Abstract
Abiotic stresses such as drought, salinity, and low temperature have drastic effects on plant growth and development. However, the molecular mechanisms regulating biochemical and physiological changes in response to stresses are not well understood. Protein kinases are major signal transduction factors among the reported molecular mechanisms mediating acclimation to environmental changes. Protein kinase ABC1 (activity of bc1 complex) is involved in regulating coenzyme Q biosynthesis in mitochondria in yeast (Saccharomyces cersvisiae), and in balancing oxidative stress in chloroplasts in Arabidopsis thaliana. In the current study, TaABC1 (Triticum aestivum L. activity of bc1 complex) protein kinase was localized to the cell membrane, cytoplasm, and nucleus. The effects of overexpressing TaABC1 in transgenic Arabidopsis plants on responses to drought, salt, and cold stress were further investigated. Transgenic Arabidopsis overexpressing the TaABC1 protein showed lower water loss and higher osmotic potential, photochemistry efficiency, and chlorophyll content, while cell membrane stability and controlled reactive oxygen species homeostasis were maintained. In addition, overexpression of TaABC1 increased the expression of stress-responsive genes, such as DREB1A, DREB2A, RD29A, ABF3, KIN1, CBF1, LEA, and P5CS, detected by real-time PCR analysis. The results suggest that TaABC1 overexpression enhances drought, salt, and cold stress tolerance in Arabidopsis, and imply that TaABC1 may act as a regulatory factor involved in a multiple stress response pathways.
Keywords: Abiotic stress response, biochemical character, gene expression, physiological character, protein kinases, TaABC1
Introduction
Plants, as sessile organisms, have evolved appropriate mechanisms to adapt to abiotic stresses, such as drought, high salt, and cold (Fujita et al., 2006). Plants trigger an orchestrated complex network of signal events to protect cellular activities and regulate whole plant physiological and biochemical responses when environmental stresses occur. Numerous stress-induced genes have been identified, including genes that function in initial perception and transmission of stress signals and those subsequently activated to enable the stress response (Ingram and Bartels, 1996; Shinozaki et al., 2003; Bartels and Sunkar, 2005; Yamaguchi-Shinozaki and Shinozaki, 2006). In diverse signalling pathways, protein phosphorylation is an important mechanism by which plants respond to external stimuli. Although genetic evidence shows that mitogen-activated protein kinase (MAPK) (Jonak et al., 1996; Mizoguchi et al., 1996; Munnik et al., 1999; Ichimura et al., 2000; Knight and Knight, 2001; Singh et al., 2002), calcium-dependent proten kinase (CDPK) (Ludwig et al., 2004), and SNF-1-related protein kinases (SnRKs) (Li et al., 2000; Mustilli et al., 2002; Kobayashi et al., 2004) are activated by abscisic acid (ABA) and osmotic stress, knowledge concerning components of these pathways, how the different molecules interact with each other, and where they are positioned in the complex signalling network is incomplete.
ABC1 (activity of bc1 complex) was originally identified as a multicopy suppressor of a cytochrome b mRNA translation defect in Saccharomyces cerevisiae (Bousquet et al., 1991). Mitochondrial ABC1 is essential for the proper conformation and activity of the cytochrome bc1 complex III in yeast (Brasseur et al., 1997). ABC1 was subsequently isolated as COQ8 and found to be necessary for yeast coenzyme Q (COQ) synthesis (Do et al., 2001). Several studies indicate that yeast abc1/coq8 mutants might be responsible for the respiratory deficiency caused by disruption of COQ biosynthesis (Saiki et al., 2003; Hsieh et al., 2004). Recent evidence showed that a yeast ABC1 homologue, p74, is required for COQ biosynthesis in Trypanosoma congolense (Baticados et al., 2005). Although the exact function of the ABC1/COQ8 protein is unknown, it is classified as a putative protein kinase based on the presence of kinase motifs (Leonard et al., 1998; Poon et al., 2000). CABC1 (chaperone activity of bc1 complex), a homologue of yeast ABC1 protein in humans, is involved in apoptosis through a mitochondrial pathway (Iiizumi et al., 2002). Mollet et al (2008) demonstrated that CABC1 mutations form a homogeneous group of ubiquinone deficiencies (Mollet et al., 2008). Plant ABC1 genes are poorly characterized. In Arabidopsis, 17 genes contain typical ABC1 motifs, but only two genes are described (Cardazzo et al., 1998; Jasinski et al., 2008). The first cloned ABC1-like protein partially restored the activity of complex III in an abc1 mutant (Cardazzo et al., 1998). Another ABC1 gene, AtOSA1 (oxidative stress-related Abc1-like), involved in balancing oxidative stress, does not complement the phenotype of abc1 mutation in yeast (Jasinski et al., 2008).
In this study, TaABC1, a member of the ABC1 protein kinase family from wheat, was overexpressed in Arabidopsis. Subsequent evidence showed that overexpression of TaABC1 produces an enhanced tolerance to drought, salt, and cold stress in Arabidopsis, and indicated the functional diversity of the ABC1 protein kinase family.
Materials and methods
Plant materials and growth conditions
Seeds of wild-type Arabidopsis thaliana (Columbia 0 type) and transgenic Arabidopsis were surface-sterilized with 10% bleach and 0.01% Triton X-100, and washed six times with sterile water. Sterile seeds were plated on Murashige and Skoog (MS) medium plus 0.8% (w/v) agar and 3.0% (w/v) sucrose. Plates were placed in darkness for 2 d at 4 °C and then transferred to a tissue culture room at 22 °C under a 12 h light/12 h dark photoperiod. After 7 d, seedlings were potted in soil mix (1:1 vermiculite:humus) and placed in a climate chamber at 22 °C, 70% relative humidity, and 150 μmol m−2 s−1 with a 12 h light/12 h darkness photoperiod.
Localization of TaABC1–green fluorescent protein (GFP) fusion protein
The coding sequence of TaABC1 was amplified with two primers (5′-GGT CTC AAG CTT ATG CCG CTG CCG CTG G-3′; BsaI site in bold and HindIII site underlined) and (5′-GGT CTC CTC GAG AAA CAA CCT TCT AAG GAA ACT TCT-3′; BsaI site in bold and BamHI site underlined). The PCR product was subcloned into the pJIT163 vector to generate pJIT163-TaABC1-GFP containing a TaABC1-GFP fusion construct under control of the cauliflower mosaic virus (CaMV) 35S promoter. The construct was further confirmed by sequencing and used for transformation of onion (Allium cepa) epidermal cells by biolistic bombardment with a GeneGun (Bio-Rad Helios™) according to the instruction manual (helium pressure, 150–300 psi). The transformed cells were incubated in MS medium at 28 °C for 36–48 h and then observed with a laser scanning confocal microscope (Leika TCS-NT).
