Summary
When iron is scarce, Bacillus subtilis expresses genes involved in the synthesis and uptake of the siderophore bacillibactin (BB) and uptake systems to pirate other microbial siderophores. Here, we demonstrate that transcriptional induction of the feuABCybbA operon, encoding the Fe-BB uptake system, is mediated by Btr (formerly YbbB) which is encoded by the immediately upstream gene. Btr contains an AraC-type DNA binding domain fused to a substrate binding protein (SBP) domain related to FeuA, the SBP for Fe-BB uptake. When cells are iron-limited, the Fur-mediated repression of btr is relieved and Btr binds to a conserved direct repeat sequence adjacent to feuA to activate transcription. If BB is present, Btr further activates feuA expression. Btr binds with high affinity to both apo-BB and Fe-BB and the resulting complex displays a significantly increased efficacy as a transcriptional activator relative to Btr alone. Btr can also activate transcription in response to the structurally similar siderophore enterobactin, although genetic analyses indicate that the two siderophores make distinct interactions with the Btr substrate binding domain. Thus, the FeuABC transporter is optimally expressed under conditions of iron starvation, when Fur-mediated repression is relieved, and in the presence of its cognate substrate.
Keywords: Bacillibactin, AraC family, regulation, ABC transporter, evolution
Introduction
Under iron-limited growth conditions microorganisms produce small iron chelating molecules named siderophores (Andrews et al., 2003). Siderophores chelate ferric ions with high affinity and the complex is imported through specific uptake systems (Neilands, 1993). Genes involved in the uptake of ferri-siderophore complexes are frequently subject to two levels of regulation: repression by the iron-sensing Fur protein and substrate induction mediated by an activator protein whose activity is controlled by the cognate siderophore (Andrews et al., 2003).
Bacillus subtilis produces a catecholate trilactone siderophore, bacillibactin (BB), under conditions of iron limitation (May et al., 2001). BB synthesis requires the products of the dhb operon and the Sfp phosphopantetheinyl transferase, encoded by a gene that is mutant (sfp0) in most laboratory strains of B. subtilis 168. Ferric-BB (Fe-BB) binds with high affinity to the FeuA substrate binding protein (SBP) (Miethke et al., 2006) and the complex is transported through the FeuBC integral membrane proteins of this ABC-type transporter (Ollinger et al., 2006). After transport, the iron is released through cleavage of the siderophore by the BesA esterase (Miethke et al., 2006).
In addition to the synthesis and uptake of BB, B. subtilis can acquire iron siderophores (xenosiderophores) produced by other microorganisms. These additional uptake systems include ABC transporters specific for ferric-citrate, ferrioxamine, ferrichrome, schizokinen and arthrobactin (Ollinger et al., 2006). In the absence of an available siderophore, iron uptake is critically dependent upon a recently identified elemental iron uptake system (encoded by the ywbLMN genes) (Ollinger et al., 2006).
B. subtilis Fur regulates the expression of iron uptake systems in response to iron availability. When intracellular iron is sufficient, iron-loaded Fur represses transcription of Fur-regulated genes (Baichoo and Helmann, 2002). Altogether, Fur regulates at least 40 genes of which approximately half encode siderophore and elemental iron uptake systems (Baichoo et al., 2002; Ollinger et al., 2006). Among the other Fur regulated genes, btr (formerly ybbB) encodes a predicted AraC-type regulator and is adjacent to the feuABC genes encoding the BB uptake system (Baichoo et al., 2002). Fur was previously shown to bind the btr promoter region and expression of btr is ~10-fold elevated by either the iron chelator 2,2′-dipyridyl or in a fur mutant (Baichoo et al., 2002).
In the present study, we show that btr (BB transport regulator) encodes an activator that mediates the BB-inducible expression of the FeuABC uptake system. Btr has a novel architecture with an amino-terminal AraC-type DNA-binding domain and a carboxy-terminal siderophore binding domain homologous to the FeuA SBP. Btr binds to a conserved direct repeat in the feuA promoter region and activates transcription. Btr is essential for the expression of the feuABCybbA operon and therefore for BB uptake.
Results
Btr encodes an unusual AraC family member
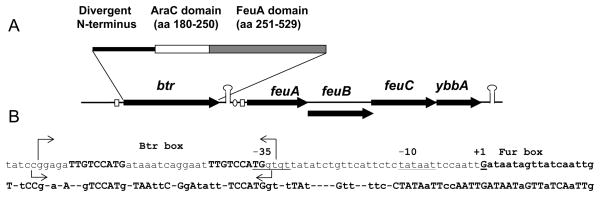
Btr is negatively regulated by Fur in the presence of sufficient iron as observed in transcriptome analyses (Baichoo et al., 2002). The btr (formerly ybbB) gene is located immediately upstream of the feuABCybbA operon which encodes the BB uptake system FeuABC and a putative esterase, YbbA (Fig. 1). Conserved domain analyses indicate that Btr is an unusually large AraC-family regulator with an N-terminal DNA-binding domain and a C-terminal domain most similar to siderophore-binding proteins such as B. subtilis FhuD (25% identical over 140 aa) and FeuA (23% identical over 193 aa) (Ollinger et al., 2006; Schneider and Hantke, 1993) and E. coli FhuD (Koster and Braun, 1990) (Fig. 1 and supplemental Fig. S1). Apparent orthologs of Btr were found in a variety of Bacillus spp. and were in each case encoded adjacent to a predicted ferri-siderophore uptake system (Fig. 1B and supplemental Fig. S2).
Figure 1. Genomic context of Btr and features of the feuABCybbA regulatory region.
A. Schematic representation of the gene organization of btr and the feuABCybbA operon. Conserved domains within the Btr protein are indicated.
B. Promoter region of the feuABCybbA operon. In the upper sequence, the transcription start site (+1) is shown as an upper case G (as determined by 5′-RACE), the −10 region is underlined, and the Fur and Btr boxes are shown in bold (in lower and upper case, respectively). Arrows indicate the regions protected by Btr in DNase I footprinting assays on the upper and the lower stand. The start codon for feuA is 11 nt downstream of the sequence shown. The lower sequence summarizes conservation of the feuA promoter region in strains with Btr orthologs (B. licheniformis, B. clausii and B. amyloliquefaciens). Upper case letters are 100% conserved, lower case letters are 75% conserved, and dashes indicate 50% or less. The regions of highest sequence conservation correspond to the recognition sites for Btr, Fur, and RNA polymerase.
