Abstract
In this study, the transcriptional state and distribution of RNA polymerase II, pre-mRNA splicing machinery elements, and rRNA transcripts were investigated in the sperm cells of Hyacinthus orientalis L. during in vitro pollen tube growth. During the second pollen mitosis, no nascent transcripts were observed in the area of the dividing generative cell, whereas the splicing factors were present and their pools were divided between newly formed sperm cells. Just after their origin, the sperm cells were shown to synthesize new RNA, although at a markedly lower level than the vegetative nucleus. The occurrence of RNA synthesis was accompanied by the presence of RNA polymerase II and a rich pool of splicing machinery elements. Differences in the spatial pattern of pre-mRNA splicing factors localization reflect different levels of RNA synthesis in the vegetative nucleus and sperm nuclei. In the vegetative nucleus, they were localized homogenously, whereas in the sperm nuclei a mainly speckled pattern of small nuclear RNA with a trimethylguanosine cap (TMG snRNA) and SC35 protein distribution was observed. As pollen tube growth proceeded, inhibition of RNA synthesis in the sperm nuclei was observed, which was accompanied by a gradual elimination of the splicing factors. In addition, analysis of rRNA localization indicated that the sperm nuclei are likely to synthesize some pool of rRNA at the later steps of pollen tube. It is proposed that the described changes in the nuclear activity of H. orientalis sperm cells reflect their maturation process during pollen tube growth, and that mature sperm cells do not carry into the zygote the nascent transcripts or the splicing machinery elements.
Keywords: Bicellular pollen, Hyacinthus orientalis L., pollen tube, pre-mRNA splicing, rRNA, sperm cells
Introduction
The mature pollen grain of angiosperms represents a unique structure with a key function in sexual plant reproduction: development and delivery of male gametes (sperm cells) into the embryo sac during pollination. This function is accomplished by the pollen tube, which germinates from the pollen grain and grows through the pistil tissues until it reaches the embryo sac. As the tip of the tube reaches the embryo sac, two sperm cells are released and double fertilization occurs. One of the sperm cells undergoes fusion with the egg cell to produce the embryo of a new plant, while the second cell fuses with the central cell to produce a nutritive endosperm. Depending on the timing of generative cell mitosis, two sperm cells may be formed during pollen development in the anther (tricellular pollen grains) or during pollen tube growth (bicellular pollen grains). Thus, in species producing bicellular pollen grain, the grain must be cultured in order to grow pollen tubes, trigger mitosis, and obtain sperm cells (Russell, 1991).
The structural features of plant sperm cells have been well characterized and can be found elsewhere (Hoefert, 1969; Haskel and Rogers, 1985; Corriveau and Coleman, 1988; Russel, 1991). In most flowering plants, the sperm cells are isomorphic, express a similar gene pool, and share an equal ability to fertilize the egg cell (Berger et al., 2008; Ingouff et al., 2009). However, little information is available on the molecular bases of their biology, including both cytoplasmic and nuclear events. The presence of nascent transcripts was confirmed in the sperm cell nuclei of rice germinating pollen grains (Haskell and Rogers, 1985). The group of Zhang et al. (1993) showed ongoing RNA and protein synthesis in the sperm cells isolated from the mature pollen grains of maize during their in vitro culture. However, in both studies, the types of RNA synthesized by the sperm nuclei were not determined. Whether RNA synthesis is related to the timing of sperm cell formation is still a matter of controversy. From recent transcriptomic studies, it is known that mature Arabidopsis thaliana pollen grains contain sperm cell-specific transcripts, including mRNAs encoding cell cycle progression proteins, and molecules involved in nuclear metabolism and protein degradation (Becker et al., 2003; Pina et al., 2005; Borges et al., 2008; Bayer et al., 2009).
Given all these data, it can be seen that nearly all of the current knowledge on plant sperm cells is limited to mature tricellular pollen grains. Information related to the sperm cells formed during pollen tube growth is very sparse. The classical experiments described by Mascarenhas (1975) showed that the early steps of pollen tube growth are transcriptionally independent. However, the division of generative cells into sperm cells was abolished after inhibition of RNA synthesis by actinomycin D (Mascarenhas, 1975). This suggests that the restarting of RNA synthesis during pollen tube growth can be essential for the proper formation of male gametes, at least in species with bicellular pollen grains. Indeed, the transcriptional activity of vegetative and generative nuclei was indicated in the growing pollen tubes of Tradescantia paludosa, Nicotiana tabacum, Lilium longiflorum (Mascarenhas, 1975), and, more recently, in the growing pollen tubes of Hyacinthus orientalis (Zienkiewicz et al., 2008a). Apart from the early studies of Reynolds and Raghavan (1982) who reported that the sperm cells of Hyoscyamus niger are transcriptionally silent, there is no information on RNA synthesis in the sperm cells of species producing bicellular pollen grains.
Sperm cells are shown to undergo a complete cell cycle. In A. thaliana, the sperm nuclei are in S-phase during anthesis and pass into G2 phase just before pollination (Friedman, 1999). In turn, the bicellular pollen grains of tobacco are released from the anthers with generative cells containing 2C. After their mitotic division in the pollen tube, the resulting sperm cells have 1C DNA contents (Tian et al., 2005). As the sperm cells are deposited in the degenerated synergid, their DNA content is between 1C and 2C. Thus, the authors suggest that receptive gametes are those that pass through the protracted S-phase and reach the G2 phase (Tian et al., 2005).
The key nuclear steps of protein-encoding gene expression are transcription and nascent transcript processing. Pre-mRNA transcript is synthesized by the RNA polymerase II complex. The largest subunit of the RNA polymerase II core enzyme contains a unique C-terminal repeat domain (CTD). The function of this domain is closely associated with its phosphorylation state (Lee and Young, 2000). To date, it is known that the CTD exists in two phosphorylation states—hypophosphorylated (Pol IIA) and hyperphosphorylated (Pol IIO). Pol IIA is found in initiation complexes, whereas Pol IIO is present in elongating polymerase complexes. The phosphorylated CTD was also shown to recruit pre-mRNA processing factors, including splicing machinery molecules (Lee and Young, 2000; Reed, 2003; Kornblihtt et al., 2004; Bentley, 2005). Newly formed pre-mRNA undergoes processing, including a splicing reaction during which the introns are removed and the exons are joined. This reaction occurs in large ribonucleic complexes, called spliceosomes, composed of five classes of small ribonucleoproteins (RNPs) (U1, U2, U4/U6, and U5 snRNPs) as well as numerous non-RNA splicing factors. Each splicing snRNP molecule contains a proper snRNA chain with a trimethylguanosine (TMG) cap on the 5' end and an Sm protein core (Will and Lührmann, 2001; Yong et al., 2004). Among non-RNP splicing factors, a large group belonging to the SR (serine–arginine rich) protein family was identified. The SR proteins function in the formation of a catalytically active spliceosome by multiple protein–protein and protein–RNA interactions (Graveley, 2000). One of the members of this family is the SC35 protein, which is capable of reactivating the splicing reaction by binding to specific pre-mRNA sequences (Will and Lührmann, 1997).
Transcription of rDNA genes by RNA polymerase I results in the formation of the rRNA primary transcript (pre-rRNA). Each pre-rRNA molecule contains three of the four rRNAs found in the ribosomes: the 18S small subunit (SSU), the 26S large subunit (LSU), and the 5.8S rRNAs. The pre-rRNA molecule is flanked at both ends by external transcribed spacers (ETSs) and the rRNAs present in the primary transcript are separated by sequences termed internal transcribed spacers (ITSs). The mature rRNAs are released from the primary transcript by several cleavage reactions catalysed by a class of small RNAs—snoRNAs and by additional nucleolar proteins (Brown and Shaw, 1998).
Here, the in situ organization of the key nuclear steps of gene expression in sperm cells of H. orientalis was analysed, with special emphasis on transcription and nascent transcript processing. Previously, it had been shown that despite the fact that the mature pollen grains of H. orienalis contains a high amount of long-lived mRNA (Zienkiewicz et al., 2006), the restarting of RNA synthesis occurs during the early steps of pollen tube growth and is accompanied by the presence of pre-mRNA splicing factors in both of the pollen nuclei (Zienkiewicz et al., 2006, 2008a, b, c). Changes in RNA synthesis were accompanied by a significant redistribution of splicing machinery elements in the vegetative and the generative nuclei of growing pollen tube.
