Abstract
Genes that enable crops to limit Na+ accumulation in shoot tissues represent potential sources of salinity tolerance for breeding. In barley, the HvNax4 locus lowered shoot Na+ content by between 12% and 59% (g−1 DW), or not at all, depending on the growth conditions in hydroponics and a range of soil types, indicating a strong influence of environment on expression. HvNax4 was fine-mapped on the long arm of barley chromosome 1H. Corresponding intervals of ∼200 kb, containing a total of 34 predicted genes, were defined in the sequenced rice and Brachypodium genomes. HvCBL4, a close barley homologue of the SOS3 salinity tolerance gene of Arabidopsis, co-segregated with HvNax4. No difference in HvCBL4 mRNA expression was detected between the mapping parents. However, genomic and cDNA sequences of the HvCBL4 alleles were obtained, revealing a single Ala111Thr amino acid substitution difference in the encoded proteins. The known crystal structure of SOS3 was used as a template to obtain molecular models of the barley proteins, resulting in structures very similar to that of SOS3. The position in SOS3 corresponding to the barley substitution does not participate directly in Ca2+ binding, post-translational modifications or interaction with the SOS2 signalling partner. However, Thr111 but not Ala111 forms a predicted hydrogen bond with a neighbouring α-helix, which has potential implications for the overall structure and function of the barley protein. HvCBL4 therefore represents a candidate for HvNax4 that warrants further investigation.
Keywords: Barley, Brachypodium, calcineurin-B like, genetic mapping, protein modelling, QTL, rice, salinity tolerance, sodium exclusion, SOS3
Introduction
The impacts of salinity on plant growth arise through the effects of dehydration (osmotic toxicity) and interference with cellular metabolism caused by high levels of Na+ in the cytoplasm (ion-specific toxicity) (Munns and Tester, 2008). Na+ can inhibit K+ uptake (Rains and Epstein, 1965), and in the cytoplasm, Na+ readily displaces K+ in many enzymes that require K+ as a co-factor for their activity (Tester and Davenport, 2003). Therefore, genes that can either help limit net Na+ uptake into the shoots or that enable a high cytoplasmic K+/Na+ ratio to be maintained are regarded as potential sources of salinity tolerance. Knowledge of chromosome regions controlling Na+ exclusion and other salinity tolerance-related traits in cultivated cereals and their near-relatives has been reviewed (Colmer et al., 2006; Genc et al., 2010; Shavrukov et al., 2010).
Various transporters and regulatory factors are known to influence net Na+ uptake in plants (reviewed by Plett and Møller, 2010), and some of these have been shown (or implicated) to govern natural genetic variation for Na+ accumulation. Member(s) of the HKT family of Na+ transporters and Na+/K+ symporters control variation for Na+ accumulation at the SKC1 locus of rice (Oryza sativa L.) (Ren et al., 2005), and are strong candidates for Na+ exclusion genes at the Nax1, Nax2, and Kna1 loci of wheat (Triticum spp.) (Huang et al., 2006; Byrt et al., 2007) and at a QTL in Arabidopsis thaliana (L.) Heynh. (Rus et al., 2006). A vacuolar H+-pyrophosphatase gene was identified at the HvNax3 Na+ exclusion locus of barley (Hordeum vulgare L.) and represents a plausible candidate for the controlling gene (Shavrukov et al., 2010). Vacuolar H+-pyrophosphatase contributes to the electrochemical gradient for H+ across the tonoplast, which can be used to power the NHX Na+/H+ antiporter. This process sequesters Na+ in the vacuole and can prevent Na+ from reaching toxic concentrations in the cytosol. In Arabidopsis, salt overly-sensitive (sos) mutants have enabled the identification of a salinity tolerance mechanism comprising the SOS1 plasma membrane H+/Na+ antiporter and its regulators—the SOS2 kinase and the SOS3 calcineurin-B like protein (Liu and Zhu, 1998; Halfter et al., 2000; Liu et al., 2000; Shi et al., 2000; Guo et al., 2001, 2004; Qiu et al., 2002; Quintero et al., 2002). However, to our knowledge, SOS genes have not been reported to control any natural variation in salinity tolerance.
A recent study in barley focusing mainly on the genetics of Zn2+ accumulation identified a locus on the long arm of barley chromosome 1H with a large effect on shoot Na+ accumulation (Lonergan et al., 2009). In the doubled-haploid (DH) mapping population made from a cross between the Australian variety Clipper and the Algerian landrace Sahara 3771, the locus controlled 79% of the variation in the trait and a ∼2-fold average difference in shoot Na+ concentration. In the current study, the phenotype of the locus (named HvNax4) has been characterized in more detail, showing that its expression is strongly dependent on growth conditions. Fine-mapping identified a co-segregating gene (HvCBL4) with close similarity to Arabidopsis SOS3. HvCBL4 was investigated as a candidate for the HvNax4 gene by mRNA expression profiling and sequencing and, in addition, by constructing the molecular model of HvCBL4 based on the known crystal structure of SOS3 from Arabidopsis.
Materials and methods
QTL analysis
QTL analysis of shoot Na+ and K+ accumulation was performed in the Clipper×Sahara 3771 mapping population of 146 DH lines, using the accumulated marker dataset (Jefferies et al., 1999; Karakousis et al., 2003; P Langridge, M. Pallotta, unpublished data), and shoot elemental content data obtained by inductively coupled plasma atomic emission spectrometry (ICPAES) (Zarcinas and Cartwright, 1987) in four experiments previously performed at the Waite Campus of the University of Adelaide.
Experiment 1. This represents the study Lonergan et al. (2009) undertook to measure mineral nutrient accumulation in plants at the vegetative developmental stage. The soil base was a calcareous aeolian sand (67% CaCO3, pH 8.6) collected at Wangary (South Australia). It was supplemented with nutrients as described by Lonergan et al. (2009), including NaCl at 4.16 mg kg−1 dry soil. Plants were raised in a growth chamber with a 15/10 °C 10/14 h day/night cycle and whole shoots of 4-week-old plants harvested for analysis.
Experiment 2. This study (Y Zhu, S Smith, unpublished data) used the low phosphorus calcareous soil described by Zhu et al. (2003) and was originally designed to investigate phosphorus use efficiency. A base of grey calcareous soil (pH 8.7; 25 mg kg−1 NaHCO3-extractable P, 38.2% CaCO3), collected from Minnipa, South Australia, was supplemented with nutrients as described by Zhu et al. (2003). These supplements did not include any phosphorus or sodium. Plants were grown in a growth chamber and whole shoots harvested at 5.5 weeks.
Experiment 3. This was the dataset Jefferies et al. (1999) generated for boron tolerance studies. A soil of silty clay loam texture was taken from the 10 cm top layer of a red-brown earth at Glenthorne Research Farm, O'Halloran Hill, South Australia (Paull et al., 1988), and supplemented with a high level of boron (0.1 g kg−1 soil) but no sodium. In 2008, an analysis of the same box of soil (CSBP, Perth, Australia) showed it to have a pH of 6.6, a conductivity (1:5 soil:water extract) of 1.33 dS m−1, and Na+ at 3.53 mEq per 100 g dry soil (0.81 g kg−1 dry soil). Plants were grown in a 25-cm deep box in a greenhouse, and shoots harvested at 5 weeks.
Experiment 4. This experiment was originally designed to study the potential effects of a combined boron and salt stress in hydroponics (M Quinn, unpublished data). Plants were grown on a wire mesh that was level with the surface of the aerated hydroponics solution, in a growth chamber, with 12/12 h day/night cycle and a constant temperature of 20 °C. The Hydrogro hydroponic nutrient solution (The Gro-Shop, Adelaide, Australia) was made by diluting stocks A and B 250-fold and does not contain sodium. When plants were 16 days old, boron (100 mM) and NaCl (50 mM) was added to the solution, and 2 d later NaCl was increased to 100 mM. After a further 4 d, whole shoots were harvested (when plants were 3 weeks old and-1 day old).
Single point linkage analysis (P <0.001) was performed using Map Manager QTX version 0.30 software (Manly et al., 2001). A minimum logarithm of the odds (LOD) score of 2.0 was used to define significant marker–trait associations.
HvNax4 fine mapping
Additional markers were obtained using previously described RFLP markers from the HvNax4 chromosome region, by converting published RFLP markers from the region into PCR markers, and by making new PCR markers from genes predicted to be in the region through co-linearity with the rice genome sequence, following established methods (Chen et al., 2009a). Primers and enzymes used for PCR markers are provided in Supplementary Table S1 at JXB online. Most of the genes from the corresponding rice interval (21–28 Mb of rice chromosome 5 sequence NC_008398.1) had homologues on rice chromosome 1 (22.43–37.58 Mb of NC_008394.1), consistent with a known duplication involving these genomic regions which predates the evolutionary divergence of the Triticeae and rice (Salse et al., 2008; data not shown). Hence, care was taken to base PCR marker primer sequences on barley ESTs which, in rice, most closely matched the gene copies on chromosome 5. The markers were scored in the Clipper×Sahara 3771 DH lines that were recombinant for the HvNax4 marker interval. The HvNax4 locus was located to a single point on the genetic map by comparing shoot [Na+] values for the DH recombinants with the phenotype distributions of the non-recombinants, in the manner described by Schnurbusch et al. (2007).