Btr is essential for growth under conditions of severe iron starvation
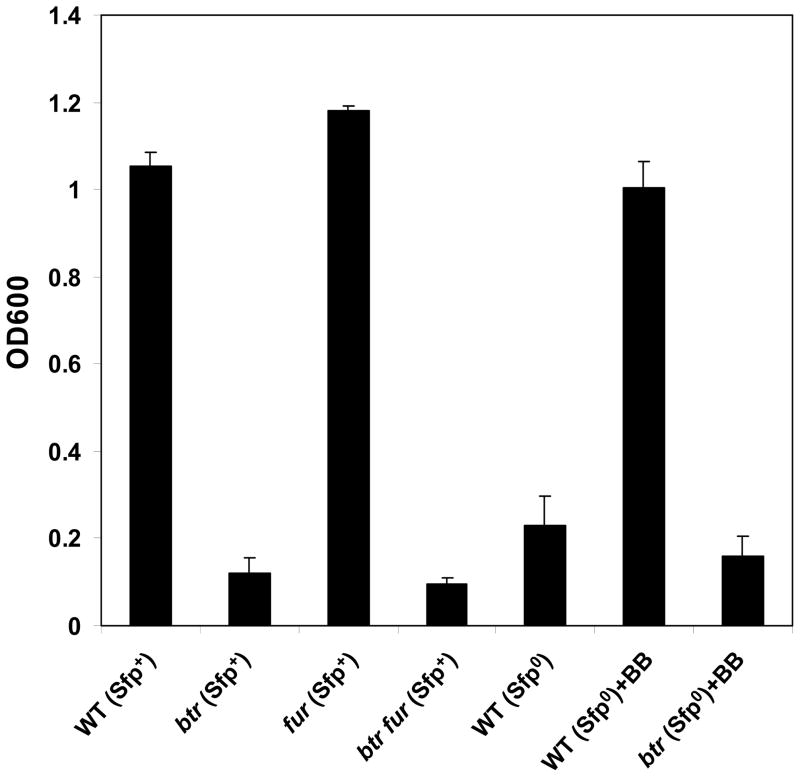
A btr null mutant was constructed in B. subtilis 168 in both BB producing (sfp+) and non-producing (sfp0) strains. Under iron starvation conditions, strains lacking functional Sfp (sfp0) secrete the BB precursor dihydroxybenzoic acid (DHBA) and its glycine conjugate (DHBG; also known as Itoic acid; Ito and Neilands, 1958). In the presence of the strong iron chelator ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDHA; pFe = 26.9), cells are unable to grow unless they can synthesize or are provided with a high affinity siderophore such as BB (pFe = 33.1) (Ollinger et al., 2006). The BB producing strain grew to high cell density even in the presence of EDDHA, as did the isogenic fur mutant. In contrast, neither the btr mutant nor the btr fur double mutant were able to grow (Fig. 2).
Figure 2.
Siderophore-dependent growth of BB-producing (sfp+) and non-producing (sfp0) strains in FS-MM as measured after 18 h. The BB-producing strains were grown in the presence of 15 μM EDDHA, and the non-producing strains were grown with 10 μM EDDHA. 5 μM exogenous BB was added where indicated (+BB).
We hypothesized that the inability of the btr mutant to grow in the presence of EDDHA was due to a lack of synthesis of the BB uptake system encoded by the adjacent feuABCybbA operon. Indeed, BB levels in the supernatant fraction from cells grown in Fe-starvation minimal medium (FS-MM) were slightly higher in the sfp+ btr strain (~450 nM BB/OD600) as compared to the sfp+ strain (~350 nM BB/OD600). The ability of the btr mutant to synthesize normal levels of BB (in the sfp+ strain), together with the inability of exogenous BB to stimulate growth of the sfp0 btr mutant (Fig. 2), suggests that Btr is required for BB transport but not for synthesis.
Btr is essential for siderophore-stimulated expression of the BB uptake system
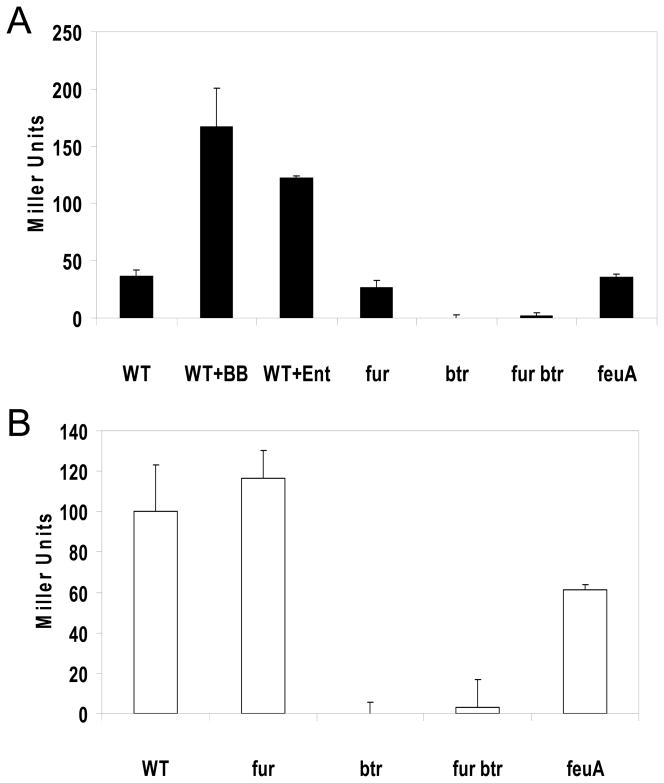
The feuABCybbA operon is under Fur control and is derepressed under iron-limiting conditions (Baichoo et al., 2002). Thus, the feuA promoter is about equally active in both wild-type and isogenic fur mutants grown in FS-MM (Fig. 3A). Significantly, feuA expression was elevated several-fold by addition of BB (Fig. 3A) or in strains able to synthesize BB (Fig. 3B). The btr mutation completely eliminated expression from the feuA promoter, both in wild-type and fur mutant cells (Fig. 3). This indicates that Btr is a positive regulator of the feuA promoter and is required for both basal (in the absence of BB) and siderophore-induced promoter activity. We speculated that the Btr-dependent basal promoter activity might be due to the presence of the siderophore precursors DHBA and DHBG. This appears unlikely, however, since a dhbA mutant (unable to produce either BB or its precursors) still expressed feuA at a comparable level to the DHBA producing strain (data not shown). In addition to BB, the structurally similar catecholate siderophore enterobactin (Ent) also activated the feuA promoter (Fig. 3A) and this was also Btr-dependent (data not shown). Like Fe-BB, Fe-Ent is transported predominantly through FeuABC (Dertz et al., 2006b; Ollinger et al., 2006).
