SUMMARY
Light and brassinosteroid (BR) antagonistically regulate the developmental switch from etiolation in the dark to photomorphogenesis in the light in plants. Here we identify GATA2 as a key transcriptional regulator that mediates the crosstalk between BR- and light-signaling pathways. Overexpression of GATA2 causes constitutive photomorphogenesis in the dark, whereas suppression of GATA2 reduces photomorphogenesis caused by light, BR deficiency, or the constitutive photomorphogenesis mutant cop1. Genome profiling and chromatin immunoprecipitation experiments show that GATA2 directly regulates genes that respond to both light and BR. BR represses GATA2 transcription through the BR-activated transcription factor BZR1, whereas light causes accumulation of GATA2 protein and feedback inhibition of GATA2 transcription. Dark-induced proteasomal degradation of GATA2 is dependent on the COP1 E3 ubiquitin ligase, and COP1 can ubiquitinate GATA2 in vitro. This study illustrates a molecular framework for antagonistic regulation of gene expression and seedling photomorphogenesis by BR and light.
INTRODUCTION
Light and brassinosteroid (BR) are key signals that determine the development program of young seedlings. To reach the surface of soil, seedlings that germinate in the dark undergo skotomorphogenesis, exhibiting elongated hypocotyls, small and folded cotyledons with undifferentiated chloroplasts, and repression of light-induced genes. Exposure to light causes a developmental switch from skotomorphogenesis to photomorphogenesis, resulting in short hypocotyls, open and expanded cotyledons, and differentiation of chloroplast (Wei and Deng, 1996). Genetic studies have identified many components that mediate this developmental switch by light. Two classes of photoreceptors, phytochrome and cryptochrome, perceiving red/far-red and blue light respectively, play major roles in promoting photomorphogenesis. A group of proteins termed CONSTITUTIVE PHOTOMORPHOGENIC/DE-ETIOLATED/FUSCA (COP/DET/FUS), which are components of the ubiquitination system or COP9 signalosome, are central repressors of photomorphogenesis (Deng et al., 1991; Wei and Deng, 1996). Several classes of transcription factors, such as the b-zip protein LONG HYPOCOTYL 5 (HY5) and the Phytochrome Interacting Factor (PIF) family of b-HLH proteins, directly regulate light-responsive gene expression and are degraded by the ubiquitin system in a light-dependent manner (Chen et al., 2004; Leivar et al., 2008; Ma et al., 2002; von Arnim et al., 1997; Wang et al., 2001). Through these components, light turn on a transcription program that supports photomorphogenic development (Chen et al., 2004; Jiao et al., 2007).
In addition to these light-signaling components, BR also plays a key role in photomorphogenesis. BR deficient mutants show typical de-etiolation phenotypes in the dark with elevated expression of many light-induced genes (Chory et al., 1991; Li et al., 1996; Song et al., 2009; Szekeres et al., 1996). While light inhibits hypocotyl elongation and promotes chlorophyll accumulation, BR promotes hypocotyl elongation and reduces chlorophyll level. BR is perceived by the cell surface receptor kinase BRI1 and downstream signal transduction activates the BZR family transcription factors (Gendron and Wang, 2007), which mediate BR-responsive gene expression (He et al., 2005). Recent studies have established a complete BR signal transduction pathway from the BRI1 to the BZR transcription factors (Kim et al., 2009; Tang et al., 2010; Kim and Wang, 2010). Activation of BZR1 and BZR2 is essential for skotomorphogenesis, as the constitutive photomorphogenesis phenotype of BR-deficient or insensitive mutants are suppressed by the dominant bzr1-1D and bes1-D mutations, which cause constitutive activation of BR-responsive gene expression (Wang et al., 2002; Yin et al., 2002). It has been proposed that light might inhibit BR synthesis or signaling to inhibit skotomorphogenesis and promote photomorphogenesis (Kang et al., 2001). However, no significant difference in BR level was observed between dark-grown and light-grown plants (Symons et al., 2008). On the other hand, physiological studies of BR-deficient Arabidopsis suggested that BR regulates phytochrome- and cryptochrome-mediated responses (Luccioni et al., 2002; Neff et al., 1999). The molecular mechanism of such BR-light interactions has remained unclear.