In this study, the nuclear metabolism of hyacinth sperm cells was analysed during 36 h of in vitro pollen tube growth. This period of cultivation closely mimics the time needed for in vivo pistil penetration by the pollen tube and fertilization in H. orientalis. The present results are the first to illustrate the spatial and temporal pattern of RNA synthesis and the organization of the molecules involved in key steps of gene expression in the sperm cells formed during pollen tube growth.
Materials and methods
Plant material
Hyacinthus orientalis L. (a commercial cultivar grown at room temperature at the Institute of General and Molecular Biology, Nicolaus Copernicus University, Toruń, Poland) pollen tubes growing in vitro were used in the investigation.
Preparation of material
Freshly collected mature pollen grains of H. orientalis were used for germination. Before placing the pollen grains, the Brewbaker and Kwak (1963) medium with 10% (w/v) polyethylene glycol 4000 was modified by adding pistils from the pollinated flowers. Pistils were collected 8 h after pollination. Five symmetrically cut pistils (10 parts) were added to each 5 ml of the medium and gently squeezed. The pollen grains were placed on the surface of this medium and lightly homogenized using a pipette. Cultivation was carried out at 25 °C in the dark. For immunolocalization of Pol IIO, Pol IIA, TMG snRNA, and SC35 protein, the pollen tubes were collected after 8, 12, 24, and 36 h of growth. The medium (with the pistils) was replaced every 8 h during pollen tube cultivation. Just before fixation and after manual removal of the pistils, the pollen tubes, and the germination medium were transferred into tubes (50 ml) and gently centrifuged to remove excess medium. The pollen tubes were fixed in a mixture of 4% (w/v) paraformaldehyde and 0.5% (v/v) glutaraldehyde prepared in phosphate-buffered saline (PBS), pH 7.2, overnight at 4 °C. The material was then enzymatically digested as previously described (Zienkiewicz et al., 2006). All the steps of the digestion procedure were carried out in tubes (50 ml) with gentle centrifugation between each step. These semi-protoplasts of pollen tubes were stored at 4 °C.
For total protein and RNA isolation, the mature, hydrated (1 h of hydration in a humid chamber at 30 °C), and germinating (1, 3, 6, 12, 24, and 36 h) pollen grains were collected. Germination was performed as described above. The samples were frozen in liquid nitrogen and stored at −80 °C. In experiments with cycloheximide, after 3 h (sample 6hCX) or 9 h (sample 12hCX) of pollen tube growth, cycloheximide (Sigma-Aldrich, St. Louis, USA) was added to the medium to a final concentration of 400 μg ml−1. Cultivation on the medium with cycloheximide was continued for 3 h and the pollen tubes were collected and stored as described above.
Transcription analysis
For in vitro transcription investigations, the pollen grains were germinated in the same way as described above, but after 8, 12, 24, 32, and 36 h of growth the pollen tubes were transferred from the standard medium containing fragmented pistils to a medium containing 0.01% (v/v) Triton X-100 for 10 min (with fragmented pistils), and then to a medium containing 20 mM 5-bromouracil for 45 min. As a control, the pollen tubes were first transferred onto a medium containing actinomycin D (25 g ml−1) for 1 h, and then to a medium containing both actinomycin D (25 g ml−1) and 20 mM 5-bromouracil for 45 min after 8, 12, 24, 32, and 36 h of growth. The material was then washed three times with fresh medium for 5 min each and then fixed. Pollen tube fixation and enzymatic digestion were carried out as previously described. The prepared material was stored at 4 °C.
Immunolocalization experiments
All immunocytochemical reactions with the material obtained in vitro were carried out in 1.5 ml Eppendorf tubes. The samples were gently centrifuged between each step of the procedure and placed on microscope slides covered with Biobond (British Biocell International, Cardiff, UK). TMG snRNA and SC35 protein were detected by incubating with the primary antibodies mouse anti-TMG (Calbiochem, Bad Soden, Germany) and mouse anti-SC35 (Sigma-Aldrich), respectively, as previously described (Zienkiewicz et al., 2006). Pol IIA and Pol IIO were localized using H14 and H5 IgM monoclonal mouse antibodies (Sigma-Aldrich), respectively. Anti-mouse IgM antibodies labelled with TRIC (Sigma-Aldrich) were used as secondary antibodies.
Incorporated 5-bromouracil was detected by incubating the samples with the primary monoclonal antibody anti-BrdUTP (F. Hoffmann-LaRoche Ltd, Rotkrenz, Switzerland) diluted 1:100 in 1% (w/v) bovine serum albumin (BSA) prepared in PBS, pH 7.2, overnight at 4 °C, and with the secondary goat anti-mouse antibody Alexa Fluor 488 diluted 1:500 in 1% (w/v) BSA in PBS for 1 h at 37 °C.
Control reactions were performed without the primary antibodies. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; Fluka).
In situ hybridization
Pre-rRNA was detected using a DNA oligonucleotide probe complementary to the ITS1 of pre-rRNA (5′-Cy3-ACG GGT TCG GGA TCG TCC GTT CGG G-3′) (www.oligo.pl, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland). 26S rRNA was detected using the DNA oligonucleotide probe complementary to the 26S rRNA sequence fragment (5′-Cy3-CAC GGA ATA AGT AAA ATA ACG TTA AAA GTA G-3′). U2 snRNA (see the Supplementary data available at JXB online) was detected using the oligonucleotide probe TOR50, which is complementary to the first 20 nucleotides of U2 snRNA (Department of Bioorganic Chemistry, Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences). All these probes were chemically labelled with Cy3 fluorochrome at the 5′ end (IBB PAN, Warsaw, Poland). For hybridization, the probes were resuspended in the hybridization buffer [30% (v/v) formamide, 4× SSC, 5× Denhardt's buffer, 1 mM EDTA, 50 mM phosphate buffer, and water (H2O)] from 100 pmol ml−1 to 200 pmol ml−1. Hybridization was performed overnight at 37 °C. In the control reactions, pollen tubes were incubated with the hybridization buffer, with omission of the probes. The positive control to ITS1 hybridization was performed as described above on the somatic tissue of the young H. orientalis anther.
Microscopy
The samples were analysed with a Nikon Eclipse TE 300 confocal laser scanning inverted microscope. The results were recorded using an argon-ion laser emitting light with a wavelength of 488 nm (blue excitation and green fluorescence) and a helium–neon laser emitting light with a wavelength of 543 nm (green excitation and red fluorescence). A mid pinhole, long exposure time (75 s), and 60× (numerical aperture: 1.4) Plan Apochromat DIC H oil immersion lens was used. Pairs of images were collected simultaneously in the green (Alexa 488 fluorescence) and red (autofluorescence) channels. The three-dimensional optical sections were acquired at 1 μm step intervals. The final images represent projection of an image stack. For image processing and analysis, the EZ 2000 Viewer software package (Nikon Europe BV, Badhoevedorp, The Netherlands) was used. For DAPI staining, an inverted Nikon Eclipse TE 80i fluorescence microscope, equipped with a mercury lamp, a UV-2EC UV narrow band filter, and a DXM 1200 FX digital camera, was used.
Total RNA analysis
Frozen samples (100 mg) were ground in liquid nitrogen using a mortar and pestle. Total RNA was isolated from the frozen powder using an RNeasy Plant Mini Kit (Qiagen, Syngen Biotech, Wrocław, Poland) according to the manufacturer's instructions. The total RNA was treated with DNase I (RNase-free) (Fermentas, Burlington, Canada). RNA samples of 1 μg were loaded on a 1.2% agarose gel and stained with ethidium bromide (Sigma-Aldrich). The gels were visualized using the DNR Bio-Imaging System (DNR Bio-Imaging Systems Ltd, Jerusalem, Israel).
Total protein extraction, SDS–PAGE, and content measurement
In each case, the proteins were isolated from 500 mg of mature (MP), hydrated (HP), and germinating pollen grains (1, 3, 6, 12, 24, and 36 h) and from pollen tubes treated with cycloheximide (6hCX and 12hCX). The frozen material was ground into a fine powder in liquid nitrogen. For total protein isolation, the ground material was resuspended in extraction buffer [100 mM TRIS-HCl buffer (pH 7.5), 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulphonyl fluoride (protease inhibitor cocktail); Sigma-Aldrich] by gentle shaking and clarified by centrifugation at 16 000 g for 30 min at 4 °C.
