Abstract
After fertilization, filial grain organs are surrounded by the maternal nucellus embedded within the integuments and pericarp. Rapid early endosperm growth must be coordinated with maternal tissue development. Parameters of maternal tissue growth and development were analysed during early endosperm formation. In the pericarp, cell proliferation is accomplished around the time of fertilization, followed by cell elongation predominantly in longitudinal directions. The rapid cell expansion coincides with endosperm cellularization. Distribution of TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick end labelling)-positive nuclei reveals distinct patterns starting in the nucellus at anthesis and followed later by the inner cell rows of the pericarp, then spreading to the whole pericarp. The pattern suggests timely and spatially regulated programmed cell death (PCD) processes in maternal seed tissues. When the endosperm is coenocytic, PCD events are only observed within the nucellus. Thereby, remobilization of nucellar storage compounds by PCD could nourish the early developing endosperm when functional interconnections are absent between maternal and filial seed organs. Specific proteases promote PCD events. Characterization of the barley vacuolar processing enzyme (VPE) gene family identified seven gene members specifically expressed in the developing grain. HvVPE2a (known as nucellain) together with closely similar HvVPE2b and HvVPE2d might be involved in nucellar PCD. HvVPE4 is strongly cell specific for pericarp parenchyma. Correlative evidence suggests that HvVPE4 plays a role in PCD events in the pericarp. Possible functions of PCD in the maternal tissues imply a potential nutritive role or the relief of a physical restraint for endosperm growth. PCD could also activate post-phloem transport functions.
Keywords: Barley, cell expansion, grain development, maternal tissues, programmed cell death
Introduction
In angiosperms, the filial seed organs (embryo and endosperm) develop within maternal tissues. During early development of barley (Hordeum vulgare L.) seeds, the diploid maternal tissues represent the bulk of the grain and consist of the nucellus, nucellar projection, inner and outer integuments, and pericarp. Vascular bundles are embedded in the pericarp. The nucellus degenerates shortly after flowering, except adjacent to the main vascular bundle, where it forms the nucellar projection, which functions as transfer tissue supplying assimilates to the endosperm. There is evidence that nucellar development and assimilate transfer involve programmed cell death (PCD; Radchuk et al., 2006; Thiel et al., 2009). Likewise, the seed coat in dicotyledonous legumes functions as the maternal conduit for the flow of assimilates to the embryo (Weber et al., 1995; Patrick and Offler, 2001). The developmentally regulated degradation of the innermost cell layer, probably by PCD, initiates the storage phase in Vicia faba seeds and is accompanied by a switch from hexose to sucrose due to loss of extracellular invertase expressed in this layer (Weber et al., 1995).
PCD is a genetically regulated process essential for development, integrity, and response to the environment. It is involved in deleting structures when unneeded, such as the embryo suspensor (Lombardi et al., 2007), thus controlling cell number and tissue size. In monocot seeds, PCD mediates degeneration of the endosperm (Olsen, 2004), aleurone depletion in germinating seeds (Pennel and Lamb, 1999), and disintegration in the nucellar projection (Radchuk et al., 2006). In dicot seeds, PCD is observed in the seed coat, suspensor, and endosperm (Lombardi et al., 2007).
In apoptosis-like PCD, nuclei are the first targets of degradation, demonstrated by nuclear shrinkage and fragmentation, chromatin condensation, and DNA laddering. DNA fragments can be cytochemically determined by the terminal deoxynucleotidyl transferase-mediated dUTP nick end labelling (TUNEL) of the 3′-OH groups (Gavireli et al., 1992). Fragmentation of DNA, detected by gel electrophoresis and TUNEL assay, indicated the presence of PCD in wheat (Triticum aestivum L.) pericarp and nucellus already at anthesis (Dominguez et al., 2001; Zhou et al., 2009).
Several groups of cysteine proteases have been shown to be involved in plant PCD. Metacaspases are only distantly related to animal caspases, lack caspase activity, and cleave after arginine and lysine residues but not aspartate (Vercammen et al., 2007). However, at least some metacaspases can be functional homologues of caspases (Bozhkov et al., 2005). Other plant proteases possess caspase-like activity and are shown to be frequently involved in plant PCD. These include vacuolar processing enzyme (VPE), also called legumain (Hara-Nishimura et al., 2005; Hatsugai et al., 2006), subtilisin (Coffeen and Wolpert, 2004), and phytaspase (Chichkova et al., 2010). Arabidopsis thaliana has four VPE genes, αVPE, βVPE, γVPE, and δVPE. Two are preferentially expressed in vegetative tissues and two are seed specific. Quadruple knockout mutants lack any caspase-1 activity, indicating the absence of other related proteases (Kuroyanagi et al., 2005). Vegetative αVPE and γVPE are involved in plant cell death during leaf senescence, lateral root formation, and diverse stresses (Kinoshita et al., 1999; Hara-Nishimura and Maeshima, 2000). Seed-specific βVPE is expressed during later seed development and is essential for processing of storage proteins in cotyledons (Shimada et al., 2003). The seed-specific δVPE is expressed in two inner cell layers of the early seed coat of Arabidopsis (Nakaune et al., 2005). This tissue undergoes PCD at early stages, thereby reducing its thickness by >50%. In a δvpe-deficient mutant, PCD is delayed and the seed coat remains thick throughout development (Nakaune et al., 2005) showing that δVPE is responsible for the PCD in Arabidopsis seed coats. Probably, VPEs in barley seeds are also involved in PCD of pericarp tissues. Nucellain, a barley homologue of VPE, is specifically expressed in nucellar tissues of the developing barley grain (Linnestad et al., 1998). However, detailed analysis of other seed-specific barley VPEs has not been performed.
Filial seed organs grow and expand rapidly within the maternal pericarp. Grains reach their maximal length ∼10 days after flowering (DAF). Thereby, the caryopsis volume increases ∼10-fold. This change can be traced back to the pronounced growth of the filial tissues (Gubatz et al., 2007). It is unclear how the pericarp copes with rapid growth of the endosperm. Here parameters of pericarp growth and development during the early stage of endosperm formation have been analysed. Cell divisions in the pericarp already decreased at 2 DAF. Thereafter the pericarp enlarges by cell expansion. Concurrently PCD occurs, beginning within the nucellus and followed later by the pericarp. The characterization of a gene family encoding VPEs identified HvVPE4 as potentially playing a role in PCD, specifically in pericarp cells. The functional significance of PCD events in the pericarp for grain development is discussed.
