) ±SE. Full information on the phosphoproteins including their classification, peptide sequences, and peak list is given in Supplementary Tables S2 and S4 at JXB online.
Calcium signalling in regulation of early cytokinin response phosphoproteins
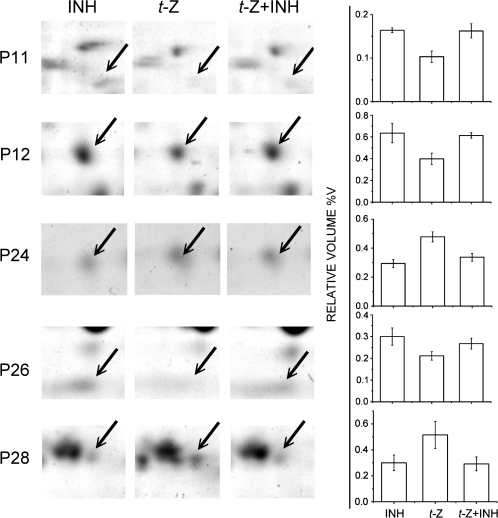
Recognition of ERD14 (P10, P26) and COR47 (P23), in which phosphorylation status and Ca2+ binding are reportedly interlinked, as early cytokinin response phosphoproteins suggested a molecular link between cytokinin action and calcium signalling. To test the involvement of calcium signalling in early phosphoproteome regulation by cytokinin, 7-day-old Arabidopsis seedlings were treated with 5 μM t-Z in the presence and absence of a calcium channel blocker (30 μM D600) and a competitive inhibitor of calcium uptake (60 μM LaCl3) for 15 min, and phosphoproteome alterations were analysed as outlined above. This resulted in identification of five phosphoproteins in which regulation by t-Z was lost in the presence of the calcium signalling inhibitors (Fig. 2), while the remaining pattern of phosphoprotein regulation remained unaltered compared with data given in Table 2.
Fig. 2.
Effect of calcium signalling inhibitors on regulation by cytokinin of early cytokinin response phosphoproteins. (A) Selected regions of 2D gels showing early cytokinin response phosphoproteins (indicated by arrows) whose regulation by 15 min treatment with 5 μM t-Z in Arabidopsis seedlings (t-Z) was abolished when calcium signalling inhibitors 30 μM D600 and 60 μM LaCl3 were administered simultaneously with 5 μM t-Z (t-Z + INH). Control samples were treated with the inhibitors only (INH). For details see Materials and methods. Spot numbers as in Fig. 1C and Table 2. (B) Relative volumes of the individual spots.
Comparison of proteome and phosphoproteome data sets
The sets of differentially expressed proteins and phosphoproteins (Fig. 1) included seven overlapping proteins: At3g09200 (T34, P22, P31), At3g16420 (T38, P19), At4g24190 (T2, P2), At5g14740 (T42, P15), At5g60640 (T11, P11), AtCg00120 (T12, T16, P12), and AtCg00490 (T13, T18, P13). However, the apparent pI and molecular mass values were indistinguishable in proteome and phosphoproteome maps for only one of them, endoplasmin homologue (At4g24190), indicating that the proteins displayed in the protein and phosphoprotein maps may be subjected to distinct PTMs affecting their apparent pI, molecular mass, or both.
Classification of identified proteins and phosphoproteins
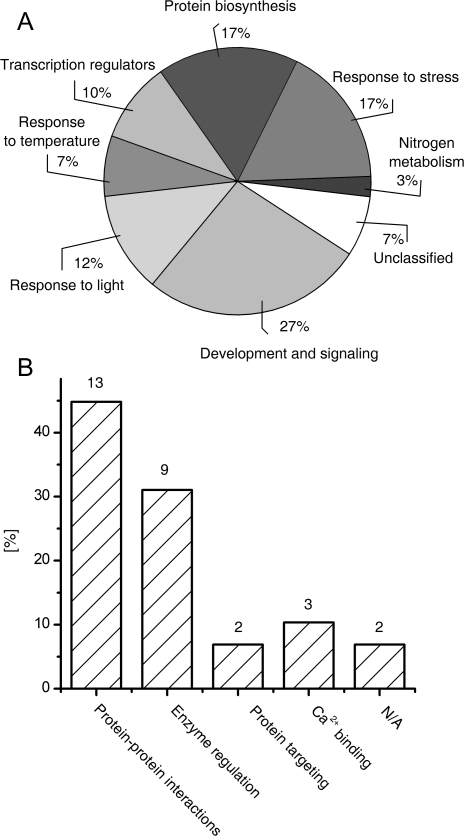
Identified proteins were categorized using criteria described by Bevan et al. (1998). As shown in Fig. 3A, significant fractions of the phosphoproteins are involved in responses to environmental (stress, light, and temperature) stimuli (36%), and signalling and development pathways (27%). Other highly represented functional categories are protein biosynthesis (17%) and transcription regulators (10%). Given previously reported data on the functional significance of phosphorylation of the phosphoproteins identified here, the present data set also indicates that phosphorylation regulation by cytokinins might mainly regulate protein–protein and protein–ligand/substrate interactions (Fig. 3B).
Fig. 3.
(A) Classification of the early cytokinin response phosphoproteins according to their cellular functions (Bevan et al., 1998) and (B) molecular processes reportedly controlled by their phosphorylation as deduced from database entries and literature review.