SDS–PAGE was performed according to Laemmli (1970). A 5 μl aliquot of protein extract per sample was loaded on 12% (w/v) polyacrylamide gels with 4% stacking gels using a Mini-Protean III apparatus (Bio-Rad). After electrophoresis, the gels were stained with Coomassie Brilliant Blue R250 according to the standard procedure.
The protein concentration was estimated in 1 μl of each sample by using the 2D Quant kit (Amersham Biosciences, USA) according to the manufacturer's instructions.
Results
Transcriptional activity of pollen tube nuclei
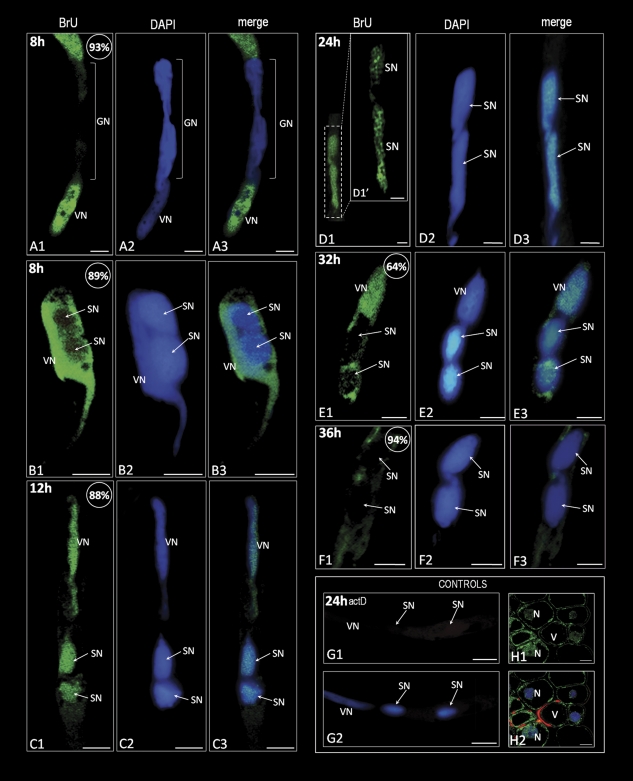
The presence of 5-bromouracil in the growing pollen tubes of H. orientalis was analysed after 8, 12, 24, 32, and 36 h of in vitro cultivation (Fig. 1). The results obtained revealed the presence of nascent transcripts as well as changes in their levels and distribution pattern during the course of pollen tube growth. After 8 h of cultivation, pollen tubes containing generative cells, dividing generative cells (Fig. 1A), or sperm cells (Fig. 1B) were observed. During generative cell division, the incorporated 5-bromouracil was observed mainly in the pollen tube cytoplasm (Fig. 1A), whereas in the area of the dividing generative cell no labelling was observed. After sperm cell formation (Fig. 1B), many pollen tubes showed a physical relationship between the sperm cells and the vegetative nucleus [termed a male germ unit (MGU)]. In such an organized structure, the most intense fluorescence was observed in the vegetative nucleus. In both of the sperm nuclei, the extent of labelling was significantly low and observed in the form of small fluorescent spots (Fig. 1B). After 12 h (Fig. 1C) and 24 h (Fig. 1D) of pollen tube growth, the incorporated 5-bromouracil was detected in the vegetative nucleus and in both of the sperm nuclei at similar levels. A less intense signal was observed in the pollen tube cytoplasm. Between 24 h and 36 h of pollen tube growth, significant changes in the levels and localization of the incorporated 5-bromouracil were seen in comparison with the previous steps of pollen tube growth. The highest signal indicating nascent transcripts was observed in the vegetative nucleus and pollen tube cytoplasm (Figs. 1E, F). In both of the sperm cells, a gradual decrease occurred in the labelling. In these nuclei, the nascent transcripts were localized in the form of a few, irregular fluorescent spots (Fig. 1E). After 36 h of pollen tube growth, no signal was detected in sperm cells (Fig. 1F). A control reaction performed on pollen tubes treated with actinomycin D showed no labelling (Fig. 1G). As a positive control, the incorporation of 5-bromouracil in the anther wall of parenchyma cells of young H. orientalis was analysed (Fig. 1H). The labelling was found in all the observed nuclei and cytoplasm. No signal was detected in the vacuoles. These results indicated that the sperm cells activate RNA synthesis soon after their formation. Ongoing transcription was observed until the final steps of pollen tube growth when inhibition of RNA synthesis occurred in both of the sperm cells. The vegetative nucleus is transcriptionally active during the whole culture period.
Fig. 1.
Localization of the incorporated 5-bromouracil (green colour) in hyacinth pollen tubes. (A1–A3) A pollen tube after 8 h of growth containing the dividing generative cell. A strong signal is present in the vegetative nucleus and in the pollen tube cytoplasm. The area of the dividing generative nucleus is completely devoid of fluorescence. (B1–B3) A pollen tube soon after sperm cell formation. The vegetative nucleus exhibits a significantly stronger signal in comparison with the sperm cells. In the sperm nuclei, a few spots of the signal are present. (C1–C3) After 12 h of pollen tube growth, the sperm nuclei exhibit a signal intensity comparable with that observed in the vegetative nucleus. (D1–D3) In both sperm nuclei of the pollen tube growing for 24 h, the signal corresponding to nascent transcripts is localized in the form of numerous irregular fluorescent clusters. (E1–E3) A pollen tube growing for 32 h. Strong homogenous fluorescence is visible in the vegetative nucleus as well as in the pollen tube cytoplasm. In the area of the sperm nuclei, only a few spots of weak signal are present. (F1–F3) No labelling can be observed in the sperm nuclei after 36 h of pollen tube growth. Weak fluorescence is present in the pollen tube cytoplasm. (G1 and G2) Control reaction performed on pollen tubes treated with actinomycin D. (H1 and H2) Positive control showing the presence of nascent transcripts in the cells of the young anther wall. An intense signal is present in the nuclei and cytoplasm of the parenchyma cells, whereas the vacuoles are devoid of green fluorescence; values presented in the upper right corner of the figures indicate the percentage of pollen tubes exhibiting the labelling pattern shown on the image. Z-series images corresponding to the whole volume of growing pollen tubes from three independent experiments (n=50) were captured and used for statistical analysis. VN, vegetative nucleus; GN, generative nucleus; SN, sperm nucleus; N, nucleus; V, vacuole; bars=10 μm.
Levels of rRNA and its localization during hyacinth pollen tube growth
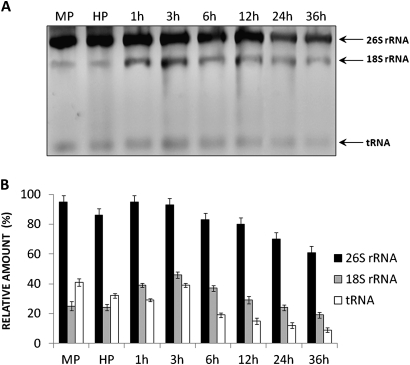
To analyse whether rRNA is present in the pool of the newly synthesized transcripts during pollen tube growth, the RNA profile of the growing pollen tube was investigated (Fig. 2) and a series of fluorescence in situ hybridization (FISH) experiments were performed (Fig. 3). Well-distinguishable pools of 26S and 18S rRNA (Fig. 2A) were observed in the profile of the total RNA of the growing pollen tube. The level of both of the rRNA classes stored in the mature pollen grain did not change significantly during its hydration (Fig. 2B). As the pollen grains started to germinate, a slight increase in 26S and 18S rRNA levels was observed during the first 3 h of pollen tube growth. Further steps of pollen tube growth were accompanied by a gradual decrease in the rRNA pool, with a minimum level observed at the last analysed stage of pollen tube elongation (Fig. 2B).
Fig. 2.
Profile of total RNA in the growing pollen tubes of hyacinth. (A) Ethidium bromide-stained gel of total RNA isolated from pollen at maturity (MP), after hydration (H), and after 1, 3, 6, 12, 24, and 36 h of in vitro germination. Well-distinguishable bands corresponding to 26S rRNA and 18S rRNA as well as to tRNA are indicated with arrows. (B) Densitometric data corresponding to the 26S rRNA (black bars), 18S rRNA (grey bars), and tRNA (white bars) from A. The values represent the average of thee replicate experiments; the error bars indicate 1 SEM.