Materials and methods
Plant material
Barley plants (H. vulgare L. cv. Barke) were cultivated and the material was collected as described earlier (Sreenivasulu et al., 2006). The developing seeds were harvested from the mid-region of the ear at 2 d intervals starting from anthesis until 24 DAF. Pericarp tissues were hand-separated under a light microscope.
For cytological studies, seeds from different developmental stages were fixed in 50% (v/v) ethanol, 5% (v/v) acetic acid, and 3.7% (w/v) formaldehyde overnight at 4 0C, dehydrated, and embedded in paraffin. Cross-sections (12 μm) of the same seed were prepared, mounted on silane-coated slides (Sigma-Aldrich, Germany), and used as for both TUNEL and in situ hybridization assays.
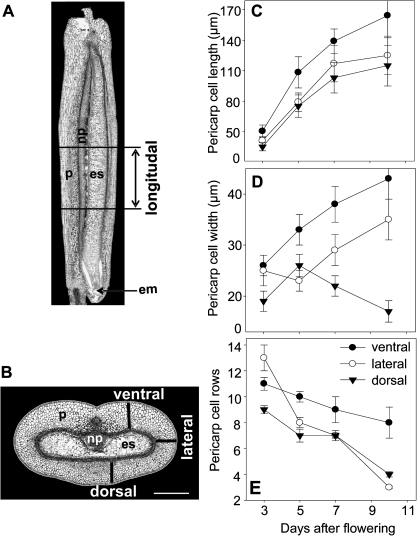
Cross-sections and longitude sections of five grains for each developmental stage (3, 5, 7, and 10 DAF) were used to measure cell number, cell width, and cell length in different regions of a grain (Fig. 1A, B). Only the middle parts of grains were analysed. Cell number and cell width were calculated in ventral, dorsal, and lateral regions of the pericarp on cross-sections, whereas cell length was measured on longitude sections, which were made in the same regions as labelled on the cross-sections (Fig. 1). Between 35 and 50 cells were measured from each of the defined regions.
Fig. 1.
Number of rows of cells, cell length, and cell width of the cells in the pericarp at different developmental stages. Pericarp cell length was calculated in ventral, dorsal, and lateral regions of longitudinal sections (A) of barley grains. Cell row number and cell width were estimated in ventral, dorsal, and lateral regions of grain cross-sections as shown by black bars (B). Cell length (C), cell width (D), and number of cell rows (D) in the developing pericarp. The values are given as means ±SE. White bars=400 μm. em, embryo; es, endosperm; np, nucellar projection; p, pericarp.
DNA isolation and electrophoresis
Genomic DNA was isolated from pericarp tissues and leaves using the DNeasy plant mini kit (Qiagen, Germany). A 2 μg aliquot of DNA from each sample was separated on a 0.7% (w/v) agarose gel at 35 V for 20 h and stained with ethidium bromide.
Identification of genes and evaluation of expression profiles from macro array analyses
To identify genes involved in the cell cycle, cytokinesis, and cell expansion, as well genes encoding VPEs, expressed sequence tag (EST) similarity searches were performed with publicly available sequences by BLASTN, and ESTs with a high percentage identity were selected. To define gene family members, Arabidopsis thaliana and Oryza sativa genome sequence annotations were used (Vandepoele et al., 2002, 2005; Guo et al., 2007). The longest VPE cDNA clones were sequenced using primers generated from the flanking plasmid sequences as well as gene-specific primers (Metabion, Germany). Sequence data were processed using the Lasergene software (DNAstar, USA). The VPE sequences have been submitted to the GenBank/EMBL data libraries under accession numbers FR696360–FR696366.
Gene expression profiles were extracted from early published transcript analysis of barley seed development by using high density macro arrays with 12 000 cDNA sequences from the developing grains. The analysis of mRNA expression was repeated twice with independently grown biological material (Sreenivasulu et al., 2006).
Real-time RT-PCR
Total RNA was extracted from tissue samples using the Gentra RNA isolation kit (Biozym, Germany), cleaned with the RNeasy RNA Isolation kit (Qiagen, Germany), and treated with RNase-free TURBO™ DNase (Ambion, USA). After purification, 8 μg of RNA were used in a reverse transcriptase reaction with SuperScript™ III reverse transcriptase (Invitrogen, Germany) using oligo(dT) primers.
Custom-made primers (Metabion, Germany) for real-time PCRs were designed by Lasergene software (DNASTAR, USA) using the following criteria (Verdier et al., 2008): Tm of 60±2 °C, PCR amplicon length between 60 bp and 450 bp, primer length of 20±5 bp, and a guanine–cytosine content between 45% and 55%. The primers were predominantly selected at the 3′ ends to obtain a high specificity of transcript sequences. The genes and their primer sequences are listed in Table 1. PCRs were performed in an ABI PRISM® 7900 HT sequence detection system (Applied Biosystems, USA) with a reaction volume and thermal profile as described by Verdier et al. (2008). Data analysis was performed using SDS 2.2.1 software (Applied Biosystems, USA).
Table 1.
