Abstract
Climacteric and non-climacteric fruits have traditionally been viewed as representing two distinct programmes of ripening associated with differential respiration and ethylene hormone effects. In climacteric fruits, such as tomato and banana, the ripening process is marked by increased respiration and is induced and co-ordinated by ethylene, while in non-climacteric fruits, such as strawberry and grape, it is controlled by an ethylene-independent process with little change in respiration rate. The two contrasting mechanisms, however, both lead to texture, colour, and flavour changes that probably reflect some common programmes of regulatory control. It has been shown that a SEPALLATA(SEP)4-like gene is necessary for normal ripening in tomato. It has been demonstrated here that silencing a fruit-related SEP1/2-like (FaMADS9) gene in strawberry leads to the inhibition of normal development and ripening in the petal, achene, and receptacle tissues. In addition, analysis of transcriptome profiles reveals pleiotropic effects of FaMADS9 on fruit development and ripening-related gene expression. It is concluded that SEP genes play a central role in the developmental regulation of ripening in both climacteric and non-climacteric fruits. These findings provide important information to extend the molecular control of ripening in a non-climacteric fruit beyond the limited genetic and cultural options currently available.
Keywords: Fragaria, fruit development, ripening, SEPALLATA genes, strawberry
Introduction
Fruits are reproductive organs unique to the Angiosperms that have evolved to promote seed dispersal. They are also one of the most important components of the human diet. Classically, two broad classes of fruits have been recognized. In climacteric fruits, for example, tomato (Solanum lycopersicum), banana (Musa acuminata), and melon (Cucumis melo), the onset of ripening is marked by increased ethylene synthesis and respiration. In non-climacteric fruits, for example, strawberry (Fragaria×ananassa), grape (Vitis vinifera),and citrus, these changes are not apparent (Giovannoni, 2004; Seymour et al., 1993). However, these distinct developmental programmes usually result in similar ripening-related changes including a loss of chlorophyll, the accumulation of carotenoids or anthocyanins, the production of volatiles, and cell wall disassembly.
The role of ethylene in inducing ripening in climacteric fruits has received substantial attention. The pathway for ethylene biosynthesis is now well established and many steps in the ethylene signal transduction pathway have been revealed (Alexander and Grierson, 2002). In stark contrast, almost nothing is known about the molecular regulation of ripening in non-climacteric fruits (Giovannoni, 2004).
Studies on non-ripening mutants of tomato suggest that certain hitherto unsuspected developmental cues may control ripening. In tomato, the ripening inhibitor (rin) mutation exhibits pleiotropic effects in the fruits that fail to soften, lack a respiratory climacteric and associated ethylene evolution, pigment accumulation, and flavour or aroma development (Robinson and Tomes, 1968). Positional cloning has revealed that a SEPALLATA (SEP)-like MADS-box transcription factor (LeMADS-RIN), resides at the rin locus and the gene is expressed primarily in fruit tissues (Vrebalov et al., 2002). The LeMADS-RIN protein contains concensus binding sites characteristic of the SEP proteins and in vitro recognizes similar DNA-binding sites to these transcription factors (Ito et al., 2008). However, in Arabidopsis SEP genes are commonly associated with the regulationof floral organ identity suggesting that, in tomato fruit, LeMADS-RIN has evolved a different biological function. In strawberry, a non-climacteric fruit, it is shown that silencing a SEP1/2 class gene in transgenic plants, which is expressed during normal fruit development and ripening, delays and modifies these important processes.
Materials and methods
Phylogenetic analysis
A phylogenetic tree was constructed based on predicted full-length protein sequences using MegAlign software (DNAStar) and aligned by ClustalW.
Transgenic strawberry lines
A 552bp antisense fragment from the 5′ end of FaMADS9 cDNA, the putative strawberry orthologue (GenBank AF484683) of LeMADS-RIN described previously (Vrebalov et al., 2002), was cloned in front of the CaMV35S promoter at the KpnI/BamHI multiple cloning site of the pBINPLUS1 plant transformation vector (Van Engelen et al., 1995). Leaf discs of the cultivar Calypso were transformed and kanamycin-resistant plants selected as described previously by Woolley et al.(2001).
Tissue sampling
Individual receptacles (minus achenes) were powdered in liquid N2 and subsamples were taken for the determination of chlorophyll, anthocyanin, and RIN expression.
Determination of chlorophyll, anthocyanin, and fruit firmness
Chlorophyll and total anthocyanins were extracted and assayed essentially as described before (Hiscox and Israelstam, 1979; Given et al., 1988b). Fruit texture measurements were made using a penetrometer (Woolley et al., 2001) and firmness was assessed as the maximum force (g) recorded at the yield point as the probe travelled 5mm into the fruit.
Isolation of nucleic acids
Total nucleic acids were isolated from fruit and leaf tissues based on a modified CTAB method (Chang et al., 1993). Total nucleic acids were precipitated from the CTAB extract with isopropanol and further purified with SSTE buffer before separating total RNA from DNA with 2M LiCl.
5′ and 3′ RACE of full-length FaMADS4 cDNA
Total RNA isolated from cultivar Calypso ripe fruit was the template for 5′ and 3′ RACE using the GeneRacer Kit (Invitrogen, Paisley, UK) essentially according to the manufacturer's protocol. For the reverse transcription step, Superscript II reverse transcriptase (Invitrogen) was used with the anchored RT primer: 5′-GCGAGCACAGAATTAATACGACTCACTATAGG(T)12VN-3′. Gene-specific forward and reverse primers were designed from the predicted F. nubicola cDNA sequence and nested PCRs performed with KOD Hot Start polymerase (Invitrogen) together with GeneRacer adapter primers. Amplicons were either sequenced directly or cloned into the pJET1.2 vector (Fermentas, York, UK) for sequencing.