Mapping was also performed using an HvNax4-segregating F2 population, made by crossing DH lines DNA83 and DNA89 from the Clipper×Sahara 3771 population. Screening for recombinants was undertaken using the multiplex assay for HvNax4-flanking PCR markers ABC257 and BCD304 (see Supplemetary Table S1 and Supplementary Fig. S1 at JXB online). Genomic DNA of the 125 F2 individuals was kindly provided by T Sutton and A Hay. F2 recombinants were scored for intervening markers and for the HvNax4 locus by measuring shoot Na+ accumulation in progeny F3 families (see below). Selected markers were also used to score F3 individuals, to allow comparisons of Na+ accumulation in plants containing the three different combinations of the two chromosomes derived from the F2 plant. This approach enables robust genotyping of single recombinants at subtle QTL loci (Chen et al., 2009b; Shavrukov et al., 2010). Shoot Na+ data were subjected to ANOVA using the Genstat software package (NAG, Oxford, UK). Within each F3 family, plants homozygous for recombinant and non-recombinant chromosomes were compared to determine whether or not there was segregation for Na+ accumulation controlled by the target chromosome region. Where the F value was significant (P <0.05), the LSD test was used to compare the means. Graphic visualization of marker and trait locus scores was used to establish locus order. Recombination fractions were converted to cM using Kosambi's mapping function (Kosambi, 1944).
F3 plants (48 per family) were scored for Na+ accumulation using the same box of soil that Jefferies et al. (1999) used for boron tolerance studies. At the time of the experiment in 2008, an analysis of the soil was performed (see Experiment 3 description). Seeds were sown 2–3 cm deep and 2 cm apart and the soil watered every second day. Four weeks after sowing, a 2-cm length of the second leaf of each plant was used to extract DNA. The remaining shoot material was harvested, oven-dried at 80 °C for 48 h, weighed, digested in 50 ml Falcon tubes containing 10–20 ml of 1.0% HNO3 at 80 °C in heating blocks for 4 h, and analysed for Na+ content using a flame photometer (model 4200, Sherwood, UK), alongside standards of known concentrations.
Gene annotation and genomic comparisons
Homologues in rice were located by BLASTn searches of the ‘genomes (chromosome)’ database at NCBI (http://www.ncbi.nlm.nih.gov/). The MSU Osa1 gene annotations (Yuan et al., 2005) were used to identify rice genes, either via the genome browser at http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/ or by BLASTn searches at http://blast.jcvi.org/euk-blast/index.cgi?project=osa1. Likewise, Brachypodium distachyon (L.) genes were identified using the genome browser at http://www.brachybase.org/cgi-bin/gbrowse/brachy8/ or by using BLASTn searches of the 8× assembly sequence at http://www.brachybase.org/blast/. Several annotated genes were not described because they showed no clear nucleotide or protein similarity to any other species (Os05g45779, Os05g45940, Bradi2g18560), because they appeared to represent pseudogene fragments of another gene in the interval (Bradi2g18610), or because they were related to retrotransposons. Arabidopsis protein homologies were searched at TAIR (http://www.arabidopsis.org/Blast/).
HvCBL4 sequencing
Barley ESTs were only available for the 3′ half of the HvCBL4 coding sequence (AV913727 and AV921332). Five-prime random amplification of cDNA ends (5′-RACE), RT-PCR, and genomic fragment amplification were used to obtain the full-length cDNA and genomic coding sequences of HvCBL4 (GenBank HM175878 to HM175881), using primers listed in Supplementary Table S2 at JXB online. Protocols were as described by Collins et al. (2008), except that the SMART II A oligonucleotide used in 5′-RACE was made entirely of RNA. RACE and RT-PCR were performed using total root RNA extracted from 3-week-old hydroponically grown plants. The entire ORF from each parent was finally RT-PCR amplified for sequencing from oligo-dT primed cDNA, using the primers CBL-21 and CBL-6. The final genomic sequence was obtained from two overlapping genomic PCR products, amplified using primers CBL-21 with CBL-18, and CBL-1 with CBL-2.
Expression analysis
A set of 24 BC1F2-derived inbred lines were developed in the Clipper background for HvNax4-candidate expression analysis and for future physiological and agronomic studies. The Clipper×Sahara 3771 DH line DNA44 carrying a Sahara 3771 HvNax4 allele was crossed with Clipper and an F1 plant backcrossed with Clipper. A BC1F1 plant containing the Sahara 3771 HvNax4 allele was selected using the ABC257–BCD304 multiplex marker assay and used to derive a BC1F2 family. BC1F2 plants homozygous for contrasting HvNax4 alleles (12 for each allele) were selected using the multiplexed markers and independently used to generate BC1F2:6 bulked seed by successive rounds of multiple plant (random) selection and selfing in the greenhouse and field. Based on the genome-wide marker dataset (Karakousis et al., 2003), line DNA44 derived around 65% of its genome from Clipper. Therefore, the BC1F2-derived lines were expected to have derived around 91% of their genomes from Clipper. They superficially resembled Clipper in appearance.
Five BC1F2 derived lines carrying the Clipper HvNax4 allele and four carrying the Sahara 3771 allele were used to compare mRNA expression of the Clipper and Sahara 3771 derived HvCBL4 alleles. Plants were grown in a greenhouse during September in the supported hydroponics system described by Shavrukov et al. (2010). Seeds were germinated on filter paper in Petri dishes and, after 4 d, seedlings of similar sizes were transferred to hydroponics. After growing plants without salt stress for 10 d, NaCl was added to the growth solution twice daily, at 10.00 h and 14.00 h, in increments of 25 mM, until a concentration of 150 mM was reached after 3 d. Supplemental CaCl2 was added with the NaCl to maintain a constant Ca2+ activity in the solution. The appropriate amount was calculated using the Visual MINTEQ computer program (Department of Land and Water Resources Engineering, Stockholm, Sweden). Whole root systems were harvested immediately prior to the first salt addition and, at the same time of day, 3 d and 5 d thereafter. Per line and time point, roots of three seedlings were bulked together to make one sample.
Quantitative RT-PCR was performed using an oligo-dT primed first strand cDNA with the primers CBL-7F and CBL-12 (see Supplementary Table S2 at JXB online), according to the methods of Burton et al. (2004), except that half-scale reactions and an RG 6000 real-time thermal cycler (Corbett Research, Australia) were used. The PCR program comprised 3 min at 95 °C, followed by 45 cycles of 1 s at 95 °C, 1 s at 55 °C, and 30 s at 72 °C, followed by a single step for 15 s at the optimal acquisition temperature (82 °C for the HvCBL4 product). PCRs were performed in triplicate to obtain an average value for each cDNA/primer combination. Normalizsation was performed according to Vandesompele et al. (2002), using amplification from barley control genes encoding glyceraldehyde-3-phosphate dehydrogenase, α-tubulin, heat shock protein 70, and cyclophilin, using primers described by Burton et al. (2004).
HvCBL4 protein sequence analysis and construction of molecular models
The Structure Prediction Meta-Server (Ginalski et al., 2003), SeqAlert (Bioinformatics and Biological Computing, Weizmann Institute of Science, Israel), 3D-PSSM Server (Imperial College of Science, Technology and Medicine, London, UK), and the Protein Data Bank (http://www.rcsb.org/pdb/) were mined to identify the most suitable three-dimensional (3D) structural template that could be used for homology (comparative) modelling of the target HvCBL4 proteins. The crystal structures of free SOS3 in complex with four Ca2+ ions (Sánchez-Barrena et al., 2005) (PDB accession 1V1G) and the SOS2–SOS3 complex (Sánchez-Barrena et al., 2007) (PDB accession 2EHB) were identified to be the best templates. The 1V1G structure was used as a template to construct 3D molecular models of the HvCBL4 proteins including four Ca2+ ions, using the Modeller 9v7 software (Sali and Blundell, 1993; Sánchez and Sali, 1998). Modelling was performed on a LINUX workstation, running a Ubuntu 8.1 operating system. Final 3D molecular models were selected from 50 models as having the most favourable parameters of the Modeller 9v7 objective function (Sali and Blundell, 1993) and Discrete Optimized Protein Energy (DOPE) scores (Eswar et al., 2006). The overall G-factors and the stereochemical quality of the final models were calculated using PROCHECK (Laskowski et al., 1993). The z-score values were calculated using ProsaIIv3 (Sippl, 1993). The DeepView program (Guex and Peitsch, 1997) was used in determining the RMSD values in the Cα positions between the models and the 1V1G template. Electrostatic potentials were calculated with the Adaptive Poisson-Boltzmann Solver (the dielectric constants of solvent and protein were 80 and 2, respectively) (http://apbs.sourceforge.net/) implemented in PyMol as a plugin, and mapped on protein molecular surfaces that were generated with a probe radius of 1.4 Å. PyMol (http://www.pymol.org) was used to generate molecular graphics.
The protein sequence alignment was made in ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and shading applied using Boxshade 3.2 (http://www.ch.embnet.org/software/BOX_form.html).