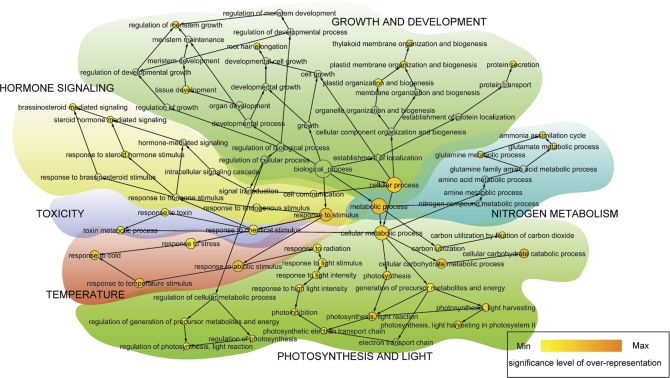
To gain deeper insights into biological processes in which the differentially regulated proteins are involved, the gene ontology (GO) of proteins showing significant (P <0.05), ≥1.4-fold changes in expression between cytokinin-challenged and control samples was analysed. The results were visualized using BiNGO, a graphical tool enabling GO classes in clustered data to be highlighted (http://www.psb.ugent.be/cbd/papers/BiNGO/) (Fig. 4). The GO categories that were identified as being significantly over-represented were ‘Growth and development’ (including subclasses such as ‘Thylakoid development’ and ’Protein transport’), ‘Nitrogen metabolism’, ‘Hormone signalling’, ‘Photosynthesis and light’ (e.g. ‘Response to light stimulus’, ‘Response to light intensity’, and ‘Photosynthesis–light reaction and light harvesting’), and ‘Toxicity’. Further, ‘Temperature response’ was the only down-regulated GO category.
Fig. 4.
Gene ontology (GO) analysis of the early cytokinin response proteins in Arabidopsis (performed in Cytoscape using BiNGO plugin version 2.3). GO categories that were significantly over-represented among the differentially expressed proteins were identified. The yellow to orange colour of the circles indicates the level of significance of over-represented categories (P=0.05, yellow; P=10−7, orange). The size of the circles is proportional to the number of proteins in each category. Links with low significance were removed manually to reduce complexity of the image.
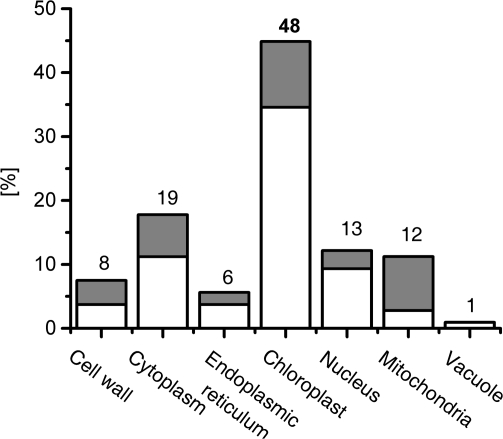
The subcellular location of each identified protein was determined according to the TAIR database (http://www.arabidopsis.org), and the results are summarized in Fig. 5. The largest group of proteins was localized to chloroplasts (52%), followed by the cytoplasm (17%) and mitochondria (14%). Among phosphoproteins, 31, 25, and 19% were localized to chloroplasts, the nucleus, and cytoplasm, respectively.
Fig. 5.
Subcellular distribution of the early cytokinin response proteins (white) and phosphoproteins (grey) according to the UniProt database (http://www.uniprot.org). The numbers above the columns represent sums of the identified proteins and phosphoproteins located in the respective cellular compartment.
Discussion
Proteomic and phosphoproteomic effects of cytokinin treatment on Arabidopsis seedlings were analysed to identify early cytokinin response proteins and phosphoproteins in order to elucidate molecular mechanisms involved in cytokinin action. The identified proteins and phosphoproteins represent a snapshot of early links in various well known cytokinin-regulated signalling circuits and cellular processes. The results also indicate as yet unrecognized links between temperature, calcium, and cytokinin signalling. Comparative analysis revealed differences in both the potency of the four representative cytokinins to trigger the responses, and the contributions of specific cytokinin receptors to phosphoproteomic responses to t-Z treatment.
Phosphoproteome isolation and analysis
A previously established procedure for isolating mammalian and yeast phosphoproteins by affinity chromatography prior to further analysis by 2-DE and MS (Makrantoni et al., 2005) was optimized and employed in this study. The procedure is specific for proteins phosphorylated at serine or threonine residues. Thus, it is capable of detecting most phosphorylated proteins in eukaryotic cells since their pSer:pThr:pTyr and pHis/pTyr ratios are typically 1800:200:1 and 10–100:1, respectively (Klumpp and Krieglstein, 2002; Laugesen et al., 2004). The procedure is reportedly reliable for plant phosphoproteome analysis (Laugesen et al., 2006; Meimoun et al., 2007), and sample preparation for the 2-DE stage of the protocol has been further improved. All proteins identified on the phosphoproteome map show the predicted phosphorylation sites in NetPhos (Blom et al., 1999). Most are already included in the PhosPhAt database (Heazlewood et al., 2008), and the possibility of PTM by phosphorylation of the others has been previously documented (Table 2). Here, phosphorylation for 60S acidic ribosomal protein P0-2 (P22) and endoplasmin homologue (SHD; P2) was confirmed directly, and it was shown that all but two differentially regulated spots are stained by the phosphoprotein-specific stain Phos-Tag (Supplementary Fig. S3 at JXB online). Further, phosphorylation-dependent shifts in apparent (SDS–PAGE) molecular masses of COR47 and ERD14 have been reported (Alsheikh et al., 2005), and increases in apparent molecular masses of these proteins after cytokinin stimulation (P10, P23) were observed.