Gene expression assays for FaMADS9
Expression of the strawberry FaMADS9 gene in receptacle tissue was determined by absolute quantification using a two-step real-time PCR method. Total RNA from each tissue was initially reverse transcribed using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's protocol but without added dithiothreitol. Synthesis of first strand cDNA was primed with the gene-specific primer: 5′-ATCCTTAGTAAACCAGTCTTT-3′. Gene expression was analysed by real-time PCR using the 7900HT Fast Real-Time PCR System (Applied Biosystems, Warrington, UK). The PCR reaction (10μl) contained 5μl qPCRMastermix Plus for SYBR Green (Eurogentec, Southampton, UK) with a passive reference dye, first strand cDNA template synthesized from 20ng total RNA, and 200nM of each primer. Absolute standards were prepared from a pGEMT (Promega, Southampton, UK) plasmid containing the FaMADS9 cDNA. Following an initial denaturation of 95oC for 1min 30s, the thermal cycling programme was 40 cycles of 95°C for 30s, annealing at 60 °C for 30s, and extension at 72 °C for 30s. Three technical replicates were run for each of the standards and the unknowns and three fruits were sampled at each stage from each line. The Ct value for each QRT-PCR was determined and a standard curve used to calculate absolute amounts of target cDNA. Results were expressed as the number of transcript copies per microgram total RNA. Forward and reverse primers for the strawberry FaMADS9gene were FaMADS9 QRT-PCR F3: 5′-AGCCAACAGAGATTTGAAAACGAAG-3′ and FaMADS9 QRT-PCR R3: 5′-GACACCACAGCATTGTACCCTATTTG-3′. The primers span an intron and prevent any detectable amplification from genomic DNA (up to 50ng). For sensitive detection of strawberry FaMADS9 expression in petals and achenes a nested real-time PCR method was used. The primary PCR contained 200nM of the forward and reverse primers FaMADS9 QRT-PCR F3 and FaMADS9 QRT-PCR R3 in a 20μl reaction containing 10μl Megamix Blue Double PCR mixture (Microzone, HaywardsHeath, UK) and 20ng first strand cDNA. The reaction was initially denatured at 95 °C for 1min 30s, followed by thermal cycling for 15 cycles of 95 °C for 30s, annealing at 60 °C for 30s, and extension at 72 °C for 30s. Template from the primary reaction (1μl) was used in the secondary real-time PCR containing the nested primers FaMADS9 QRT-PCR F4: 5′-CAGAGATTTGAAAACGAAGTTGGATG-3′ and FaMADS9 QRT-PCR R4: 5′-TGTTGAGTTCCATATAGCATAGTCTGGTG-3′.
Gene expression assays for FaMADS4
To quantify the low expression of FaMADS4 in fruit tissue, a nested real-time PCR method similar to that for FaMADS9 was used. The primary PCR contained 200nM of the forward and reverse primers FaMADS4 QRT-PCR F1:5′-CTACAACGCACGCAGAGACG-3′and FaMADS4 QRT-PCR R1:5′-GTCGCATTGTAAACGCTCAA-3′ in a 10μl reaction containing 5μl Megamix Blue Double PCR mixture (Microzone) and 10ng first strand cDNA. Each primer spanned an intron–exon border. The reaction was initially denatured at 95 °C for 1min 50s, followed by thermal cycling for 15 cycles of 95 °C for 10s, annealing at 55 °C for 30s, and extension at 72 °C for 20s. Template from the primary reaction (0.5μl) was used in the secondary real-time PCR containing 200nM of the inner (nested) primers FaMADS4 QRT-PCR F2: 5′-AGCTTCAGCAGCTTGAGAAT-3′ and FaMADS4 QRT-PCR R2: 5′-CTTCATTACTCTCTTCCAACTTCA-3′.This reaction (10μl) contained 5μl Mesa Green qPCRMastermix Plus for SYBR assay (Eurogentec) with passive reference dye. Absolute standards were prepared from sequenced purified products of a PCR with FaMADS4 QRT-PCR F1/R1 primers. Following an initial activation of 95°C for 5min, the thermal cycling programme was 40 cycles of 95 °C for 15s, annealing at 55 °C for 20s, and extension at 72 °C for 40s.
Gene expression assays for all other genes
A two-step real-time PCR method similar to that for FaMADS9 was used for determining the expression of other genes in receptacle tissue, except that first strand cDNA synthesis, primed with oligo dT12–18, cDNA template from 10ng of RNA, was used and absolute standards were purified RT-PCR products. The following genes (GenBank accession numbers in parentheses) were assayed: AGL6-like MADS-box gene (DV440581), SEP3-like MADS-box gene (DV440160), SHATTERPROOF2-like MADS-box gene (CO380891), AGAMOUS-like MADS-box gene (AF168468), endo-1,4-β-D-glucanase (AF041405), chalcone synthase (AY997297), flavanone-3-hydroxylase (AY691919), and quinoneoxidoreductase (AY158836). Primers are described in Supplementary Table S3 at JXB online.