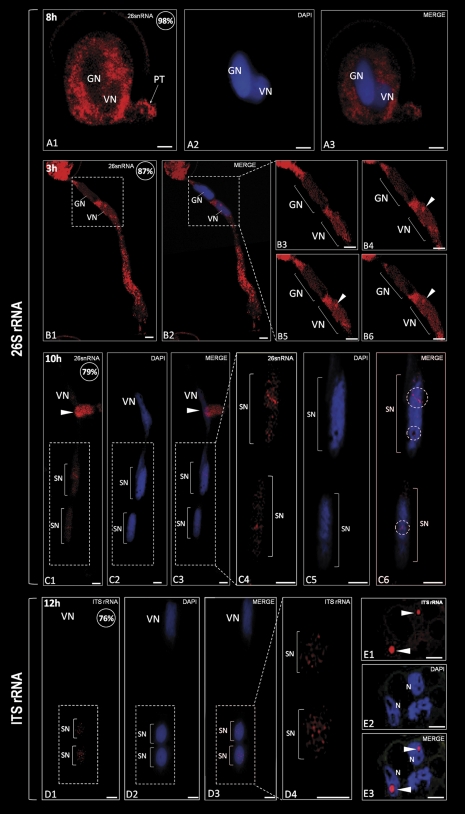
Fig. 3.
Localization of rRNA (red colour) in hyacinth pollen tubes by FISH. (A1–A3) In the young pollen tube, the signal corresponding to 26S rRNA is observed only in the cytoplasm. Accumulation of fluorescence occurs inside the pollen grain and at the tip of the small pollen tube (arrow). Pollen nuclei do not show any labelling. (B1 and B2) After 3 h of pollen tube growth, the strongest signal is present in the pollen tube cytoplasm. At this time, the fluorescence corresponding to 26S rRNA appears in the vegetative nucleus, whereas the generative nucleus does not show any fluorescence. (B3–B6) On the consecutive optical sections of both pollen tube nuclei (area marked with the dashed line on B1 and B2), accumulation of the signal in the vegetative nucleus is observed in a round-shaped fluorescent cluster (arrowhead). (C1–C3) After sperm cell formation, 26S rRNA appears to be located in both sperm nuclei and the vegetative nucleus. The arrowhead indicates the fragment of the other pollen tube. (C4–C6) Magnification of the pollen tube zone containing sperm cells (area marked with the dashed line on C1). In both sperm nuclei, the signal is present in the form of irregular fluorescent clusters, which appear to occupy nuclear areas devoid of DNA (C6; circles). (D1–D4) The fluorescence corresponding to immature rRNAs is observed only in the sperm nuclei where it is visible as numerous spots of different sizes and shapes (D4). (E1-E3) A positive control performed on the somatic tissues of hyacinth anther revealed accumulation of ITS containing rRNAs exclusively in the nucleoli of the parenchyma cell nuclei (arrowheads); values presented in the upper right corner of the figures indicate the percentage of pollen tubes exhibiting the labelling pattern showed on the image. Z-series images corresponding to the whole volume of growing pollen tubes from three independent experiments (n=50) were captured and used for statistical analysis. VN, vegetative nucleus; GN, generative nucleus; SN, sperm nucleus; N, nucleus; bars=10 μm.
In situ localization of 26S rRNA at the beginning of pollen grain germination showed its presence exclusively in the cytoplasm of the pollen grain and the emerging pollen tube (Fig. 3A). As germination proceeded, the signal corresponding to 26S rRNA was abundantly localized in the cytoplasm of the proximal area of the pollen tube as well as the pollen grain (Fig. 3B). The hybridization signal was observed in the vegetative nucleus where the accumulation of labelling was observed in the form of a round-shaped fluorescent cluster (Fig 3B3–B6; arrowhead). A less intense signal was also uniformly localized throughout the vegetative nucleoplasm (Fig. 3B). In the generative nucleus, no labelling was detected. After sperm cell formation (Fig. 3C), a decrease in the cytoplasmic pool of 26S rRNA was observed in comparison with the pre-mitotic steps of pollen tube growth. A nuclear hybridization signal corresponding to 26S rRNA was detected in the vegetative nucleus and both of the sperm nuclei (Fig. 3C). The labelling in the sperm nuclei was observed in the form of numerous irregular fluorescent clusters, probably localized between the DNA clumps (Fig. 3C4–C6; circles).
Detection of immature rRNA transcripts (Fig. 3D) revealed their presence in both of the sperm nuclei at the 12th hour of pollen tube growth. A hybridization signal was observed as numerous small fluorescent spots spread throughout the sperm nucleoplasm. Fluorescent labelling was seen in the pollen tube cytoplasm (Fig. 3D). The last analysed steps of pollen tube growth (between 24 h and 36 h) were accompanied by a complete lack of signal in the sperm nuclei (not shown). The positive control, performed on the somatic tissue of the young hyacinth anther, showed strong accumulation of ITS rRNA in the nucleoli of the parenchyma cells (Fig. 3E). The data indicate that both of the sperm cells contained pre-mature and mature rRNAs during their life cycle. During further steps of pollen tube growth, the pool of rRNA in the sperm cells gradually decreases until there is a complete lack at the final steps of culture.
Protein content in the growing pollen tube of hyacinth
Protein content was measured at different stages of H. orientalis pollen tube growth (Fig. 4). Mature and hydrated pollen grains were shown to contain the highest levels of proteins. Early steps of pollen tube growth were accompanied by a gradual and strong decrease in the total protein pool. A significant increase in the protein content occurred between the third and sixth hour of pollen grain germination. During the further steps of pollen tube elongation, a continuous reduction in the total protein pool was seen, with the lowest levels observed at the last analysed step of culture.
Fig. 4.
Analysis of protein contents during H. orientalis pollen tube growth. (A) Coomassie blue-stained gel of total proteins from pollen grains at maturity (MP), after hydration (H), and after 1, 3, 6, 12, 24, and 36 h of in vitro germination. Lanes marked as 6hCHX and 12hCHX indicate pollen tubes treated for 3 h with cycloheximide added to the medium at the third and ninth hour of growth, respectively. Lanes 6hC and 12hC indicate control pollen tubes growing for 6 h and 12 h, respectively. (B) Protein concentration (μg) in 1 μl of extracts isolated from the mature, hydrated, and germinating pollen grains of hyacinth. The temporal variants of pollen tube growth are the same as shown in A. Grey blocks of the graph correspond to the pollen tubes treated with cycloheximide. The values represent the average of three replicate experiments; the error bars indicate 1 SEM.
To investigate the intensity of protein synthesis before and after sperm cell formation, H. orientalis pollen tubes were treated with cycloheximide. A translation inhibitor was added to the medium at the third and ninth hour of pollen tube growth and the incubation was continued for 3 h. The treatment caused a significant depletion in the protein content, in comparison wiith the pollen tubes growing on control medium (Fig. 4B; grey bars). The reduction in the total protein pool was >30% and ∼15% in the pollen tubes growing for 3 h and 9 h, respectively, before cycloheximide treatment. At the morphological level, cycloheximide treatment caused strong inhibition of pollen tube growth, arrest of sperm cell formation, and defects in the pollen tube structure (Supplementary Fig. S1 at JXB online). These results indicate intense protein synthesis at the time of sperm cell formation. After the second pollen mitosis, the rates of ongoing translation were low and were followed by a continuous decrease in the total protein content during the further steps of pollen tube growth.
Distribution of the initiating and elongating RNA polymerase II forms
Before the mitotic division of the generative cell, the hypophosphorylated form of RNA polymerase II (Pol IIA) was localized in both of the nuclei present in the pollen tube (Fig. 5A). In the vegetative nucleus, a strong labelling was uniformly distributed throughout the area of the nucleoplasm, whereas in the generative nucleus less intense fluorescence was localized in the form of a few, irregular spots (Fig. 5A). After sperm cell formation, the initiating form of Pol II was present in the vegetative nucleus as well as in the sperm nuclei (Fig. 5B, and Supplementary Fig. S5 at JXB online). The distribution pattern of labelling was similar in all the observed nuclei because irregular spots of fluorescence were spread throughout their area. The later steps of pollen tube growth were accompanied by a lack of Pol IIA in both of the sperm nuclei (Fig. 5C). In the vegetative nucleus, relatively strong labelling was still observed. No signal was observed in the pollen tubes after the control reaction (Fig. 5D).