Primers used in real-time RT-PCR analyses
| Gene | Primer name | Sequence | Localization | Product size |
| HvVPE1 | VPE1_3u | 5′-CCGAAAGGCTGCAATCAACCAA-3′ | 1110–1175 | 65 |
| VPE1_3r | 5′-GCAGCTCGTACCTCCTCCACAAGA-3′ | |||
| HvVPE2a | VPE2a_u | 5′-TGCGCTGCAGTACACGGAA-3′ | 1243–1300 | 57 |
| VPE2a_r | 5′-TCTAGCTAGCTAGGAACCTCCG-3′ | |||
| HvVPE2b | VPE2b_u | 5′-CAGCGCTTGCAACGGCTACGA-3′ | 1431–1503 | 72 |
| VPE2b_r | 5′-TGCAAGCGGATCAGGGCTGTG-3′ | |||
| HvVPE2c | VPE2c_u | 5′-GCGTCTCTGAGGCCCAAATGA-3′ | 1468–1519 | 51 |
| VPE2c_r | 5′-TTATAACCGCCGCAAGCACTGAT-3′ | |||
| HvVPE2d | VPE2d_u | 5′-GCTGCCTTTGCCCATCCTG-3′ | 1050–1141 | 91 |
| VPE2d_r | 5′-TCCCCCGTTTAACTGCTCATACTT-3′ | |||
| HvVPE3 | VPE3_3u | 5′-TGCTTGAACGAGCCTTAGGTGAAT-3′ | 1656–1771 | 115 |
| VPE3_3r | 5′-ATATTTGTTGCCGGGGAGAGTTCT-3′ | |||
| HvVPE4 | VPE4_2u | 5′-ATCGGCGGCCTCCTGTTCG-3′ | 1193–1386 | 193 |
| VPE4_2r | 5′-CGCCGGCGTTGCAGATGTT-3′ | |||
| VPE4_3u | 5′-CGGCCCGGACGACCACATC-3′ | 469–602 | 133 | |
| VPE4_3r | 5′-CCCGCCGGCGTGCTTCTT-3′ | |||
| TEF1 α-subunita | HZ42K12_u | 5′-GACCGCCGCCGCCTCAT-3′ | 4–452 | 448 |
| HZ42K12_r | 5′-GATACCAGCCTCAAAACCACCAGT-3′ | |||
| Actinb | Hvactin_u | 5′-GGTAGGGATGGGGCAGAAGG-3′ | 157–72 | 415 |
| Hvactin_r | 5′-ACCAGCGAGATCCAAACGAAGAA-3′ |
The absence of genomic DNA contamination was confirmed by real-time PCR using non-reverse-transcribed RNA. The efficiency of cDNA synthesis was estimated by real-time reverse transcription-PCR (RT-PCR) using the VPE4_2 and VPE4_3 primer pairs located on the 3′ and 5′ ends of the HvVPE4 gene, respectively. CT values using these primer pairs were almost identical (data not shown), confirming the high efficiency of cDNA synthesis. The efficiencies of PCRs were estimated using the LinRegPCR software (Ramakers et al., 2003). Melting curve analysis was performed for all PCR products to confirm the occurrence of specific amplification peaks and the absence of primer dimer formsation (data not shown). Expression data of the HvVPE genes were normalized by subtracting the mean reference genes (translation elongation factor 1 α-subunit) CT value from the mean CT value (ΔCT) for each cDNA. The normalized ΔCT values from two biological with two technical repetitions were averaged and coefficients of variation (CV%) were calculated. The mean ΔCT values, for which CV% did not exceed 3%, were used for further ΔΔCT calculation. ΔΔCT values were calculated in comparison with the lowest signal value observed in a tissue for each gene. The fold differences were calculated using the formula ratio=2– ΔΔCT.
TUNEL assay
In situ detection of DNA fragmentation was carried out with the TUNEL method essentially following the protocol of the in situ Cell Death Detection kit (Roche Diagnostics, Germany). Briefly, prepared seed sections were de-waxed, rehydrated, and treated with proteinase K, followed by incubation in a mixture of fluorescein-labelled deoxynucleotides and TdT (TUNEL mix) for 60 min at 37 °C. The fluorescein fluorescence of nuclei with fragmented DNA was observed using a microscope with blue light excitation. Controls were performed in which TdT was omitted. For positive control of the reaction, the sections were treated with DNase (1500 U ml−1) prior to performing labelling with the TUNEL mix.
In situ hybridization
Grain sections, prepared as above described, were de-waxed, rehydrated, and treated with 2 μg ml−1 proteinase K for 30 min at 37 °C. Finally, tissue sections were dehydrated and dried before applying the hybridization solution. Hybridization and immunological detection were performed after Drea et al. (2005) with 1 ng μl−1 digoxigenin-labelled sense and antisense RNA probes for HvVPE4 cDNA synthesized using T3 or T7 RNA polymerase (Roche, Germany). Hybridization signals were detected by alkaline phosphatase-conjugated anti-digoxigenin antibody and visualized with 4-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate (Roche, Germany).
Results
Cell number, cell width, and cell length during pericarp growth
Between anthesis and 10 DAF the length of the barley caryopsis increases >5-fold. Thus, the growth of the maternal pericarp has to keep pace with that of the endosperm. To monitor the changes in volume of the pericarp, the cell length, cell width, and the number of rows of cells in the dorsal, lateral, and ventral regions of the pericarp were measured on the basis of longitudinal (Fig. 1A) and cross-sections (Fig. 1B) from the middle part of grains between 3 and 10 DAF. Pericarp cells elongate 2- to 3-fold in length, especially in the ventral region, followed by the lateral and dorsal areas (Fig. 1C). Cell widths increase in the ventral region by 2-fold and in the lateral region by 1.5-fold with minor changes in the dorsal region (Fig. 1D). The number of pericarp cells between the inner integument and epidermis decreases only slightly in the ventral area (0.5-fold) but in a more pronounced way in the dorsal (2-fold) and lateral (4-fold) regions (Fig. 1E). The data suggest that pericarp growth between 3 and 10 DAF occurs predominantly by elongation in the longitudinal direction. In addition, the numbers of rows of cells decreased in all areas but the decrease was more pronounced in lateral and dorsal regions.
Expression analysis of genes involved in the cell cycle, cytokinesis, and cell expansion
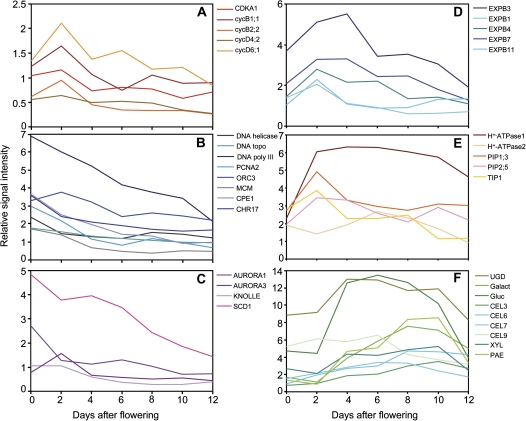
From a previous gene expression study of the barley pericarp based on a 12k macro array (Sreenivasulu et al., 2006), expression profiles of genes involved in cell cycle regulation, DNA replication, repair, and chromatin remodelling, as well as mitosis and cytokinesis were extracted (Supplementary Table S1 available at JXB online). Cell cycle regulation genes such as cyclin-dependent kinase A and cyclin isoforms show maximal expression at 2 DAF (Fig. 2A). Genes involved in DNA replication, repair, and chromatin remodelling have their highest expression even earlier than 2 DAF (Fig. 2B). Similarly, expression of genes associated with mitosis and cytokinesis peaks at or before 2 DAF (Fig. 2C).
Fig. 2.