Figure 3. Btr is required for expression of the feuABCyybA operon.
A. Expression from the feuA promoter region was assayed using an feuA-lacZ transcriptional fusion. The BB non-producing strains (sfp0) were grown in FS-MM and induced by addition of either bacillibactin (BB) or enterobactin (Ent) at 5 μM. Btr was required both for the basal level expression observed in this iron starvation medium and for the siderophore-induced expression.
B. Elevated expression is observed in BB producing strains, even in the absence of added BB.
BB is synthesized by the nonribosomal peptide synthetase complex encoded by the dhb operon, exported from the cell by an unknown mechanism, and then the Fe-BB complex is internalized by the FeuABC transporter and cleaved by the BesA esterase to release iron (Miethke et al., 2006). To determine if induction of feuA expression is only responsive to Fe-BB taken up from the medium, we measured feuA promoter activity in a strain that can produce BB, but is defective in uptake (sfp+ feuA). Expression was slightly reduced relative to transport competent strains, but was reproducibly elevated relative to cells unable to synthesize BB (Fig. 3B vs. 3A; last column). This suggests that BB can activate the expression of its cognate uptake system independent of import into the cell, and therefore presumably prior to export. Since the BB exporter is not yet identified, it was not possible to test the effect on induction of blocking export. Mutations of the besA esterase or the putative esterase encoded in the feuA operon (ybbA) did not significantly affect Btr-dependent activation of the feuA operon (data not shown).
Btr binds to the feuA regulatory region
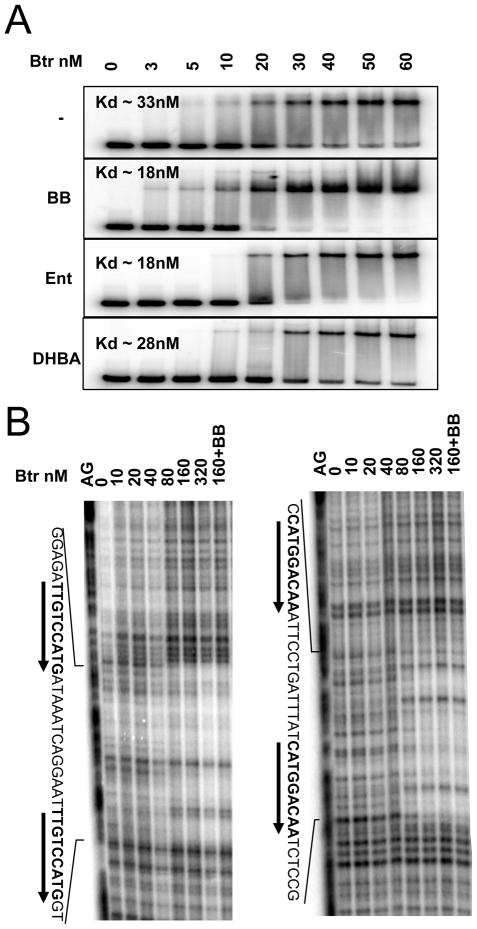
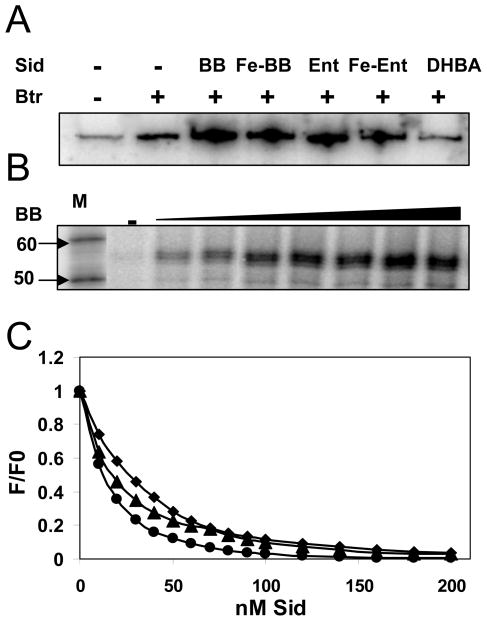
Purified Btr bound in vitro to the feuA regulatory region in electrophoretic mobility shift assays (EMSA) with an apparent Kd of ~33 nM (Fig 4A). The affinity of Btr for the feuA promoter region was enhanced ~2-fold by either BB or Ent, but not by DHBA. The Btr binding site overlaps the −35 element of the feuA promoter, as shown by DNase I footprinting (Fig. 4B). The 38 bp protected region contains a perfect direct repeat (TTGTCCATG), with a 13 base spacer (Fig. 1B and Fig. 4B). Incubation of Btr with BB prior to the footprinting reaction did not obviously change the location of the binding site or the cleavage pattern (Fig 4B).
Figure 4. Btr binds tightly and specifically to the feuA regulatory region.
A. EMSA analysis of Btr binding in the absence or presence of siderophores. Measured DNA- binding affinities (Kd) are indicated in the insets.
B. DNase I footprinting of Btr binding to the feuA regulatory region on the sense (left) and anti-sense (right) strands. Btr binds to a direct repeat overlapping the −35 region (Fig. 1B). Protected regions were indexed against the A+G chemical sequencing ladder which was itself compared against dideoxy-sequencing reactions (data not shown).