Based on previous reports, the changes in phosphoprotein levels detected here may reflect phosphorylation events mainly involved in regulation of protein–protein or protein–ligand/substrate interactions. Modulation of enzyme activity and specificity by phosphorylation has been shown for histone deacetylase (Pflum et al., 2001), protein disulphide isomerase (Guthapfel et al., 1996), β-carbonic anhydrase (Church et al., 1980), Rubisco (Aggarwal et al., 1993), RNA helicase (Yang et al., 2005), and ATP synthase (Murtazina et al., 2001). Phosphorylation also reportedly mediates association and assembly of protein complexes of Rubisco (Aggarwal et al., 1993), 60S acidic ribosomal protein P0-2 (Naranda and Ballesta, 1991), endoplasmin (Brunati et al., 2000), tubulins (Blume et al., 2009), proteasome subunits (Umeda et al., 1997), and heat shock proteins (Picard, 2002). Down-regulation of eIF2 (P14), and up-regulation of eIF5A-2 and probable elongation factor 1-γ1 (P28, P9) are consistent with previously reported stimulation of proteosynthesis by cytokinins as eIF2 reportedly inhibits initiation of proteosynthesis in its phosphorylated form (Zhang et al., 2008) while phosphorylation of eIF5 and EF-1 reportedly stimulates formation of initiation complexes (Majumdar et al., 2002) and enhances proteosynthesis (Belle et al., 1995), respectively. Further, phosphorylation reportedly promotes NAP1 import into the nucleus (Calvert et al., 2008), while HMGB1 requires phosphorylation for export from the nucleus (Youn and Shin, 2006). Finally, several phosphoproteins reportedly related to other signalling pathways were identified that may be involved in as yet unrecognized branches of cytokinin signalling and/or as molecular players in cross-talk between cytokinins and other stimuli. For example, PATL-2 (P1) is reportedly involved in membrane trafficking events (Peterman et al., 2004) and its phosphorylation has been localized to its phosphoinositide-binding pocket (Jones et al., 2009). This is consistent with recent indications of a role for intracellular trafficking in cytokinin signalling (Dortay et al., 2008).
Temperature perception
A novel theme highlighted by the present analysis is differential regulation by cytokinins of proteins and phosphoproteins reportedly involved in responses to high and low temperatures (Nylander et al., 2001; Sung et al., 2001; Bae et al., 2003; Goulas et al., 2006; Lim et al., 2006; Sasaki et al., 2007). The proteins include heat shock cognate 70 kDa protein 1, fructose-1,6-bisphosphatase, phosphoribulokinase, phosphoglycerate kinase, 60S acidic ribosomal protein P0-2, putative ribonucleoprotein At2g37220, and cold shock domain protein 2, and the phosphoproteins comprise heat shock protein 81-2, 60S acidic ribosomal protein P0-2, and dehydrins COR47 and ERD14. Interestingly, all the proteins were down-regulated by cytokinins, suggesting there may be shared components of cytokinin and temperature signalling pathways. In plants, mechanisms underlying temperature perception are poorly understood (Penfield, 2008). However, a two-component signalling pathway is known to act in temperature perception in cyanobacteria (e.g. Synechocystis), with a histidine kinase perceiving and relaying temperature signals (Suzuki et al., 2000), and a two-component signalling pathway is the main known component of cytokinin signalling chains in plants. Accordingly, a role for cytokinins in responses to cold stress was recently deduced from the apparent attenuation of cytokinin signalling under cold stress (Argueso et al., 2009). Decreases in levels of endogenous cytokinins in heat shock-treated plants have also been documented (Hare et al., 1997), and Burkhanova et al. (2001) found that responses to BA were enhanced in heat-shocked Arabidopsis thaliana, prompting speculation that heat shock proteins may be involved in cytokinin signalling.
Chloroplast biogenesis and function
The differentially regulated proteins and phosphoproteins detected in the presented experiments include a remarkably high percentage of proteins located to chloroplasts—45%—compared with 7.9% chloroplast proteins predicted for the whole genome (Bevan et al., 1998). They reflect most processes involved in chloroplast biogenesis and function, including: mRNA processing; protein biosynthesis, folding, and degradation; light and dark reactions of photosynthesis; carbon utilization; carbohydrate metabolism; glycolysis; fatty acid biosynthesis; and stress responses. Both the number and variety of functions in which they are implicated substantially exceed estimates obtained in two previous proteomic analyses of cytokinin action. In Physcomitrella patens, four early response phosphoproteins located to chloroplasts were identified following short cytokinin treatment (Heintz et al., 2006), and 10 up-regulated chloroplast proteins were found when effects of continuously increasing endogenous cytokinin levels in dark-grown Arabidopsis seedlings were investigated (Lochmanová et al., 2008). For two of the Physcomitrella phosphoproteins (Rubisco large subunit and carbonic anhydrase) functional matches are present in our data set. The high number of early cytokinin response proteins and phosphoproteins located in chloroplasts might indicate an as yet uncharacterized direct branch in cytokinin signalling responsible for cytokinin action in chloroplasts. Consistently, 10 interactors of cytokinin receptors have been located to chloroplasts (Dortay et al., 2008), and one of them, Rubisco small chain 1A (At1g67090), was identified here as a constituent of the differentially regulated spot P13. Brenner et al. (2005) have proposed that rapid transfer of cytokinin signals to plastids, or direct perception and interpretation of the signals by the plastids, may explain the fast regulation of five chloroplast transcripts by cytokinin treatment. Interestingly, the chloroplast cytokinin pool has been found to be dynamic (Benková et al., 1999), and compartmentation into chloroplasts of some cytokinin biosynthesis and metabolism pathways has been reported (Brzobohatý et al., 1993; Kristoffersen et al., 2000; Takei et al., 2004; Kiran et al., 2006).