Results
The HvNax4 QTL
The three pot experiments with the Clipper×Sahara 3771 DH mapping population each revealed a single highly significant (LOD >3.0) QTL governing shoot [Na+], which was near the end of the long arm of chromosome 1H, most highly associated with the clustered markers ABC257, cMWG733, BCD808a, and CDO669b. The Clipper allele conferred lower [Na+]. The QTL detected in experiments 1, 2, and 3 had respective LOD scores of 39.1, 27.5, and 13.3, and controlled 84%, 83%, and 35% of the total variation in [Na+] as determined by marker regression analysis. The QTL was not detected in the hydroponics dataset (experiment 4), even at a level of moderate significance (LOD >2.0). However, this was not simply due to mislabelling, because the boron accumulation QTL on chromosome 4H that was known to segregate in this population (Jefferies et al., 1999; Sutton et al., 2007) was strongly manifested in this dataset (LOD 11.5; most highly associated with marker: HvBot1). ICPAES data for other elements were available for experiments 1 and 3. Despite the fact that Na+ exclusion loci often exert an inverse effect on K+ and Na+ concentrations, no K+ QTL was detected in either experiment, on chromosome 1H or elsewhere in the genome. Surprisingly, in experiment 1 (only) a QTL for Mg2+ accumulation was detected at the same marker position as the Na+ QTL. The LOD was 4.8 and the Sahara 3771 allele conferred lower [Mg2+]. Data for shoot dry weight were available for experiments 1, 2, and 3, and for root dry weight for experiment 2, but these data did not reveal QTL on 1H. The locus on 1HL was named HvNax4 (for Hordeum vulgare Na+ exclusion 4), to distinguish it from the durum wheat Nax1 and Nax2 loci, which do not reside on chromosome regions corresponding to barley 1HL (Lindsay et al., 2004; Huang et al., 2006; Byrt et al., 2007), and the HvNax3 locus on barley chromosome 7H (Shavrukov et al., 2010).
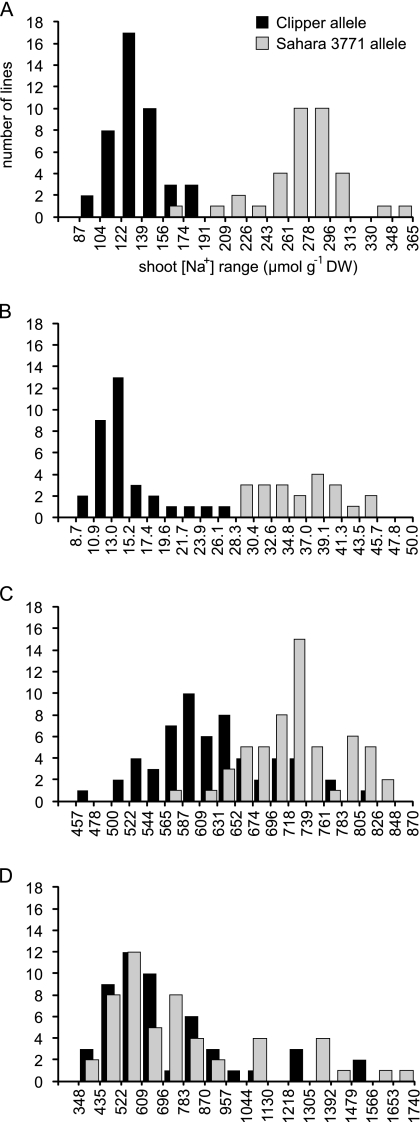
Distributions of shoot [Na+] values in DH lines carrying the Clipper or Sahara 3771 alleles of HvNax4 were examined in lines that were non-recombinant for the QTL marker interval ABC152–7SGlob, and are shown in Fig. 1. The Clipper allele reduced Na+ levels by an average of 50%, 59%, and 12% in experiments 1, 2, and 3, respectively (Table 1). Overall shoot [Na+] values in lines containing the Clipper or Sahara 3771 allele varied widely between the experiments, from 14.9 and 36.2 μmol g−1 DW, respectively, in experiment 2, to 716 and 773 μmol g−1 DW, respectively, in experiment 4 (Table 1).
Fig. 1.
Frequency distributions for shoot [Na+], in Clipper×Sahara 3771 DH lines carrying the Clipper (black) or Sahara 3771 (grey) alleles of HvNax4, in experiment 1 (A), 2 (B), 3 (C), and 4 (D).
Table 1.
Mean shoot [Na+] for Clipper×Sahara 3771 DH lines carrying alternate HvNax4 alleles
| Experiment | Substrate and [Na+] | HvNax4 allele | Mean shoot [Na+] ±SEM (μmol g−1 DW) | Difference in shoot [Na+] associated with HvNax4a |
| 1 | Calcareous aeolian sand; | Clipper | 136±3 | 50%*** |
| 1.64 mg kg−1 dry soil | Sahara 3771 | 270±5 | ||
| 2 | P-deficient grey calcareous | Clipper | 14.9±0.6 | 59%*** |
| soil; [Na+] unknown | Sahara 3771 | 36.2±1.0 | ||
| 3 | B-toxic red-brown earth; | Clipper | 628±8 | 12%*** |
| 0.81 g kg−1 dry soil | Sahara 3771 | 713±7 | ||
| 4 | Aerated hydroponics; | Clipper | 716±40 | 7% (ns at P=0.01) |
| 100 mM | Sahara 3771 | 773±46 |
*** Differences significant at P=0.001.
HvNax4 fine mapping
Initial BLAST searches using sequences of RFLP probes mapped to the HvNax4 region of barley 1HL identified a related genomic region on the long arm of rice chromosome 5, consistent with the known relationship between these two chromosome regions (Stein et al., 2007). The co-linearity with rice was used for the targeted generation of 15 gene-based PCR markers in the HvNax4 region (see Supplementary Table S1 at JXB online). Fragments from 10 other genes were amplified and sequenced from Clipper and Sahara 3771, but these could not be converted to markers due to the absence of polymorphism (data not shown). In addition, five gene-based RFLP markers previously mapped to the HvNax4 region, either in the Clipper×Sahara 3771 population (ABC257, BCD808=BCD265, and BCD304) or in other populations of barley and wheat (BCD1930 and cMWG733; Dubcovsky et al., 1995) were converted to PCR assays (see Supplementary Table S1 at JXB online). During PCR marker development, 42,901 bp of sequence from 30 genes was compared between the parents, revealing a total of 111 SNPs and 12 insertion/deletions (data not shown), giving an average frequency of one polymorphism every 349 bp.
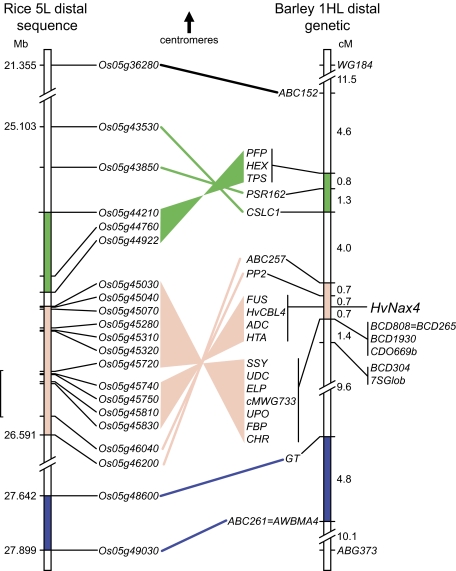
The RFLP markers ABC152 and PSR162, mapped to wheat 1L (Dubcovsky et al., 1995; Quarrie et al 2005), were scored in the entire Clipper×Sahara 3771 DH population as RFLPs. In addition, the 20 new PCR markers were scored in all 21 Clipper×Sahara 3771 DH lines that were recombinant for the HvNax4 interval ABC152–7SGlob, giving rise to an improved map (Fig. 2). Differences in gene order between rice and barley suggested the presence of at least two inversion differences (Fig. 2).
Fig. 2.
Rice–barley comparative map. Lines connect putatively orthologous genes in rice and barley, with colour/shading indicating chromosomal segments separated by ancestral rice–barley inversion breakpoints. Sequences of markers on the right showed strong similarity to rice chromosomes other than chromosome 5, except for the wheat genomic RFLP probe WG184, for which there was no significant match in rice. BCD808=BCD265 and ABC261=AWBMA4 represent homologous RFLP probes. Positions on rice chromosome 5 refer to the sequence NC_008398.1. The bracket on the left indicates the deduced rice HvNax4 interval.
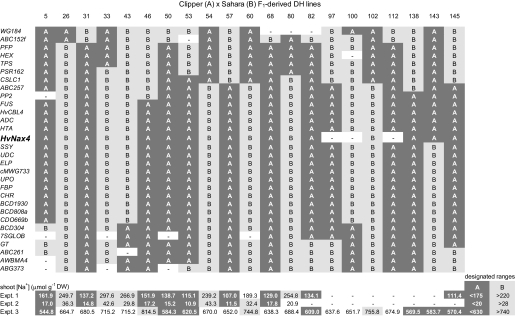
The frequency distributions in Fig. 1 were used subjectively to designate [Na+] ranges that could be used to predict the HvNax4 allele type in experiments 1, 2, and 3. These ranges were used to allocate HvNax4 genotypes to the lines recombinant for the HvNax4 interval ABC152–7SGlob, enabling HvNax4 to be defined as a single point locus on the map (Fig. 3). In the DH lines, HvNax4 co-segregated with the markers FUS, HvCBL4, ADC, and HTA and was separated by one recombination event from the next closest marker(s) on either side (Figs 2, 3).
Fig. 3.
Mapping with DH lines. Recombinants for the HvNax4 interval were assigned HvNax4 genotypes, based on the [Na+] ranges for line carrying Clipper (A) or Sahara 3771 (B) alleles defined by the frequency distributions in Fig. 1. Shading indicates [Na+] values falling within either of the designated ranges (bottom right). Line 46 likely carries the Clipper allele because it was designated Clipper in two out of the three experiments and in the experiments that gave the clearest definition (1 and 2).