Fig. 5.
Localization of hypo- (Pol IIA) and hyperphosphorylated (Pol IIO) forms of RNA polymerase II (red colour) in hyacinth pollen tubes. (A1–A3) In the pollen tube growing for 8 h, the labelling indicating Pol IIA is present exclusively in the area of the vegetative and generative nuclei. In the latter nucleus, only some irregular fluorescence clusters are present. In the vegetative nucleus, the labelling is much more intense and uniformly distributed throughout the nucleoplasm. (B1–B3) After generative cell division, both the sperm nuclei exhibit the presence of irregular fluorescent spots corresponding to Pol IIA. The magnification of the area marked with the dashed line in B1 is shown in Supplementary Fig. S5 at JXB online. (C1-C3) In the pollen tube growing for 36 h, both sperm cells show no labelling. A strong signal is present in the vegetative nucleus. (D1 and D2). Control reaction by omitting the primary anti-Pol IIA antibody. (E1–E3) Before generative cell division, the strong and uniformly distributed fluorescence indicating Pol IIO is located in the vegetative nucleus. In the generative nucleus, numerous clusters of less intense signal are present. (F1–F3) After second pollen mitosis, the signal is observed in the vegetative and sperm nuclei, wherein numerous spots of fluorescence are present. (G1–G3) Later steps of pollen tube growth were accompanied by a lack of the signal in the sperm nuclei and its reduction in the vegetative nucleus. (H1 and H2) Control reaction conducted by omitting incubation with the anti-Pol IIO antibody; values presented in the upper right corner of the figures indicate the percentage of pollen tubes exhibiting the labelling pattern showed on the image. Z-series images corresponding to the whole volume of growing pollen tubes from three independent experiments (n=50) were captured and used for statistical analysis; VN, vegetative nucleus; GN, generative nucleus; SN, sperm nucleus; bars=10 μm.
Before sperm cell formation, the hyperphosphorylated form Pol IIO was detected in the vegetative and generative nuclei (Fig. 5E). Both of the nuclei strongly differed in Pol IIO localization pattern. In the vegetative nucleus, an intense and uniformly distributed fluorescence was observed, whereas in the generative nucleus only irregular spots of labelling were present (Fig. 5E). After generative cell division (Fig. 5F), the elongating form of Pol II was localized in the vegetative nucleus as well as in both of the sperm cells. In the vegetative nucleus, a very intense labelling was uniformly distributed throughout the area of the nucleoplasm. In the sperm nuclei, only single spots of fluorescence were observed (Fig. 5F). After 36 h of pollen tube growth, Pol IIO was detected only in the vegetative nucleus, whereas both of the sperm cells were devoid of any labelling (Fig. 5G). The signal present in the vegetative nucleus was significantly weaker than at the previous step of pollen tube growth and was visible in the form of numerous irregular spots spread throughout the nucleoplasm. No labelling was observed in the control performed by omitting the primary H14 antibody (Fig. 5H). These data indicate that the pre-mRNA transcriptional machinery is present in both of the sperm cells, however at a lower level than in the vegetative nucleus. The final steps of pollen tube growth were accompanied by the elimination of the initiating and elongating forms of Pol II from both sperm cells.
Localization of the pre-mRNA splicing machinery elements
In order to assess whether molecules involved in pre-mRNA processing are present in the sperm cells formed during H. orientalis pollen tube growth, immunolocalization of the TMG cap-containing snRNAs and of the SC35 splicing factor was performed. Both the antigens were present in the H. orientalis sperm nuclei, although their levels and localization patterns changed significantly during the examined steps of pollen tube growth (Fig 6).
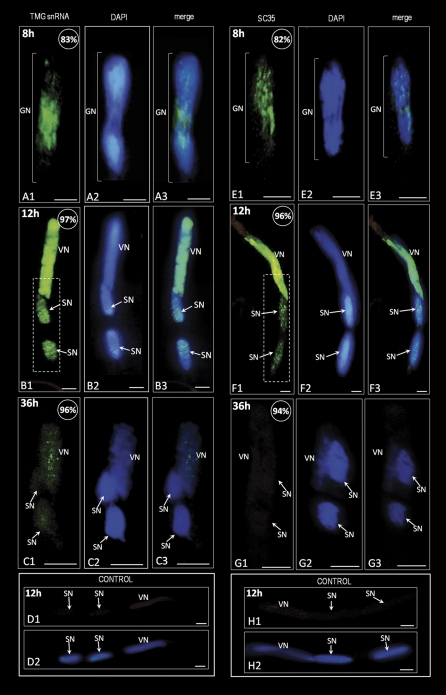
Fig. 6.
Immunolocalization of pre-mRNA splicing machinery elements (TMG snRNA and SC35 protein; green colour) in hyacinth pollen tubes. (A1–A3) A dividing generative cell. The signal appears to be located between two groups of daughter chromosomes. (B1–B3) After 12 h of pollen tube growth, the fluorescence corresponding to TMG snRNAs is present in two sperm nuclei. The signal is visible as numerous irregular fluorescent spots spread throughout the sperm nucleoplasm. In the vegetative nucleus, strong and uniform fluorescence is observed. Magnification of the area marked with a dashed line on B1 is shown in Supplementary Fig. S6 at JXB online. (C1–C3) A pollen tube growing for 36 h. A weak signal is observed only in the vegetative nucleus, whereas both sperm nuclei are almost completely devoid of labelling. (D1 and D2) Control reaction by omitting the primary anti-TMG antibody. (E1–E3) A generative cell during mitosis. The signal indicating the SC35 splicing factor is located in the area between the newly forming sperm nuclei. (F1–F3) After 12 h of pollen tube growth, the signal is present in the vegetative nucleus as well as in both sperm nuclei. In the vegetative nucleus, intense and uniformly distributed fluorescence is observed. Sperm nuclei exhibit low intensity staining, localized mainly in the form of numerous irregular fluorescent clusters. Magnification of the area marked with the dashed line on F1 is shown in Supplementary Fig. S7. (G1–G3) A pollen tube growing for 36 h. Lack of the signal corresponding to SC35 protein is observed in the sperm nuclei and in the vegetative nucleus. (H1 and H2) Control reaction by omitting incubation with the primary anti-SC35 antibody. The values presented in the upper right corner of the figures indicate the percentage of pollen tubes exhibiting the labelling pattern shown in the image. Z-series images corresponding to the whole volume of growing pollen tubes from three independent experiments (n=50) were captured and used for statistical analysis; VN, vegetative nucleus; GN, generative nucleus; SN, sperm nucleus; bars=10 μm.
After 8 h of H. orientalis pollen tube growth, TMG snRNAs were present in both vegetative and generative nuclei. In the vegetative nucleus, a strong homogenous signal was present, whereas in the generative nucleoplasm the fluorescence was visible in the form of numerous spots (Supplementary Fig. S2 at JXB online). During generative cell division, an accumulation of fluorescence was observed, indicating that the TMG snRNAs were localized in the area between the two groups of daughter chromosomes (Fig. 6A). After completion of division, the TMG snRNAs were localized in the vegetative nucleus as well as in both the newly formed sperm nuclei (Fig. 6B, and Supplementary Figs S4, S6 at JXB online). However, the labelling present in the sperm nuclei was lower than in the vegetative nucleus and visible as numerous small spots of bright fluorescence (Fig. 6B). In the vegetative nucleus, the TMG snRNAs were uniformly distributed throughout the area of the nucleoplasm. Between 24 h and 36 h of culture, a strong decrease in the TMG snRNA pool was observed in all the pollen tube nuclei (Fig. 6C). Both sperm nuclei exhibited complete lack of labelling, whereas in the vegetative nucleus only a few spots of weak fluorescence were present. The control reaction performed by omitting the anti-TMG antbody showed no labelling in the pollen tube growing for 12 h (Fig. 6D).