Cross-species arrays
Microarray experiments were conducted to quantify transcriptional differences between fruit from two transgenic strawberry lines (MADS A and MADS I) and wild-type plants. Since no large-scale strawberry microarrays are currently available, a cross-species experiment was performed using the Affymetrix Arabidopsis thaliana ATH1-121051 GeneChip® array (ATH1 GeneChip®; Hammond et al., 2005, 2006). Briefly, genomic DNA isolated from strawberry was biotinylated and hybridized to an ATH1 GeneChip® array as described previously (Hammond et al., 2005). The ATH1 GeneChip® array was then scanned and a gDNA hybridization cell intensity file generated (.CEL). Probe-pairs from the gDNA CEL file were selected for subsequent transcriptome analysis of the strawberry fruits using a parser written in Perl (www.perl.com; Hammond et al., 2005) to generate custom probe mask files. The Perl script selected probe-pairs in which the PM probe has agDNA hybridization intensity greater than a user-defined threshold, and hence at the defined intensity, probe-pairs with a lower value are defined as having poor hybridization between the strawberry gDNA and the Arabidopsis ATH1 GeneChip probe and would not be representative of the gene. These probes are not useful for transcriptional analyses. A gDNA-based probe-selection at a hybridization intensity threshold of 250 was selected for analysis of transcriptional data. This threshold was selected as optimal, based on (i) the retention of 81% of probe-sets with over 84% represented by two or more probe-pairs (see Supplementary Fig. S1A at JXB online) and (ii) the maximal detection of significantly (P<0.05) differentially expressed genes between the MADS A and MADS I transgenic lines and wild-type (see Supplementary Fig.S1B at JXB online).
Total RNA was extracted from red ripe receptacle tissue (minus achenes) of MADS A, MADS I, and wild-type plants as described above. Three replicates, consisting of tissue from single fruits were performed. Total RNA was labelled and hybridized to AffymetrixArabidopsis thaliana GeneChip arrays using standard protocols (Affymetrix, Santa Clara, CA, USA). GeneChip arrays were scanned using an Affymetrix 3000 GeneArray scanner and cell intensity files (.cel files) were generated. The RNA.cel files were loaded into GeneSpring GX (Agilent Technologies, Palo Alto, CA, USA) using the RMA normalization algorithm and the probe mask file generated from the gDNA hybridization (Hammond et al., 2005). Initially, genes significantly (P<0.05) differentially expressed between either of the transgenic lines and the wildtype by more than 1.3-fold were identified using a one-way ANOVA with a Benjamini and Hochberg False Discovery Rate of 5% to correct for multiple testing (see Supplementary Table S1 at JXB online). Hierarchical clustering was performed using a Pearson correlation similarity measure in GeneSpring GX (Agilent Technologies). Common genes significantly differentially expressed between both FaMADS9 transgenic lines and the wild-type were identified by combining the two data sets and performing the one-way ANOVA as above (see Supplementary Table S2 at JXB online).
Results
Phylogenetic analysis of strawberry FaMADS9
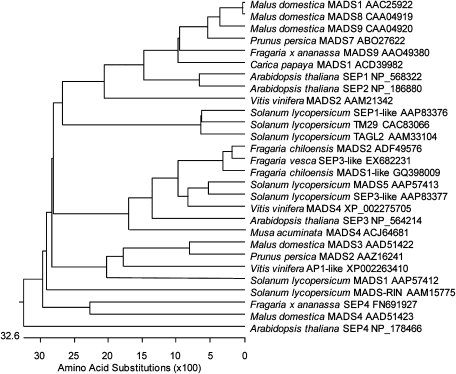
The relationship between FaMADS9, isolated by Vrebalov et al. (2002), and other SEP proteins in selected fleshy fruit species as compared with the SEP1/SEP2/SEP3/SEP4 clade of Arabidopsis, a dry-fruited species, was analysed by constructing a phylogenetic tree (Fig. 1). The phylogram suggests that FaMADS9 belongs to the SEP1/2 clade. By contrast, the tomato LeMADS-RIN is more closely aligned with SEP4. Strawberry belongs to the Rosaceae family and FaMADS9 is most similar to the SEP1/2 proteins MdMADS1, 8, and 9 and PpMADS7 in the related apple (MalusXdomestica Borkh.) and peach (Prunus persica) species, respectively. A new SEP gene (FaMADS4) has also been isolated from strawberry (described below and included in Fig. 1) encoding a protein that is more closely related to SEP4 and LeMADS-RIN than FaMADS9.However, this gene was expressed at a very low level in ripening fruit and was not investigated further. A SEP4-like homologue, MdMADS4, is also present in apple.
Fig. 1.
Phylogenetic analysis of SEPALLATA proteins in selected fruit species. A phylogenetic tree of full-length protein sequences from apple (Malus domestica), Arabidopsis, banana (Musa acuminata), grape (Vitis vinifera), papaya (Carica papaya), peach (Prunus persica), strawberry (Fragaria×ananassa), and tomato (Solanum lycopersicum) was constructed using MegAlign software (DNAStar) after aligning by ClustalW.
Down-regulation of a fruit-related SEP1/2 gene in strawberry alters normal fruit development and ripening
To examine the function of FaMADS9,transgenic strawberry plants were producedcontaining a 552 bp fragment of the FaMADS9 gene sequence inthe antisense orientation ectopically expressed using the cauliflower mosaic virus (CaMV) 35S promoter. Ten independent T0 plants were generated in tissue culture and three lines gave plants with markedly altered flower and fruit phenotypes. The primary transformants were vegetatively propagated from runners (stolons) and these clones were then used for further analysis.