Chromatin remodelling and nuclear proteins
Novel insights into possible involvement of cytokinins in chromatin remodelling were obtained by identifying HMGB1, histone deacetylase HDT2, and possibly NAP1;1 as early cytokinin response phosphoproteins. Interestingly, cytokinin response genes have been found to be up-regulated in hmgb1 knockout mutants (Lildballe et al., 2008), while NAP1;2 protein reportedly interacts with ARR7 (Dortay et al., 2008), and NAP1 proteins are positive regulators in the ABA signalling pathway (Liu et al., 2009). The expression level of genes involved in chromatin remodelling has been found to change after just 120 min cytokinin treatment (Brenner et al., 2005), clearly demonstrating the power of proteomic profiling for identifying primary events in cytokinin action.
Cytokinin and cytokinin receptor specificity in eliciting proteomic/phosphoproteomic alterations
The present experiments showed that the representatives of the four cytokinin classes had largely similar qualitative proteomic and phosphoproteomic effects, although the extent of up- or down-regulation varied for most proteins and phosphoproteins. The varying degree of responsiveness might reflect the molecular basis of differential activities of different cytokinin classes previously observed in various physiological experiments (e.g. Mok et al., 1978; Sujatha and Reddy, 1998; Lexa et al., 2003; Hradilová et al., 2007), and is consistent with distinct specificities of the individual cytokinin receptors for individual cytokinin moieties (Spíchal et al., 2004; Yonekura-Sakakibara et al., 2004; Romanov et al., 2006). Generally, the highest degree of differential regulation of proteins and phosphoproteins was elicited by BA and TDZ (reportedly the most potent cytokinin in activation of the cytokinin primary response gene ARR5 in Arabidopsis; Spíchal et al., 2004). The present study is the first global comparison of the effects of all four classes of cytokinins. However, Rashotte et al. (2003) compared the effects of BA and t-Z on expression profiles in Arabidopsis and found that while both cytokinins up-regulated largely overlapping sets of genes, far fewer genes were found to be down-regulated by t-Z than by BA, concluding that some of these genes may be specifically down-regulated by BA or not actually regulated by cytokinin.
The profiling of t-Z action in the cytokinin receptor double mutants ahk2ahk3, ahk2cre1, and ahk3cre1 provides first insights into the specificity of outputs of specific cytokinin receptors at the proteome level. Most of the identified phosphoproteins were found to be differentially regulated primarily by a single cytokinin receptor. However, indications of inter-receptor cooperation were seen for some of the differentially regulated phosphoproteins. Detection of a significant number of phosphoproteins regulated by two individual cytokinin receptors was consistent with reportedly overlapping functions of the cytokinin receptors (Inoue et al., 2001; Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). Defining the output of cytokinin receptors as the number of phosphoproteins they apparently regulate, their output decreases in the order AHK3>AHK4/CRE1/WOL1>>AHK2. This observation is consistent with degrees of involvement of the cytokinin receptors in various biological processes previously found in morphological, physiological, and molecular analysis of cytokinin receptor mutants (Riefler et al., 2006).
Calcium signalling
The present work provides the first molecular link between cytokinin action and signalling pathways involving modulation of free Ca2+ levels, showing that early cytokinin response phosphoproteins include ERD14 and COR47, for which correlations between phosphorylation status and Ca2+ binding have been demonstrated (Alsheikh et al., 2005). In addition, it is shown that inhibition of calcium signalling abolishes cytokinin regulation of several phosphoproteins, further supporting the interlinking of cytokinin and calcium signalling. In planta, inhibition of calcium signalling disrupts cytokinin-induced bud formation in the moss Funaria (Saunders and Helper, 1983). Ca2+ signalling is reportedly involved in the transduction of diverse abiotic, biotic, and developmental stimuli including temperature and plant hormones (Sanders et al., 2002). In this context, increases in the phosphorylation of SHD and HSP 81-2 (P2, P3), proteins related to the hsp-90 family, whose members are known to be autophoshorylated in the presence of Ca2+ (Csermely and Kahn, 1991), were also observed. Further, Ca2+-mediated signalling may represent a rapid mechanism of transmitting cytokinin signals into chloroplasts. It has long been established that chloroplast-localized physiological processes are subject to regulation by Ca2+ and, accordingly, a Ca2+-sensing receptor has been localized to the chloroplast and found to modulate cytoplasmic Ca2+ concentrations (Nomura et al., 2008; Weinl et al., 2008).