F2 analysis
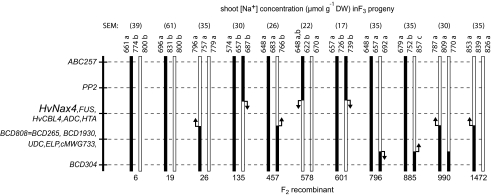
Further mapping was undertaken using the DNA83×DNA89 F2 population. The multiplex assay for the HvNax4-flanking markers ABC257 and BCD304 (see Supplementary Fig. S1 at JXB online) was used to screen 125 F2 individuals, identifying eight single recombinants and one double recombinant (plant 990). The recombinants were scored with ten of the intervening PCR markers, and their HvNax4 genotypes were determined by assaying shoot Na+ accumulation in progeny F3 individuals grown in the glasshouse in the soil from Glenthorne Research Farm. The analysis (Fig. 4) also included two non-recombinant F2 plants heterozygous for both flanking markers (plants 6 and 19) and their F3 progeny families, which were used as HvNax4-segregating controls.
Fig. 4.
Mapping with recombinant DNA83×DNA89 F2 plants and their F3 progeny. Markers on the left were scored in the F2 recombinants, which defined the illustrated chromosome types (black=Clipper derived; white=Sahara 3771 derived). F3 plants from each family were grown in soil, scored individually for marker ABC257 (families 6, 19, 457, 578, 796, and 885), BCD304 (families 6, 19, 26, 135, 601, and 1472) or cMWG733 (family 990), and shoot [Na+]. For each F3 family, mean [Na+] for the two homozygous classes (outer numbers) and heterozygotes (middle number) are presented, together with the family standard error of the means (SEM). Within each family, means that were not significantly different at P=0.05 are denoted by the same letters. Inferred HvNax4 position relative to each recombination point is indicated by an arrow. In control families 6 and 19, individuals found to be recombinant between ABC257 and BCD304 were excluded from the analysis. Analysis of each F3 family was based on 5–16 plants of each homozygote class and 7–24 plants of each heterozygous class.
Comparisons between sibling F3 homozygotes provided a consensus position for the HvNax4 locus, co-segregating with the markers FUS, HvCBL4, ADC, and HTA (Fig. 4). Only the data from family 796 were not entirely consistent with this location. Differences in average shoot [Na+] values between the genotypic classes were in the directions expected if HvNax4 was segregating in this family (and if HvNax4 was located in the consensus position), however, these differences were not significant. The HvNax4 position indicated by the F3 analysis was the same as the one provided by the DH analysis (Figs 2, 3), indicating that this locus position was accurate.
In HvNax4-segregating families, significant differences in shoot [Na+] values between the heterozygote and homozygote F3 classes were suggestive of the Clipper HvNax4 Na+ exclusion allele being recessive (families 6, 19, 135, 601), dominant (family 457) or incompletely dominant (family 885). However, in all HvNax4-segregating families except family 19, the trend was for the heterozygotes to be intermediate. Overall, these data indicate that the HvNax4 exclusion/accumulation alleles control shoot Na+ accumulation in an incompletely dominant fashion.
In the HvNax4-segregating F3 families, individuals homozygous for the Clipper or Sahara 3771 HvNax4 allele accumulated an average of 652 and 761 μmol g−1 DW Na+, respectively. The difference in shoot Na+ accumulation due to HvNax4 (14%) is similar to the 12% observed for the DHs in experiment 3 (Table 1; Jefferies et al., 1999), which was undertaken in the same box of soil as the F3 analysis, albeit with a slightly later shoot sampling time (at 5 weeks instead of 4 weeks).
Genes in the corresponding rice and Brachypodium intervals
If the chromosome segment in the immediate vicinity of HvNax4 only underwent a single genomic inversion since the evolutionary divergence of rice from barley, as depicted in Fig. 2, then the smallest corresponding rice HvNax4 interval can be defined as the 211 kb between the SSY (Os05g45720) and PP2 (Os05g46040) genes. BLAST searches of the Brachypodium genomic sequence identified a corresponding interval of 198 kb on Brachypodium chromosome 2 (6.45 Mb to 16.65 Mb in the Bd2 8× assembly), consistent with the known relationships between the rice, Triticeae and Brachypodium genomes (Vogel et al., 2010). Of the 28 predicted genes in the rice interval, 24 have putative orthologues in the Brachypodium interval, present in the same order as in rice, while the Brachypodium interval contains six genes with no orthologue in the rice interval (Table 2). Therefore, rice and Brachypodium together identify 34 genes with potential to be located in the SSY–PP2 interval of barley. These genes include FUS, HvCBL4, ADC, and HTA, which were mapped in barley and found to co-segregate with HvNax4 (Fig. 3, 4).
Table 2.
Gene content of HvNax4 intervals in rice and Brachypodium, as defined by the HvNax4 flanking gene markers SSY and PP2
| Barley marker | Rice gene | Brachypodium gene | Annotation/Predicted function |
| SSY | Os05g45720 | Bradi2g18810 | Starch synthase, putative, expressed (starch synthesis) |
| Os02g09450 (non-syntenic copy on rice chromosome 2) | Bradi2g18800 | Glycerophosphoryl diester phosphodiesterase family protein, putative, expressed (energy production and conversion) | |
| Os05g45730 | Bradi2g18790 | NADH:ubiquinone oxidoreductase, putative, expressed (energy production and conversion) | |
| HTA | Os05g45740 | Bradi2g18780 | Mitochondrial ATP synthase g subunit family protein, putative, expressed (proton transport) |
| ADC | Os05g45750 | Bradi2g18770 (browser) Bradi2g18767 (BLAST) | ATP-dependent Clp protease ATP-binding subunit clpX, putative, expressed (various functions such as chaperones, proteases, helicases) |
| Os05g45760 | Bradi2g18760 | Protein with PPR domain, putative, expressed (protein interaction domain, various functions) | |
| Os05g45770 | Bradi2g18750 | Expressed protein (unknown function) | |
| HvCBL4 | Os05g45810 (OsCBL4) | Bradi2g18740 | Calcineurin B-like protein 4, putative, expressed (signal transduction) |
| Os05g45820 | Bradi2g18735 (BLAST only) | Heavy metal-associated (HMA) domain containing protein, expressed (transporter of heavy metal cations) | |
| FUS | Os05g45830 | Bradi2g18730 | Expressed protein, with homology to CASC3 domain (mRNA regulation) |
| Os07g41190 (non-syntenic copy on rice chromosome 7) | Bradi2g18710 | WD40 domain containing protein (protein interaction, various functions including signal transduction, pre-mRNA processing, and cytoskeleton assembly) | |
| Os05g45840 | No clear homologue | GTPase-activating protein, putative, expressed (intracellular trafficking and secretion) | |
| Os05g45850 | Bradi2g18720 (browser) Bradi2g18717 (BLAST) | Protein with PPR domain, expressed (protein interaction domain, various functions) | |
| Os05g45860 | Bradi2g18700 | Glycoside hydrolase family 17 member, putative, expressed (carbohydrate metabolism) | |
| Os05g45870 and Os05g45875 (joined by full-length cDNAs) | No clear homologue | F-box family protein, expressed (ubiquitin-mediated protein degradation) | |
| Os05g45880 | Bradi2g18690 | Choline kinase, putative, expressed (lipid metabolism) | |
| Os05g45890 | Bradi2g18680 (browser) Bradi2g18677 (BLAST) | tRNA His guanylyltransferase family protein, expressed (tRNA His maturation, protein translation) | |
| Os05g45900 | Bradi2g18670 | Inositol polyphosphate 5-phosphatase, expressed (hydrolyse 5-phosphates from a variety of myo-inositol phosphate and phosphoinositide phosphate substrates, signalling) | |
| Os05g45910 | Bradi2g18660 | Conserved hypothetical protein | |
| Os05g45920 | Bradi2g18650 | S1 RNA binding domain containing protein, expressed (protein translation) | |
| Os05g45930 | Bradi2g18640 | IQ calmodulin-binding motif family protein, putative, expressed (various functions, regulated by calcium signalling) | |
| Os05g45950 | Bradi2g18630 | Outer mitochondrial membrane voltage-dependent anion channel, putative, expressed (metabolite transport) | |
| No clear homologue | Bradi2g18620 | Hydrophobic protein from soybean (HPS)-like subfamily protein (unknown function) | |
| No clear homologue | Bradi2g18600 | Hydrophobic protein from soybean (HPS)-like subfamily protein (unknown function) | |
| No clear homologue | Bradi2g18590 | Hypothetical protein (unknown function) | |
| No clear homologue | Bradi2g18580 | F-box family protein, expressed (ubiquitin-mediated protein degradation) | |
| Os05g45954 | Bradi2g18570 | Dual-AP2-domain-containing transcription factor, expressed (development) | |
| Os05g45960 | Bradi2g18550 | Delta(1)-pyrroline-5-carboxylate dehydrogenase (P5CDH), putative, expressed (mitochondrial enzyme involved in proline degradation) | |
| Os05g45980 | No clear homologue | MAP65/ASE1 family microtubule associated protein (cell division, cytoskeleton function) | |
| Os05g45990 | No clear homologue | F-box family protein, expressed (ubiquitin-mediated protein degradation) | |
| Os05g46000 | Bradi2g18540 | Ras-related GTPase protein, putative, expressed (signalling, intracellular trafficking) | |
| Os05g46020 | Bradi2g18530 | WRKY transcription factor, expressed (various functions) | |
| Os05g46030 | Bradi2g18520 | Myosin head family protein, expressed (organelle trafficking, development) | |
| PP2 | Os05g46040 | Bradi2g18510 | Protein phosphatase 2C (PP2C) type serine/threonine phosphatase, putative, expressed (signalling) |
OsCBL4, corresponding to the HvCBL4 marker, encodes a calcineurin-B like protein with close similarity to the salt overly-sensitive 3 (SOS3) gene that contributes to salinity tolerance and limitation of Na+ uptake in wild-type Arabidopsis (Liu and Zhu, 1997). In rice, OsCBL4 and two other CBLs (OsCBL7 and OsCBL8) are the proteins that are most similar to SOS3 (Fig. 5; Martínez-Atienza et al., 2007). OsCBL4 can complement the sos3-1 mutation in Arabidopsis (Martínez-Atienza et al., 2007) and its product locates to the plasma membrane (Hwang et al., 2005), which is also the site of SOS3 function (Ishitani et al., 2000; Quintero et al., 2002). ZmCBL4, another plasma membrane-localized CBL from maize (Zhao et al., 2009; Fig. 5) is most similar to OsCBL4 in rice, and can also complement the Arabidopsis sos3-1 mutant (Wang et al., 2007).