Transcript profiling of selected genes involved in the cell cycle (A), DNA replication (B), cell division and cytokinesis (C), as well as in cell expansion (D–F) consisting of expansins (D), plasma membrane H+-ATPase and aquaporins (E), and enzymes for cell wall modification (F) in the developing barley pericarp. Transcript levels were extracted from cDNA macro array data (Sreenivasulu et al., 2006). The values were calculated as the means of two independent experiments and are shown as relative signal intensities. CDKA, cyclin-dependent kinase A; CEL, endo-1,4-β-glucanase; CHR17, DNA-dependent ATPase; CPE, SMC2-like condensing protein; cyc, cyclin; DNA topo, DNA topoisomerase; DNA poly III, DNA polymerase III; EXPB, β-expansin; Galact, α-galactosidase; Gluc, α-glucosidase; MCM, minichromosome maintenance family protein; ORC, origin recognition complex; PAE, pectin acetylesterase; PCNA, proliferating cell nuclear antigen; PIP, plasma membrane intrinsic protein; TIP, tonoplast intrinsic protein; SCD, stomatal cytokinesis-defective; UGD, UDP-D-glucuronate decarboxylase; XYL, α-D-xylosidase.
Cell expansion determines organ size and occurs by water uptake into the vacuole and surface expansion of the cell wall (Szymanski and Cosgrove, 2009). H+-ATPases acidify cell walls, which activates expansins, leading to cell wall synthesis and cell expansion (Cosgrove, 2000). From the microarray data set, gene expression profiles of expansins, H+-ATPases, and enzymes of cell wall biosynthesis were extracted (Supplementary Table S1). In the barley pericarp, five expansin genes have their highest expression between 3 and 4 DAF (Fig. 2D). H+-ATPase 1 shows high gene expression between 2 and 10 DAF (Fig. 1D). Accordingly, gene expression of nine genes involved in cell wall biosynthesis shows a broad maximum between 3 and 10 DAF (Fig. 2E).
Such a pattern of gene expression suggests that during pericarp growth, cell proliferation events occur very early, between 0 and 2 DAF, whereas cell expansion and cell wall synthesis happen later (from 3 to 10 DAF), which is in accordance with the increasing cell volume during that period (Fig. 1C, D).
DNA degradation in the growing barley pericarp
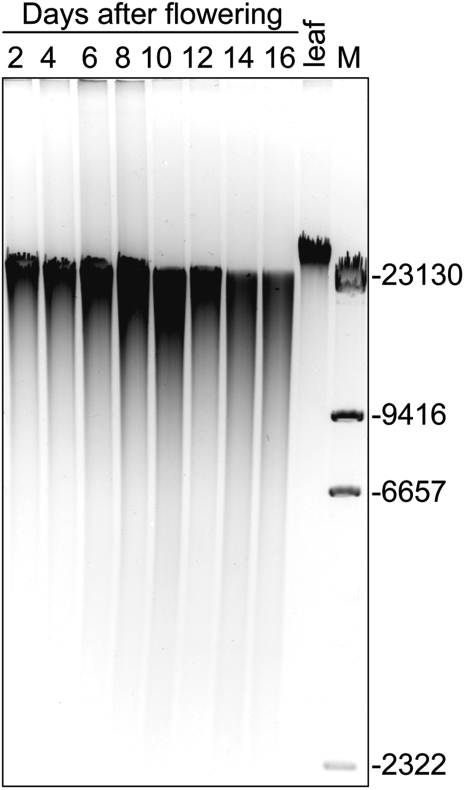
The fact that cell row numbers in the pericarp decrease between 3 and 10 DAF suggests the PCD process is occurring, which often is accompanied by fragmentation and degradation of nuclear DNA. Therefore, total DNA was isolated from barley pericarp from 2 to 16 DAF and separated by DNA gel electrophoresis. In comparison with the sample isolated from leaves, a smear was detected for the DNA of all pericarp samples, which increased with ageing (Fig. 3). Such a pattern indicates DNA degradation throughout pericarp development. However, there is no clear laddering of DNA as found previously for maternal tissues of wheat (Dominguez et al., 2001), possibly because of different DNA extraction methods.
Fig. 3.
DNA fragmentation in the pericarp during barley grain development. The DNA was isolated from the pericarp fraction of developing grains harvested at 2 d intervals and from young, fully expanded leaves, and was separated in an agarose gel.
Localization of PCD in the developing barley grains by TUNEL assay
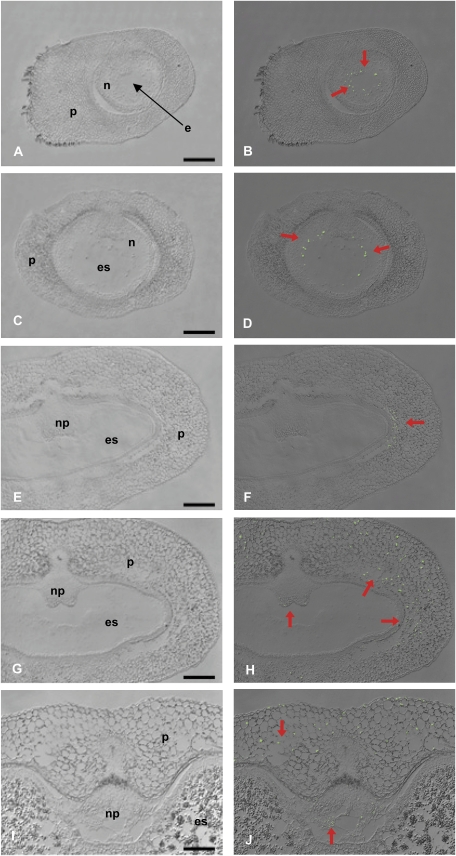
PCD is evidenced by loss of nuclei, which can be analysed by TUNEL staining that labels 3′-OH ends produced as a result of DNA fragmentation (Fig. 4B, D, F, H, and J). Figure 4A, C, E, G, and I show bright field microscopic sections. At anthesis, nuclei of the inner cell layers of the nucellus are labelled (Fig. 4B). At 2 DAF, TUNEL-positive nuclei spread outwards following disintegration of highly vacuolated cells in the nucellus (Fig. 4D). Only occasional labelling of nuclei in the pericarp is detected at this stage. However, at 4 DAF, TUNEL-positive nuclei are located in the inner pericarp layers, especially in dorsal regions (Fig. 4F), but also in other parts of the pericarp. At 6 DAF, TUNEL-labelled nuclei also appear in dorsal and lateral regions of the pericarp (Fig. 4H) as well as in ventral areas (Fig. 4J). Some TUNEL-positive nuclei are present in cells at the margins of the nucellar projection between 6 and 10 DAF (Fig. 4J). No signal is detected in endosperm cells between anthesis and 10 DAF. TUNEL labelling was absent from control sections when the TdT enzyme had been omitted. Almost all nuclei are labelled in positive controls, treated with DNase prior to TUNEL assay, demonstrating the validity of the procedure (data not shown).