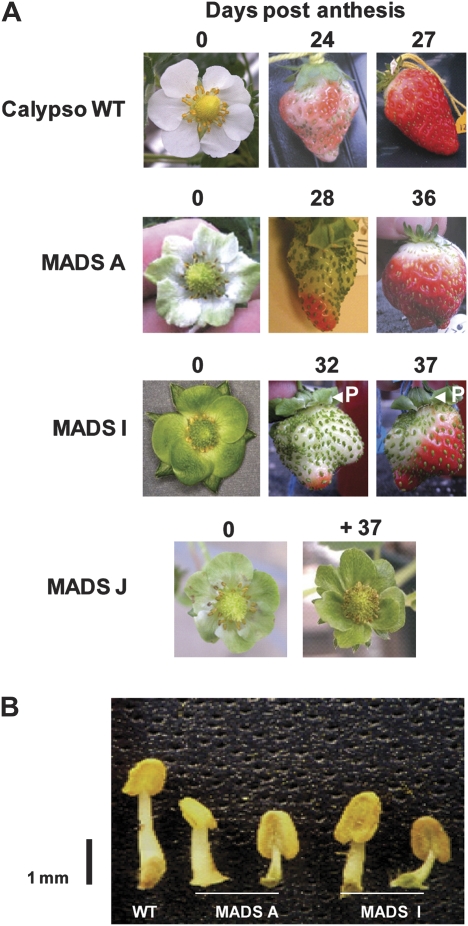
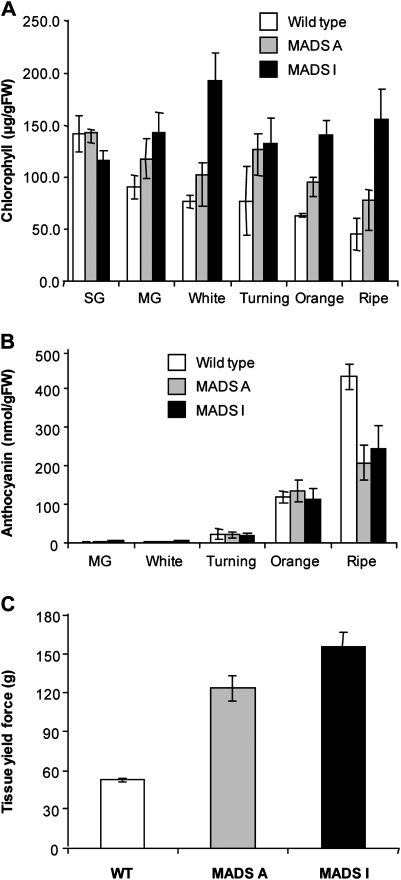
Clones from all three transgenic lines (A, I, and J) produced flowers with petals that exhibited increasing green colouration in the order A<I<J compared with the normally white petals of the untransformed plants (Fig.2A). In normal plants, the petals abscise 2–3d after anthesis. Abscission of petals in the transgenic plants was either delayed (A) or was totally abolished (I and J).Line J exhibited such a severe developmental phenotype that the fruit failed to advance beyond the immature stage and were not analysed further. In lines A and I, it was observed that self-pollination occurred much less readily than in the wild-type controls and hand pollination was necessary for optimum fruit expansion. This was consistent with altered anther development and pollen release observed in the flowers from these plants (Fig.2B). The antisense lines A and I achieved normal fruit expansion with no significant differences in morphology prior to ripening, but showed altered ripening in both receptacle and achenes. The fruit of the strawberry is a false fruit, the fleshy part being derived from the receptacle while the true fruits ‘achenes’ are embedded in the surface of the receptacle. The onset of ripening, observed as an increase in the production of red anthocyanin pigments in the receptacle, was delayed by around 10d compared with wild-type plants.Ripe fruits sampled from the transgenic lines were on average 11d (A) and 12d (I) older than the wild-type controlsto achieve comparable developmental stages. Unlike wild-type fruits, where ripening is normally initiated throughout the receptacle, anthocyanin accumulation in the transgenic lines was often observed first at the distal end before it progressed towards the calyx (Fig.2A). Chlorophyll was retained in the achenes of line I, which remained green with indications of a similar trend in A (Fig.3A). By contrast, there were no differences in chlorophyll content in receptacle tissue between the transgenic and wild-type receptacles (data not shown). Examination by light microscopy did not reveal any significant structural differences in the achenes from these transgenic lines. Although anthocyanin production increased in the developing receptacles of line A and I, it never attained the levels found in wild-type fruits (Fig.3B). Anthocyanin content in the achenes was not measured due to insufficient tissue for reliable determination. The fruits from linesA and I retained a significantly firmer texture than wild-type controls, as determined by measuring tissue yield force at the stage where the fruits were red ripe and ready for consumption (Fig.3C).
Fig. 2.
A MADS-box gene affects flower and fruit development in strawberry.(A) Altered development of flowers and fruit in untransformed (WT) strawberry and three independent lines (MADS A, MADS I, and MADS J) transformed with an antisense strawberry FaMADS9 transgene sequence. P, attached petals.(B) Anthers in two independent transformed lines (MADS A and MADS I).
Fig. 3.
Repression of strawberry FaMADS9 alters normal ripening.(A) Chlorophyll content in achenes; (B) anthocyanin content in receptacle tissue of three independent fruits sampled at each stage of development; (C) firmness of receptacle tissue. Measurements were made on 43, 35, and 30 ripe fruits from untransformed, MADS A and MADS I plants, respectively. Bars represent mean values (±SEM) of untransformed wild type (white) and independent antisense transformants MADS A (grey) and MADS I (black). SG and MG are small green and mature green fruits, respectively.
Comparison of FaMADS9 target gene expression in receptacles, achenes, and petals of wild-type and antisense lines
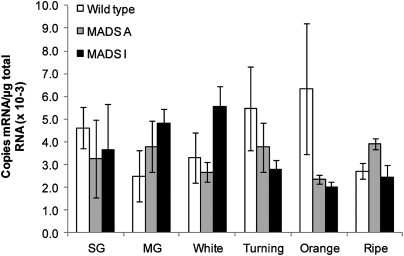
In order to determine low levels of FaMADS9 expression in achene and petal tissue a modified quantitative RT-PCR (QRT-PCR) method for single copy detection was used. In wild-type plants FaMADS9 was expressed in developing receptacles, achenes, and petals (Fig.4A–C). The fruit and petal phenotypes of the antisenselines described above were associated with a significant reduction in FaMADS9 expression in these tissues (Fig.4A–C). The petals showed a significant reduction in the level of expression of the gene even though it was an order of magnitude lower in this tissue than in achenes or receptacles (Fig.4C). Furthermore, line I, which had the more severe petal and fruit phenotypes, almost always displayed the lowest levels of FaMADS9 expression (Fig.4A–C).