Conclusion
In conclusion, a novel proteome- and phosphoproteome-wide view of changes in abundance of proteins and phosphoproteins is presented that might be functionally relevant for the many biological processes regulated by cytokinins. Importantly, the results indicate as yet unrecognized links between temperature, calcium, and cytokinin signalling. Interlinking of cytokinin and calcium signalling is further supported by loss of cytokinin regulation of several phosphoproteins following inhibition of calcium signalling. Rapid regulation of a number of chloroplast phosphoproteins suggests a currently uncharacterized direct signalling chain responsible for cytokinin action in chloroplast. Comparative analysis of the representatives of the four cytokinin classes revealed largely similar regulation patterns in the 7-day-old Arabidopsis seedlings. First insights into the specificity of cytokinin receptors on phosphoproteomic effects were obtained from analysis of cytokinin action in the set of cytokinin receptor double mutants. The presented data provide a new framework for further detailed investigations, using, for example, mutants and transgenic plants, of molecular mechanisms involved in cytokinin action. Further identification of kinase(s) and phosphatase(s) involved in phosphorylation and dephosphorylation events triggered by cytokinins and elucidation of their relationships to cytokinin receptors are important challenges for future work. Interestingly, two protein kinases have been identified as AHK2 and AHK4 interactors, and a phosphatase as an ARR4 interactor (Dortay et al., 2008).
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Schematic representation of the experimental design.
Figure S2. Confirmation of peptide phosphorylation.
Figure S3. Detection of phosphoproteins isolated using the PhosphoProtein Purification Kit and separated by 2-DE.
Table S1. Early cytokinin response proteins of Arabidopsis (an extended version of Table 1)
Table S2. Early cytokinin response phosphoproteins of Arabidopsis (an extended version of Table 2)
Table S3. Detailed MS information on the early cytokinin response proteins of Arabidopsis.
Table S4. Detailed MS information on the early cytokinin response phosphoproteins of Arabidopsis.
Acknowledgments
We thank Professor Thomas Schmülling for ahk2ahk3, ahk2cre1, and ahk3cre1 seeds, and Dr Přemysl Souček for providing us with unpublished ARR3 and ARR5 expression data. This work was supported by grants LC06034 and 1M06030 (Ministry of Education of the Czech Republic), IAA600040701 (Grant Agency of the Academy of Sciences of the Czech Republic), GACR 206/09/2062 (Grant Agency of the Czech Republic), and AV0Z50040507, AV0Z50040702 and AV0Z40310501 (Academy of Sciences of the Czech Republic).
References
- Aggarwal KK, Saluja D, Sachar RC. Phosphorylation of rubisco in Cicer arietinum: non-phosphoprotein nature of rubisco in Nicotiana tabacum. Phytochemistry. 1993;34:329–335. [Google Scholar]
- Alsheikh MK, Svensson JT, Randall SK. Phosphorylation regulated ion-binding is a property shared by the acidic subclass dehydrins. Plant, Cell and Environment. 2005;28:1114–1122. [Google Scholar]
- Argueso CT, Ferreira FJ, Kieber JJ. Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant, Cell and Environment. 2009;32:1147–1160. doi: 10.1111/j.1365-3040.2009.01940.x. [DOI] [PubMed] [Google Scholar]
- Bae MS, Cho EJ, Choi E, Park OK. Analysis of the Arabidopsis nuclear proteome and its response to cold stress. The Plant Journal. 2003;36:652–663. doi: 10.1046/j.1365-313x.2003.01907.x. [DOI] [PubMed] [Google Scholar]
- Benková E, Witters E, Van Dongen W, Kolář J, Motyka V, Brzobohatý B, Van Onckelen HA, Macháčková I. Cytokinins in tobacco and wheat chloroplasts: occurrence and changes due to light/dark treatment. Plant Physiology. 1999;121:245–251. doi: 10.1104/pp.121.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle R, Minella O, Cormier P, Morales J, Poulhe R, Mulner-Lorillon O. Phosphorylation of elongation factor-1 (EF-1) by cdc2 kinase. Progress in Cell Cycle Research. 1995;1:265–270. doi: 10.1007/978-1-4615-1809-9_21. [DOI] [PubMed] [Google Scholar]
- Bevan M, Bancroft I, Bent E, et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. Journal of Molecular Biology. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Blume YB, Lloyd CW, Yemets AI. Plant tubulin phosphorylation and its role in cell cycle progression. In: Blume Y, Baird W, Yemets A, Breviario D, editors. The plant cytoskeleton: a key tool for agro-biotechnology. Vol. III. Dordrecht, The Netherlands: Springer; 2009. pp. 145–159. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T. Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. The Plant Journal. 2005;44:314–333. doi: 10.1111/j.1365-313X.2005.02530.x. [DOI] [PubMed] [Google Scholar]
- Brunati AM, Contri A, Muenchbach M, James P, Marin O, Pinna LA. Grp94 (endoplasmin) co-purifies with and is phosphorylated by golgi apparatus casein kinase. FEBS Letters. 2000;471:151–155. doi: 10.1016/s0014-5793(00)01378-8. [DOI] [PubMed] [Google Scholar]