Fig. 4.
General view on cross-sections of barley caryopses at 0 (A), 2 (C), 4 (E), 6 (G), and 10 DAF (I) and localization of nuclear DNA fragmentation detected by the TUNEL assay at 0 (B), 2 (D), 4 (F), 6 (H), and 10 DAF (J). TUNEL-positive nuclei are visualized as green signals and indicated by red arrows. e, embryo sac; es, endosperm; n, nucellus; np, nucellar projection; p, pericarp. Bars=200 μm.
The data show that the distribution of TUNEL-positive nuclei in the pericarp reveals a distinct pattern starting in the nucellus between 0 and 2 DAF, spreading to the inner cell rows of the pericarp at 4 DAF and to the whole pericarp later on. Such a pattern suggests timely and spatially regulated PCD processes during pericarp growth.
Identification and gene expression of vacuolar processing enzymes in barley
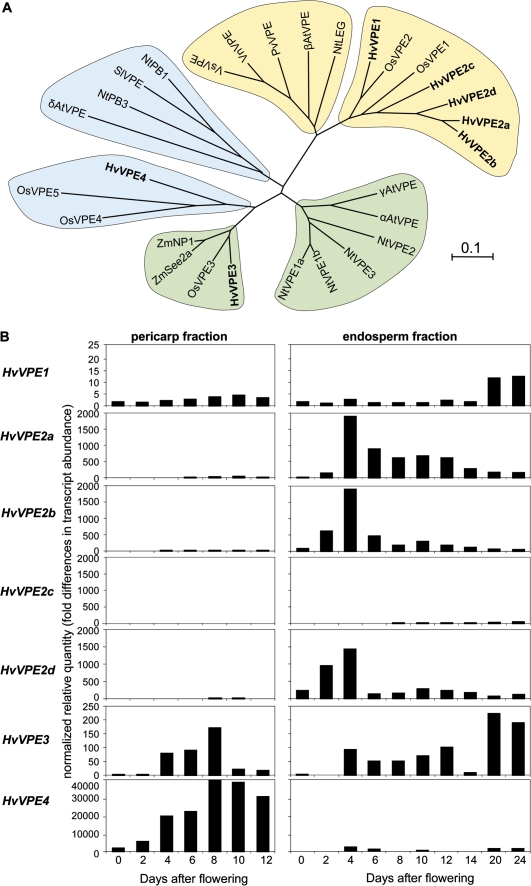
A previous gene expression study detected up-regulated genes in the pericarp potentially related to PCD (Sreenivasulu et al., 2006), especially proteases and VPEs. Some VPEs have been identified as higher plant caspases with caspase-1-like activity (Hara-Nishimura et al., 2005). Seven barley cDNAs encoding putative VPEs were identified from available databases (Table 2, Fig. 5A). The HvVPE2a cDNA is identical to the previously described nucellain (Linnestad et al., 1998). Three additional cDNAs (HvVPE2b, HvVPE2c, and HvVPE2d) have high homology to the known HvVPE2a sequence (76.4–94.4% identity at the amino acid level). The corresponding proteins and HvVPE1 group together with the seed-type VPEs (Fig. 5A). HvVPE3, identified from an EST collection from vegetative barley tissues, belongs to the monocot subgroup of vegetative-type VPEs. HvVPE4 groups with two rice VPEs and is weakly similar to the PCD-type of VPEs, including Arabidopsis δVPE with proven caspase-1-like activity (Nakaune et al., 2005), tobacco NtPB3, and tomato VPE (Fig. 5A).
Table 2.
List of VPE genes in barley
| Gene | Reference EST (full length or partial sequence) | Predicted protein localization (probability) | Length of signal peptide (amino acids) | Alternative name, highest BLAST hit (% identity) | Reference |
| HvVPE1 | HS03M20 (full) | Vacuole (0.755) | 21 | Legumain 1 (100%), (CAQ00096) | Martinez and Diaz (2008) |
| HvVPE2a | HB03N17 (partial) | ND | Nucellain (100%), (AAD04882) | Linnestad et al. (1998) | |
| HvVPE2b | HZ61D22 (full) | Apoplast (0.78) | 27 | Legumain 2 (97.5%), (CAQ00097) | Martinez and Diaz (2008) |
| HvVPE2c | HT01N09 (full) | Vacuole (0.633) | 28 | Legumain 3 (79.3%), (CAQ00097) | Martinez and Diaz (2008) |
| HvVPE2d | HY07P11 (full) | Vacuole (0.446) or apoplast (0.437) | 26 | Legumain 3 (99.0%), (CAQ00097) | Martinez and Diaz (2008) |
| HvVPE3 | TC156635 (full) | Apoplast (0.576) | 36 | Legumain 4 (100%), (CAQ00099) | Martinez and Diaz (2008) |
| HvVPE4 | HZ45O04 (full) | Apoplast (0.82) | 24 | Legumain 5 (100%), (CAQ00099) | Martinez and Diaz (2008) |
Fig. 5.
Barley VPEs and their expression in developing grains. (A) Unrooted phylogenetic tree of plant VPEs drawn with the ClustalW and Mega4 software. The horizontal scale represents the evolutionary distance expressed as a number of substitutions per amino acid. Barley VPEs are shown in bold. VPEs are separated into a vegetative (green), a seed type (yellow), and a type potentially related to PCD (blue). Two subgroups comprising VPEs from monocot and eudicot plants correspondingly are distinguishable in each type. GenBank accession numbers of the sequences are as follows: Arabidopsis thaliana, αAtVPE (At2g25940), βAtVPE (At1g62710), γAtVPE (At4g32940), δAtVPE (At3g20210); Nicotiana tabacum, NtLEG (CAB42651), NtPB1 (CAB42650), NtPB3 (CAE84598), NtVPE1a (BAC54827), NtVPE1b (BAC54828), NtVPE2 (BAC54829), NtVPE3 (BAC54830); Oryza sativa, OsVPE1 (Os04g0537900), OsVPE2 (Os02g0644000), OsVPE3 (Os01g0559600), OsVPE4 (Os05g0593900), OsVPE5 (Os05g0593900); Phaseolus vulgaris, PvVPE (O24326); Solanum lycopersicum SlVPE (CAB51545); Vicia narbonensis VnVPE (CAB16318); Vicia sativa, VsVPE (CAA07639); Zea mays, ZmNP1 (AAD04883), ZmSee2a (CAB64544). (B) Transcript profiling of the HvVPE genes in pericarp (left) and endosperm fractions (right) of developing barley grains determined by real-time quantitative RT-PCR analysis.