Fig. 5.
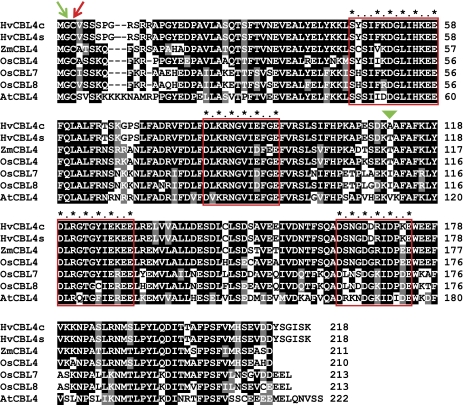
Alignment of barley HvCBL4 proteins from Clipper (c) and Sahara 3771 (s), maize ZmCBL4 (Wang et al., 2007), rice OsCBL4, OsCBL7, and OsCBL8 (Hwang et al., 2005) and Arabidopsis AtCBL4 (SOS3). Dashes represent gaps introduced into the alignment. Shading indicates >50% amino acid sequence identity (black) or similarity (grey). EF hand motifs are boxed and calcium binding residues marked with asterisks (X, Y, Z, –Y, –X, –Z residues, respectively). Residues with predicted N-myristoylation or S-acylation are indicated by green and red arrows, respectively. The single amino acid residue difference between HvCBL4c and HvCBL4s is marked by the green triangle.
The product of the PP2 gene most closely matches four proteins in Arabidopsis including ABI2, which is a negative regulator of the salinity tolerance mechanism involving SOS3 (Ohta et al., 2003). However, the barley PP2 marker was separated from HvNax4 by four recombination events (Figs 3, 4), making this gene an unlikely candidate for HvNax4.
At least four other genes from the rice/Brachypodium HvNax4 intervals have potential roles in salinity tolerance. Os05g45820 encodes a protein containing a heavy metal-associated (HMA) domain. Proteins of this class are known to transport heavy metals such as copper, cadmium, cobalt, and zinc, although they have not yet been reported to transport sodium. However, TargetP analysis (http://www.cbs.dtu.dk/services/TargetP-1.0/) suggested a mitochondrial location (score 0.85) for this rice protein. The Os05g45880 product is most similar to three choline kinases in Arabidopsis, encoded by At1g74320, At4g09760, and At1g71697. Salt stress increases the abundance of transcripts from these genes and stimulates choline kinase activity, which may impact on salinity tolerance by causing membrane structure remodelling, or by producing lipid messengers or compatible solutes (Tasseva et al., 2004). The Os05g45960 product is most similar in Arabidopsis to the Δ(1)-pyrroline-5-carboxylate dehydrogenase encoded by At5g62530. Mutants of At5g62530 are salinity-tolerant and contain elevated levels of the compatible osmolyte proline (Borsani et al., 2007). Os05g46020 encodes a WRKY transcription factor which is transcriptionally up-regulated by salinity stress (Ramamoorthy et al., 2008). Over-expression of at least some salt-inducible WRKYs in Arabidopsis has been shown to result in salinity tolerance (Jiang and Deyholos, 2009).
The HvCBL4 gene
The HvCBL4 gene was regarded to be the best candidate for HvNax4 and was investigated further. ESTs covering the 5′ end of HvCBL4 were not available, so the complete 5′ end of the transcript sequence was obtained using RACE. Recovery of the full HvCBL4 genomic sequence revealed the gene structure (see Supplementary Fig. S2 at JXB online). Compared with the rice orthologue OsCBL4 (consensus of NM_001062683.1 and CI639696), the 5′ UTR of HvCBL4 is 154 bp longer, and contains an additional intron. The rice gene also contained an additional intron in the region corresponding to exon four of HvCBL4, but otherwise the positions of introns in the two genes were conserved. Comparison of Clipper and Sahara 3771 HvCBL4 sequences revealed 14 single nucleotide substitutions and three small insertion/deletion differences (see Supplementary Fig. S2 at JXB online). Four of the former were located in coding-exon sequence, and only one results in a difference between the predicted Clipper and Sahara 3771 products (Alanine or Threonine at position 111, respectively) (Fig. 5).
HvCBL4 protein and structural modelling
In Arabidopsis, SOS1, SOS2, and SOS3 contribute to salinity tolerance (Liu and Zhu, 1998; Liu et al., 2000; Shi et al., 2000). The SOS3 protein contains four Ca2+ binding domains (EF hands) that are thought to enable sensing of transient Ca2+ signals generated by salt stress (Liu and Zhu, 1998). A physical interaction between SOS3 and the SOS2 serine–threonine protein kinase activates the kinase and localizes SOS2 to the plasma membrane. At this intracellular location, the SOS2–SOS3 complex phosphorylates and activates the plasma membrane SOS1 Na+/H+ antiporter in a Ca2+-dependent manner, leading to restoration of ion homeostasis (Halfter et al., 2000; Guo et al., 2001, 2004; Qiu et al., 2002; Quintero et al., 2002). SOS3 can form Ca2+-stabilized dimers which may be important for in planta function (Sánchez-Barrena et al., 2005). Crystal structures of the SOS3 dimer with Ca2+ and of the SOS2–SOS3 complex have been elucidated (Sánchez-Barrena et al., 2005, 2007), providing the opportunity to construct molecular models of the barley HvCBL4 protein and explore possible consequences of the Ala111Thr substitution.
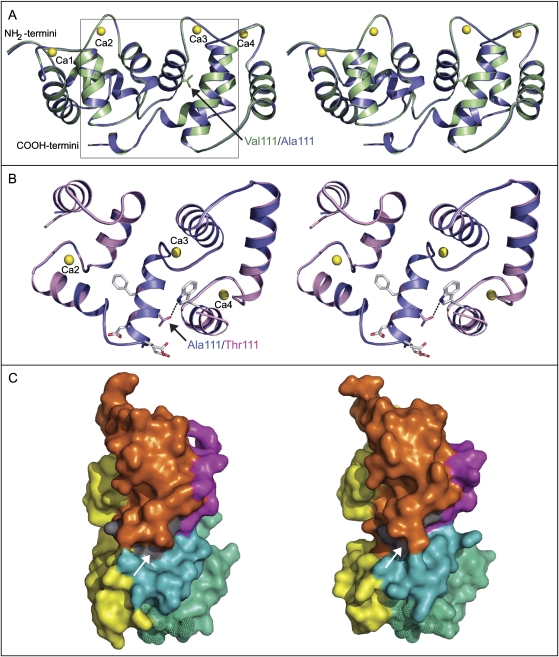
The sequences of the HvCBL4 proteins were subjected to protein modelling using the SOS3 structure (PDB accession 1V1G; Sánchez-Barrena et al., 2005) as a template. The shape and distribution of the secondary structure elements in the modelled barley proteins corresponded closely to those of SOS3 (Fig. 6A). Overall G-factors (Laskowski et al., 1993) for 1V1G and the Clipper and Sahara 3771 models were 0.33, 0.10, and 0.08, respectively, while RMSD (root-mean-square deviation) values in the Cα positions with respect to 1V1G were 0.21 Å and 0.26 Å, respectively. The stereochemical quality and overall G-factors showed that 92–93% of the residues were in positions that were most favoured, 7–8% were in allowed regions and 0% in disallowed regions. The z-scores (Sippl, 1993), which reflect combined statistical potential energy for 1V1G and the barley Clipper and Sahara 3771 models, were –9.21, –9.18, and –9.21, respectively. The SOS3 protein structure therefore served as an excellent template for modelling of the barley HvCBL4 proteins, and the resulting models can be regarded as highly reliable.
Fig. 6.
3D structure of SOS3 and molecular models of HvCBL4. (A), Stereoview of ribbon representations of the superposed Arabidopsis SOS3 structure (green) and the Clipper HvCBL4 model (steel blue), showing the disposition of secondary structure elements. The four bound Ca2+ ions are shown as yellow spheres. Side chains of Val111 (SOS3) and Ala111 (Clipper HvCBL4) are shown in sticks and are arrowed. (B), Stereoview of partial ribbon representations of the superimposed Clipper (steel blue) and Sahara 3771 (pink) HvCBL4 models, detailing the region containing the Ala111Thr substitution. It is an enlargement of the boxed section in (A) and is rotated 45° North with respect to (A). Amino acid residue Thr111 (cpk green colour) in Sahara 3771 forms a hydrogen bond with Trp163 (dashed line). Side chains of residues surrounding the polymorphic site are included to indicate local environment. The last 12 (C-terminal) residues are omitted. (C), Surface representations of SOS3 (left) and the Clipper HvCBL4 model (right), rotated by about 90° North and 90° clockwise with respect to (A). Arrows (and grey) highlight residues 110–112 of HvCBL4 containing the polymorphic site, and the corresponding residues in SOS3. Yellow indicates EF hand domains, cyan and green-cyan indicate the two sections of the molecule that would move apart to expose a hydrophobic SOS2-binding crevice, magenta indicates the 12 C-terminal residues, and orange indicates the remainder of the surface. The dimerization surface in each protein is dotted.