Transformation of TaABC1 in Arabidopsis
To produce 35S-TaABC1 plants, a 1444 bp fragment containing the coding sequence of TaABC1 was amplified using two primers (5′-GGT ACC AGG CAG GGG GGC ATC-3′; the KpnI site underlined) and (5′-GGA TCC AAA CAA CCT TCT AAG GAA ACT TCT-3′; the BamHI site underlined). The PCR product was subcloned into the vector pPZP211 (Hajdukiewicz et al., 1994), in which transgene expression is under control of the CaMV 35S promoter. Transformation of Arabidopsis was performed by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens strain GV3101. Phenotypic analyses were performed on T3 or T4 homozygous lines.
Drought, salt, and cold tolerance assays in transgenic Arabidopsis
Drought tolerance assays were performed on seedlings. 35S-TaABC1, wild-type, and vector control seeds were germinated on MS medium. One-week-old seedlings were planted in identical pots containing mixed soil (1:1 vermiculite:humus) and well watered. The seedlings were cultured in a greenhouse (22 °C, 70% humidity, 150 μmol m−2 s−1, 12 h light/12 h dark cycle) without watering.
Salt tolerance assays were conducted at the seedling stage on plates and in soil. For the salt tolerance assay on plates, wild-type, 35S-TaABC1, and vector control seeds were germinated on MS medium. Five days after germination, seedlings from each line were carefully transferred to new MS media supplemented with different concentrations of NaCl. After 10 d growth in the treatment media, plants with no green or dead cotyledons were scored. Arabidopsis seedlings were cultured as described above for the salt tolerance assay in soil. Water was withheld for 4 weeks and plants were then well irrigated with NaCl solution (350 mM) applied at the bottom of the pots. When the soil was completely saturated with salt solution, free NaCl solution was removed and the plants were cultured under normal conditions. Survival rates were recorded 2 weeks later.
Cold tolerance assays were carried out on seedlings. Normally cultured Arabidopsis seedlings (4 weeks old) were stressed in a –8 °C freezer for 2 h, and subsequently cultured under normal growing conditions. Survival rates were scored after 6 d. All abiotic stress tolerance experiments were carried out in triplicate.
Cell membrane stability
Plant cell membrane stability indices (MSIs) were determined with a conductivity meter (DDS-1, YSI); MSI (%)=(1–initial electrical conductivity/electrical conductivity after boiling)×100. Twenty 8-day-old seedlings (grown on MS medium, 0.8% agar) were transferred to a horizontal screen; seedling roots were completely submerged in NaCl solution (200 mM). When signs of stress began to appear on the control plants, seedlings were removed and immediately thoroughly rinsed with double-distilled water (ddH2O) prior to immersion in 20 ml of ddH2O at room temperature. After 2 h the initial conductivities of the solutions were recorded. The samples were then boiled for 30 min, cooled to room temperature, and the final conductivities were measured.
Water loss determination
Water loss was measured using 10 plants each of transgenic and control plants. Four-week-old plants were detached from roots and weighed immediately [fresh weight (FW)], and then the plants were left on the laboratory bench (humidity, 45–50%, 20–22 °C) and weighed at designated time intervals. The proportions of FW loss were calculated relative to the initial plant weights. The plants were finally dried for 24 h at 80 °C to a constant dry weight (DW). Relative water contents (RWCs) were measured according to the formula: RWC (%)=(desiccated weight–DW)/(FW–DW).
Osmotic potential (OP)
OP was measured with a Micro-Osmometer (Fiske® Model 210, Fiske® Associates). Measurements were taken in the freezing point mode at room temperature. Four 4-week-old plants of each line were collected as a sample, which was finely ground using a mortar and pestle before being transferred to a microcentrifuge tube. The supernatant sap was obtained after centrifuging at 12 000 rpm for 10 min at room temperature. Tests were done in triplicate.
Photochemistry efficiency assays
Photochemistry efficiency was measured with a portable modulated chlorophyll fluorometer (OS-30p, Opti-Sciences, USA). After fully expanded leaves were allowed to dark-adapt for 60 min, the maximum photochemistry efficiency of photosystem II (PSII) (Fv/Fm) measurements were taken to assess changes in the primary photochemical reactions of photosynthetic potential at an early stage of drought stress. Three measurements were made for each plant, and 20 plants were used for transgenic and control lines.
Chlorophyll (Chl) content assay
Leaf Chl content measurements were performed as described by Hiscox and Israelstam (1979); that is, by measuring absorbance at 663 nm, and 645 nm of Chl extracted in dimethylsulphoxide (DMSO) using a spectrophotometer (SmartSpec™ 3000, Bio-Rad, USA). Transgenic and control plants were treated with 350 mM NaCl and measurements were made 24 h before and after the treatment.
Detection of reactive oxygen species (ROS)
For superoxide detection, detached fully expanded rosette leaves from 4-week-old transgenic and wild-type plants were harvested. Some of these leaves were incubated with 100 mM NaCl for 0.5/1.5 h, and others were incubated with ddH2O. These samples were vacuum-infiltrated with 0.1 mg ml−1 nitroblue tetrazolium (NBT) in 25 mM HEPES buffer, pH 7.6. Samples were incubated at room temperature in the dark for 2 h. To remove Chl, stained samples were transferred to 80% ethanol and incubated at 70 °C for 10 min. Twenty leaves of each line were treated and measured for each treatment.
Gene expression analysis
Twelve-day-old seedlings grown on MS medium were treated, or not treated, with polyethylene glycol (PEG)-6000 (25.4%, –1.4 MPa). Total RNA was extracted using Trizol reagent and treated with RNase-free DNase. For real-time quantitative PCR, 2 μg of total RNA was used for retrotranscription with a SuperScript First-Strand Synthesis System kit (Invitrogen). Quantitative expression assays were performed with the SYBR Green Master Mix kit and an ABI 7300 sequence detection system according to the manufacturer's protocol (Applied Biosystem). RT-PCR conditions were as follows: 5 min at 95 °C, and 40 cycles of 15 s at 95 °C, 30 s at 60 °C, 31 s at 72 °C. Samples were run in triplicate on each 96-well plate and independent PCRs were repeated at least three times. The relative quantification method (ΔΔCT) was used to evaluate quantitative variation between replicates (Zhang et al., 2008), and the Actin gene was used as an internal control to normalize all data. The primer pairs used for real-time PCR are listed in Supplementary Table S1 available at JXB online.
Results
Sequence analysis, phylogenetic tree, and subcellular localization of TaABC1
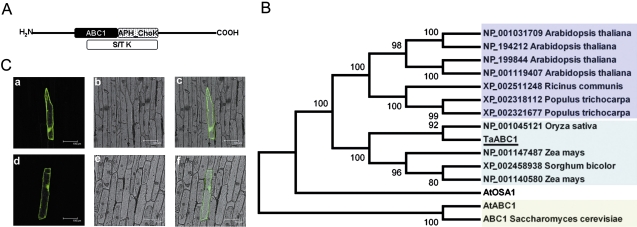
Using the Conserved Domain Database search engine at the National Center for Biotechnology Information (Marchler-Bauer et al., 2009), a conserved region of ∼120 amino acids that is characteristic of the so-called ABC1 protein family (Fig. 1A) was detected in the putative TaABC1 protein sequence (HM773264). In addition, an aminoglycoside phosphotransferase choline kinase (APH_ChoK) domain was also detected within the putative protein sequence of TaABC1 (Fig. 1A).