Btr activates transcription in response to siderophores
Btr is sufficient to activate transcription of feuA in vitro. In the absence of Btr, there was a low level of specific transcript in run-off transcription assays (Fig. 5A, lane 1). Addition of Btr alone led to a slight, but reproducible increase in transcription (lane 2). In the presence of Btr and a cognate siderophore, there was an ~3-fold increase in the yield of the run-off transcript (lanes 3–6). Consistent with the in vivo results, DHBA itself was unable to activate transcription (lane 7). In titration experiments using BB, high levels of transcription activation were achieved with concentrations in the 100–200 nanomolar range (Fig. 5B). This suggests that Btr binds BB with comparable avidity as FeuA itself (Miethke et al., 2006).
Figure 5. Siderophore-dependent induction of feuA transcription is mediated by Btr.
A. In vitro transcription from the feuA promoter is weakly stimulated by Btr (400 nM; second lane) and strongly stimulated by Btr in the presence of the siderophores (Sid), BB or Ent.
B. Btr-dependent transcriptional activation is highly sensitive to BB (increasing concentrations from 50–600 nM). RNA size markers (50 and 60 nt) are indicated to the left.
C. Siderophores bind tightly to Btr as monitored by fluorescence emission quenching (F/Fo). Siderophores (Sid) included BB (◆), Fe-BB (●), and Fe-Ent (▲) and were added at the indicated concentrations. Apparent Kd values were determined using non-linear regression analysis as ~27, 15, and 20 nM, respectively.
Btr binds bacillibactin with high affinity
It has been previously shown that Fe-BB, and to a lesser extent BB, quenches the intrinsic fluorescence of the FeuA SBP (Miethke et al., 2006). When the FeuA-like SBP domain of Btr was modeled using as a template the known structure of E. coli FhuD (Clarke et al., 2000; Clarke et al., 2002) the presumed siderophore-binding cleft contained 3 conserved Trp residues, including one also present in FeuA (Supp. Figs. S1-S3). Nearly complete quenching of Btr Trp fluorescence was observed in the presence of BB, Fe-BB, or Fe-Ent. The highest affinity binding was observed for Fe-BB (Fig. 5C). Indeed, Btr bound to both BB (Kd ~27 nM) and Fe-BB (Kd ~15 nM) with higher affinity than that reported for the extracellular Fe-BB receptor lipoprotein FeuA (Kd of ~190 nM for BB and 57 nM for Fe-BB; Miethke et al., 2006).
Recognition of Fe-BB and Fe-Ent can be genetically separated
The strong fluorescence quenching upon binding of siderophores is consistent with an interaction in the Trp-rich substrate binding cleft of the FeuA-like domain. The ability of both Fe-BB and Fe-Ent to be transported by the same ABC transporter, and to interact productively with the same regulatory protein (Btr), is consistent with the fact that both are cyclic, trimeric lactones. However, these two complexes have opposite chirality in solution (∧ for Fe-BB and Δ for Fe-Ent; Bluhm et al., 2002). In addition, BB is based on a cyclic trimeric lactone ring of L-Thr residues and there is a Gly spacer between the Thr and 2,3-dihydroxybenzoate moieties. In contrast, Ent is based on a trimeric L-Ser lactone ring. These differences suggest that Fe-BB and Fe-Ent may make distinct sets of contacts with Btr.
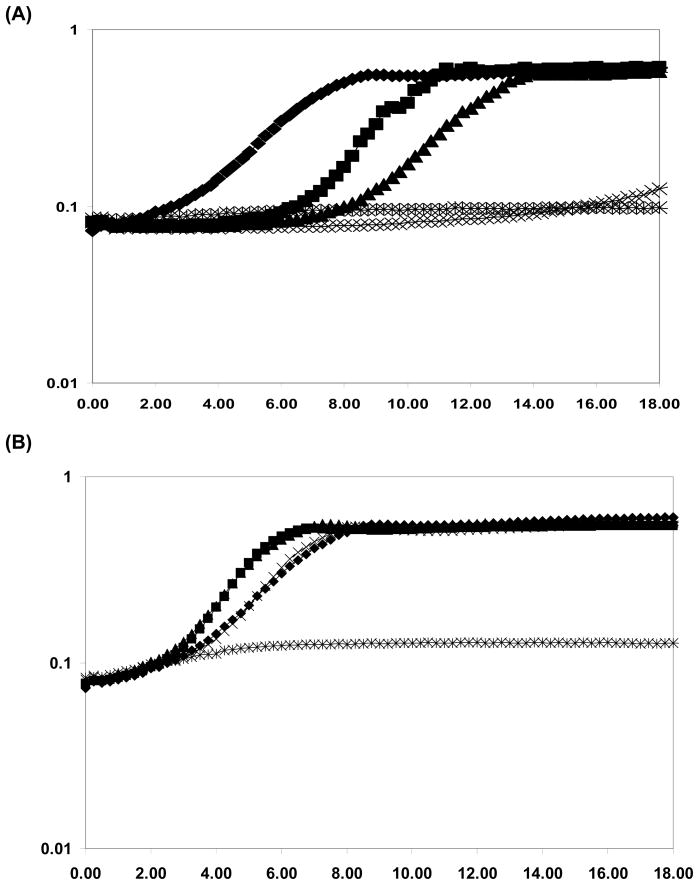
To test this hypothesis we generated a series of mutant proteins with one more of the three conserved Trp residues substituted by Ala. Growth experiments demonstrate that the btr W385A mutant strain retained the ability to use Fe-Ent as an iron source, but lost the ability to grow in the presence of Fe-BB (Fig. 6). In contrast, the btr W296A mutant strain retained the ability to grow with both siderophores, although at a reduced rate in the case of Fe-BB. The inability of the btr W385A mutant strain to grow in the presence of Fe-BB is likely due to ineffective activation of the feuA transport operon: an inference supported by measurements of feuA promoter activity under these conditions. In FS-MM, wild-type Btr efficiently activated a feuA-lacZ fusion in response to either Fe-BB (187 Miller units) or Fe-Ent (265 Miller units). Similarly, the Btr W296A mutant protein responded to both Fe-BB and Fe-Ent (151 and 136 Miller units, respectively). In contrast, the Btr W385A responded well to Fe-Ent (137 Miller units), but not to Fe-BB (41 Miller units).