- Brzobohatý B, Moore I, Kristoffersen P, Bako L, Campos N, Schell J, Palme K. Release of active cytokinin by a β-glucosidase localized to the maize root meristem. Science. 1993;262:1051–1054. doi: 10.1126/science.8235622. [DOI] [PubMed] [Google Scholar]
- Brzobohatý B, Moore I, Palme K. Cytokinin metabolism: implications for regulation of plant growth and development. Plant Molecular Biology. 1994;26:1483–1497. doi: 10.1007/BF00016486. [DOI] [PubMed] [Google Scholar]
- Burkhanova EA, Mikulovich TP, Kudryakova NV, Kukina IM, Smith AR, Hall MA, Kulaeva ON. Heat shock pre-treatment enhances the response of Arabidopsis thaliana leaves and Cucurbita pepo cotyledons to benzyladenine. Plant Growth Regulation. 2001;33:195–198. [Google Scholar]
- Calvert ME, Keck KM, Ptak C, Shabanowitz J, Hunt DF, Pemberton LF. Phosphorylation by casein kinase 2 regulates Nap1 localization and function. Molecular and Cellular Biology. 2008;28:1313–1325. doi: 10.1128/MCB.01035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Hwang I. Cytokinin: perception, signal transduction, and role in plant growth and development. Journal of Plant Biology. 2007;50:98–108. [Google Scholar]
- Church GA, Kimelberg HK, Sapirstein VS. Stimulation of carbonic anhydrase activity and phosphorylation in primary astroglial cultures by norepinephrine. Journal of Neurochemistry. 1980;34:873–879. doi: 10.1111/j.1471-4159.1980.tb09660.x. [DOI] [PubMed] [Google Scholar]
- Csermely P, Kahn CR. The 90-kDa heat shock protein (hsp-90) possesses an ATP binding site and autophosphorylating activity. Journal of Biological Chemistry. 1991;266:4943–4950. [PubMed] [Google Scholar]
- Damerval C, De Vienne D, Zivy M, Thiellement H. Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis. 1986;7:52–54. [Google Scholar]
- Dortay H, Gruhn N, Pfeifer A, Schwerdtner M, Schmülling T, Heyl A. Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. Journal of Proteome Research. 2008;7:3649–3660. doi: 10.1021/pr0703831. [DOI] [PubMed] [Google Scholar]
- Ferreira FJ, Kieber JJ. Cytokinin signaling. Current Opinion in Plant Biology. 2005;8:518–525. doi: 10.1016/j.pbi.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Goulas E, Schubert M, Kieselbach T, Kleczkowski LA, Gardeström P, Schröder W, Hurry V. The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. The Plant Journal. 2006;47:720–734. doi: 10.1111/j.1365-313X.2006.02821.x. [DOI] [PubMed] [Google Scholar]
- Guthapfel R, Gueguen P, Quemeneur E. ATP binding and hydrolysis by the multifunctional protein disulfide isomerase. Journal of Biological Chemistry. 1996;271:2663–2666. doi: 10.1074/jbc.271.5.2663. [DOI] [PubMed] [Google Scholar]
- Hare PD, Cress WA, van Staden J. The involvement of cytokinins in plant responses to environmental stress. Plant Growth Regulation. 1997;23:79–103. [Google Scholar]
- Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, Schulze WX. Phosphat: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Research. 2008;36:D1015–D1021. doi: 10.1093/nar/gkm812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz D, Erxleben A, High AA, Wurtz V, Reski R, Van Dorsselaer A, Sarnighausen E. Rapid alteration of the phosphoproteome in the Moss physcomitrella patens after cytokinin treatment. Journal of Proteome Research. 2006;5:2283–2293. doi: 10.1021/pr060152e. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, et al. In planta functions of the Arabidopsis cytokinin receptor family. Proceedings of the National Academy of Sciences, USA. 2004;101:8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S, Ikeda Y, Morgante M, Wang X, Zuo J, Hanafey MK, Gaasterland T, Tingey SV, Chua N. Monitoring genome-wide changes in gene expression in response to endogenous cytokinin reveals targets in Arabidopsis thaliana. FEBS Letters. 2003;554:373–380. doi: 10.1016/s0014-5793(03)01194-3. [DOI] [PubMed] [Google Scholar]
- Hradilová J, Malbeck J, Brzobohatý B. Cytokoinin regulation of gene expression in the AHP gene family in Arabidopsis thaliana. Journal of Plant Growth Regulation. 2007;26:229–244. [Google Scholar]
- Hradilová J, Řehulka P, Řehulková H, Vrbová M, Griga M, Brzobohatý B. Comparative analysis of proteomic changes in contrasting flax cultivars upon cadmium exposure. Electrophoresis. 2010;31:421–431. doi: 10.1002/elps.200900477. [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Jones AME, MacLean D, Studholme DJ, Serna-Sanz A, Andreasson E, Rathjen JP, Peck SC. Phosphoproteomic analysis of nuclei-enriched fractions from Arabidopsis thaliana. Journal of Proteomics. 2009;72:439–451. doi: 10.1016/j.jprot.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Kakimoto T. Biosynthesis of cytokinins. Journal of Plant Research. 2003;116:233–239. doi: 10.1007/s10265-003-0095-5. [DOI] [PubMed] [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T. Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His→Asp phosphorelay circuitry. Plant and Cell Physiology. 2005;46:339–355. doi: 10.1093/pcp/pci033. [DOI] [PubMed] [Google Scholar]