Fig. 1.
Structure, localization, and homological analysis of TaABC1. (A) Schematic illustration of the TaABC1 protein structure. Identified domains are depicted as follows: black box, ABC1; dotted box, aminoglycoside phosphotransferase, choline kinase (APH_ChoK) domain; white box, Ser/Thr protein kinase (S/T K). (B) Phylogenetic tree of ABC1 proteins. The tree was constructed with the MEGA 4.1 program; bootstrap values are in percentages. (C) Subcellular localization of TaABC1 protein by GFP fusion expression in onion epidermal cells. The GFP control or TaABC1-GFP fusion was driven by the CaMV 35S promoter. Cells were bombarded with DNA-coated gold particles and analysed by fluorescence microscopy. Photographs were taken after 16 h of incubation. Panels from left to right: images of GFP control (a) or TaABC1-GFP (d) in dark field, in bright field (b and e), and the combined images (c and f). (This figure is available in colour at JXB online.)
A phylogenetic tree was constructed based on the putative amino acid sequences of TaABC1, some plant ABC1 proteins, and yeast ABC1 protein (Fig. 1B). TaABC1 was clustered in the same monophyletic group as putative ABC1 proteins from rice, sorghum, and maize. In addition, TaABC1 protein showed >59.6% identity with putative ABC1 proteins from Ricinuss communis, Populus tichocarpa, and A. thaliana, but <30% identity with AtOSA1, AtABC1, and yeast ABC1. Furthermore, the results indicated that AtOSA1 protein did not group with AtABC1 and yeast ABC1 protein, but rather it formed a separate branch.
To examine its subcellular localization, TaABC1 was fused in-frame to the GFP gene under control of the CaMV 35S promoter. The constructs TaABC1–GFP and GFP were introduced into onion epidermal cells by particle bombardment. As shown in Fig. 1C, the TaABC1–GFP was localized to the cell membrane, cytoplasm, and nucleus.
Salt and osmotic responses of TaABC1-overexpressing Arabidopsis plants
Six T4 homozygous transgenic lines were randomly selected for detection of gene expression. Further phenotypic analyses were performed on three lines (OE1, OE4, and OE6) which possess a higher expression level of TaABC1 (Supplementary Fig. S1 at JXB online).
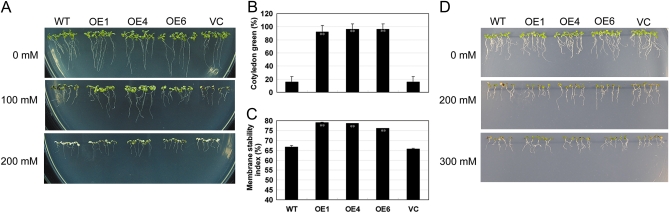
Salts inhibit seedling growth in a concentration-dependent manner (Xiong et al., 2002). Previous studies showed that TaABC1 was a salt-induced gene (Wang et al., 2007), but the role of TaABC1 in response to salt was not clear. The 35S-TaABC1 Arabidopsis plants and the two controls were grown as described in the Materials and methods. Differences were observed at the young seedling stage. Cotyledon greening and expansion, as well as root growth of the control plants, were inhibited at the seedling stage on plates containing 100 mM NaCl (Fig. 2A). When the NaCl concentration was increased to 200 mM, the growth of control plants was strongly inhibited and <20% of seedlings had small green cotyledons, whereas TaABC1 transgenic seedlings were still green (Fig. 2B). In the salinity stress tests, a significant difference was observed in terms of cell membrane stability between 8-day-old TaABC1 transgenic and control plants. After the 200 mM NaCl treatment was applied for 5 h, the control plants began to show symptoms of salt stress, whereas no signs of stress were shown on TaABC1 plants. MSI determinations showed that the cell membrane stability of TaABC1 plants was 12–16% higher than that of the control plants (Fig. 2C), and the difference was highly significant (P <0.01).
Fig. 2.
Salt and osmotic tolerance of 35S-TaABC1, wild-type (WT), and vector control (VC) plants. (A) NaCl effects on newly germinated seedling growth. 35S-TaABC1 (lines OE1, OE4, and OE6), WT, and VC seeds were planted on MS medium for germination. Five days after germination, seedlings from each line were carefully transferred to a new MS medium containing 100 mM and 200 mM NaCl. Photographs of representative seedlings of 35S-TaABC1, WT, and VC plants were taken after 10 d of growth in the treatment medium. (B) NaCl dose-response analysis of post-germinative growth (green cotyledons). Results were scored at 10 d on MS medium containing 200 mM NaCl. Values are the mean ±SD (triplicate measurements; n=15). (C) Cell membrane stability measurement of 35S-TaABC1, WT, and VC plants under 200 mM NaCl treatment. Values are the mean ±SD (triplicate measurements; n=20). (D) Mannitol effects on newly germinated seedling growth. 35S-TaABC1, WT, and VC seeds were germinated for 5 d on MS medium and then transferred to a new MS medium containing 200 mM and 300 mM manntiol for an additional 10 d of growth. Representative seedlings are shown. (This figure is available in colour at JXB online.)
To determine further if TaABC1 was involved in the response to salt stress or general osmotic effect stress, 5-day-old seedlings grown on MS medium were transferred to MS medium with 200 mM and 300 mM mannitol (an osmotic agent) for 10 d. The growth of control plants on plates containing different concentrations of mannitol was slightly inhibited compared with 35S-TaABC1 plants (Fig. 2D), suggesting that TaABC1 is involved in a general osmotic response to salt.
Drought response of TaABC1-overexpressing Arabidopsis plants
To characterize the performance of 35S-TaABC1 plants under drought stress, 7-day-old 35S-TaABC1, wild-type, and vector control plants were grown in soil for an additional 4 weeks (initial stage of drought stress). Thereafter, plants were not watered for 15 d (late stage of drought stress) to induce drought stress. The plants were then rewatered after 3 d (Fig. 3A). Before rewatering, the wilting levels of wild-type plants and vector control plants were more obvious than those of the 35S-TaABC1 plants. After rewatering, 49–80% of 35S-TaABC1 plants survived, whereas the corresponding survival rates were 25% for wild-type plants and 11% for vector control plants (Fig. 3B).
Fig. 3.