Figure 6. Genetic separation of BB and Ent recognition by Btr.
A. Growth curves of the btr mutant strain complemented with various btr alleles. The btr mutant strain is unable to grow in FS-MM containing 10 μM EDDHA+ 5 μM BB (✳) whereas the btr mutant complemented with the FLAG-tagged wild type Btr grows well under these conditions (□). Note that if EDDHA and BB are omitted from the growth medium, the complemented strain also grows well (◆). Strains complemented with btr W296A(▲), but not those complemented with btr W385A (×), grow in the presence of 10 μM EDDHA+ 5 μM BB.
B. Growth curves of the btr mutant strain complemented with various btr alleles are shown as in panel A but in medium containing 5 μM Ent instead of BB. Note that in this medium both the btr W296A and btr W385A alleles support robust growth.
Discussion
In response to iron deprivation, many bacteria synthesize siderophores together with their cognate uptake systems (Andrews et al., 2003; Moore and Helmann, 2005). In several well characterized systems, the cell has the potential to synthesize several different siderophores that may differ both in chemical properties and affinity for ferric iron. The ability of a siderophore to promote iron nutrition depends not only on its ability to access environmental iron, but also its ability to deliver iron to the producer cell. Delivery can be compromised by competition from other organisms since many bacteria can efficiently internalize ferri-siderophore complexes that they themselves do not synthesize. In addition, in the mammalian host siderocalin binds tightly to many catecholate siderophores thereby rendering them unable to promote bacterial growth (Goetz et al., 2002). To optimize their ability to obtain environmental iron, some bacteria have evolved systems to prioritize the production and transport of specific siderophores. As a result, many siderophore systems are regulated at two different levels: iron-dependent repression mediated by the ferric uptake regulator (Fur) and siderophore-specific induction (Braun and Mahren, 2005; Brickman et al., 2007; Visca et al., 2002).
At least two distinct types of regulatory systems have been described that mediate substrate (siderophore)-induction. In the first class, siderophore systems are induced in response to the extracellular ferri-siderophore complexes sensed by a cell surface (outer membrane) receptor that then triggers activation of an alternative σ factor of the extracytoplasmic function (ECF) family (Helmann, 2002). Well characterized examples include the induction of ferric citrate uptake in E. coli (Van Hove et al., 1990) and pyoverdine uptake in Pseudomonas spp. (for a recent review see Visca et al., 2002). In E. coli, induction involves the interaction between the FecA receptor and the FecR membrane protein which in turn regulates the ECF σ FecI. In general, these systems respond selectively to the bound ferri-siderophore complex rather than apo-siderophore. Thus, the further induction of receptor synthesis responds to the ability of the siderophore to successfully scavenge iron and bring it to the receptor.
The second class involves the “iron subfamily” of AraC-type transcription activators. In these systems, an AraC-type regulator mediates the siderophore-dependent induction of the uptake system and, in some cases, of the corresponding biosynthetic operon. Siderophore-mediated regulation by AraC-type transcription factors has been best documented in Yersinia pestis (YbtA), P. aeruginosa (PchR), and Bordetella spp. (AlcR and BfeR) and similar regulators have been described in other Pseudomonads (PdtC, QbsA), Sinorhizobium meliloti (RhrA) and Vibrio vulnificans (DesR) (Anderson and Armstrong, 2004; Beaumont et al., 1998; Fetherston et al., 1996; Heinrichs and Poole, 1993; Lynch et al., 2001; Pelludat et al., 1998). The few AraC family regulators studied to date appear to respond to a broad range of chemical inducers. For example, BfeR responds to the neuroendocrine catecholamines epinephrine, norepinephrine, and dopamine (Anderson and Armstrong, 2006).
The molecular basis of siderophore recognition has yet to be defined for any of the iron-subfamily AraC regulators. In all previously described examples, the regulator contains a C-terminal AraC DNA-binding domain and a more divergent N-terminal region likely to mediate inducer recognition (Brickman and Armstrong, 2002). In general, there has been little detailed biochemical study of these siderophore-sensing regulators and the inducer binding sites have not been defined.
Here we describe Btr, the first reported intracellular siderophore sensor from a Gram-positive bacterium. Btr is encoded adjacent to the bacillibactin transport system and appears to have evolved by duplication and appropriation of the gene encoding the feuA-encoded SBP to encode a siderophore-specific sensing domain appended to an AraC-type DNA-binding domain. As a result, Btr is significantly longer (529 amino acids) than previously described iron-subfamily regulators (250–300 aa). Thus, Btr represents a novel solution to the problem of coupling siderophore recognition with transcription activation in a one-component regulator (Ulrich et al., 2005). Orthologs of Btr are present in several different Bacilli and sequence alignments of the Btr orthologs reveal high levels of similarity in both the SBP and DNA-binding domains (from 44%–65% identity; Supp. Fig. S2). In contrast, the N terminal region has diverged significantly (< 23% identity). This region, proposed to function in inducer recognition in other siderophore-sensing AraC family members (Gallegos et al., 1997), has been functionally replaced by the C-terminal BB binding domain.
Btr binds to a conserved direct repeat element (ttgTCCATG -N13-ttgTCCATG; Fig. 1B) upstream of the feuA promoter. Searches of the B. subtilis 168 genome failed to identify additional candidate Btr-binding sites, suggesting that the feuABCybbA operon may be the only target for Btr regulation. Btr binds tightly to this DNA regulatory site in both the absence and presence of BB. Indeed, derepression of Btr upon iron starvation leads to the basal expression of the FeuABC transport system and thereby primes the cell to be able to import Fe-BB (or Fe-Ent) present in the environment. The import of either of these siderophores can trigger further induction of the FeuABC transport system, even if the cell is itself unable to synthesize BB (as in sfp0 strains). In vitro studies confirm that both apo- and Fe-BB stimulate transcription. The FeuABC transporter is dispensable for induction in strains that produce BB which suggests that apo-BB is also sufficient for induction in vivo. These results lead us to suggest that the binding of BB (or Fe-BB) to the SBP domain of Btr alters the protein conformation to enhance DNA-binding and to more efficiently activate transcription.