EF hands of CBLs often vary from a canonical 12 amino acid residue EF hand structural motif (Lewit-Bentley and Réty, 2000; Batistič and Kudla, 2009). As in SOS3, the HvCBL4 proteins contain an insertion of two amino acid residues between the first and second (X and Y) Ca2+ binding position in the first EF-hand (Fig. 5). Furthermore, Ca2+ binding carboxylic groups present at the Y position in the canonical motif are absent from some of the EF hands (first three EF hands of HvCBL4; Fig. 5). The four EF-hands of SOS3 bind Ca2+, despite these variations (Sánchez-Barrena et al., 2005). Molecular modelling indicated that binding of Ca2+ ions to all four EF hands of HvCBL4 was mediated by close interactions with backbone carbonyl groups, or with acidic, hydroxy, and amino side-chain groups (data not shown).
SOS3 is N-myristoylated at the Gly2 residue, and this modification is essential for its salinity tolerance function and its ability to recruit SOS2 to the plasma membrane (Ishitani et al., 2000). The MYR Predictor tool (http://mendel.imp.ac.at/myristate/SUPLpredictor.htm; Maurer-Stroh et al., 2002) predicted that structural features of residues 2–18 of HvCBL4 were favourable for recognition by myristoyl CoA:protein N-myristoyltransferase, and it suggested that Gly2 would be N-myristoylated. For both versions of the protein, the overall confidence score was 2.262, and the probability of a false positive prediction was 2.75e−04. N-myristoylation sites were also predicted at the conserved Gly2 in the other CBL proteins shown in Fig. 5. Another lipid modification (S-acylation) occurs at Cys3 of a related Arabidopsis CBL (CBL1) and is required for its plasma membrane localisation (Batistič et al., 2008). Cysteine occurs at this position in HvCBL4 and other closely related CBLs (Fig. 5), suggesting that they may also be S-acylated.
SOS3 and the HvCBL protein models have a two-lobed architecture comprising two globular domains (each with two EF-hands), connected by a short linker region of around six residues (Fig. 6A; Sánchez-Barrena et al., 2005). In SOS3, opposing sections of each lobe move apart to expose a hydrophobic cleft which binds SOS2 (Fig. 6C). The Ala111/Thr111 substitution of HvCBL4 is located on an α-helix adjacent to the linker, and is not within the hydrophobic cleft that would be expected to interact with a SOS2-type protein (Fig. 6A–C). The substitution is also not within the region corresponding to the homodimerization interaction interface of SOS3, located at one end of the molecule (Fig. 6C). However, Thr111 (but not Ala111) is predicted to form a hydrogen bond with Trp163 from the neighbouring α-helix within the C-terminal globular domain (Fig. 6B), and this bond could influence structural packing and stability of the barley proteins.
HvCBL4 expression
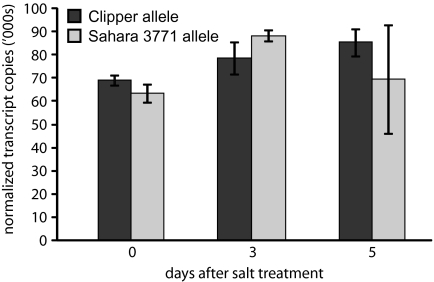
Hydroponics did not allow expression of HvNax4-controlled Na+ exclusion differences in experiment 4 (Fig. 7). However, a supported hydroponics system was used for the transcript analysis because it could provide clean samples of root tissue suitable for RNA extraction and it could be used to analyse constitutive expression levels of the HvCBL4 alleles. BC1F2-derived lines that were mostly Clipper in their genetic background were grown under saline (150 mM) conditions and the expression of Clipper and Sahara 3771 HvCBL4 alleles was measured by quantitative RT-PCR, at 0, 3, or 5 d after salt addition. No expression difference between the alleles was detected (Fig. 7). There was also no detectable induction of HvCBL4 expression by salt treatment. The latter observation corresponds with reports that SOS3 and OsCBL4 were non-inducible by NaCl (Quan et al., 2007; Gu et al., 2008), but contrasts with a finding that ZmCBL4 was induced by NaCl in maize roots (Wang et al 2007).
Fig. 7.
HvCBL4 transcript analysis. Quantitative RT-PCR was used to monitor HvCBL4 transcript abundance in roots of BC1F2 derived lines, homozygous for HvNax4 alleles from Clipper (five lines) or Sahara 3771 (four lines), grown in supported hydroponics containing 150 mM NaCl. Means ±SEM are shown, with individual BC1F2 derived lines being treated as replicates for each allele and time point.
Discussion
HvNax4 chromosome location
This current work reports the fine mapping, candidate gene identification, and phenotypic characterization of the barley Na+ accumulation locus on the long arm of barley chromosome 1H that was initially discovered by QTL mapping in the Clipper×Sahara 3771 cross by Lonergan et al. (2009). In a barley Harrington×TR306 cross, Mano and Takeda (1997) detected a QTL for salinity tolerance at germination, positioned just proximally to the ABC261 marker on 1H, which corresponds closely to the position of HvNax4 (Fig. 2), although they did not measure Na+ accumulation. In a CM72×Gairdner barley cross, Xue et al. (2009) detected a 1H QTL (qSPP1s) controlling spike number per plant in saline field soil. Comparisons via a map with common markers (http://www.triticarte.com.au/) indicated that the QTL was close to HvNax4 (data not shown). However, they measured Na+ accumulation and it did not map to this QTL. In rice, Prasad et al. (2000) detected QTL in the vicinity of cDNA probes RZ70 (Os05g41550) and RZ225 (Os05g47446) on the long arm of chromosome 5, for vigour and dry matter accumulation, in seedlings grown in saline filter paper culture. This corresponds to the HvNax4 interval in rice (Fig. 2), suggesting that rice may possess a functional orthologue of the HvNax4 gene.
HvNax4 phenotype
Levels of shoot Na+ in plants carrying either allele of HvNax4 varied by over 20-fold between the four initial experiments, increasing through experiments 2, 1, 3, and 4 (Table 1), despite a similar plant age at sampling (3–5 weeks), suggesting that there were differences across the experiments in salinity stress and/or physiological factors contributing to net Na+ uptake. Perhaps more interesting was the fact that HvNax4 effects also varied widely across experiments, from a 59% in shoot [Na+] in experiment 2 to no significant effect in hydroponics (experiment 4) (Table 1). The extent of the genetic effect decreased through experiments 2, 1, 3, and 4, and was therefore negatively correlated with overall shoot [Na+]. However, in supported hydroponics, HvNax4 still had no detectable impact when NaCl was reduced to 1 mM in the growth solution and Na+ concentrations in plant shoots were only around 39 μmol g−1 DW (J Rivandi, unpublished data). These observations indicate that there was some characteristic of these hydroponics experiments other than external [Na+] that prevented genotypic discrimination. Further work is therefore needed to identify the environmental factor(s) necessary to manifest the HvNax4 Na+ exclusion phenotype.
HvNax4 had no measurable effect on plant biomass where this was tested (experiments 1, 2, and 3), even at the highest of Na+ accumulation levels, which was encountered in experiment 3 (628 and 713 μmol g−1 DW for plants with the Clipper and Sahara 3771 HvNax4 allele, respectively). James et al. (2006a) reported that patches of Na+-induced leaf cell death in barley occurred with overall leaf Na+ content in the range of 1,100–1,900 μmol g−1 DW. However, they observed no overall correlation between symptoms and leaf [Na+] between barley genotypes, which was proposed to be due to variation in mechanisms that enable leaf tissue to tolerate high levels of accumulated Na+. Such mechanisms appeared to involve the abililty of tolerant barley genotypes to maintain high cytoplasmic [K+] when [Na+] was also high (James et al., 2006a). Therefore, a failure to observe a growth effect of HvNax4 in the current study may have been due to a high level of tissue tolerance in the Clipper and Sahara 3771 parents, insufficient accumulation levels, or other factors such as exposure to stress for an insufficient duration. Added to the environmental sensitivity of expression of the HvNax4-controlled Na+ exclusion trait, these considerations emphasize the need for grain yield testing in the field to gauge the potential agronomic value of the HvNax4. Accordingly, the BC1F2 derived lines carrying Clipper or Sahara 3771 HvNax4 alleles are currently being trialled at saline field sites. Should HvNax4 prove to offer a yield advantage, the PCR markers generated in this study could be deployed by barley breeders for improving varieties.
A common feature of Na+ exclusion loci is the inverse effect they have on Na+ and K+ accumulation, for example, at wheat loci Nax1, Nax2, and Kna1 (Ren et al., 2005; James et al., 2006b; Dubcovsky et al., 1996), rice SKC1, and Arabidopsis SOS2 and SOS3 (Liu and Zhu, 1997; Zhu et al., 1998; Yang et al., 2009). These K+ effects appear to be an indirect result of altered Na+ status in the tissues, as the cereal loci encode members of the HKT family of Na+ transporters or Na+/K+ symporters (Horie et al., 2001; Mäser et al., 2002; Garciadeblás et al., 2003), while SOS2 and SOS3 directly regulate the activity of the SOS1 Na+/H+ antiporter (Quintero et al., 2002). By contrast, K+ content was not significantly altered by HvNax3 (Shavrukov et al., 2010) or HvNax4, suggesting that transport functions controlled by these barley genes may differ somehow (for example, site of action) from those of the aforementioned loci, in such a way as to avoid triggering changes in K+ transport.