Responses to drought of 35S-TaABC1, wild-type (WT), and vector control (VC) plants. (A) Drought tolerance assays of 35S-TaABC1 (lines OE1, OE2, OE4, and OE6), WT, and VC plants. One-week-old 35S-TaABC1, WT, and VC plants, were transplanted to soil for an additional 4 weeks (initial stage of drought stress). Thereafter, plants continued to be not watered for 15 d (late stage of drought stress) to induce drought stress. Photographs of representative seedlings of 35S-TaABC1, WT, and VC plants were taken at the initial stage of drought stress, late stage of drought stress, and 3 d after rewatering. (B) Quantitative analysis of survival of 35S-TaABC1 (lines OE1, OE2, OE4, and OE6), WT, and VC plants 3 d after rewatering. Values are the mean ±SD (triplicate measurements; n=20 plants). (C) Water loss rates of 35S-TaABC1 (lines OE1, OE2, OE4, and OE6), WT, and VC plants. Detached leaves from 4-week-old plants grown on soil were left on the laboratory bench (humidity, 45–50%, 20–22 °C) and weighed at various time points after detachment. The proportions of fresh weight loss were calculated relative to the initial plant weights. Values are mean ±SD (n=10 plants). (D) Relative water contents of 35S-TaABC1 (lines OE1, OE2, OE4, and OE6), WT, and VC plants. Detached leaves from 4-week-old plants grown on soil were left on the laboratory bench (humidity, 45–50%, 20–22 °C) and weighed at various time points after detachment. The plants were finally dried for 24 h at 80°C to a constant dry weight (DW). Relative water contents were measured according to the formula: RWC (%)=(desiccated weight–DW)/(FW–DW). Values are mean ±SD (n=10 plants). (E) Osmotic potential (OP) measurement of 35S-TaABC1 (lines OE1, OE2, OE4, and OE6), WT, and VC plants. Values are the mean ±SD (triplicate measurements; n=20 plants). (F) Photochemical efficiency measurement of 35S-TaABC1 (lines OE1, OE2, OE4, and OE6), WT, and VC plants. One-week-old 35S-TaABC1, WT, and VC plants, were transplanted to soil for an additional 22 d and 28 d (normal conditions and moderate drought stress conditions) prior to measuring photochemical efficiencies (Fv/Fm). Values are the mean ±SD (triplicate measurements; n=20 plants). (This figure is available in colour at JXB online.)
To assess the water retention ability of 35S-TaABC1 plants, FW losses in detached rosette leaves were measured. The FW loss of detached rosette leaves in 35S-TaABC1 transgenic plants was <38%, as opposed to 49% and 50% for wild-type and vector control plants, respectively (Fig. 3C). The final RWCs of the 35S-TaABC1 plants were significantly higher than those of the control plants (Fig. 3D). This suggests that the 35S-TaABC1 plants possess higher water retention ability than wild-type and vector control plants.
An OP assay was undertaken to assess the osmotic adjustment ability of 35S-TaABC1 plants. The OP of 35S-TaABC1 transgenic plants was significantly higher than those of control plants, and there was no significant difference between the two controls (Fig. 3E). Overexpression of TaABC1 apparently led to enhanced OP.
To evaluate further the photosynthetic activity of 35S-TaABC1 plants, 1-week-old 35S-TaABC1, wild-type, and vector control plants were transplanted to soil and grown for an additional 22 d and 28 d (approximately normal conditions and moderate drought stress conditions, respectively), and the maximum photochemistry efficiencies of PSII (Fv/Fm) were measured. Under normal conditions, there were no evident differences between 35S-TaABC1 plants and the controls in Fv/Fm ratio. Under moderate drought stress conditions, Chl fluorescence values showed that the Fv/Fm ratios of the 35S-TaABC1 plants were significantly higher than those of the control plants (P <0.01) (Fig. 3F).
Salt response of TaABC1-overexpressing Arabidopsis plants
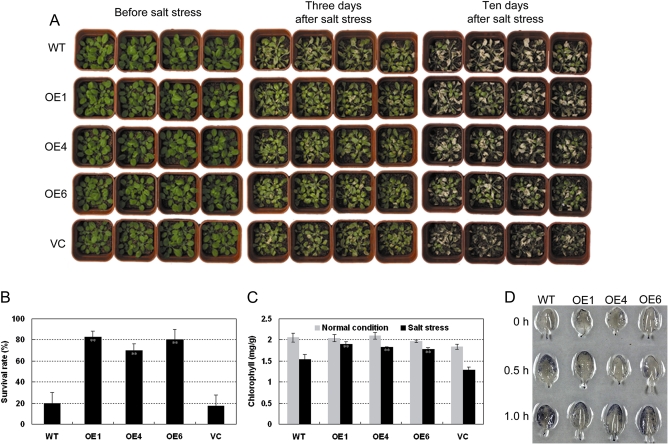
To examine further salt stress responses of TaABC1 transgenics, 35S-TaABC1 and control plants were grown in potted soil for 4 weeks, and then soaked with 350 mM NaCl solution. About 3 d after NaCl treatment, the control plants withered, whereas the leaves of transgenic plants were still green. Ten days later, signs of salt stress were evident; 35S-TaABC1 plants were much less affected than the control plants (Fig. 4A). The survival rates of TaABC1 transgenics were much higher than those of the controls (Fig. 4B).
Fig. 4.
Responses to salt of 35S-TaABC1, wild-type (WT), and vector control (VC) plants. (A) Four-week-old 35S-TaABC1 (lines OE1, OE4, and OE6), WT, and VC plants were irrigated with NaCl solution (350 mM); free NaCl solution was removed and the plants were cultured under normal conditions. Photographs of representative seedlings were taken 3 d and 10 d after treatment. (B) Quantitative analysis of 35S-TaABC1 (lines OE1, OE4, and OE6), WT, and VC plant survival 10 d after 350 mM NaCl treatment. Values are the mean ±SD (triplicate measurements; n=20 plants). (C) Chl contents in leaves before or after 350 mM NaCl treatment for 18 h. Values are the mean ±SD of triplicate measurements. (D) ROS production in leaves of 35S-TaABC1 and WT plants. Four-week-old detached leaves were stained with nitroblue tetrazolium (NBT). The photographs are representative leaves from three independent experiments. (This figure is available in colour at JXB online.)
The Chl contents of 35S-TaABC1 and control plants were measured before and after 350 mM NaCl treatment. There were no significant differences in Chl content between 35S-TaABC1 and control plants under normal conditions but, 18 h after 350 mM NaCl treatment, Chl contents of all plants were reduced and leaf Chl contents of the control plants decreased more rapidly than those of the 35S-TaABC1 plants (P <0.01) (Fig. 4C). These results indicated that the retention of stay-green features in 35S-TaABC1 plants was superior to that of the control plants.
Salt stress also leads to an accumulation of high levels of ROS (Van Camp et al., 1996; Borsani et al., 2001). The presence of ROS was further examined with NBT staining for superoxide in the 35S-TaABC1 and control plants with or without salt stress. Under non-salt stress conditions, 35S-TaABC1 and control leaves showed slight NBT staining, indicative of low levels of superoxide (Fig. 4D). However, under stress conditions, the staining levels of control leaves were much higher than in 35S-TaABC1 leaves (Fig. 4D) showing that 35S-TaABC1 plants scavenged superoxide more effectively than the controls.