Fig. 4.
FaMADS9 gene expression in strawberry receptacles, achenes and petals.(A) Receptacle tissue; (B) achenes; (C) petals. Expression of an endogenous RIN-like gene was determined by absolute QRT-PCR. Bars indicate the mean values (±SEM) of three fruits sampled from each line (see Fig. 2) at different stages of development. Data for wild-type petals only extends to day 2 as from this point these petals abscised.
Effect of repressing FaMADS9 on the programme of ripening-related gene expression in strawberry
To assess downstream gene expression effects in the FaMADS9 transgenic lines, cross-species microarray experiments were performed. The Affymetrix Arabidopsis thaliana ATH1-121051 GeneChip® array was used, since comprehensive public strawberry array resources were not available. Although FaMADS9 cDNA was isolated using a tomato probe and tomato oligo microarrays are available,the choice was made to use the array from Arabidopsis because this species is phylogenetically more closely related to strawberry than tomato (Judd et al., 1999) and this array is more comprehensive than that of tomato. A mixed physical and bioinformatic method was used to optimize the analysis of the transcriptional data (Hammond et al., 2005, 2006). Briefly, genomic DNA (gDNA) from strawberry was hybridized onto the ATH1 GeneChip array and a parser script, written in Perl, was used to remove probe-pairs whose perfect match (PM) probe signal intensity value was below a user-defined gDNA hybridization intensity threshold. The optimum gDNA hybridization intensity threshold was selected based on the retention of probe-sets and maximal detection of differentially expressed transcripts (see Supplementary Fig.S1 at JXB online). Thus, only probes with good homology (gDNA hybridization signal intensity ≥250) to strawberry gDNA were used to analyse the transcriptional data.
Hierarchical clustering of genes significantly (P<0.05) differentially expressed in red ripe receptacle tissue between the lines A and I and the wild-type, identified groups of genes with similar expression profiles (see Supplementary Fig.S2 and Supplementary Table S1 at JXB online). Down-regulated transcripts in clusters I and II included those with homology to Arabidopsis AGL (AGL5; At2g42830), MYB (At5g12870 and At5g55020), F-box (At1g64290, At2g17020, and At3g06380), and bHLH (At4g36540) transcription factor families. Also present in these clusters were transcripts with homology to Arabidopsis cell wall-related genes (pectin methylesterase inhibitor, At5g62360; cellulase, At1g70710; pectate lyase, At4g13710; and pectinesterase, At5g49180) and an ACC oxidase (At2g19590) gene. From the Gene Ontology (GO) annotations associated with the homologous Arabidopsis transcripts, several groups of genes were identified that were significantly (P<0.05) over-represented in Clusters I and II, including those involved in the ubiquitin cycle (GO:6512), ovule and carpel development (GO:48481 and GO:48440), and signal transduction (GO:7165). Transcripts with elevated levels in both transgenic lines (cluster V) had homology to members of the Arabidopsis F-box (At2g15640), bZIP (At1g68640), and MYB (At1g72650 and At2g01060) transcription factor families, as well as two AGL (AGL16; At3g57230 and AGL24; At4g24540) and two NAM genes (At1g12260 and At3g04060). Genes up-regulated in A or I lines (clusters III and IV, respectively) included those with homology to Arabidopsis CLAVATA1 (At1g75820) and pectinesterase 2 (At1g53830).
To distinguish genes differentially expressed in both FaMADS9 transgenic lines, the data sets from each were combined and compared with the wild-type. In total, 87 genes were identified, 38 with higher and 49 with lower transcript abundance in the transgenic lines compared with the wild-type (see Supplementary Table S2 at JXB online). These included transcripts with homology to Arabidopsis AGL5, three F-box genes (At1g64290, At2g15640, and At2g17020), an endo-1,4-β-D-glucanase (At1g70710), and a homeobox gene (At1g05230).
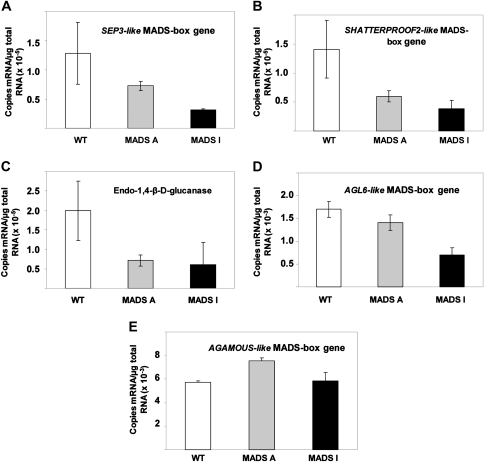
Based on the microarray analyses, a number of genes whose expression was down-regulated in the FaMADS9-silenced lines were selected for further expression analysis by QRT-PCR (Fig.5). Strawberry genes whose sequences were available in public databases were identified from the corresponding Arabidopsis probes. Three genes that were identified as being down-regulated in both of the transgenic lines MADS A and MADS I were a SEP3-like MADS-box gene (GenBank DV440160; Fig.5A), a SHATTERPROOF2-like MADS-box gene (GenBank CO380891; Fig.5B), and an endo-1,4-β-D-glucanase (GenBank AF041405; Fig.5C). The QRT-PCR analyses confirmed that all these genes were repressed in the red ripe transgenic fruit.The expression of two other genes related to FaMADS9, an AGL6-like MADS-box gene (GenBank DV440581) and an AGAMOUS-like MADS-box gene (GenBank AF168468) were also investigated. The AGL6-like MADS-box gene was repressed in red ripe receptacle tissue of both transgenic lines in the order MADS I>MADS A>WT whereas expression of the AGAMOUS-like MADS-box gene did not show a trend related to the phenotypic differences of these plants (Fig. 5E,F, respectively). In addition, the expression of two genes related to anthocyanin biosynthesis, chalcone synthase and flavanone-3-hydroxylase, and a gene involved in flavour biogenesis, quinoneoxidoreductase (GenBank accessions AY997297, AY691919, and AY158836, respectively) were also assayed by QRT-PCR. Although the expression of these genes is known to be enhanced during the ripening of normal fruit, they were not identified from microarray data as having altered expression in the transgenic lines, and this was confirmed by QRT-PCR (see Supplementary Fig.S3 at JXB online).