Before the second pollen mitosis, fluorescence indicating the presence of SC35 protein was observed in both pollen grain nuclei (Supplementary Fig. S3 at JXB online). In the vegetative nucleus, a strong and homogeneous labelling was observed, whereas in the generative nucleus the SC35 protein was localized in the form of numerous spots. During generative cell division (Fig. 6E), the labelling was observed mainly between the condensed chromosomes as well as in the area between the two forming daughter nuclei. In the pollen tube growing for 12 h, a strong difference in the SC35 protein distribution pattern was seen between the vegetative nucleus and the sperm nuclei (Fig. 6F). In the vegetative nucleus, an intense and uniform fluorescence covered the whole area of the nucleoplasm, whereas in the sperm nuclei the labelling was observed as numerous irregular fluorescent spots (Fig. 6F, and Supplementary Fig. S7 at JXB online). After 36 h of culture, the SC35 protein pool was not detected in the vegetative nucleus or in either of the sperm nuclei (Fig. 6G). No labelling could be observed in the nuclei of the pollen tube grown for 12 h after the control reaction performed with omission of anti-SC35 antibody (Fig. 6H). These results indicate that pre-mRNA splicing machinery elements are present in the sperm cells. At least some part of their pool is delivered into the sperm cells by the generative cell during its mitosis. The later steps of a sperm cell's life cycle are accompanied by the elimination of pre-mRNA splicing machinery elements from both of the sperm nuclei.
Discussion
Plant male gametes were initially thought to be metabolically silent and fully dependent on the vegetative cell and/or the pollen tube (Mascarenhas, 1975; McCormick, 1993). However, the successful exploration of a single cell transcriptome in recent years uncovered the fact that sperm cells contain their own pool of genes, transcripts, and promoters (Borges et al., 2008; Singh et al., 2008; Bayer et al., 2009; Wei et al., 2010). Most of the data concerning sperm cell-specific genes, their expression, and possible functions come from studies on A. thaliana, which produces tricellular pollen grains at anthesis. To date, there is almost no information about the organization of gene expression in the sperm nuclei formed during the germination of bicellular pollen grains. Recently, Tian et al. (2005) analysed the nuclear behaviour of tobacco sperm cells formed during pollen tube growth, however only in the context of the cell cycle. The data presented here constitute, to the best of our knowledge, the first report regarding the spatial and temporal organization of RNA synthesis and the RNA processing machinery throughout late microgametogenesis in a species producing bicellular pollen grains.
Highly condensed chromatin and a small amount of cytoplasm in the sperm cells suggest their very low or zero transcriptional activity (McCormick, 1993). Investigations of RNA synthesis in the sperm cells reported to date gave ambiguous results. In the germinating tricellular pollen grains of Secale cereale, 5-H3-labelled uridine was incorporated into the vegetative nucleus and both of the sperm nuclei (Haskell and Rogers, 1985). In turn, a complete lack of ongoing RNA synthesis was observed in the sperm nuclei of H. niger, which produces bicellular pollen grains at anthesis (Reynolds and Raghavan, 1982). The data presented here clearly indicate that activation of RNA synthesis occurs in newly formed sperm cells of hyacinth, although the levels of 5-bromouracil incorporated into the sperm cells were much lower than those in the vegetative nucleus. Similar to the generative cell (Blomstedt et al., 1996), the occurrence of RNA synthesis in H. orientalis sperm cells may indicate their at least partial metabolic autonomy from the pollen tube. On the other hand, recent findings of Slotkin et al. (2009) strongly suggest that the vegetative nucleus plays a significant role in the regulation of gene expression in sperm cells. The authors showed that in the A. thaliana mature pollen, retrotransposons [transposable elements (TEs)] are activated only in the vegetative nucleus and produce small interfering RNAs (siRNAs), which in turn are transported via pollen cytoplasm to the sperm cells. There, siRNAs mediate silencing of TEs in the sperm cells, avoiding TE activation in germ cells and thus preventing aberrant transposition events that may induce genomic rearrangements which could be transmitted to the next generation (Slotkin et al., 2009).
In the transcriptome of A. thaliana sperm cells, several transcription factors were found, among which the MYB family members were highly expressed (Borges et al., 2008). These molecules play regulatory roles in the key developmental processes in plants such as cell cycle, proliferation, or biotic stress (Martin and Paz-Ares, 1997). Moreover, some of the sperm-specific promoters identified in A. thaliana were shown to possess binding sites for transcription factors (Borges et al., 2008). The presence of transcription regulators in the sperm cells of the mature pollen grains strongly suggests that they activate RNA synthesis at further steps of pollen tube growth. Thus, the present results provide strong evidence for the activation of transcriptional activity in sperm cells during pollen tube growth, as previously suggested by others (Zhang et al., 1993).
The ongoing RNA synthesis in the H. orientalis sperm nuclei observed soon after their formation strongly suggests that the sperm cells progress at least through the G1 phase of the cell cycle. In turn, the transcriptional silence observed at further steps of pollen tube growth might reflect the arrest of the nuclei in the S-phase or the end of the sperm cell cycle, when they reach G2 phase. Such temporal changes in the transcriptional activity seem to resemble the nuclear activity observed during the interphase of somatic cell growth (Bryant and Francis, 1985). In most of bi- and tricellular pollen grains examined so far, the sperm cells do not progress to the S-phase and fuse with the target cells of the embryo sac in the G1 phase (Bino et al., 1990; Mogensen and Holm, 1995; Williams and Friedman, 2002, 2004). However, the sperm cells of A. thaliana (Friedman, 1999) and tobacco (Tian et al., 2005) were shown to accomplish a complete cell cycle during pollen tube elongation. Therefore, it seems that cell cycle coordination in the sperm cells may be a species-specific feature. More detailed studies on the DNA content are needed to address this hypothesis in H. orientalis.
Although transcriptomic studies enable the exploration of the mRNA pool in a single cell, little attention has been paid to other RNA types. Analysis of the total RNA profile of the growing H. orientalis pollen tube showed that the rRNA pool seemed to be temporally correlated with the total protein content after sperm cell formation. It can be assumed that rRNA expression levels in the growing pollen tube reflect its requirement for ribosomal units catalysing translation. Using the FISH technique, it was shown that in the growing H. orientalis pollen tube, rRNA appears not only in the vegetative nucleus and pollen tube cytoplasm but also in both sperm nuclei. Immature (ITS rRNA) and mature rRNAs (26S rRNA) appeared in the sperm nuclei soon after the completion of generative cell division and were observed until the final steps of microgametogenesis. These findings strongly support the existence of rRNA synthesis in the plant sperm cells. Interestingly, no accumulation of rRNA transcripts in the nucleoli-like structure was observed in the H. orientalis sperm nuclei. Unlike a typical interphase nucleus, the sperm nuclei exhibited high levels of chromatin condensation and a small interchromatin area (Hoefert, 1969). By analogy to animal sperm cells (Czaker, 1984; Nadel et al., 1995), it is proposed that rRNA gene clusters could be separated in the H. orientalis sperm nucleus, and they are, therefore, not organized into a nucleolus or nucleolus-like structures. These findings also showed that rRNA synthesis in the sperm nuclei might occur at very low levels, compared with that observed in the vegetative nucleus. This suggests that the sperm cells may require fewer ribosomal subunits than the pollen tube. Inhibition of protein synthesis with cycloheximide in the elongating pollen tube of H. orientalis showed that protein synthesis rates are much higher during the early steps of pollen grain germination. After sperm cell formation, mRNA translation also occurred, although at a lower level. It cannot be excluded that some pool of the new proteins is synthesized in the sperm cells. This assumption is in agreement with previous findings showing that the translation of some sperm transcripts occurs in the sperm cells of tricellular pollen grains (Singh et al., 2008; Russel et al., 2010).
The presence of pre-mRNA splicing machinery elements and both the forms of RNA Pol II temporally overlaps with the detectable RNA transcription in the H. orientalis sperm nuclei. The expected decrease in the total RNA synthesis rates at the later steps of pollen tube growth was accompanied by a gradual elimination of molecules involved in the transcription and processing of pre-mRNA. This strongly suggests that some part of the synthesized RNA is of the mRNA type. Moreover, the spatial distribution of RNA Pol II and splicing factors in the sperm nuclei differs significantly from that observed in the vegetative nucleus. Both forms of RNA Pol II and the splicing machinery elements were localized mainly in the form of irregular clusters spread throughout the sperm nucleoplasm. As has been shown previously, such a distribution pattern was characteristic for both pollen grain nuclei of H. orientalis at the time of low transcriptional activity and high chromatin condensation (Zienkiewicz et al., 2006, 2008a, b, c). The strong relationship between th e transcriptional activity and spatial distribution of the gene expression machinery has been well documented in plant and animal cells (Carmo-Fonseca et al., 1992; Misteli, 2000). Thus, we are convinced that such a localization pattern of the examined nuclear antigens reflects the differences in RNA synthesis intensity between the sperm and vegetative nuclei.