Cold response of TaABC1-overexpressing Arabidopsis plants
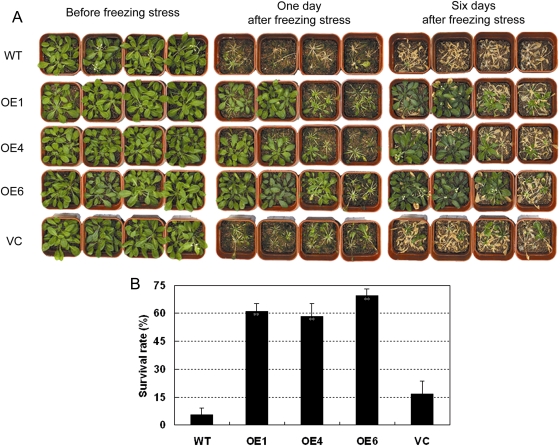
Because TaABC1 is also induced by cold (Wang et al., 2007), it was expected that 35S-TaABC1 plants would have altered responses to cold. Four-week-old 35S-TaABC1 and control plants were exposed to –8 °C for 1.5 h. After 1 of recovery under normal conditions, most 35S-TaABC1 plants were phenotypically green, whereas most of the control plants were dead (Fig. 5A); 6–16% of the control plants and 58–69% of the 35S-TaABC1 plants grew normally after recovery (Fig. 5B). These data suggest that TaABC1-overexpressing plants have increased tolerance to freezing stress.
Fig. 5.
Responses of 35S-TaABC1, wild-type (WT), and vector control (VC) plants to cold. (A) Four-week-old 35S-TaABC1 (lines OE1, OE4, and OE6), WT, and VC plants were cold stressed at –8 °C for 2 h and then transferred to normal conditions for recovery. Photographs of representative seedlings of 35S-TaABC1, WT, and VC plants were taken after 1 d and 6 d of recovery. (B) Survival rates were determined as the number of visibly green plants after rehydration. Values are the mean ±SD (triplicate measurements; n=15 plants). (This figure is available in colour at JXB online.)
Expression of stress-responsive genes in TaABC1-overexpressing Arabidopsis plants
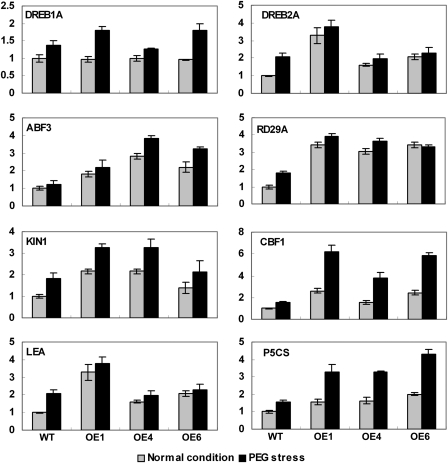
To elucidate the molecular mechanism of TaABC1 in osmotic response, the expression of stress-responsive genes identified in different regulated pathways was monitored by real-time PCR in transgenic Arabidopsis. Under normal conditions, the expression of DREB2A, RD29A, ABF3, KIN1, DREB1B/CBF1, LEA, and P5CS in TaABC1-overexpressing plants was substantially increased compared with wild-type plants, whereas there was no significant induction of expression of DREB1A/CBF3 in either transgenic or wild-type plants (Fig. 6). However, with PEG treatment for 2 h, the expressions of the tested marker genes, including DREB1A/CBF3, DREB2A, RD29A, ABF3, KIN1, DREB1B/CBF1, LEA, and P5CS, was up-regulated in both wild-type and transgenic plants. Thus, overexpression of TaABC1 increases expression of stress-responsive genes in Arabidopsis.
Fig. 6.
Expression of stress-responsive genes in 35S-TaABC1 and wild-type plants induced by PEG (–1.4 MPa) using real-time PCR. Total RNA was extracted from 2-week-old plants grown under normal or PEG treatment for 2 h. Transcript levels were measured by real-time RT-PCR of DREB1A, DREB2A, RD29A, ABF3, KIN1, CBF1, LEA, and P5CS under normal conditions (grey bars) or PEG treatment for 2 h (black bars). Actin was used as an internal control. Data represent the mean ±SD of three replicates.
Discussion
Plants have acquired an ability to survive stress conditions by developing highly organized signalling networks, in which protein kinase phosphorylation is one of the central signalling events that occur in response to environmental stress (Ichimura et al., 2000). The effects of overexpression of TaABC1, a wheat ABC1 protein kinase, in Arabidopsis were investigated and it was found that it induced various stress-responsive genes involved in stress signalling pathways. The transgenic plants had increased tolerance to drought, salt, and cold stress.
TaABC1-encoded protein kinase localizes to the cell membrane, cytoplasm, and nucleus
Phylogenetic analysis revealed that AtOSA1 does not group with AtABC1 (Fig. 1B), in agreement with Jasinski et al. (2008). The present studies also showed that TaABC1 does not cluster with AtABC1 or AtOSA1; rather, it clustered with putative ABC1 proteins from rice, sorghum, and maize. TaABC1 protein showed >59.6% identities with putative ABC1 proteins from R. communis, P. tichocarpa, and A. thaliana. It is therefore speculated that although ABC1 family proteins possess the conserved ABC1 domain, they also contain other kinase domains, partially explaining why they share high identity levels, but cluster into different groups.
Only two ABC1 genes are characterized amongst 17 genes containing typical ABC1 motifs in Arabidopsis (Cardazzo et al., 1998; Jasinski et al., 2008). The first representative ABC1 in plants (AtABC1, At4g01660) is a structural and functional homologue of yeast ABC1, and it allows partial restoration of the complex III activity of a yeast abc1 mutant, suggesting subcellular localization of AtABC1 in mitochondria (Bousquet et al., 1991; Cardazzo et al., 1998). However, AtOSA1, another ABC1 gene in Arabidopsis, is located in chloroplasts as a further factor playing a role in the balance of oxidative stress (Jasinski et al., 2008). TaABC1 protein was present in the cell membrane, cytoplasm, and nucleus (Fig. 1C). These localizations are different from those reported for AtABC1 and AtOSA1. This further confirms the view that ABC1 family proteins contain the conserved ABC1 domain, but have different kinase domains that lead to different localizations of ABC1 proteins and presumably the diverse functions they perform. Protein phosphorylation, catalysed by protein kinases, is one of the major post-translational modifications involved in the signal transduction pathway. It is generally acknowledged that signal transduction occurs at the plasma membrane level, in the cytosol, and at the transcriptional level (Hardie, 1999; Olsen et al., 2006; Afzal et al., 2008; Colcombet and Hirt, 2008; de la Fuente van Bentem and Hirt, 2009). However, strictly nucleus-localized and nucleus–cytosol-localized protein kinases are attracting increasing attention in signalling networks (Pandey et al., 2002; Cheong et al., 2003; Dammann et al., 2003; Riera et al., 2004;Choi et al., 2005; Raichaudhuri et al., 2006; Vert and Chory, 2006; Salinas et al., 2006; Takahashi et al., 2007; Yoo et al., 2008). Therefore, some researchers assume that nucleocytoplasmic trafficking machinery probably controls cytosolic translocation of an activated form, or activation in the nucleus through promoting interactions with their substrates under specific cellular conditions (Dahan et al., 2010).