The HvCBL4 candidate gene
Proteins in rice or maize corresponding most closely to Arabidopsis SOS1, SOS2, and SOS3 (OsSOS1, OsCIPK24, and OsCBL4/ZmCBL4) can substitute for the salinity tolerance functions of the respective Arabidopsis proteins, either in Arabidopsis or yeast, suggesting that cereals contain SOS-like machinery with potential to contribute to salinity tolerance (Martínez-Atienza et al., 2007; Wang et al., 2007). Consistent with this proposal, HvCBL4, the positional orthologue of OsCBL4 (Fig. 2; Table 2), formed 3D molecular models which were very similar overall to the known SOS3 crystal structure (Sánchez-Barrena et al., 2005). Furthermore, it was suggested that HvCBL4 has EF-hand motifs capable of binding Ca2+, and that Cys2 would be N-myristoylated, which are features required for SOS3 functionality.
The Ala111Thr substitution between the Clipper and Sahara 3771 HvCBL4 proteins is located outside of regions expected to participate directly in homodimerization, Ca2+ binding, N-myristoylation, S-acylation, or binding to a SOS2 homologue protein. The amino acid residue at this position is also not well conserved among other CBLs (Fig. 5), and there are rice CBLs that have either Ala or Thr at this position, namely OsCBL5 and OsCBL4, respectively (Hwang et al., 2005). Nonetheless, the substitution is a non-conservative one, in that Ala is hydrophobic and Thr polar. Furthermore, Thr111 (but not Ala111) could potentially form a hydrogen bond with Trp163 from an adjacent α-helix (Fig. 6B). Conceivably, local change in electrostatic and dipole characteristics of the protein (data not shown) or alterations to the stability or dynamics of the entire α-helical bundle structure caused by the substitution could impact on functional characteristics of the protein, such as affinity for Ca2+ or a SOS2 counterpart. These possibilities could be tested using in vitro and in vivo approaches previously applied to investigate aspects of SOS3 function (Halfter et al., 2000; Ishitani et al., 2000; Qiu et al., 2002; Quintero et al., 2002; Guo et al., 2004; Sánchez-Barrena et al., 2005).
No difference in the constitutive mRNA expression levels of the Clipper and Sahara 3771 HvCBL4 alleles was detected in supported hydroponics (Fig. 7). However, in experiment 4 undertaken in hydroponics, HvNax4 did not exert a significant genotypic effect on Na+ accumulation. Therefore, it is possible that conditions that manifest HvNax4 effects on Na+ may also be required for differential expression of HvNax4 alleles, if such an mRNA expression difference is the basis for the phenotypic variation at this locus.
While HvCBL4 co-segregated with the HvNax4 locus and is similar to the SOS3 salinity tolerance gene, decisive proof that HvCBL4 is the HvNax4 gene is currently lacking. HKT genes mapping to the wheat Nax1 and Nax2 Na+ exclusion loci are regarded as strong candidates for the controlling genes, because the respective HKT genes were found to have no expression, or no genomic copy at all, in susceptible genotypes (Huang et al., 2006; Byrt et al., 2007). By contrast, natural variation in HKT genes residing at the rice SKC1 locus and at a Na+ accumulation locus in Arabidopsis appears to be more subtle, being based on a quantitative difference in activity of the encoded transporter, or a quantitative difference in mRNA expression level, respectively (Ren et al., 2005; Rus et al., 2006). Further work is required to identify whether there are subtle differences between the activities of the Clipper and Sahara 3771 HvCBL4 alleles, or to reveal an alternative gene that could be a stronger candidate for HvNax4.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Multiplex marker.
Supplementary Fig. S2. Sequence of Clipper and Sahara 3771 HvCBL4 genes.
Supplementary Table S1. PCR markers.
Supplementary Table S2. Primers used in HvCBL4 gene sequencing and Q-PCR.
Acknowledgments
We are grateful to Paul Lonergan, Yuangwan Zhu, Michael Quinn, Stephen Jefferies, Peter Langridge, Andrew Barr, Robin Graham, Susan Barker, Sally Smith, Stewart Coventry, and Jason Eglinton for generous access to unpublished ICPAES data, and to Yuri Shavrukov, Alison Hay, and Neil Shirley for technical assistance. This work was supported by the ARC, GRDC, and the South Australian Government.
Glossary
Abbreviations
- HvNax4
Hordeum vulgare Na+ exclusion locus 4
- SOS
SALT OVERLY SENSITIVE
- RT-PCR
reverse transcription-polymerase chain reaction
- QTL
quantitative trait locus/loci
- CBL
calcineurin-B like
- HKT
high-affinity K+ transporter
- 3D
three-dimensional
References
- Batistič O, Kudla J. Plant calcineurin B-like proteins and their interacting protein kinases. Biochimica et Biophysica Acta. 2009;1793:985–992. doi: 10.1016/j.bbamcr.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Batistič O, Sorek N, Schültke S, Yalovsky S, Kudla J. Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. The Plant Cell. 2008;20:1346–1362. doi: 10.1105/tpc.108.058123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu J-K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2007;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB. The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiology. 2004;134:224–236. doi: 10.1104/pp.103.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrt CS, Platten JD, Spielmeyer W, James RA, Lagudah ES, Dennis ES, Tester M, Munns R. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiology. 2007;143:1918–1928. doi: 10.1104/pp.106.093476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Baumann U, Fincher GB, Collins NC. Flt-2L, a locus in barley controlling flowering time, spike density, and plant height. Functional and Integrative Genomics. 2009b;9:243–254. doi: 10.1007/s10142-009-0114-2. [DOI] [PubMed] [Google Scholar]
- Chen A, Brûlé-Babel A, Baumann U, Collins NC. Structure–function analysis of the barley genome: the gene-rich region of chromosome 2HL. Functional and Integrative Genomics. 2009a;9:67–79. doi: 10.1007/s10142-008-0099-2. [DOI] [PubMed] [Google Scholar]
- Collins NC, Shirley NJ, Saeed M, Pallotta M, Gustafson JP. An ALMT1 gene cluster controlling aluminum tolerance at the Alt4 locus of rye (Secale cereale L.) Genetics. 2008;179:669–682. doi: 10.1534/genetics.107.083451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer TD, Flowers TJ, Munns R. Use of wild relatives to improve salt tolerance in wheat. Journal of Experimental Botany. 2006;57:1059–1078. doi: 10.1093/jxb/erj124. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Luo M-C, Dvořák J. Differentiation between homoeologous chromosomes 1A of wheat and 1Am of Triticum monococcum and its recognition by the wheat Ph1 locus. Proceedings of the National Academy of Sciences, USA. 1995;92:6645–6649. doi: 10.1073/pnas.92.14.6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Santa María G, Epstein E, Luo M-C, Dvořák J. Mapping of the K+/Na+discrimination locus Kna1 in wheat. Theoretical and Applied Genetics. 1996;92:448–454. doi: 10.1007/BF00223692. [DOI] [PubMed] [Google Scholar]
- Eswar N, Marti-Renom MA, Webb B, Madhusudhan MS, Eramian D, Shen M, Pieper U, Sali A. Current Protocols in Bioinformatics. Supplement 15. 2006. Comparative protein structure modeling using Modeller. 5.6.1–5.6.30. John Wiley & Sons Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciadeblás B, Senn ME, Bañuelos MA, Rodríguez-Navarro A. Sodium transport and HKT transporters: the rice model. The Plant Journal. 2003;34:788–801. doi: 10.1046/j.1365-313x.2003.01764.x. [DOI] [PubMed] [Google Scholar]
- Genc Y, Oldach K, Verbyla AP, Lott G, Hassan M, Tester M, Wallwork H, McDonald GK. Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theoretical and Applied Genetics. 2010;121:877–894. doi: 10.1007/s00122-010-1357-y. [DOI] [PubMed] [Google Scholar]
- Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- Gu Z, Ma B, Jiang Y, Chen Z, Su X, Zhang H. Expression analysis of the calcineurin B-like gene family in rice (Oryza sativa L.) under environmental stresses. Gene. 2008;415:1–12. doi: 10.1016/j.gene.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu J-K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. The Plant Cell. 2001;13:1383–1399. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Qiu Q-S, Quintero FJ, Pardo JM, Ohta M, Zhang C, Schumaker KS, Zhu J-K. Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. The Plant Cell. 2004;16:435–449. doi: 10.1105/tpc.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu J-K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proceedings of the National Academy of Sciences, USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. The Plant Journal. 2001;27:129–138. doi: 10.1046/j.1365-313x.2001.01077.x. [DOI] [PubMed] [Google Scholar]
- Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, Dennis ES, Munns R. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiology. 2006;142:1718–1727. doi: 10.1104/pp.106.088864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y-s, Bethke PC, Cheong YH, Chang H-S, Zhu T, Jones RL. A gibberellin-regulated calcineurin B in rice localizes to the tonoplast and is implicated in vacuole function. Plant Physiology. 2005;138:1347–1358. doi: 10.1104/pp.105.062703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim C-S, Shi W, Zhu J-K. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. The Plant Cell. 2000;12:1667–1677. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RA, Davenport RJ, Munns R. Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiology. 2006b;142:1537–1547. doi: 10.1104/pp.106.086538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RA, Munns R, von Caemmerer S, Trejo C, Miller C, Condon AG. Photosynthetic capacity is related to the cellular and subcellular partitioning of Na+, K+ and Cl– in salt-affected barley and durum wheat. Plant, Cell and Environment. 2006a;29:2185–2197. doi: 10.1111/j.1365-3040.2006.01592.x. [DOI] [PubMed] [Google Scholar]