Fig. 5.
Expression of selected strawberry genes determined by absolute QRT-PCR. (A) SEP3-like MADS-box gene (DV440160); (B) SHATTERPROOF2-like MADS-box gene (CO380891); (C) endo-1,4-β-D-glucanase (AF041405); (D) AGL6-like MADS-box gene (DV440581); (E) AGAMOUS-like MADS-box gene (AF168468).Bars represent mean values (±SEM) of three fruits sampled from each line (see Fig. 2).
Cloning and expression analysis of FaMADS4
The sequence of a putative orthologue of SEP4 has been retrieved from a WGS sequence (kindly provided by DJ Sargent, unpublished data) of Fragaria nubicola cultivar ‘601’, a diploid species of strawberry, using tBlastN. From this information primers were designed to clone a full-length 985 bp cDNA from the octaploid Fragaria ananassa (GenBank accession FN691927). This cDNA encodes a protein with 51% identity over 268 amino acids to LeMADS-RIN, and which amongst Arabidopsis MADS-box proteins has the highest similarity to the SEP4 transcription factor (45% identity over 280 amino acids). The corresponding gene has been named FaMADS4.
In WT fruit the FaMADS4 gene is expressed at levels <1% of those of FaMADS9 and shows a modest increase in expression from the mature green to the orange stage (Fig.6). Expression of FaMADS4 was not substantially altered in the transgenic fruits down-regulated by the FaMADS9 construct, although the expression was reduced at the orange stage.
Fig. 6.
Expression of FaMADS4 in strawberry receptacle. Expression of a SEP4-like gene was determined by absolute QRT-PCR. Bars indicate the mean values (±SEM) of three fruits sampled from each line (see Fig. 2) at different stages of development.
Discussion
Fleshy fruits have traditionally been placed in one of two classes dependent on their ripening behaviour. In climacteric fruits, such as tomato and banana, ripening is associated with a burst of respiration and is initiated and co-ordinated by ethylene. By contrast, ripening in non-climacteric fruits such as strawberry and grape appears to be an ethylene-independent process (Giovannoni, 2004). Fruit in both classes, however, show changes in colour, texture, and flavour that probably reflect similar programmes of gene expression (Fei et al., 2004).
MADS-box transcription factors are expressed in a range of species with fleshy fruits including tomato, banana, apple, grape, and strawberry (Vrebalov et al., 2002; Fei et al., 2004; Malcomber and Kellogg, 2005). In tomato it has been shown that a SEP4-like gene (LeMADS-RIN) is necessary for ripening (Vrebalov et al., 2002). At the N-terminal region SEP proteins have highly conserved MADS-box/MEF2 and K-box domains involved in transcription factor binding and protein–protein interactions, respectively.The C-terminal region, however, is highly variable and confers important functional properties on the protein. This region in LeMADS-RIN is essential for normal fruit development in tomato and its disruption accounts for ripening inhibition in the rin mutant (Ito et al., 2008). It has been demonstrated here that a gene of the SEP1/2 subfamily can modulate the development and ripening of strawberry receptacle, achene, and petal tissues. Like the SEP4 LeMADS-RIN gene in tomato, suppression of the strawberry SEP1/2 FaMADS9 gene delays normal ripening. By contrast, silencing of TM29, a SEP1/2-like gene, in tomato produced parthenocarpic fruit and was not reported to affect ripening (Ampomah-Dwamena et al., 2002). Genes of the SEP3 clade are relatively highly expressed in banana fruit, but functional studies showed they were not able to complement ripening in the tomato rin mutant (Elitzur et al., 2010). These observations indicate that different classes of SEP genes can have non-redundant and specialized functions in fleshy fruit species.
In the rin tomato mutant, where the functional MADS-box transcript is abolished, the fruit show complete inhibition of ripening in the homozygous state, but in heterozygous individuals delayed ripening is apparent, presumably due to the presence of lower than normal levels of LeMADS-RIN transcript. This latter situation is probably analogous to our antisense transgenic strawberry fruit exhibiting moderate levels of FaMADS9 expression. However, in contrast to tomato where a SEP4-like gene is involved in the control of ripening, severe repression of the FaMADS9 SEP1/2-like gene in strawberry results in a complete inhibition of receptacle development. The expression pattern of FaMADS9 is consistent with its involvement in both fruit development and ripening.
We wanted to investigate further if a ripening-related homologue of the tomato SEP4 LeMADS-RIN gene could be found in strawberry. No SEP4 genes have been identified from strawberry fruit EST libraries in the public databases. However, recent information from high throughput genomic sequencing of Fragaria nubicola, a diploid species, has allowed us to clone a SEP4-like gene (FaMADS4) from Fragaria ananassa (cv. Calypso), an octoploid species. However, expression analysis revealed very low levels of transcript in ripening fruits, and no consistent relationship between mRNA levels of the strawberry SEP4 and inhibition of fruit development or ripening in the transgenic lines. The relatively low expression of the FaMADS4 gene may explain why FaMADS9 was the only cDNA isolated from a strawberry ripe fruit cDNA library using the tomato LeMADS-RIN probe, and also why our microarray data did not show any evidence of differential SEP4 expression between the wild type and transgenic lines.