Overexpression of TaABC1 enhances tolerance to abiotic stress in Arabidopsis
Dehydration, salinity and low temperature stresses in plants lead to membrane disorganization, inhibition of photosynthesis, and generation of ROS (Hasegawa et al., 2000).
Maintenance of membrane integrity and stability is thought to be a major component of environmental stress tolerance (Levitt, 1980). Cell membrane stability is positively correlated with various physiological and biochemical parameters conditioning responses to environmental stresses such as changes in OP, leaf rolling index, and/or leaf RWC (Munns, 2002). The cell MSI under environmental stress can be easily estimated by measurements of electrolyte leakage from cells. In this study, the MSI of 35S-TaABC1 plants under osmotic stress was higher than that of control plants (Fig. 2C). Because cell membrane stability is positively related to OP and RWC, the RWCs and OP of detached leaves were further tested. The RWCs of 35S-TaABC1 plants were higher than those of the control plants (Fig. 3D). A similar result was obtained for OP. The OP of 35S-TaABC1 plants was significantly higher than that of the control plants under well-watered conditions (Fig. 3E). These results indicated that TaABC1-overexpressing transgenic Arabidopsis had higher water retention and osmotic adjustment abilities, and thus increased tolerance to abiotic stresses.
Abiotic stresses provoke oxidative damage to photosynthetic proteins and pigments. The photochemical efficiency of PSII (Fv/Fm) and Chl content have been used as physiological senescence markers (Krause and Weis, 1991; Franke and Schreiber, 2007; Lim et al., 2007; Schippers et al., 2008; Osakabe et al., 2010). The effect of drought and salt stress on the levels of these senescence markers in 35S-TaABC1 and control plants was investigated. The drought and salt treatments caused severe damage to photochemical efficiency and Chl contents of wild-type plants (Figs 3F, 4C). ROS homeostasis is maintained by production of ROS, scavenging enzymes and non-enzymatic antioxidants (Mustilli et al., 2002; Mittler et al., 2004; Davletova et al., 2005). Superoxides in 35S-TaABC1 and wild-type plants were measured by NBT staining under salt stress conditions. It was found that transgenic plants overexpressing TaABC1 had enhanced tolerance to oxidative stress compared with wild-type plants (Fig. 4D). These results confirmed that overexpression of TaABC1 reduced the levels of damage to photosynthetic proteins and pigments, thereby increasing stress tolerance levels in transgenic Arabidopsis.
Generally, drought tolerance accompanies hypersensitivity to ABA treatments during seed germination and early seedling development (Hu et al., 2006; Ko et al., 2006; Zhang et al., 2007; Osakabe et al., 2010). However, in the system used here, enhanced tolerance to stress is not accompanied by sensitivity of seed germination and early seedling development to ABA in overexpressed TaABC1 transgenic plants (data not shown). The understanding is that ABA dependence might not be the major mode by which TaABC1 is involved in drought, salt, and cold stress signalling pathways. It is possible that expression of related genes in other signal pathways increases the OP and reduces the levels of damage to photosynthetic proteins and pigments to enhance multistress tolerance. Under stress conditions, enhancement of OP leads to a decrease of water loss, an increase in RWC, maintenance of regular cell turgor, and avoidance of damage to cell membranes. The higher OP probably also prevents entry of harmful ions, and relieves ion damage to cell membranes; and, moreover, it commonly lowers freezing points in plant cells. In addition, the levels of damage to photosynthetic proteins and pigments decline, thereby maintaining life under stress conditions. Finally, overexpression of TaABC1 conferred tolerance to drought, salt, and cold.
TaABC1 enhances multiple stress-responsive genes
Overexpression of genes such as DREB1A/CBF3, DREB2A, RD29A, ABF3, DREB1B/CBF1, and P5CS conferred stress tolerance in transgenic plants (Yamaguchi-Shinozaki and Shinozaki, 1994; Kishor et al., 1995; Jaglo-Ottosen et al., 1998; Liu et al., 1998; Kasuga et al., 1999; Kang et al., 2002; Dubouzet et al., 2003; Sakuma et al., 2006). To identify whether TaABC1 affects the expression of stress-related genes, eight marker genes involved in the stress response were compared in TaABC1-overexpressing and wild-type plants. Overexpression of DREB1/CBF in transgenic plants increased tolerance to freezing, drought, and salt stresses (Liu et al., 1998; Kasuga et al., 1999; Dubouzet et al., 2003). Ito et al. (2006) showed that overexpression of OsDREB1 also improved drought and chilling tolerance in rice (Ito et al., 2006). However, the results failed to show that DREB1A/CBF3 underwent increased expression in 35S-TaABC1 plants under normal conditions (Fig. 6), suggesting that there may be other stress pathways involved in TaABC1-mediated stress tolerance.
The RD29A gene is a drought-, cold-, and ABA-inducible gene with a dehydration-responsive element (DRE) and an ABA-reaponsive element (ABRE) present in its promoter region (Shinozaki and Yamaguchi-Shinozaki, 1997). Overexpression of constitutively active DREB2A resulted in significant drought stress tolerance, but only slight freezing tolerance in transgenic Arabidopsis plants (Sakuma et al., 2006). ABF3, KIN1, and DREB1B/CBF1 are thought to be involved in different stress regulation pathways (Yamaguchi-Shinozaki and Shinozaki, 2006). The high transcript levels of DREB2A, RD29A, ABF3, KIN1, and DREB1B/CBF1 in 35S-TaABC1 plants suggest that TaABC1 may act upstream of these genes in stress tolerance and is therefore involved in a cross-talk between the complex networks of stress-responsive genes.
Late embryogenesis abundant (LEA) proteins and Δ1-pyrroline-5-carboxylate synthetase (P5CS) function in the protection of cells during osmotic stress (Taji et al., 2002; Battaglia et al., 2008). The present study showed that the expression levels of LEA and P5CS were up-regulated in 35S-TaABC1 plants under normal and stress conditions (Fig. 6). This is consistent with higher OP in TaABC1 plants.
The expression of DREB1A/CBF3 showed no clear changes in 35S-TaABC1 plants, but DREB1B/CBF1, DREB2A, and ABF3 underwent increased expression under normal conditions. This demonstrated that TaABC1 was possibly more dependent on the DREB1B/CBF1, DREB2A, and ABF3 stress pathways. Moreover, it seems likely that cross-talk among different stress signalling pathways occurs in TaABC1-induced stress responses.