Analyses of light-responsive promoters have identified a number of light-response promoter elements (LREs), including the G-box, GATA and GT1 motifs (Terzaghi and Cashmore, 1995). It has been suggested that combinations of LREs, rather than individual elements, confer proper light-responsiveness to a promoter (Puente et al., 1996; Terzaghi and Cashmore, 1995). For example, the combination of G-box with GATA element is critical for promoter activation in response to the signals from multiple photoreceptors as well as for repression by the COP/DET system (Chattopadhyay et al., 1998b). Most of the light-signaling transcription factors identified so far bind to the G-box (Liu et al., 2008; Jiao et al., 2007). The transcription factor that regulates light-responsive genes through the essential GATA element has not been identified in plants (Arguello-Astorga and Herrera-Estrella, 1998; Chattopadhyay et al., 1998b; Jiao et al., 2007; Terzaghi and Cashmore, 1995). In fungi, such as Neurospora, two GATA-type factors bind to GATA element and regulate gene expression in response to light signal (Scazzocchio, 2000). It has long been proposed that members of the Arabidopsis GATA family of transcription factors might play a similar role (Jeong and Shih, 2003; Manfield et al., 2007), however, genetic evidence for this hypothesis is absent.
In this study, we identify a GATA-type transcription factor (GATA2) as a junction between light and BR pathways. Overexpression and loss-of-function experiments demonstrate that GATA2 is a major positive regulator of photomorphogenesis that mediates a gene expression profile with significant overlap to those caused by light treatment or BR-deficiency. BR-activated BZR1 directly represses GATA2 transcription, whereas light signaling stabilizes the GATA2 protein, likely by inhibiting a COP1-dependent degradation process. The results demonstrate that GATA2 is not only a key light-signaling transcription factor but also a junction for the crosstalk between the BR and light-signaling pathways. The results support a mode of BR-light antagonism through transcriptional and posttranslational regulation of common transcription factors.
RESULTS
GATA2 is a positive regulator of photomorphogenesis
The suppression of the photomorphogenesis phenotype of bri1 by the bzr1-1D mutation suggests that BR inhibits photomorphogenesis through BZR1 and its downstream target genes. Based on BR responsive expression and the presence of BR-response elements in their promoters (He et al., 2005), two BR-repressed genes encoding GATA-type transcription factors, GATA2 and GATA4 were considered putative target genes of BZR1. Since previous studies of GATA sequence in light-responsive promoter implicated unknown GATA factors in light-responsive gene expression (Chattopadhyay et al., 1998b), we tested whether GATA2 and GATA4 play a role in light- or BR-regulated gene expression and photomorphogenesis.
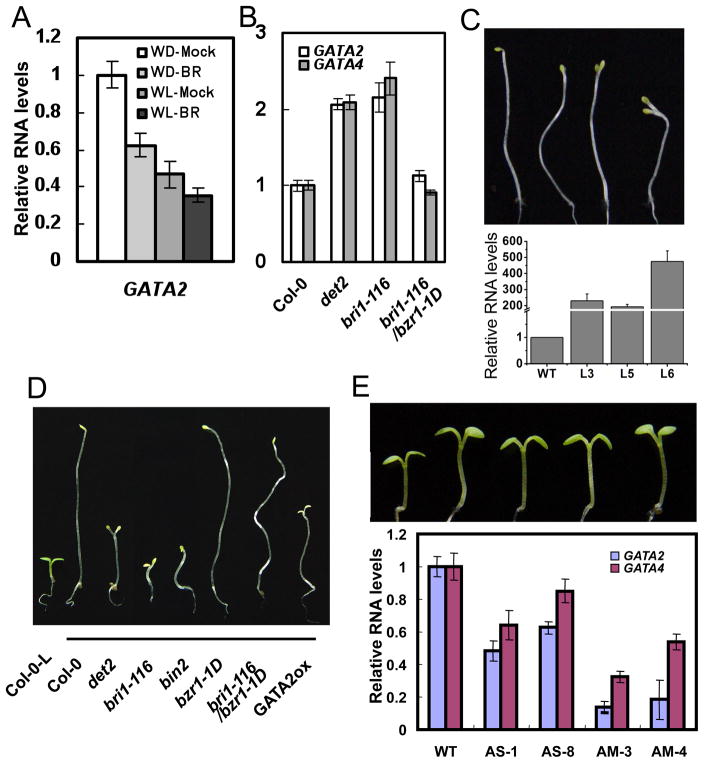
GATA2 and GATA4 are two closest members of the subfamily I of Arabidopsis GATA factors (Reyes et al., 2004). Quantitative RT-PCR analysis confirmed that the transcript level of GATA2 is reduced by BR treatment. GATA2 is expressed at a higher level in the dark than in the light, and BR repression is also more obvious in the dark than in the light (Figure 1A). GATA2 RNA level is increased in the BR-deficient mutant det2 and BR-insensitive mutant bri1-116, but repressed by the bzr1-1D mutation (Figure 1B). A GATA2 promoter-GUS reporter gene showed strong expression in hypocotyls and petioles (Figure S1), where cell elongation is most sensitive to light and BR. GATA2 expression was also detected in root tips, the junctions of floral organs, and styles of plants grown under light (Figure S1). RT-PCR assays confirmed ubiquitous expression of GATA2 in various tissues (Figure S1I). A GATA2-YFP fusion protein is localized in the nucleus (Figure S1J–O). Such expression pattern and subcellular localization of GATA2 is consistent with a role as transcription factor for photomorphogenesis. Recent co-expression analysis has shown that GATA2 and GATA4 show strong coexpression with each other (Manfield et al., 2007).