The gene expression patterns of VPE genes were determined in pericarp and endosperm fractions of the barley grains between anthesis and 24 DAF by quantitative real-time PCR. HvVPE1 transcripts are present at basal levels in both pericarp and endosperm fractions, with increased abundance in the maturing endosperm (Fig. 5B). Real-time RT-PCR analysis distinguishes the gene expression patterns of HvVPE2a–HvVPE2d despite their high similarity. Transcripts levels of HvVPE2a–HvVPE2d genes are very low in the pericarp. In endosperm fractions, mRNA levels of HvVPE2a, HvVPE2b, and HvVPE2d peak at 4 DAF and decline afterwards. Contents of HvVPE2c mRNA are low throughout development. The HvVPE3 gene expression is detected in pericarp and endosperm. Its mRNA levels in pericarp peak between 4 and 10 DAF, while in endosperm they increase from 4 DAF until maturation. In contrast, HvVPE4 transcript levels accumulate specifically and abundantly in the pericarp, with a maximum between 8 and 10 DAF (Fig. 5B).
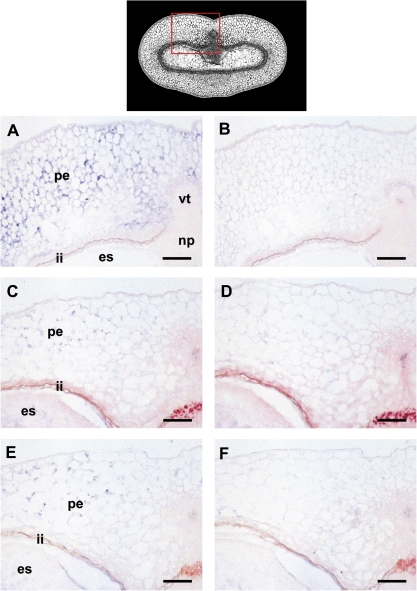
The data suggest a specific role for HvVPE4 in the pericarp in comparison with the other isoforms. Cell type-specific expression of the HvVPE4 gene was analysed by in situ hybridization. HvVPE4 mRNA signals are detected exclusively in the pericarp at 6, 8, and 10 DAF (Fig. 6A, C, E). A slight gradient in transcript abundance is present, increasing from the inner to the outer cell layers of the pericarp. No signals are present in the endosperm, integuments, nucellar projection, and vascular bundle.
Fig. 6.
Localization of HvVPE4 transcripts in developing grains at 6 (A), 8 (C), and 10 DAF (E) by in situ hybridization with an antisense probe. The same regions were also hybridized with a sense probe (B, D, and F) as a negative control. The top panel shows a median transverse section of a developing caryopsis with the region shown in A–F (red box). es, endosperm; ii, integuments; np, nucellar projection, pe, pericarp, vt, vascular bundle. Bars=100 μm.
Discussion
After fertilization, the filial organs of barley grains are surrounded by the maternal nucellus, which itself is embedded within the integuments and the pericarp. Until 10 DAF, the barley grain increases >5-fold in length. Therefore, the rapid growth of the endosperm during cellularization has to be coordinated with development of maternal tissues. The aim of the study was to analyse parameters of the development of the parts of the maternal grain during early stages of endosperm formation. Evidence is provided that cell divisions in the pericarp already decreased at 2 DAF, and thereafter the pericarp enlarges by cell expansion. At the same time PCD occurs, beginning within the nucellus and spreading later to the pericarp. Characterization of a gene family encoding VPEs identified HvVEP4 as potentially playing a role in PCD, specifically in pericarp cells.
The expression analysis of genes related to the cell cycle, DNA replication, mitosis, and cytokinesis clearly shows that cell proliferation events occur very early in the pericarp, with their maximum at 2 DAF, or even before, indicating that cell division activity in the pericarp is almost complete at around the time of fertilization. The pericarp grows predominantly by cell elongation in the longitudinal direction, with the highest rates between 3 and 6 DAF and lower rates between 6 and 10 DAF (Fig. 1C). Remarkably, pericarp cells in the ventral region elongate more, followed by the lateral and dorsal areas. The ventral region contains the main vascular bundle from which assimilates are unloaded into the surrounding pericarp. The increase in pericarp cell width is much less pronounced compared with elongation, but again the highest rates occurred in the ventral area (Fig. 1D). The increase in pericarp cell elongation between 3 and 5 DAF is accompanied by gene expression related to cell expansion (Fig. 2D, E) and cell wall synthesis (Fig. 2F). In summary, it is concluded that cell proliferation is almost completed by around 2 DAF in the pericarp. Later, the pericarp grows mainly by cell elongation predominantly in the longitudinal direction.
Remarkably, the number of rows of cells in the pericarp decreases steadily, most markedly in lateral and dorsal regions. Such a loss of cells is indicative of PCD. This assumption is further strengthened by the observed DNA degradation within the pericarp between 2 and 10 DAF (Fig. 3). The hallmark of apoptosis-like PCD is degradation of the nuclei (Reape and McCabe, 2008). TUNEL staining (Fig. 4) shows such DNA fragmentation in the maternal tissues. The distribution of TUNEL-positive nuclei shows that PCD occurs sequentially. Degradation of nuclei begins in the nucellus at anthesis, is followed by the inner cell rows of the pericarp at 4 DAF, and spreads to the whole pericarp later on. Such a pattern suggests timely and spatially regulated PCD processes during growth of maternal seed tissues. TUNEL-positive nuclei were also detected in the nucellus and pericarp of developing wheat grains (Domingues et al., 2001; Zhou et al., 2009).