More detailed analyses of the images obtained with the confocal microscope showed that in most cases, the fluorescence corresponding to the examined antigens was localized between the dense DNA clumps brightly stained by DAPI. In most eukaryotic cells, mRNA synthesis and processing were shown to occur in the inter- and/or perichromatin areas of the nucleus (Lamond and Spector, 2003; Spector, 2003). Ultrastructural observations of the actively transcribing generative cell nucleus of the H. orientalis pollen grain showed that both forms of RNA Pol II as well as TMG snRNAs were mainly localized in the interchromatin areas, between the condensed chromatin clumps (Zienkiewicz et al., 2008c). Thus, a similar localization of RNA Pol II and splicing factors in the sperm nuclei, which are actively synthesizing mRNAs, is suggested. However, the correspondence of these nuclear areas with interchromatin domains needs to be proved by further ultrastructural studies.
One of the most remarkable features of the distribution of splicing factors observed during H. orientalis microgametogenesis was their distribution during generative cell mitosis. It was shown that TMG snRNAs and SC35 protein are likely to be divided equally between the forming sperm cells. Several lines of evidence suggest that the molecules involved in pre-mRNA splicing are inherited by the daughter cells, at least in somatic cells. In the case of HeLa cells, when mitosis is completed, the splicing machinery elements are imported into the nucleus just after reconstruction of the nuclear envelope (Prasanth et al., 2003). This fact suggests that H. orientalis sperm cells might not synthesize their own pool of splicing factors, and that processing of the pre-mRNAs synthesized during the life cycle of sperm cells might be catalysed by the splicing machinery derived from the generative cell. To date, there is only one report documenting the presence of SR splicing factor in the sperm cells of A. thaliana mature pollen grains (Fang et al., 2004). However, these authors did not discuss its origin, distribution, or possible role in the nuclei of the sperm cells.
The later steps of male gametogenesis in H. orientalis were accompanied by silencing of the transcriptional activity and elimination of the molecules involved in pre-mRNA synthesis and processing from the sperm cells. If it is assumed that the in vitro cultivation system mimics the in vivo conditions, it could be possible that from 24 h to 36 h of pollen tube growth in the pistil, just before fertilization, the sperm nuclei of H. orientalis become transcriptionally inactive. Therefore, it could be that in H. orientalis, some part of the mRNA pool needed for proper functioning of sperm cells during fertilization is synthesized during pollen tube growth. Such sperm cell-specific transcripts were, for example, found in A. thaliana. The mRNA encoding short suspensor protein (SSP) was shown to be present in the sperm cells; however, it did not undergo translation (Bayer et al., 2009). The SSP protein is synthesized soon after fertilization is completed and it acts as the key regulator of Arabidopsis zygote and embryo development (Bayer et al. 2009). Several reports also indicate that some transcripts found in the sperm cells might be delivered by the generative cell during its division. In the generative and sperm cells of L. longiflorum, Xu et al. (1999) showed the presence of mRNA encoding LGC1 protein. This protein was exclusively localized in the membrane of the sperm cells; thus, these authors suggested its possible role in sperm–egg cell fusion during fertilization (Xu et al., 1999). A similar function was confirmed for the GCS1 gene in A. thaliana (Mori et al., 2006). In turn, the AtMGH3 gene encoding histone H3 was shown to be transcribed in both generative and sperm cells. This protein probably controls chromatin remodelling and transcription during sperm cell development (Okada et al., 2005).
Conclusion
This study has shown the following characters of the sperm cells of H. orientalis: (i) they activate their transcriptional activity during pollen tube growth; (ii) they synthesize rRNA; (iii) they possess pre-mRNA transcription and processing machineries; and (iv) they undergo gradual inactivation of nuclear activity at the final steps of pollen tube growth. All these events are interpreted as a complete course of the H. orientalis sperm cell maturation process. The lack of RNA synthesis and removal of the molecules involved in its processing during the later steps of microgametogenesis suggest that mature sperm cells of H. orientalis do not carry these nuclear components into the target cells of the female gametophyte during fertilization. This is in agreement with previous observations showing that just after fertilization, activation of the zygote genome occurs in H. orientalis L., including rapid RNA synthesis and biogenesis of splicing machinery elements (Pięciński et al., 2008). To sum up, to our knowledge, this is the first characterization of the key nuclear steps of gene expression in the sperm cells formed during bicellular pollen grain germination. Further experiments are ongoing to investigate the sperm cell physiology and interactions at the molecular level, not only by using in vitro systems but also during pollen tube growth in the pistil.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Effect of cycloheximide on hyacinth pollen tube morphology and behaviour of the nuclei. (a, b, and c, transmitted light images; a′, b′, and c′, fluorescence images of pollen tubes shown ina, b, and c stained with propidium iodide.) In comparison with the control pollen tubes (a, a′), the treatment with cycloheximide after 3 h (b, b′) and 9 h (c, c′) of pollen tube growth caused numerous structural and physiological abnormalities. Strong inhibition of growth and shape deformations are observed. Disturbances in movement of the nuclei and arrest of second pollen mitosis occurred in all the pollen tubes growing on cycloheximide-containing medium. VN, vegetative nucleus; GN, generative nucleus; in a, a′ bars=50 μm and in b, b′ and c, c′ bars=10 μm.
Figure S2. TMG snRNA localization in hyacinth pollen tube before generative cell division (8 h of growth). The intense and homogeneous signal is present in the vegetative nucleus. In the generative nucleus, numerous fluorescent spots are present. VN, vegetative nucleus; GN, generative nucleus; bars=10 μm.
Figure S3. Immunolocalization of SC35 splicing factor in hyacinth pollen tube before the second pollen mitosis (8 h of growth). The fluorescence is present in the vegetative and generative nuclei. A strong and uniformly distributed signal is observed in the vegetative nucleoplasm, whereas in the generative nucleus numerous fluorescent spots are present. VN, vegetative nucleus; GN, generative nucleus; bars=10 μm.
Figure S4. Detection of U2 snRNA in hyacinth pollen tube by FISH. In both the sperm nuclei and the vegetative nucleus, an intense signal is present. In the pollen tube cytoplasm, labelling is observed. VN, vegetative nucleus; SN, sperm nucleus; bars=10 μm.
Figure S5. Magnification of the area marked with the dashed line in Fig. 5B1. VN, vegetative nucleus; SN, sperm nucleus; bars=10 μm.
Figure S6. Magnification of the area marked with the dashed line in Fig. 6B1. VN, vegetative nucleus; SN, sperm nucleus; bars=10 μm.
Figure S7. Magnification of the area marked with the dashed line in Fig. 6F1. VN, vegetative nucleus; SN, sperm nucleus; bars=10 μm.
Acknowledgments
This work was supported by the grant of Polish Ministry of Science and Higher education no. n N303 290434.