Based on various analyses, it is speculated that the higher tolerance of TaABC1 plants to drought, salt, and cold stresses may be due to the combined effects of up-regulated expression of stress-responsive genes, increased OP, and decreased damage to photosynthetic proteins and pigments.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Expression levels of TaABC1 in different transgenic Arabidopsis lines.
Table S1. Primer pairs used in quantitative real-time PCR.
Acknowledgments
We are grateful to Professor Robert A. McIntosh (Plant Breeding Institute, University of Sydney, NSW, Australia) for revising the manuscript. This work was supported by the National Basic Research Program of China (2010CB951501) and the National High Technology Research and Development Program of China (2006AA100201).
Glossary
Abbreviations
- ABA
abscisic acid
- ABF3
abscisic acid-responsive elements-binding factor 3
- ABRE
ABA-responsive element
- CBF1
C-repeat/DRE-binding factor 1
- Chl
chlorophyll
- DRE
dehydration-responsive element
- PEG
polyethylene glycol
- RD29A
responsive to dessication 29A
References
- Afzal AJ, Wood AJ, Lightfoot DA. Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Molecular Plant-Microbe Interactions. 2008;21:507–517. doi: 10.1094/MPMI-21-5-0507. [DOI] [PubMed] [Google Scholar]
- Bartels D, Sunkar R. Drought and salt tolerance in plants. Critical Reviews in Plant Science. 2005;24:23–58. [Google Scholar]
- Baticados W, Witola W, Inoue N, Kim J, Kuboki N, Xuan X, Yokoyama N, Sugimoto C. Expression of a gene encoding Trypanosoma congolense putative Abc1 family protein is developmentally regulated. Journal of Veterinary Medical Science. 2005;67:157–164. doi: 10.1292/jvms.67.157. [DOI] [PubMed] [Google Scholar]
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. The enigmatic LEA proteins and other hydrophilins. Plant Physiology. 2008;148:6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Valpuesta V, Botella MA. Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiology. 2001;126:1024–1030. doi: 10.1104/pp.126.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet I, Dujardin G, Slonimski P. ABC1, a novel yeast nuclear gene, has a dual function in mitochondria: it suppresses a cytochrome b mRNA translation defect and is essential for the electron transfer in the bc 1 complex. EMBO Journal. 1991;10:2023–2031. doi: 10.1002/j.1460-2075.1991.tb07732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasseur G, Tron G, Dujardin G, Slonimski PP, Brivet-Chevillotte P. The nuclear ABC1 gene is essential for the correct conformation and functioning of the cytochrome bc1 complex and the neighbouring complexes II and IV in the mitochondrial respiratory chain. European Journal of Biochemistry. 1997;246:103–111. doi: 10.1111/j.1432-1033.1997.t01-1-00103.x. [DOI] [PubMed] [Google Scholar]
- Cardazzo B, Hamel P, Sakamoto W, Wintz H, Dujardin G. Isolation of an Arabidopsis thaliana cDNA by complementation of a yeast abc1 deletion mutant deficient in complex III respiratory activity. Gene. 1998;221:117–125. doi: 10.1016/s0378-1119(98)00417-x. [DOI] [PubMed] [Google Scholar]
- Cheong YH, Moon BC, Kim JK, et al. BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiology. 2003;132:1961–1972. doi: 10.1104/pp.103.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HI, Park HJ, Park JH, Kim S, Im MY, Seo HH, Kim YW, Hwang I, Kim SY. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiology. 2005;139:1750–1761. doi: 10.1104/pp.105.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochemical Journal. 2008;413:217–226. doi: 10.1042/BJ20080625. [DOI] [PubMed] [Google Scholar]
- Dahan J, Wendehenne D, Ranjeva R, Pugin A, Bourque S. Nuclear protein kinases: still enigmatic components in plant cell signalling. New Phytologist. 2010;185:355–368. doi: 10.1111/j.1469-8137.2009.03085.x. [DOI] [PubMed] [Google Scholar]
- Dammann C, Ichida A, Hong B, Romanowsky SM, Hrabak EM, Harmon AC, Pickard BG, Harper JF. Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiology. 2003;132:1840–1848. doi: 10.1104/pp.103.020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver D, Coutu J, Shulaev V, Schlauch K, Mittler R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. The Plant Cell. 2005;17:268–281. doi: 10.1105/tpc.104.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Hirt H. Protein tyrosine phosphorylation in plants: more abundant than expected? Trends in Plant Science. 2009;14:71–76. doi: 10.1016/j.tplants.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Do T, Hsu A, Jonassen T, Lee P, Clarke C. A defect in coenzyme Q biosynthesis is responsible for the respiratory deficiency in Saccharomyces cerevisiae abc1 mutants. Journal of Biological Chemistry. 2001;276:18161–18168. doi: 10.1074/jbc.M100952200. [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Franke R, Schreiber L. Suberin—a biopolyester forming apoplastic plant interfaces. Current Opinion in Plant Biology. 2007;10:252–259. doi: 10.1016/j.pbi.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion in Plant Biology. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Molecular Biology. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Hardie DG. Plant protein serine/threonine kinase: classification and functions. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:97–131. doi: 10.1146/annurev.arplant.50.1.97. [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- Hiscox J, Israelstam G. A method for the extraction of chlorophyll from leaf tissue without maceration. Canadian Journal of Botany. 1979;57:1332–1334. [Google Scholar]
- Hsieh E, Dinoso J, Clarke C. A tRNATRP gene mediates the suppression of cbs2-223 previously attributed to ABC1/COQ8. Biochemical and Biophysical Research Communications. 2004;317:648–653. doi: 10.1016/j.bbrc.2004.03.096. [DOI] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences, USA. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. The Plant Journal. 2000;24:655–665. doi: 10.1046/j.1365-313x.2000.00913.x. [DOI] [PubMed] [Google Scholar]
- Iiizumi M, Arakawa H, Mori T, Ando A, Nakamura Y. Isolation of a novel gene, CABC1, encoding a mitochondrial protein that is highly homologous to yeast activity of bc1 complex. Cancer Research. 2002;62:1246–1250. [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant and Cell Physiology. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Jasinski M, Sudre D, Schansker G, Schellenberg M, Constant S, Martinoia E, Bovet L. AtOSA1, a member of the Abc1-like family, as a new factor in cadmium and oxidative stress response. Plant Physiology. 2008;147:719–731. doi: 10.1104/pp.107.110247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Kiegerl S, Ligterink W, Barker PJ, Huskisson NS, Hirt H. Stress signaling in plants: a mitogen-activated protein kinase pathway is activated by cold and drought. Proceedings of the National Academy of Sciences, USA. 1996;93:11274–11279. doi: 10.1073/pnas.93.20.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. The Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnology. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kishor P, Hong Z, Miao GH, Hu C, Verma D. Overexpression of [delta]-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiology. 1995;108:1387–1394. doi: 10.1104/pp.108.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Knight MR. Abiotic stress signalling pathways: specificity and cross-talk. Trends in Plant Science. 2001;6:262–267. doi: 10.1016/s1360-1385(01)01946-x. [DOI] [PubMed] [Google Scholar]