Figure 1. GATA2 is a positive regulator of photomorphogenesis.
(A), BR treatment reduces GATA2 RNA level. Arabidopsis seedlings grown in the dark (WD) or light (WL) for five days were treated with mock solution or 100 nM 24-epibrassinolide (BR) for 3 hours and the expression of GATA2 was analyzed by qRT-PCR. (B) qRT-PCR analysis of GATA2 and GATA4 RNA levels in 5-day-old dark-grown wild type (Col-0), det2, bri1-116, and bri1-116 bzr1-1D. (C) Dark-grown phenotypes of three GATA-ox lines. Lower panels show qRT-PCR of GATA2 expression (see also Figure S1P, S1R). (D) Phenotypes of light-grown (first on left) or dark-grown seedlings of wild type (Col-0), BR mutants and a representative GATA2-ox transgenic line 6. (E) Phenotypes of antisense (AS) or artificial-microRNA (AM) transgenic Arabidopsis seedlings with reduced levels of GATA2 and GATA4 (see also Figure S2A for quantitation data). Lower panel shows qRT-PCR analysis of GATA2 and GATA4 in these transgenic seedlings. All error bars are standard deviation (SD).
To investigate the function of these GATA factors, we generated transgenic plants over expressing GATA2 and GATA4 under the control of the cauliflower mosaic virus 35S promoter (GATA-ox). Of five GATA2-ox transgenic lines four lines exhibited obvious short hypocotyls and open cotyledons in the dark, similar to the BR-deficient or insensitive mutants (Figure 1C, 1D and S1P–Q). Similarly five of ten GATA4-ox lines also showed shorter hypocotyl phenotypes, however the overall phenotypes were weaker than the GATA2-ox lines (Figure S1R). We further generated GATA2 antisense (GATA-AS) and artificial microRNA (GATA-AM) transgenic plants. The constructs contain conserved sequence and are expected to also suppress GATA4. Many GATA-AS and GATA-AM lines showed long hypocotyl phenotypes in the light (Figure 1E and S2A), but not in the dark (Figure S2G). These results demonstrate that GATA2 plays an important role in promoting photomorphogenesis, and GATA4 is likely to play a similar but less prominent role.