The possibility was considered that the antisense configuration of the FaMADS9 transgene might be acting by suppressing both its own transcription and that of certain other related MADS-box genes. However, analysis of strawberry ESTs and changes in the expression of related gene sequences in our transgenic plants suggested that this was not the case. A BlastN search of approximately 21000 strawberry nucleotide sequences available in GenBank found four MADS-box gene sequences with some similarity to the 552 bp FaMADS9 transgene. These were SEP3-like (83% identity over 213 bp), AGL6-like (84% identity over 172 bp), AGAMOUS-like (89% identity over 68 bp), and SHATTERPROOF2-like (87% over 47 bp). The microarray data, later confirmed by QRT-PCR, identified that the SEP3-like and SHATTERPROOF2-like genes were down-regulated in the transgenic lines. The sensitive QRT-PCR method also detected suppression of an AGL6-like MADS-box gene in strawberry receptacles. However, repression of these genes was much less pronounced than that of FaMADS9 in this tissue. These data are entirely consistent with the remarkable specificity shown by antisense transgenes (Palecanda and Sharrock, 2001). These observations strongly suggest that the suppression of this gene alone is responsible for altering the development and ripening of strawberry fruit.
Retention of chlorophyll in the achenes of our antisense A and I lines is suggestive of the maintenance of a vegetative state and the arrest of ripening in these organs. Achenes are the true fruits of the strawberry and play a fundamental role in the growth and development of the receptacle through the action of auxin (Nitsch, 1950; Given et al., 1988a). The achenes resemble dry fruits and also undergo ripening changes during normal development including degradation of chlorophyll, accumulation of anthocyanins, and loss of water. The development and ripening of the receptacle is known to be influenced by the achenes (Given et al., 1988a). FaMADS9 may co-ordinate these events in both tissues.
The antisense transgene also altered petal development. The ‘green petal’ phenotype of FaMADS9 lines resembled the effects of silencing the SEP1/2-like geneTM29 in tomato (Ampomah-Dwamena et al., 2002). This contrasts with the lack of an observable petal phenotype in tomato when the SEP4 LeMADS-RIN was suppressed. This tomato gene is mainly expressed in fruit tissue, but like FaMADS9 is also expressed in the inflorescence (Malcomber and Kellogg, 2005). Similarly, the lack of an observable phenotype in other tissues does not exclude the possibility that these genes might be expressed in other parts of the plant. The SEP genes in Arabidopsis show considerable functional redundancy in controlling the identity of floral organs. Our findings, together with those reported for tomato, reinforce the view that SEP genes play a major role in the developmental regulation of ripening; however, they also suggest the specific role of each class of SEP gene may vary according to tissue and species (Malcomber and Kellogg, 2005; Zahn et al., 2005).
To study gene expression profiles in strawberry, as influenced by the antisense FaMADS9 transgene, an Arabidopsis microarray was used. Cross-species array experiments have now been demonstrated to provide a useful measure of the transcriptome where species-specific arrays are inadequate or not publicly available (Hammond et al., 2005). Nevertheless, while cross-species hybridizations can be optimized for utility via incorporation of appropriate controls (such as genomic DNA hybridization as described in this study) the results of the cross-species arrays should be interpreted with caution. For example, every hybridization reaction cannot be independently verified and the evolutionary distance between the species may result in cross-hybridization between members of the same gene family. However, the validation of a subset of these genes by QRT-PCR has shown this approach to be robust for the analysis of the strawberry transcriptome, even with TFs that may be expected to be transcribed at releatively low levels. Using this approach, it has been possible to investigate the expression of a wide range of genes in strawberry without any pre-selection based on limited information available in the literature and to identify novel gene expression associated with ripening and changes caused by altered FaMADS9 expression.
Our microarray experiments have revealed the pleiotropic effects of silencing the FaMADS9 transcript, including effects on the expression of both regulatory and metabolic genes. SEP proteins are known to form multimers (Malcomber and Kellogg, 2005) and it is likely that these complexes are necessary for ripening. Several MADS-box transcription factors, including those with sequence homology to Arabidopsis SEP3 and SHP 2 were among the genes repressed in the transgenic strawberry fruits. Interestingly, the SHP genes are involved in silique development and dehiscence in the dry fruits of Arabidopsis. Although there is no analogous process to dehiscence in strawberry fruits, both fruit types undergo cell separation events leading either to the parting of tissues in the dehiscence zones of siliques or softening in the fleshy receptacle. These cell wall degradation processes may be controlled directly or indirectly by this class of MADS-box genes (Ferrándiz, 2002). In the current study, the transgenic fruits showed reduced pectate lyase and endo-1,4-β-D-glucanase expression consistent with enhanced fruit firmness.Tomato also shows induction of an SHP-like gene (TAGL1) during development and ripening and, recently, TAGL1 has been shown to be involved in tomato fruit development and ripening (Vrebalov et al., 2009). By contrast, neither SEP3-like nor SEP1/2-like genes appear to have a clear fruit-related function in tomato (Ampomah-Dwamena et al., 2002; Fei et al., 2004), The induction of a likely SEP3 orthologue, but not MADS-RIN or FaMADS9 homologues, has been observed in ripening grapes, another non-climacteric fruit (Fei et al., 2004).
Differences may be expected between the regulatory factors involved in controlling ripening in fruiting species given the diversity of fruit forms in the Angiosperms. However, our work provides evidence that MADS-box genes are common features of the ripening regulatory network in both climacteric and non-climacteric fruits. The challenge will be to determine how these master switches are modulated by hormones such as auxin and ethylene, in addition to their links to the downstream effectors bringing about changes in fruit colour, texture, and flavour.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Validation of gDNA test for microarray.
Supplementary Fig. S2. Gene expression profiles in wild-type and FaMADS9 transgenic strawberry fruit.