- Jefferies SP, Barr AR, Karakousis A, Kretschmer JM, Manning S, Chalmers KJ, Nelson JC, Islam AKMR, Langridge P. Mapping of chromosome regions conferring boron toxicity tolerance in barley (Hordeum vulgare L.) Theoretical and Applied Genetics. 1999;98:1293–1303. [Google Scholar]
- Jiang Y, Deyholos MK. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Molecular Biology. 2009;69:91–105. doi: 10.1007/s11103-008-9408-3. [DOI] [PubMed] [Google Scholar]
- Karakousis A, Barr AR, Kretschmer JM, Manning S, Jefferies SP, Chalmers KJ, Islam AKM, Langridge P. Mapping and QTL analysis of the barley population Clipper×Sahara. Australian Journal of Agricultural Research. 2003;54:1137–1140. [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Annals of Eugenics. 1944;12:172–175. [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- Lewit-Bentley A, Réty S. EF-hand calcium-binding proteins. Current Opinion in Structural Biology. 2000;10:637–643. doi: 10.1016/s0959-440x(00)00142-1. [DOI] [PubMed] [Google Scholar]
- Lindsay MP, Lagudah ES, Hare RA, Munns R. A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Functional Plant Biology. 2004;31:1105–1114. doi: 10.1071/FP04111. [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proceedings of the National Academy of Sciences, USA. 2000;97:3730–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K. An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proceedings of the National Academy of Sciences, USA. 1997;94:14960–14964. doi: 10.1073/pnas.94.26.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- Lonergan PF, Pallotta MA, Lorimer M, Paull JG, Barker SJ, Graham RD. Multiple genetic loci for zinc uptake and distribution in barley (Hordeum vulgare) New Phytologist. 2009;184:168–179. doi: 10.1111/j.1469-8137.2009.02956.x. [DOI] [PubMed] [Google Scholar]
- Manly KF, Cudmore RH, Jr., Meer JM. Map Manager QTX, cross-platform software for genetic mapping. Mammalian Genome. 2001;12:930–932. doi: 10.1007/s00335-001-1016-3. [DOI] [PubMed] [Google Scholar]
- Mano Y, Takeda K. Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L.) Euphytica. 1997;94:263–272. [Google Scholar]
- Mäser P, Hosoo Y, Goshima S, et al. Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proceedings of the National Academy of Sciences, USA. 2002;99:6428–6433. doi: 10.1073/pnas.082123799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu J-K, Pardo JM, Quintero FJ. Conservation of the salt overly sensitive pathway in rice. Plant Physiology. 2007;143:1001–1012. doi: 10.1104/pp.106.092635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. Journal of Molecular Biology. 2002;317:541–557. doi: 10.1006/jmbi.2002.5426. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annual Review of Plant Biology. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Ohta M, Guo Y, Halfter U, Zhu J-K. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proceedings of the National Academy of Sciences, USA. 2003;100:11771–11776. doi: 10.1073/pnas.2034853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull JG, Cartwright B, Rathjen AJ. Responses of wheat and barley genotypes to toxic concentrations of boron. Euphytica. 1988;39:137–144. [Google Scholar]
- Plett DC, Møller IS. Na+ transport in glycophytic plants: what we know and what we would like to know. Plant, Cell and Environment. 2010;33:612–626. doi: 10.1111/j.1365-3040.2009.02086.x. [DOI] [PubMed] [Google Scholar]
- Prasad SR, Bagali PG, Hittalmani S, Shashidhar HE. Molecular mapping of quantitative trait loci associated with seedling tolerance to salt stress in rice (Oryza sativa L.) Current Science. 2000;78:162–164. [Google Scholar]
- Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K. Regulation of SOS1, a plasma membrane Na+/H+exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proceedings of the National Academy of Sciences, USA. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. The Plant Cell. 2007;19:1415–1431. doi: 10.1105/tpc.106.042291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrie SA, Steed A, Calestani C, et al. A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring×SQ1 and its use to compare QTLs for grain yield across a range of environments. Theoretical and Applied Genetics. 2005;110:865–880. doi: 10.1007/s00122-004-1902-7. [DOI] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu J-K, Pardo JM. Reconstruction in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proceedings of the National Academy of Sciences, USA. 2002;99:9061–9066. doi: 10.1073/pnas.132092099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rains DW, Epstein E. Transport of sodium in plant tissue. Science. 1965;148:1611. doi: 10.1126/science.148.3677.1611. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy R, Jiang S-Y, Kumar N, Venkatesh PN, Ramachandran S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant and Cell Physiology. 2008;49:865–879. doi: 10.1093/pcp/pcn061. [DOI] [PubMed] [Google Scholar]
- Ren Z-H, Gao J-P, Li L-G, Cai X-L, Huang W, Chao D-Y, Zhu M-Z, Wang Z-Y, Luan S, Lin H-X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nature Genetics. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE. Natural variants of At HKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genetics. 2006;2:1964–1973. doi: 10.1371/journal.pgen.0020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, Calcagno T, Cooke R, Delseny M, Feuillet C. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. The Plant Cell. 2008;20:11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez R, Sali A. Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proceedings of the National Academy of Sciences, USA. 1998;95:13597–13602. doi: 10.1073/pnas.95.23.13597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Barrena MJ, Fujii H, Angulo I, Martínez-Ripoll M, Zhu J-K, Albert A. The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Molecular Cell. 2007;26:427–435. doi: 10.1016/j.molcel.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Barrena MJ, Martínez-Ripoll M, Zhu J-K, Albert A. The structure of the Arabidopsis thaliana SOS3: molecular mechanism of sensing calcium for salt stress response. Journal of Molecular Biology. 2005;345:1253–1264. doi: 10.1016/j.jmb.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Schnurbusch T, Collins NC, Eastwood RF, Sutton T, Jefferies SP, Langridge P. Fine mapping and targeted SNP survey using rice-wheat gene colinearity in the region of the Bo1 boron toxicity tolerance locus of bread wheat. Theoretical and Applied Genetics. 2007;115:451–461. doi: 10.1007/s00122-007-0579-0. [DOI] [PubMed] [Google Scholar]
- Shavrukov Y, Gupta NK, Miyazaki J, Baho MN, Chalmers KJ, Tester M, Langridge P, Collins NC. HvNax3 – a locus controlling shoot sodium exclusion derived from wild barley (Hordeum vulgare ssp. spontaneum) Functional and Integrative Genomics. 2010;10:277–291. doi: 10.1007/s10142-009-0153-8. [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu J-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proceedings of the National Academy of Sciences, USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- Stein N, Prasad M, Scholz U, et al. A 1,000-loci transcript map of the barley genome: new anchoring points for integrative grass genomics. Theoretical and Applied Genetics. 2007;114:823–839. doi: 10.1007/s00122-006-0480-2. [DOI] [PubMed] [Google Scholar]
- Sutton T, Baumann U, Hayes J, et al. Boron-toxicity tolerance in barley arising from efflux transporter amplifiction. Science. 2007;318:1446–1449. doi: 10.1126/science.1146853. [DOI] [PubMed] [Google Scholar]
- Tasseva G, Richard L, Zachowski A. Regulation of phosphatidylcholine biosynthesis under salt stress involved choline kinases in Arabidopsis thaliana. FEBS Letters. 2004;566:115–120. doi: 10.1016/j.febslet.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R. Na+ tolerance and Na+ transport in higher plants. Annals of Botany. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3:1–12. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Garvin DF, Mockler TC, et al. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Wang M, Gu D, Liu T, Wang Z, Guo X, Hou W, Bai Y, Chen X, Wang G. Overexpression of a putative maize calcineurin B-like protein in Arabidopsis confers salt tolerance. Plant Molecular Biology. 2007;65:733–746. doi: 10.1007/s11103-007-9238-8. [DOI] [PubMed] [Google Scholar]
- Xue D, Huang Y, Zhang X, Wei K, Westcott S, Li C, Chen M, Zhang G, Lance R. Identification of QTLs associated with salinity tolerance at late growth stage in barley. Euphytica. 2009;169:187–196. [Google Scholar]
- Yang Q, Chen Z-Z, Zhou X-F, Yin H-B, Li X, Xin X-F, Hong X-H, Zhu J-K, Gong Z. Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Molecular Plant. 2009;2:22–31. doi: 10.1093/mp/ssn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Wang A, et al. The Institute for Genomic Research Osa1 rice genome annotation database. Plant Physiology. 2005;138:18–26. doi: 10.1104/pp.104.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarcinas BA, Cartwright B. Acid dissolution of soils and rocks for the determination of boron by inductively coupled plasma atomic emission spectrometry. Analyst. 1987;112:1107–1112. [Google Scholar]
- Zhao J, Sun Z, Zheng J, et al. Cloning and characterization of a novel CBL-interacting protein kinase from maize. Plant Molecular Biology. 2009;69:661–674. doi: 10.1007/s11103-008-9445-y. [DOI] [PubMed] [Google Scholar]
- Zhu J-K, Liu J, Xiong L. Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. The Plant Cell. 1998;10:1181–1191. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y-G, Smith FA, Smith SE. Phosphorus efficiencies and responses of barley (Hordeum vulgare L.) to arbuscular mycorrhizal fungi grown in highly calcareous soil. Mycorrhiza. 2003;13:93–100. doi: 10.1007/s00572-002-0205-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.