Glossary
Abbreviations
- BrdUTP
bromodeoxyuridine triphosphate
- DAPI
4′,6-diamidino-2-phenylindole
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- ETS
external transcribed spacer
- ITS
internal transcribed spacer
- PBS
phosphate-buffered saline
- snRNA
small nuclear RNA
- SSC
saline sodium citrate buffer
- TMG
2,2,7-trimethylguanosine
- TRIC
Texas red
References
- Bayer M, Nawy T, Giglione C, Galli M, Meinnel T, Lukowitz W. Paternal control of embryonic patterning in Arabidopsis thaliana. Science. 2009;323:1485–1488. doi: 10.1126/science.1167784. [DOI] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA. Transcriptional profiling of Arabidopsis tissues reveals the unique characteristic of the pollen transcriptome. Plant Physiology. 2003;133:713–725. doi: 10.1104/pp.103.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Current Opinion in Cell Biology. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Berger F, Hamamura Y, Ingouff M, Higashiyama T. Double fertilization—caught in the act. Trends in Plant Science. 2008;13:437–443. doi: 10.1016/j.tplants.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Bino RJ, Van Tuyl JM, De Vries JN. Flow cytometric determination of relative nuclear DNA contents in bicellulate and tricellulate pollen. Annals of Botany. 1990;65:3–8. [Google Scholar]
- Blomstedt CK, Knox RB, Singh MB. Generative cells of Lilium longiflorum possess translatable mRNA and functional protein synthesis machinery. Plant Molecular Biology. 1996;5:1083–1086. doi: 10.1007/BF00040727. [DOI] [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiology. 2008;148:1168–1181. doi: 10.1104/pp.108.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewbaker JL, Kwak BH. The essential role of calcium ions in pollen germination and pollen tube growth. American Journal of Botany. 1963;50:859–865. [Google Scholar]
- Brown JW, Shaw PJ. Small nucleolar RNAs and pre-rRNA processing in plants. The Plant Cell. 1998;10:649–657. doi: 10.1105/tpc.10.5.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JA, Francis D. The cell division cycle in plants. Cambridge: Cambridge University Press; 1985. [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. Journal of Cell Biology. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corriveau JL, Coleman AW. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. American Journal of Botany. 1988;75:1443–1458. [Google Scholar]
- Czaker R. Observations on the dynamics of argyrophilic nucleolar material in the nuclei of mice spermatids. Experientia. 1984;40:960–963. doi: 10.1007/BF01946459. [DOI] [PubMed] [Google Scholar]
- Fang Y, Hearn S, Spector DL. Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis. Molecular Biology of the Cell. 2004;15:2664–2673. doi: 10.1091/mbc.E04-02-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman WE. Expression of the cell cycle in sperm of Arabidopsis: implication for understanding patterns of gametogenesis and fertilization in plants and other eukaryotes. Development. 1999;126:1065–1075. doi: 10.1242/dev.126.5.1065. [DOI] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskell DW, Rogers OM. RNA synthesis by vegetative and sperm nuclei of trinucleate pollen. Cytologia. 1985;50:805–809. [Google Scholar]
- Hoefert LL. Fine structure of sperm cells in pollen grains of Beta. Protoplasma. 1969;68:237–240. [Google Scholar]
- Ingouff M, Sakata T, Li J, Sprunck S, Dresselhaus T, Berger F. The two male gametes share equal ability to fertilize the egg cell in Arabidopsis thaliana. Current Biology. 2009;19:R19–R20. doi: 10.1016/j.cub.2008.11.025. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nature Reviews Molecular Cell Biology. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annual Review of Genetics. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- Martin C, Paz-Ares J. MYB transcription factors in plants. Trends in Genetics. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Mascarenhas JP. The biochemistry of angiosperm pollen development. Botanical Review. 1975;41:259–314. [Google Scholar]
- McCormick S. Male gametophyte development. The Plant Cell. 1993;5:1265–1275. doi: 10.1105/tpc.5.10.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. Journal of Cell Science. 2000;113:1841–1849. doi: 10.1242/jcs.113.11.1841. [DOI] [PubMed] [Google Scholar]
- Mogensen HL, Holm PB. Dynamics of nuclear DNA quantities during zygote development in barley. The Plant Cell. 1995;7:487–494. doi: 10.1105/tpc.7.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nature Cell Biology. 2006;1:64–71. doi: 10.1038/ncb1345. [DOI] [PubMed] [Google Scholar]
- Nadel B, De Lara J, Ward WS. Structure of the rRNA genes in the hamster sperm nucleus. Journal of Andrology. 1995;16:517–522. [PubMed] [Google Scholar]
- Okada T, Bhalla PL, Singh MB. Expressed sequence tag analysis of Lilium longiflorum generative cells. Plant and Cell Physiology. 2006;47:698–705. doi: 10.1093/pcp/pcj040. [DOI] [PubMed] [Google Scholar]
- Pięciński S, Smoliński DJ, Zienkiewicz K, Bednarska E. Changes in poly (A) RNA and TMG snRNA distribution in the embryo sac of Hyacinthus orientalis L. before and after fertilization. Sexual Plant Reproduction. 2008;21:247–257. [Google Scholar]
- Pina C, Pinto F, Feijo JA, Becker JD. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiology. 2005;138:744–756. doi: 10.1104/pp.104.057935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Sacco-Bubulya PA, Prasanth SG, Spector DL. Sequential entry of components of the gene expression machinery into daughter nuclei. Molecular Biology of the Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. Coupling transcription, splicing and mRNA export. Current Opinion in Cell Biology. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- Reynolds TL, Raghavan V. An autoradiographic study of RNA synthesis during maturation and germination of pollen grains of Hyoscyamuns niger. Protoplasma. 1982;111:177–182. [Google Scholar]
- Russell SD. Isolation and characterization of sperm cells in flowering plants. Annual Review in Plant Physiology and Plant Molecular Biology. 1991;42:189–204. [Google Scholar]
- Russell SD, Gou X, Wei X, Yuan T. Male gamete biology in flowering plants. Biochemical Society Transactions. 2010;38:598–603. doi: 10.1042/BST0380598. [DOI] [PubMed] [Google Scholar]
- Singh MB, Bhalla PL, Russell SD. Molecular repertoire of flowering plant male germ cells. Sexual Plant Reproduction. 2008;21:27–36. [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzić M, Becker JD, Feijó JA, Martienssen RA. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. The dynamics of chromosome organization and gene regulation. Annual Review of Biochemistry. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- Tian HQ, Yuan T, Russell SD. Relationship between double fertilization and the cell cycle in male and female gametes of tobacco. Sexual Plant Reproduction. 2005;17:243–252. [Google Scholar]
- Wei LQ, Xu WY, Deng ZY, Su Z, Xue Y, Wang T. Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genomics. 2010;11:338. doi: 10.1186/1471-2164-11-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Protein functions in pre-mRNA splicing. Current Opinion in Cell Biology. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Current Opinion in Cell Biology. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Williams JH, Friedman WE. Identification of diploid endosperm in an early angiosperm lineage. Nature. 2002;415:522–526. doi: 10.1038/415522a. [DOI] [PubMed] [Google Scholar]
- Williams JH, Friedman WE. The four-celled female gametophyte of Illicium (Illiciaceae; Austrobaileyales): implications for understanding the origin and early evolution of monocots, eumagnoliids, and eudicots. American Journal of Botany. 2004;91:332–351. doi: 10.3732/ajb.91.3.332. [DOI] [PubMed] [Google Scholar]
- Xu H, Swoboda I, Bhalla PL, Singh MB. Male gametic cell-specific gene expression in flowering plants. Proceedings of the National Academy of Sciences, USA. 1999;96:2554–2558. doi: 10.1073/pnas.96.5.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Golembe TJ, Battle DJ, Pellizzoni L, Dreyfuss G. SnRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Molecular and Cellular Biology. 2004;24:2747–2756. doi: 10.1128/MCB.24.7.2747-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Gifford DJ, Cass DD. RNA and protein synthesis in sperm cells isolated from Zea mays L. pollen. Sexual Plant Reproduction. 1993;6:239–243. [Google Scholar]
- Zienkiewicz K, Smoliński DJ, Bednarska E. Distribution of poly(A) RNA and splicing machinery elements in mature Hyacinthus orientalis L. pollen grains and pollen tubes growing in vitro. Protoplasma. 2006;227:95–103. doi: 10.1007/s00709-005-0153-z. [DOI] [PubMed] [Google Scholar]
- Zienkiewicz K, Zienkiewicz A, Rodriguez-Garcia MI, Smoliński DJ, Świdziński M, Bednarska E. Transcriptional activity and distribution of splicing machinery elements during Hyacinthus orientalis L. pollen tube growth. Protoplasma. 2008a;233:129–139. doi: 10.1007/s00709-008-0298-7. [DOI] [PubMed] [Google Scholar]
- Zienkiewicz K, Zienkiewicz A, Smoliński DJ, Rafińska K, Świdziński M, Bednarska E. Transcriptional state and distribution of poly(A) RNA and RNA polymerase II in differentiating Hyacinthus orientalis L. pollen grains. Sexual Plant Reproduction. 2008b;21:233–245. [Google Scholar]
- Zienkiewicz K, Zienkiewicz A, Smoliński DJ, Świdziński M, Bednarska E. Intracellular organization of the pre-mRNA splicing machinery during Hyacinthus orientalis L. pollen development. Sexual Plant Reproduction. 2008c;21:217–231. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.