- Kiran NS, Polanská L, Fohlerová R, et al. Ectopic over-expression of the maize β-glucosidase Zm-p60.1 perturbs cytokinin homeostasis in transgenic tobacco. Journal of Experimental Botany. 2006;57:985–996. doi: 10.1093/jxb/erj084. [DOI] [PubMed] [Google Scholar]
- Klumpp S, Krieglstein J. Phosphorylation and dephosphorylation of histidine residues in proteins. European Journal of Biochemistry. 2002;269:1067–1071. doi: 10.1046/j.1432-1033.2002.02755.x. [DOI] [PubMed] [Google Scholar]
- Kristoffersen P, Brzobohatý B, Höhfeld I, Bako L, Melkonian M, Palme K. Developmental regulation of the maize Zm-p60.1 gene encoding a β-glucosidase located to plastids. Planta. 2000;210:407–415. doi: 10.1007/pl00008149. [DOI] [PubMed] [Google Scholar]
- Larsen MR, Sørensen GL, Fey SJ, Larsen PM, Roepstorff P. Phospho-proteomics: evaluation of the use of enzymatic de-phosphorylation and differential mass spectrometric peptide mass mapping for site specific phosphorylation assignment in proteins separated by gel electrophoresis. Proteomics. 2001;1:223–228. doi: 10.1002/1615-9861(200102)1:2<223::AID-PROT223>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Laugesen S, Bergoin A, Rossignol M. Deciphering the plant phosphoproteome: tools and strategies for a challenging task. Plant Physiology and Biochemistry. 2004;42:929–936. doi: 10.1016/j.plaphy.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Laugesen S, Messinese E, Hem S, Pichereaux C, Grat S, Ranjeva R, Rossignol M, Bono J. Phosphoproteins analysis in plants: a proteomic approach. Phytochemistry. 2006;67:2208–2214. doi: 10.1016/j.phytochem.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Lexa M, Genkov T, Malbeck J, Macháčková I, Brzobohatý B. Dynamics of endogenous cytokinin pools in tobacco seedlings: a modelling approach. Annals of Botany. 2003;91:585–597. doi: 10.1093/aob/mcg061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lildballe DL, Pedersen DS, Kalamajka R, Emmersen J, Houben A, Grasser KD. The expression level of the chromatin-associated HMGB1 protein influences growth, stress tolerance, and transcriptome in Arabidopsis. Journal of Molecular Biology. 2008;384:9–21. doi: 10.1016/j.jmb.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Lim CJ, Yang KA, Hong JK, Choi JS, Yun D, Hong JC, Chung WS, Lee SY, Cho MJ, Lim CO. Gene expression profiles during heat acclimation in Arabidopsis thaliana suspension-culture cells. Journal of Plant Research. 2006;119:373–383. doi: 10.1007/s10265-006-0285-z. [DOI] [PubMed] [Google Scholar]
- Liu Z, Gao J, Dong A, Shen W. A truncated Arabidopsis nucleosome assembly protein 1, atNAP1;3T, alters plant growth responses to abscisic acid and salt in the Atnap1;3-2 mutant. Molecular Plant. 2009;2:688–699. doi: 10.1093/mp/ssp026. [DOI] [PubMed] [Google Scholar]
- Lochmanová G, Zdráhal Z, Konečná H, Koukalová Š, Malbeck J, Souček P, Válková M, Kiran NS, Brzobohatý B. Cytokinin-induced photomorphogenesis in dark-grown Arabidopsis: a proteomic analysis. Journal of Experimental Botany. 2008;59:3705–3719. doi: 10.1093/jxb/ern220. [DOI] [PubMed] [Google Scholar]
- Majumdar R, Bandyopadhyay A, Deng H, Maitra U. Phosphorylation of mammalian translation initiation factor 5 eIF5) in vitroand in vivo. Nucleic Acids Research. 2002;30:1154–1162. doi: 10.1093/nar/30.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrantoni V, Antrobus R, Botting CH, Coote PJ. Rapid enrichment and analysis of yeast phosphoproteins using affinity chromatography, 2D-PAGE and peptide mass fingerprinting. Yeast. 2005;22:401–414. doi: 10.1002/yea.1220. [DOI] [PubMed] [Google Scholar]
- Meimoun P, Ambard-Bretteville F, Colas-des Francs-Small C, Valot B, Vidal J. Analysis of plant phosphoproteins. Analytical Biochemistry. 2007;371:238–246. doi: 10.1016/j.ab.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Miller CO, Skoog F, von Saltza MH, Strong F. Kinetin, a cell division factor from deoxyribonucleic acid. Journal of the American Chemical Society. 1955;77:1392. [Google Scholar]
- Mok D, Mok M. Cytokinin metabolism and action. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- Mok M, Mok D, Armstrong D. Differential cytokinin structure–activity relationships in Phaseolus. Plant Physiology. 1978;61:72–75. doi: 10.1104/pp.61.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtazina DA, Petukhov SP, Rubtsov AM, Storey KB, Lopina OD. Phosphorylation of the alpha-subunit of Na, K-ATPase from duck salt glands by cAMP-dependent protein kinase inhibits the enzyme activity. Biochemistry. 2001;66:865–874. doi: 10.1023/a:1011900718655. [DOI] [PubMed] [Google Scholar]
- Naranda T, Ballesta JP. Phosphorylation controls binding of acidic proteins to the ribosome. Proceedings of the National Academy of Sciences, USA. 1991;88:10563–10567. doi: 10.1073/pnas.88.23.10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. The Plant Cell. 2004;16:1365–1377. doi: 10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura H, Komori T, Kobori M, Nakahira Y, Shiina T. Evidence for chloroplast control of external Ca2+-induced cytosolic Ca2+transients and stomatal closure. The Plant Journal. 2008;53:988–998. doi: 10.1111/j.1365-313X.2007.03390.x. [DOI] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Molecular Biology. 2001;45:263–279. doi: 10.1023/a:1006469128280. [DOI] [PubMed] [Google Scholar]