PCD events in the maternal seed tissue are coordinated with endosperm development. Between anthesis and 2 DAF, when the endosperm becomes coenocytic, PCD is observed only within the nucellus as indicated by TUNEL staining. Specific proteases and/or VPEs may promote PCD events such as nucellain (identified here as HvVPE2a), which is found to be expressed in the early endosperm fraction (Fig. 4B) but localized exclusively in the nucellus (Linnestad et al., 1998). Because the nucellus is always co-isolated with the endosperm fraction (Sreenivasulu et al., 2006; Radchuk et al., 2009), the activity of HvVPE2b and HvVPE2d genes, whose sequences and expression are very similar to those of HvVPE2a, can also be expected in the nucellus. High caspase-like activities were also detected in early developing barley grains (Boren et al., 2006). The nucellus accumulates starch already at anthesis and undergoes degradation as early as 2 DAF (Radchuk et al., 2006, 2009). The remobilization of its storage compounds by regulated cell disintegration could nourish the coenocytic endosperm at a stage when functional maternal–filial connections have not yet been established (Sreenivasulu et al., 2010). This fine-tuned developmental regulation of the nucellar disintegration process and possible relationship to the nutrition of the coenocytic endosperm might regulate final endosperm cell number and could thus be an important determinant of storage capacity (Sreenivasulu et al., 2010). In addition, deteriorated nucellar cells provide space for the rapidly expanding endosperm.
The pericarp itself develops by coincidental cell expansion and PCD. Some plant VPEs are shown to be involved in PCD (Hatsugai et al., 2006). Of the three VPE genes expressed in the barley pericarp (HvVPE1, HvVPE3, and HvVPE4), only HvVPE4 is tissue specific and shows weak similarity to Arabidopsis δVPE. The Arabidopsis δVPE has caspase-1 activity, is specifically expressed in seed coat layers, and is required for PCD of these tissues (Nakaune et al., 2005). High HvVPE4 gene expression between 4 and 12 DAF (Fig. 5B) coincides with the results of TUNEL staining. In addition, in situ hybridization reveals HvVPE4 mRNA signals exclusively in the pericarp at 6, 8, and 10 DAF but not in endosperm, integuments, the nucellar projection, and the vascular bundle. This correlative evidence suggests that HvVPE4 is involved in PCD events in the pericarp. However, additional genetic and biochemical investigations are required to validate this hypothesis.
The physiological PCD function in the early pericarp of barley implies a potential nutritive role due to the associated remobilization of transient nutrient reserves from the pericarp (Radchuk et al., 2009). Starch remobilization in the pericarp coincides with PCD of the maternal grain tissues. Furthermore, the removal of cell rows could relieve a physical restraint for the growing endosperm. Finally, PCD events in the nucellus, pericarp, and nucellar projection could activate post-phloem transport functions. In the maize placenta–chalazal region, PCD coincides with endosperm cellularization and is rapidly and co-ordinately completed prior to beginning the storage phase. In this way, PCD functions as an adaptive process to facilitate the passage of solutes (Kladnik et al., 2004).
PCD events and cell expansion in the pericarp take place concurrently and are accompanied by cellularization of the endosperm (Olsen, 2004). This suggests a tight regulation of processes in the pericarp interconnected with that of the endosperm. Signals for PCD probably originate in the filial tissues. It is shown that sink demand is communicated by a turgor signal (Patrick and Offler, 2001). Thus, PCD might be initiated by osmotic/turgor changes, induced by sugar accumulation in regions where invertase is located, as has been speculated for the placenta–chalazal region of maize grains (Kladnik et al., 2004).
Supplementary data
Supplementary data are available at JXB online.
Table S1. Expression profiles of genes involved in the cell cycle, cytokinesis, and cell expansion as well as vacuolar processing enzyme genes in the developing pericarp of barley grains.
Acknowledgments
We wish to thank Angela Stegmann, Gabriele Einert, Elsa Fessel, and Uta Siebert for excellent technical assistance. This work was supported in part by the German Ministry of Education and Research within the German Plant Genome Initiative (grant GABI-sysSEED, FKZ 0315044A) and by the Deutsche Forschungsgemeinschaft (grant WE1608/2-1).
Glossary
Abbreviations
- DAF
days after flowering
- PCD
programmed cell death
- VPE
vacuolar processing enzyme
References
- Borén M, Höglund AS, Bozhkov P, Jansson C. Developmental regulation of a VEIDase caspase-like proteolytic activity in barley caryopsis. Journal of Experimental Botany. 2006;57:3747–3753. doi: 10.1093/jxb/erl136. [DOI] [PubMed] [Google Scholar]
- Bozhkov PV, Suarez MF, Filonova LH, Daniel G, Zamyatnin AA Jr, Rodriguez-Nieto S, Zhivotovsky B, Smertenko A. Cysteine protease mcII-Pa executes programmed cell death during plant embryogenesis. Proceedings of the National Academy of Sciences, USA. 2005;102:14463–14468. doi: 10.1073/pnas.0506948102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichkova NV, Shaw J, Galiullina RA, et al. Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. EMBO Journal. 2010;29:1149–1161. doi: 10.1038/emboj.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffeen WC, Wolpert TJ. Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. The Plant Cell. 2004;16:857–873. doi: 10.1105/tpc.017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Domínguez F, Moreno J, Cejudo FJ. The nucellus degenerates by a process of programmed cell death during the early stages of wheat grain development. Planta. 2001;213:352–360. doi: 10.1007/s004250000517. [DOI] [PubMed] [Google Scholar]
- Drea S, Leader DJ, Arnold BC, Shaw P, Dolan L, Doonan JH. Systematic spatial analysis of gene expression during wheat caryopsis development. The Plant Cell. 2005;17:2172–2185. doi: 10.1105/tpc.105.034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavireli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labelling of nuclear DNA fragmentation. Journal of Cell Biology. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubatz S, Dercksen VJ, Brüss C, Weschke W, Wobus U. Analysis of barley (Hordeum vulgare) grain development using three-dimensional digital models. The Plant Journal. 2007;52:779–790. doi: 10.1111/j.1365-313X.2007.03260.x. [DOI] [PubMed] [Google Scholar]