Experimental Procedures
Strain construction and growth conditions
All strains used in this study are listed in Table 1 and oligonucleotide primers are in the Supplemental Material. For selection, antibiotics were added at the following concentrations: erythromycin (1μg/ml) and lincomycin (25μg/ml) (for selecting for macrolide-lincosamide-streptogramin B (MLS) resistance), spectinomycin (100μg/ml), chloramphenicol (10μg/ml), kanamycin (15μg/ml) and neomycin (10μg/ml). Growth curves were done using a Bioscreen C MBR system for 24 hours with OD600 measurements every 10 min. Ent was purchased from EMC microcollections GmbH (Germany).
Table 1.
Strains used in the study:
| Strain | Genotype | Source or reference |
|---|---|---|
| CU1065 | trpC2 att SPβ sfp0 | Lab stock |
| HB5800 | CU1065 sfp+ | Ollinger et al., 2006 |
| HB5714 | HB5800 yuiI::spc | Ollinger et al., 2006 |
| HB8240 | HB5800 ybbA::kan | This study |
| HB8242 | HB5800 fur::kan | This study |
| HB2501 | CU1065 fur::kan | Fuangthong et al., 2002 |
| HB8246 | CU1065 btr::spc | This study |
| HB8247 | HB5800 btr::spc | This study |
| HB8248 | HB2501 btr::spc | This study |
| HB8249 | HB8242 btr::spc | This study |
| HB8254 | CU1065 SPβc2Δ2::Tn917::φ(feuA-cat-lacZ) | This study |
| HB8255 | CU1065 sfp+SPβc2Δ2::Tn917::φ (feuA-cat-lacZ) | This study |
| HB8256 | CU1065 fur::kan SPβc2Δ2::Tn917::φ (feuA- cat-lacZ) | This study |
| HB8257 | CU1065 sfp+ fur::kan SPβc2Δ2::Tn917::φ (feuA- cat-lacZ) | This study |
| HB8258 | CU1065 btr::spc SPβc2Δ2::Tn917::φ (feuA-cat- lacZ) | This study |
| HB8259 | CU1065 sfp+ btr::spc SPβc2Δ2::Tn917::φ (feuA- cat-lacZ) | This study |
| HB8260 | CU1065 fur::kan btr::spc SPβc2Δ2::Tn917::φ (feuA-cat-lacZ) | This study |
| HB8261 | CU1065 sfp+ fur::kan btr::spc SPβc2Δ2::Tn917::φ (feuA-cat-lacZ) | This study |
| HB8262 | ZB307A SPβc2Δ2::Tn917::φ (feuA-cat-lacZ) | This study |
| HB8263 | CU1065 feuA::cat | This study |
| HB8266 | CU1065 feuA::cat SPβc2Δ2::Tn917::φ (feuA- cat-lacZ) | This study |
| HB8267 | CU1065 sfp+ feuA::cat | This study |
| HB8280 | HB5800 yuiI::spc SPβc2Δ2::Tn917::φ (feuA-cat- lacZ) | This study |
| HB8281 | CU1065 sfp+ SPβc2Δ2::Tn917::φ (feuA-cat- lacZ) | This study |
| HB8282 | CU1065 sfp+ ybbA::kan SPβc2Δ2::Tn917::φ (feuA-cat-lacZ) | This study |
| HB8283 | CU1065 sfp+ feuA::catSPβc2Δ2::Tn917::φ (feuA-cat-lacZ) | This study |
| HE8242 | E. coli BL21 pLYS S contains btr in pET16b | This study |
Routine molecular biology procedures were done using E. coli DH5α for routine DNA cloning as described (Sambrook and Russel, 2001). Isolation of B. subtilis chromosomal DNA, transformation and specialized SPβ transduction were done according to Cutting and Vander Horn (Cutting and VanderHorn, 1990). Restriction enzymes, DNA ligase and DNA polymerases were used according to the manufacturer’s instructions (New England Biolabs).
B. subtilis was grown in LB or in a MOPS-based, iron-starvation minimal medium, FS-MM (Chen et al., 1993). Metals were added from filter-sterilized stocks before inoculation. Unless otherwise indicated, liquid media were inoculated from an overnight pre-culture and incubated at 37°C with shaking at 200 rpm.
Mutants in btr and ybbA were constructed using long-flanking-homology polymerase chain reaction (LFH-PCR) as described (Butcher and Helmann, 2006). To construct an feuA-lacZ transcriptional fusion the feuA regulatory region was amplified from genomic DNA by PCR and cloned as a HindIII-BamHI fragment into pJPM122 (Slack et al., 1993). The resulting construct was linearized with ScaI and transformed into ZB307A (Zuber and Losick, 1987) selecting for Neomycin resistance. An SPβ transducing lysate was prepared by heat induction and used to transduce different strain backgrounds as indicated (SI Table 1) (Slack et al., 1993). β-galactosidase activity was assayed using a modification of the procedure of Miller (Miller, 1972) as described in (Bsat et al., 1998).
Overproduction and purification of Btr
The Btr coding region was amplified from the B. subtilis CU1065 genome. The resulting fragment was digested with NdeI and BamHI and cloned into pET16B (Novagen) since preliminary studies revealed that the N-terminal His tag increased protein solubility in E. coli. After sequence confirmation, the resulting plasmid was used to transform E.coli BL21(DE3)(pLysS). A single colony was grown overnight in LB containing ampicillin (200 μg/ml). The overnight culture was used to inoculate 2 l LB medium containing ampicillin (200μg/ml), and the flask was incubated at 37°C with vigorous shaking to an OD600 of 0.6. Isopropyl-β-D-thiogalactopyranoside was added to 1 mM (final), and the cells were harvested after further incubation for 3h at 25°C.