- Penfield S. Temperature perception and signal transduction in plants. New Phytologist. 2008;179:615–628. doi: 10.1111/j.1469-8137.2008.02478.x. [DOI] [PubMed] [Google Scholar]
- Peterman TK, Ohol YM, McReynolds LJ, Luna EJ. Patellin1, a novel sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiology. 2004;136:3080–3094. doi: 10.1104/pp.104.045369. discussion 3001–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflum MK, Tong JK, Lane WS, Schreiber SL. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. Journal of Biological Chemistry. 2001;276:47733–47741. doi: 10.1074/jbc.M105590200. [DOI] [PubMed] [Google Scholar]
- Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cellular and Molecular Life Sciences. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Carson SDB, To JPC, Kieber JJ. Expression profiling of cytokinin action in Arabidopsis. Plant Physiology. 2003;132:1998–2011. doi: 10.1104/pp.103.021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. The Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov GA, Lomin SN, Schmülling T. Biochemical characteristics and ligand-binding properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. Journal of Experimental Botany. 2006;57:4051–4058. doi: 10.1093/jxb/erl179. [DOI] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF. Calcium at the crossroads of signaling. The Plant Cell. 2002;14(Supplement):S401–S417. doi: 10.1105/tpc.002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders MJ, Helper PK. Calcium antagonists and calmodulin inhibitors block cytokinin-induced bud formation in Funaria. Developmental Biology. 1983;99:41–49. doi: 10.1016/0012-1606(83)90252-x. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Kim M, Imai R. Arabidopsis cold shock domain protein2 is a RNA chaperone that is regulated by cold and developmental signals. Biochemical and Biophysical Research Communications. 2007;364:633–638. doi: 10.1016/j.bbrc.2007.10.059. [DOI] [PubMed] [Google Scholar]
- Souček P, Klíma P, Reková A, Brzobohatý B. Involvement of hormones and KNOXI genes in early Arabidopsis seedling development. Journal of Experimental Botany. 2007;58:3797–3810. doi: 10.1093/jxb/erm236. [DOI] [PubMed] [Google Scholar]
- Spíchal L, Rakova NY, Riefler M, Mizuno T, Romanov GA, Strnad M, Schmülling T. Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant and Cell Physiology. 2004;45:1299–1305. doi: 10.1093/pcp/pch132. [DOI] [PubMed] [Google Scholar]
- Strnad M. The aromatic cytokinins. Physiologia Plantarum. 1997;101:674–688. [Google Scholar]
- Sujatha M, Reddy TP. Differential cytokinin effects on the stimulation of in vitro shoot proliferation from meristematic explants of castor (Ricinus communis L.) Plant Cell Reports. 1998;17:561–566. doi: 10.1007/s002990050442. [DOI] [PubMed] [Google Scholar]
- Sung DY, Vierling E, Guy CL. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiology. 2001;126:789–800. doi: 10.1104/pp.126.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I, Los DA, Kanesaki Y, Mikami K, Murata N. The pathway for perception and transduction of low-temperature signals in Synechocystis. The EMBO Journal. 2000;19:1327–1334. doi: 10.1093/emboj/19.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant and Cell Physiology. 2004;45:1053–1062. doi: 10.1093/pcp/pch119. [DOI] [PubMed] [Google Scholar]
- Umeda M, Manabe Y, Uchimiya H. Phosphorylation of the C2 subunit of the proteasome in rice (Oryza sativa L.) FEBS Letters. 1997;403:313–317. doi: 10.1016/s0014-5793(97)00073-2. [DOI] [PubMed] [Google Scholar]
- Weinl S, Held K, Schlücking K, Steinhorst L, Kuhlgert S, Hippler M, Kudla J. A plastid protein crucial for Ca2+-regulated stomatal responses. New Phytologist. 2008;179:675–686. doi: 10.1111/j.1469-8137.2008.02492.x. [DOI] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu ZR. Phosphorylations of DEAD box p68 RNA helicase are associated with cancer development and cell proliferation. Molecular Cancer Research. 2005;3:355–363. doi: 10.1158/1541-7786.MCR-05-0022. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H. Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiology. 2004;134:1654–1661. doi: 10.1104/pp.103.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JH, Shin J- S. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. Journal of Immunology. 2006;177:7889–7897. doi: 10.4049/jimmunol.177.11.7889. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Kanyuka K, Parry MAJ, Powers SJ, Halford NG. GCN2-dependent phosphorylation of eukaryotic translation initiation factor-2alpha in Arabidopsis. Journal of Experimental Botany. 2008;59:3131–3141. doi: 10.1093/jxb/ern169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.