- Guo J, Song J, Wang F, Zhang XS. Genome-wide identification and expression analysis of rice cell cycle genes. Plant Molecular Biology. 2007;64:349–360. doi: 10.1007/s11103-007-9154-y. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Hatsugai N, Nakaune S, Kuroyanagi M, Nishimura M. Vacuolar processing enzyme: an executor of plant cell death. Current Opinion in Plant Biology. 2005;8:404–408. doi: 10.1016/j.pbi.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Maeshima M. Vacuolar processing enzymes and aquaporins. In: Robinson ADG, Rogers JC, editors. Vacuolar compartments in plants. Sheffield, UK: Sheffield Academic Press; 2000. pp. 20–42. [Google Scholar]
- Hatsugai N, Kuroyanagi M, Nishimura M, Hara-Nishimura I. A cellular suicide strategy of plants: vacuole-mediated cell death. Apoptosis. 2006;11:905–911. doi: 10.1007/s10495-006-6601-1. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Yamada K, Hiraiwa N, Nishimura M, Hara-Nishimura I. Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. The Plant Journal. 1999;19:43–53. doi: 10.1046/j.1365-313x.1999.00497.x. [DOI] [PubMed] [Google Scholar]
- Kladnik A, Chamusco K, Dermastia M, Chourey P. Evidence of programmed cell death in post-phloem transport cells of the maternal pedicel tissue in developing caryopsis of maize. Plant Physiology. 2004;136:3572–3581. doi: 10.1104/pp.104.045195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi M, Yamada K, Hatsugai N, Kondo M, Nishimura M, Hara-Nishimura I. VPE is essential for mycotoxin-induced cell death in Arabidopsis thaliana. Journal of Biological Chemistry. 2005;280:32914–32920. doi: 10.1074/jbc.M504476200. [DOI] [PubMed] [Google Scholar]
- Linnestad C, Doan DN, Brown RC, Lemmon BE, Meyer DJ, Jung R, Olsen OA. Nucellain, a barley homolog of the dicot vacuolar-processing protease, is localized in nucellar cell walls. Plant Physiology. 1998;118:1169–1180. doi: 10.1104/pp.118.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi L, Ceccarelli N, Picciarelli P, Lorenzi R. DNA degradation during programmed cell death in Phaseolus coccineus suspensor. Plant Physiology and Biochemistry. 2007;45:221–227. doi: 10.1016/j.plaphy.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Martinez M, Diaz I. The origin and evolution of plant cystatins and their target cysteine proteinases indicate a complex functional relationship. BMC Evolutionary Biology. 2008;8:198. doi: 10.1186/1471-2148-8-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaune S, Yamada K, Kondo M, Kato T, Tabata S, Nishimura M, Hara-Nishimura I. A vacuolar processing enzyme, δVPE, is involved in seed coat formation at the early stage of seed development. The Plant Cell. 2005;17:876–887. doi: 10.1105/tpc.104.026872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA. Nuclear endosperm development in cereals and Arabidopsis thaliana. The Plant Cell. 2004;16:S214–S227. doi: 10.1105/tpc.017111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick JW, Offler CE. Compartmentation of transport and transfer events in developing seeds. Journal of Experimental Botany. 2001;52:551–564. [PubMed] [Google Scholar]
- Pennel RI, Lamb C. Programmed cell death in plants. The Plant Cell. 1997;9:1157–1168. doi: 10.1105/tpc.9.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk V, Borisjuk L, Radchuk R, Steinbiss HH, Rolletschek H, Broeders S, Wobus U. Jekyll encodes a novel protein involved in the sexual reproduction of barley. The Plant Cell. 2006;18:1652–1666. doi: 10.1105/tpc.106.041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk VV, Borisjuk L, Sreenivasulu N, Merx K, Mock HP, Rolletschek H, Wobus U, Weschke W. Spatio-temporal profiling of starch biosynthesis and degradation in the developing barley grain. Plant Physiology. 2009;150:190–204. doi: 10.1104/pp.108.133520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Reape TJ, McCabe PF. Apoptotic-like programmed cell death in plants. New Phytologist. 2008;180:13–26. doi: 10.1111/j.1469-8137.2008.02549.x. [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamada K, Kataoka M, et al. Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. Journal of Biological Chemistry. 2003;278:32292–32299. doi: 10.1074/jbc.M305740200. [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Borisjuk L, Junker BH, Mock HP, Rolletschek H, Seiffert U, Weschke W, Wobus U. Barley grain development: toward an integrative view. International Review of Cell and Molecular Biology. 2010;281:49–89. doi: 10.1016/S1937-6448(10)81002-0. [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Radchuk V, Strickert M, Miersch O, Weschke W, Wobus U. Gene expression patterns reveal tissue-specific signaling networks controlling programmed cell death and ABA-regulated maturation in developing barley seeds. The Plant Journal. 2006;47:310–327. doi: 10.1111/j.1365-313X.2006.02789.x. [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Cosgrove DJ. Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Current Biology. 2009;19:R800–R811. doi: 10.1016/j.cub.2009.07.056. [DOI] [PubMed] [Google Scholar]
- Thiel J, Weier D, Sreenivasulu N, Strickert M, Weichert N, Melzer M, Czauderna T, Wobus U, Weber H, Weschke W. Different hormonal regulation of cellular differentiation and function in nucellar projection and endosperm transfer cells—a microdissection-based transcriptome study of young barley grains. Plant Physiology. 2008;148:1436–1452. doi: 10.1104/pp.108.127001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D. Genome-wide analysis of core cell cycle genes in Arabidopsis. The Plant Cell. 2002;14:903–916. doi: 10.1105/tpc.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inzé D, De Veylder L. Genome-wide identification of potential plant E2F target genes. Plant Physiology. 2005;139:316–328. doi: 10.1104/pp.105.066290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D, Declercq W, Vandenabeele P, Van Breusegem F. Are metacaspases caspases? Journal of Cell Biology. 2007;179:375–380. doi: 10.1083/jcb.200705193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J, Kakar K, Gallardo K, Signor CL, Aubert G, Schlereth A, Town CD, Udvardi MK, Thompson RD. Gene expression profiling of M. truncatula transcription factors identifies putative regulators of grain legume seed filling. Plant Molecular Biology. 2008;67:567–580. doi: 10.1007/s11103-008-9320-x. [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Heim U, Buchner P, Wobus U. Seed coat-associated invertases of fava bean control both unloading and storage functions: cloning of cDNAs and cell type-specific expression. The Plant Cell. 1995;7:1835–1846. doi: 10.1105/tpc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wang L, Li J, Song X, Yang C. Study on programmed cell death and dynamic changes of starch accumulation in pericarp cells of Triticum aestivum L. Protoplasma. 2009;236:49–58. doi: 10.1007/s00709-009-0046-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.