The cell pellet was suspended in 10 ml of 50 mM NaH2PO4, 5 mM Tris, pH 8.0, 20 mM imidazole, 2 mM DTT, 300 mM NaCl, and 5% glycerol, and the cells were broken by sonication. The soluble fraction was collected and purified using Ni-NTA beads (Qiagen) according to the manufacturer’s instruction. Samples were analyzedon 12% SDS/PAGE to identify fractions that contained Btr and dialyzed overnight against 50 mM Tris, pH 8.0, 100 mMNaCl pH 8.0, 2 mM DTT and 5% glycerol. The proteins were concentrated by ultrafiltration and injected on a Superdex 200 FPLC column using the same buffer. The fractions containing Btr were concentrated using ultra-filtration and stored at −20°C in 50 mM Tris, pH 8.0, 100 mMNaCl pH 8.0, 2 mM DTT with 50% glycerol. Note that protein could not be concentrated above 2 μM because of precipitation.
DNaseI footprinting and DNA-binding assays
The feuA promoter region was amplified from B. subtilis chromosomal DNA by PCR using a [γ-32P ATP] labeled primer, and DNaseI footprinting done as previously described (Fuangthong and Helmann, 2002). EMSAs were performed using DNA (<100 pM) essentially as described (Gaballa and Helmann, 1998) with the exception that the binding buffer was 20mM Tris pH8.0, 50 mM NaCl, 50 mM KCl, 5% glycerol 5μg/ml Salmon sperm DNA, 2mMDTT.
Determination of the feuA transcription start site
The feuA transcription start site was determined by RACE using the 5′ RACE kit from Invitrogen according to the manufacturer’s instructions.
Purification of BB
Purification of BB was done as described (Dertz et al., 2006a) with a few modifications. Briefly, B. subtilis was grown in FS-MM for 48h. The culture supernatant fraction was acidified to pH 3 with HCl and extracted 3 times with ethyl acetate (200 ml). The pooled ethyl acetate fractions were dried over NaSO4, filtered and dried using rotary evaporation. The resulting material was dissolved in a minimal volume of methanol and added drop-wise into a stirring beaker of ether. The precipitate was removed by centrifugation and the clear solution was dried and dissolved in water. The soluble fraction was loaded onto a reverse-phase C18 column that was pre-equilibrated with methanol and washed thoroughly with water. BB was eluted using 50% methanol, dried, and resuspended in water. After saturation, residual BB was removed by centrifugation and dissolved in a minimal volume of dimethyl formamide. The concentration of the BB was determined spectrophotometrically after iron addition (ε490= 4700 M−1 cm−1 for Fe-BB). Fe-BB was formed by adding equimolar amounts of FeCl3 (in 0.1 N HCl) to BB, neutralization, and removal of excess Fe by one step purification using a C18 column.
In vitro transcription
The in vitro transcription reactions were done using 50 ng (5 nM) of feuA promoter fragment PCR (300 bp) in transcription buffer (20mM Tris HCl pH 8.0, 20 mM KCl, 2%glycerol, 0.5 mM DTT, 0.1 mg/ml acetylated BSA, 20 mM MgCl2 and 10 U/reaction of RNasin, RNAse inhibitor). Btr with or without BB was added to the DNA prior to RNA polymerase and incubated on ice for 15 min. The B. subtilis RNA polymerase and σA were purified using modifications of published procedures as summarized in (Helmann, 2003). Briefly, RNA polymerase was purified from a B. subtilis strain containing a His-tagged beta-prime subunit using polymin P precipitation and elution, metal ion affinity chromatography, and a Superdex 200 size exclusion column. The purified RNA polymerase was associated with sub-stoichiometric amounts of σA. σA-saturated holoenzyme was re-constituted by mixing purified RNA polymerase with purifiedσA (1:5 molar ratio) in transcription buffer and incubating on ice for 15 min. RNA polymerase was added to a final concentration of 40 nM and incubated for 15 min at 37°C. The reaction was started by adding an NTP mixture containing 0.25 mM of ATP, CTP, GTP and 0.025 mM UTP and 2.5 μCi of [α-32P UTP]. The reactions were incubated for 30 min at 37°C. The reaction products were denatured at 90°C and separated on 6% denaturing polyacrylamide sequencing gel with a DECADE RNA marker (Ambion). In some cases, the RNA products were ethanol precipitated in the presence of sodium acetate and glycogen. The RNA was washed with 70% cold ethanol, dried and dissolved in formamide containing loading dye.
Fluorescence spectroscopy
Intrinsic fluorescence measurements were obtained using a Perkin-Elmer LS55 spectrofluorometer with a band width of 10 nm. The optimal excitation and emission wavelengths were 275 and 554 nm, respectively. Measurements were made with 50 nM of Btr in 20 mM Tris, pH 8.0, 5% glycerol, 100 mM NaCl and 2 mM DTT.
Site-directed mutagenesis of Btr
An N-terminal FLAG tag was added to Btr by PCR. In brief, the btr ORF was amplified by PCR using primers Btr-F down-FLAG (containing the FLAG tag encoding sequence) and Btr-R (see Supplemental Material for primer sequences). The btr promoter region was amplified using primers Btr-F-up and Btr-R-up-flag which is complementary to Btr-F down-flag primer. The two fragments were fused together by overlap extension PCR (Ho et al., 1989). Site directed mutagenesis for W296, W365 and W385 was carried out using a similar procedure. The PCR fragments were cloned into pDG1664 vector (Guerout-Fleury et al., 1996) and integrated into a btr mutant strain at the thrC locus by double crossover recombination. In this strain, btr (and relevant mutant variants) are expressed under control of the native promoter.