Supplementary Fig. S3. Expression of chalcone synthase, flavanone-3-hydroxylase, and quinoneoxidoreductase in strawberry receptacles determined by absolute QRT-PCR.
Supplementary Table S1. Normalized probe-set signal values of genes significantly (P<0.05) differentially (>1.3-fold) expressed in FaMADS9 transgenic fruits compared with wild-type fruits.
Supplementary Table S2. Normalized probe-set signal values of genes significantly (P<0.05) differentially (>1.3-fold) expressed in both FaMADS9 transgenic fruits compared with wild-type fruits.
Supplementary Table S3. Primer sequences for QRT-PCR of strawberry genes other than FaMADS9.
Acknowledgments
We would like to thank the Biotechnology and Biological Sciences Research Council (UK) and the Department for Environment, Food and Rural Affairs (UK) for financial support. Also Malcolm Bennett (University of Nottingham) for useful discussions during the preparation of this manuscript.
Conflict of interest statement: No conflicts declared.
References
- Alexander L, Grierson D. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. Journal of Experimental Botany. 2002;53:2039–2055. doi: 10.1093/jxb/erf072. [DOI] [PubMed] [Google Scholar]
- Ampomah-Dwamena C, Morris BA, Sutherland P, Veit B, Yao JL. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiology. 2002;130:605–617. doi: 10.1104/pp.005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Elitzur T, Vrebalov J, Giovannoni JJ, Goldschmidt EE, Friedman H. The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene. Journal of Experimental Botany. 2010;61:1523–1535. doi: 10.1093/jxb/erq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Z, Tang X, Alba RM, White RA, Ronning CM, Martin GB, Tanksley SD, Giovannoni JJ. Comprehensive EST analysis of tomato and comparative genomics of fruit ripening. The Plant Journal. 2004;40:47–59. doi: 10.1111/j.1365-313X.2004.02188.x. [DOI] [PubMed] [Google Scholar]
- Ferrándiz C. Regulation of fruit dehiscence in Arabidopsis. Journal of Experimental Botany. 2002;53:2031–2038. doi: 10.1093/jxb/erf082. [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. Genetic regulation of fruit development and ripening. The Plant Cell. 2004;16:S170–S180. doi: 10.1105/tpc.019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Given NK, Venis MA, Grierson D. Hormonal regulation of ripening in the strawberry, a non-climacteric fruit. Planta. 1988a;174:402–406. doi: 10.1007/BF00959527. [DOI] [PubMed] [Google Scholar]
- Given NK, Venis MA, Grierson D. Purification and properties of phenylalanine ammonia-lyase from strawberry fruit and its synthesis during ripening. Journal of Plant Physiology. 1988b;133:31–37. [Google Scholar]
- Hammond JP, Bowen HC, White PJ, Mills V, Pyke KA, Baker AJM, Whiting SN, May ST, Broadley MR. A comparison of the Thlaspicaerulescens and Thlaspiarvense shoot transcriptomes. New Phytologist. 2006;170:239–260. doi: 10.1111/j.1469-8137.2006.01662.x. [DOI] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, Craigon DJ, Higgins J, Emmerson Z, Townsend H, White PJ, May ST. Using genomic DNA-based probe-selection to improve the sensitivity of high-density oligonucleotide arrays when applied to heterologous species. Plant Methods. 2005;1:10. doi: 10.1186/1746-4811-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JD, Israelstam GF. A method for the extraction of chlorophyll from leaf tissue without maceration. Canadian Journal of Botany. 1979;57:1332–1334. [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. The Plant Journal. 2008:55,212–223. doi: 10.1111/j.1365-313X.2008.03491.x. [DOI] [PubMed] [Google Scholar]
- Judd WS, Campbell CS, Kellogg EA, Stevens PF. Plant systemati cs:a Phylogenetic approach. Sunderland: Mass:Sinauer Associates Inc; 1999. [Google Scholar]
- Malcomber ST, Kellogg EA. SEPALLATA gene diversification: brave new whorls. Trends in Plant Science. 2005;10:427–435. doi: 10.1016/j.tplants.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Nitsch JP. Growth and morphogenesis of the strawberry as related to auxin. American Journal of Botany. 1950;37:211–215. [Google Scholar]
- Palecanda L, Sharrock RA. Molecular and phenotypic specificity of an antisense PHYB gene in Arabidopsis. Plant Molecular Biology. 2001;46:89–97. doi: 10.1023/a:1010686805488. [DOI] [PubMed] [Google Scholar]
- Robinson R, Tomes M. Ripening inhibitor: a gene with multiple effects on ripening. Report of the Tomato Genetics Cooperative. 1968;18:36–37. [Google Scholar]
- Seymour GB, Taylor JE, Tucker GA. Biochemistry of fruit ripening. London, New York: Chapman & Hall; 1993. [Google Scholar]
- Van Engelen F, Molthoff J, Conner A, Nap J, Pereira A, Stiekema W. pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Research. 1995;4:288–290. doi: 10.1007/BF01969123. [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJM, et al. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. The Plant Cell. 2009;21:3041–3062. doi: 10.1105/tpc.109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni JJ. A MADS-box gene necessary for fruit ripening at the tomato Ripening-Inhibitor (Rin) locus. Science. 2002;296:343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- Woolley LC, James DJ, Manning K. Purification and properties of an endo-β-1,4-glucanase from strawberry and down-regulation of the corresponding gene, cel1. Planta. 2001;214:11–21. doi: 10.1007/s004250100577. [DOI] [PubMed] [Google Scholar]
- Zahn LM, Kong H, Leebens-Mach JH, Kim S, Soltis PM, Landherr LL, Soltis DE, DePamphilis CW, Ma H. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics. 2005;169:2209–2223. doi: 10.1534/genetics.104.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.