- Ko JH, Yang SH, Han KH. Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. The Plant Journal. 2006;47:343–355. doi: 10.1111/j.1365-313X.2006.02782.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. The Plant Cell. 2004;16:1163–1177. doi: 10.1105/tpc.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause G, Weis E. Chlorophyll fluorescence and photosynthesis: the basics. Annual Review of Plant Biology. 1991;42:313–349. [Google Scholar]
- Leonard C, Aravind L, Koonin E. Novel families of putative protein kinases in bacteria and archaea: evolution of the ‘eukaryotic’ protein kinase superfamily. Genome Research. 1998;8:1038–1047. doi: 10.1101/gr.8.10.1038. [DOI] [PubMed] [Google Scholar]
- Levitt J. Responses of plants to environmental stress: water, radiation, salt and other stresses. New York: Academic Press; 1980. [Google Scholar]
- Li J, Wang XQ, Watson MB, Assmann SM. Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science. 2000;287:300–303. doi: 10.1126/science.287.5451.300. [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim Y, Breeze E, et al. Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. The Plant Journal. 2007;52:1140–1153. doi: 10.1111/j.1365-313X.2007.03317.x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. The Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Romeis T, Jones J. CDPK-mediated signalling pathways: specificity and cross-talk. Journal of Experimental Botany. 2004;55:181–188. doi: 10.1093/jxb/erh008. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, et al. CDD: specific functional annotation with the conserved domain database. Nucleic Acids Research. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends in Plant Science. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K. A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1996;93:765–769. doi: 10.1073/pnas.93.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet J, Delahodde A, Serre V, Chretien D, Schlemmer D, Lombes A, Boddaert N, Desguerre I, De Lonlay P, Ogier de Baulny H. CABC1 gene mutations cause ubiquinone deficiency with cerebellar ataxia and seizures. American Journal of Human Genetics. 2008;82:623–630. doi: 10.1016/j.ajhg.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Ligterink W, Meskiene, Calderini O, Beyerly J, Musgrave A, Hirt H. Distinct osmo-sensing protein kinase pathways are involved in signalling moderate and severe hyper-osmotic stress. The Plant Journal. 1999;20:381–388. doi: 10.1046/j.1365-313x.1999.00610.x. [DOI] [PubMed] [Google Scholar]
- Munns R. Comparative physiology of salt and water stress. Plant, Cell and Environment. 2002;25:239–250. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. The Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Mizuno S, Tanaka H, Maruyama K, Osakabe K, Todaka D, Fujita Y, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K. Overproduction of the membrane-bound receptor-like protein kinase 1, RPK1, enhances abiotic stress tolerance in arabidopsis. Journal of Biological Chemistry. 2010;285:9190–9201. doi: 10.1074/jbc.M109.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Tiwari SB, Tyagi W, Reddy MK, Upadhyaya KC, Sopory SK. A Ca2+/CaM-dependent kinase from pea is stress regulated and in vitro phosphorylates a protein that binds to AtCaM5 promoter. European Journal of Biochemistry. 2002;269:3193–3204. doi: 10.1046/j.1432-1033.2002.02994.x. [DOI] [PubMed] [Google Scholar]
- Poon W, Davis D, Ha H, Jonassen T, Rather P, Clarke C. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. Journal of Bacteriology. 2000;182:5139–5146. doi: 10.1128/jb.182.18.5139-5146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichaudhuri A, Bhattacharyya R, Chaudhuri S, Chakrabarti P, Dasgupta M. Domain analysis of a groundnut calcium-dependent protein kinase: nuclear localization sequence in the junction domain is coupled with nonconsensus calcium binding domains. Journal of Biological Chemistry. 2006;281:10399–10409. doi: 10.1074/jbc.M511001200. [DOI] [PubMed] [Google Scholar]
- Riera M, Figueras M, Lopez C, Goday A, Pages M. Protein kinase CK2 modulates developmental functions of the abscisic acid responsive protein Rab17 from maize. Proceedings of the National Academy of Sciences, USA. 2004;101:9879–9884. doi: 10.1073/pnas.0306154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R, Ogiyama Y, Kainou T, Nishi T, Matsuda H, Kawamukai M. Pleiotropic phenotypes of fission yeast defective in ubiquinone-10 production. A study from the abc1Sp (coq8Sp) mutant. Biofactors. 2003;18:229–235. doi: 10.1002/biof.5520180225. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. The Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas P, Fuentes D, Vidal E, Jordana X, Echeverria M, Holuigue L. An extensive survey of CK2 alpha and beta subunits in Arabidopsis: multiple isoforms exhibit differential subcellular localization. Plant and Cell Physiology. 2006;47:1295–1308. doi: 10.1093/pcp/pcj100. [DOI] [PubMed] [Google Scholar]
- Schippers JH, Nunes-Nesi A, Apetrei R, Hille J, Fernie AR, Dijkwel PP. The Arabidopsis onset of leaf death5 mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. The Plant Cell. 2008;20:2909–2925. doi: 10.1105/tpc.107.056341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiology. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Singh K, Foley RC, Onate-Sanchez L. Transcription factors in plant defense and stress responses. Current Opinion in Plant Biology. 2002;5:430–436. doi: 10.1016/s1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. The Plant Journal. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Nasir KH, Ito A, Kanzaki H, Matsumura H, Saitoh H, Fujisawa S, Kamoun S, Terauchi R. A high-throughput screen of cell-death-inducing factors in Nicotiana benthamiana identifies a novel MAPKK that mediates INF1-induced cell death signaling and non-host resistance to Pseudomonas cichorii. The Plant Journal. 2007;49:1030–1040. doi: 10.1111/j.1365-313X.2006.03022.x. [DOI] [PubMed] [Google Scholar]
- Van Camp W, Capiau K, Van Montagu M, Inze D, Slooten L. Enhancement of oxidative stress tolerance in transgenic tobacco plants overproducing Fe-superoxide dismutase in chloroplasts. Plant Physiology. 1996;112:1703–1714. doi: 10.1104/pp.112.4.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- Wang CX, Jing RL, Mao XG, Pang XB, Liu HM, Chang XP. Cloning and expression analysis of a new stress-responsive gene TaABC1L in wheat. Acta Agronomica Sinica. 2007;33:878–884. [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. The Plant Cell. 2002;14(Suppl):S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008;451:789–795. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wollenweber B, Jiang D, Liu F, Zhao J. Water deficits and heat shock effects on photosynthesis of a transgenic Arabidopsis thaliana constitutively expressing ABP9, a bZIP transcription factor. Journal of Experimental Botany. 2008;59:839–848. doi: 10.1093/jxb/erm364. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. The Plant Cell. 2007;19:1912–1929. doi: 10.1105/tpc.106.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.