Supplementary Material
Acknowledgments
We thank Dr. K. Raymond for advice on BB purification, M. Miethke and M. Marahiel for communicating results prior to publication, and S. MacLellan for B. subtilis RNA polymerase. This work was supported by a grant from the NIH (GM-59323)
References
- Anderson MT, Armstrong SK. The BfeR regulator mediates enterobactin-inducible expression of Bordetella enterobactin utilization genes. J Bacteriol. 2004;186:7302–7311. doi: 10.1128/JB.186.21.7302-7311.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MT, Armstrong SK. The Bordetella bfe system: growth and transcriptional response to siderophores, catechols, and neuroendocrine catecholamines. J Bacteriol. 2006;188:5731–5740. doi: 10.1128/JB.00495-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Baichoo N, Helmann JD. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol. 2002;184:5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baichoo N, Wang T, Ye R, Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol. 2002;45:1613–1629. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- Beaumont FC, Kang HY, Brickman TJ, Armstrong SK. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport in Bordetella pertussis and Bordetella bronchiseptica. J Bacteriol. 1998;180:862–870. doi: 10.1128/jb.180.4.862-870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluhm ME, Kim SS, Dertz EA, Raymond KN. Corynebactin and enterobactin: related siderophores of opposite chirality. J Am Chem Soc. 2002;124:2436–2437. doi: 10.1021/ja016651s. [DOI] [PubMed] [Google Scholar]
- Braun V, Mahren S. Transmembrane transcriptional control (surface signalling) of the Escherichia coli Fec type. FEMS Microbiol Rev. 2005;29:673–684. doi: 10.1016/j.femsre.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Brickman TJ, Armstrong SK. Bordetella interspecies allelic variation in AlcR inducer requirements: identification of a critical determinant of AlcR inducer responsiveness and construction of an alcR(Con) mutant allele. J Bacteriol. 2002;184:1530–1539. doi: 10.1128/JB.184.6.1530-1539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Anderson MT, Armstrong SK. Bordetella iron transport and virulence. Biometals. 2007 doi: 10.1007/s10534-006-9031-1. [DOI] [PubMed] [Google Scholar]
- Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- Butcher BG, Helmann JD. Identification of Bacillus subtilisσW-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol Microbiol. 2006;60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- Chen L, James LP, Helmann JD. Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions. J Bacteriol. 1993;175:5428–5437. doi: 10.1128/jb.175.17.5428-5437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TE, Ku SY, Dougan DR, Vogel HJ, Tari LW. The structure of the ferric siderophore binding protein FhuD complexed with gallichrome. Nat Struct Biol. 2000;7:287–291. doi: 10.1038/74048. [DOI] [PubMed] [Google Scholar]
- Clarke TE, Rohrbach MR, Tari LW, Vogel HJ, Koster W. Ferric hydroxamate binding protein FhuD from Escherichia coli: mutants in conserved and nonconserved regions. Biometals. 2002;15:121–131. doi: 10.1023/a:1015249530156. [DOI] [PubMed] [Google Scholar]
- Cutting SM, VanderHorn PB. Molecular Biological Methods for Bacillus. Chichester: John Wiley and Sons, Ltd; 1990. Genetic Analysis. [Google Scholar]
- Dertz EA, Stintzi A, Raymond KN. Siderophore-mediated iron transport in Bacillus subtilis and Corynebacterium glutamicum. J Biol Inorg Chem. 2006a doi: 10.1007/s00775-006-0151-4. [DOI] [PubMed] [Google Scholar]
- Dertz EA, Xu J, Stintzi A, Raymond KN. Bacillibactin-mediated iron transport in Bacillus subtilis. J Am Chem Soc. 2006b;128:22–23. doi: 10.1021/ja055898c. [DOI] [PubMed] [Google Scholar]
- Fetherston JD, Bearden SW, Perry RD. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol. 1996;22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- Fuangthong M, Helmann JD. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc Natl Acad Sci U S A. 2002;99:6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Helmann JD. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Heinrichs DE, Poole K. Cloning and sequence analysis of a gene (pchR) encoding an AraC family activator of pyochelin and ferripyochelin receptor synthesis in Pseudomonas aeruginosa. J Bacteriol. 1993;175:5882–5889. doi: 10.1128/jb.175.18.5882-5889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- Helmann JD. Purification of Bacillus subtilis RNA polymerase and associated factors. Methods Enzymol. 2003;370:10–24. doi: 10.1016/S0076-6879(03)70002-0. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Ito T, Neilands JB. Products of “Low-iron Fermentation” with Bacillus subtilis: Isolation, Characterization and Synthesis of 2,3-Dihydroxybenzoylglycine. Journal of the American Chemical Society. 1958;80:4645–4647. [Google Scholar]
- Koster W, Braun V. Iron (III) hydroxamate transport into Escherichia coli. Substrate binding to the periplasmic FhuD protein. J Biol Chem. 1990;265:21407–21410. [PubMed] [Google Scholar]
- Lynch D, O’Brien J, Welch T, Clarke P, Cuiv PO, Crosa JH, O’Connell M. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J Bacteriol. 2001;183:2576–2585. doi: 10.1128/JB.183.8.2576-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JJ, Wendrich TM, Marahiel MA. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J Biol Chem. 2001;276:7209–7217. doi: 10.1074/jbc.M009140200. [DOI] [PubMed] [Google Scholar]
- Miethke M, Klotz O, Linne U, May JJ, Beckering CL, Marahiel MA. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol Microbiol. 2006 doi: 10.1111/j.1365-2958.2006.05321.x. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Moore CM, Helmann JD. Metal ion homeostasis in Bacillus subtilis. Curr Opin Microbiol. 2005;8:188–195. doi: 10.1016/j.mib.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Neilands JB. Siderophores. Arch Biochem Biophys. 1993;302:1–3. doi: 10.1006/abbi.1993.1172. [DOI] [PubMed] [Google Scholar]
- Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. Role of the Fur regulon in iron transport in Bacillus subtilis. J Bacteriol. 2006;188:3664–3673. doi: 10.1128/JB.188.10.3664-3673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelludat C, Rakin A, Jacobi CA, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- Schneider R, Hantke K. Iron-hydroxamate uptake systems in Bacillus subtilis: identification of a lipoprotein as part of a binding protein-dependent transport system. Mol Microbiol. 1993;8:111–121. doi: 10.1111/j.1365-2958.1993.tb01208.x. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Mueller JP, Sonenshein AL. Mutations that relieve nutritional repression of the Bacillus subtilis dipeptide permease operon. J Bacteriol. 1993;175:4605–4614. doi: 10.1128/jb.175.15.4605-4614.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hove B, Staudenmaier H, Braun V. Novel two-component transmembrane transcription control: regulation of iron dicitrate transport in Escherichia coli K-12. J Bacteriol. 1990;172:6749–6758. doi: 10.1128/jb.172.12.6749-6758.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visca P, Leoni L, Wilson MJ, Lamont IL. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol Microbiol. 2002;45:1177–1190. doi: 10.1046/j.1365-2958.2002.03088.x. [DOI] [PubMed] [Google Scholar]
- Zuber P, Losick R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987;169:2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.