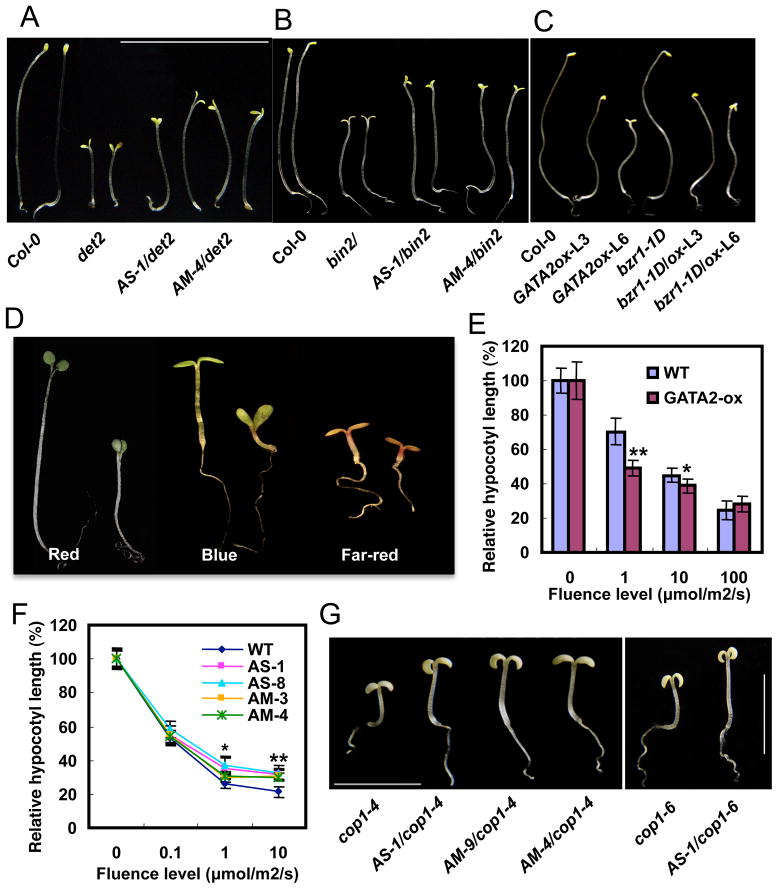
As positive regulator of photomorphogenesis, the increased expression of GATA2 in BR mutants is likely to contribute to the de-etiolation phenotypes. To determine if GATA2 play a role in BR regulation of photomorphogenesis, we crossed the GATA-AS and GATA-AM lines with the BR deficient mutant det2 and BR insensitive mutant bin2, and these plants exhibited longer hypocotyls than the det2 and bin2 single mutants (Figure 2A, 2B, S2B, and S2C). We also crossed the GATA2-ox line with bzr1-1D, which suppresses the de-etiolation phenotypes of the BR-biosynthetic or signaling mutants (Figure 1D). Seedlings homozygous for both GATA2-ox and bzr1-1D had short hypocotyls and open cotyledon in the dark, resembling the phenotype of GATA2-ox (Figure 2C, and S2D), consistent with GATA2 acting downstream of BZR1. These results support an important role of repressing GATA2 in BR inhibition of photomorphogenic development.
Figure 2. GATA2 acts downstream of both BR and light signaling pathways to promote photomorphogenesis.
(A) Phenotypes of det2 mutants crossed with the GATA2/4 antisense (AS) or artificial microRNA (AM) lines (see also Figure S2B). (B) Phenotypes of the bin2 mutant crossed with the GATA-AS and -AM lines (see also Figure S2C). (C) Phenotypes of bzr1-1D mutants crossed with GATA2-ox (see also Figure S2D). (D) GATA2-ox plants (right of each pair) have short hypocotyls than wild type (left) when grown under red (26 μmol/m2/s), blue (13 μmol/m2/s) and far-red (100 μmol/m2/s) light conditions (see also Figure S2E). (E) Relative hypocotyl lengths of GATA2-ox seedlings (L3 line) grown under various fluence rate of red light. (F) Fluence-rate response curve of hypocotyl lengths of GATA2-AS and –AM lines grown in the dark or various intensities of blue light. Error bars in E and F are SD and significant differences from WT are marked (**p<0.01, *p<0.05). (G) Phenotypes of dark-grown cop1 mutants crossed with GATA-AS or GATA-AM lines (see also quantitation data in Figure S2I). The seedlings were grown for 7 days.
Light regulates seedling development through several photoreceptor families that absorb light of distinct wavelengths. To test if GATA2 functions in any specific photoreceptor pathway, we grew the GATA2-ox and GATA-knockdown lines under monochromatic red, far-red, or blue light. The GATA2-ox plants had shorter hypocotyls and the GATA-AS or GATA-AM plants showed longer hypocotyls under all wavelengths of light but not in the dark (Figure 2D, 2E, and S2E–G), suggesting that GATA2 is likely to function downstream of all photoreceptors. Fluence-rate response analyses indicate that the GATA2-ox plants have enhanced sensitivity and the GATA-AS and AM plants have reduced sensitivity to light (Figure 2E, 2F, and S2G). To test if GATA2 is downstream of the master photomorphogenic repressor COP1 (Deng et al., 1991), we crossed the GATA-AS and GATA-AM lines into the cop1-4 and cop1-6 mutants. Knockdown of GATA partly suppressed the de-etiolation phenotypes of the cop1 mutants (Figure 2G, S2H), suggesting that GATA2 functions downstream of COP1 in the light signaling pathway.
GATA2 overexpression causes similar transcriptomic changes as light and BR deficiency
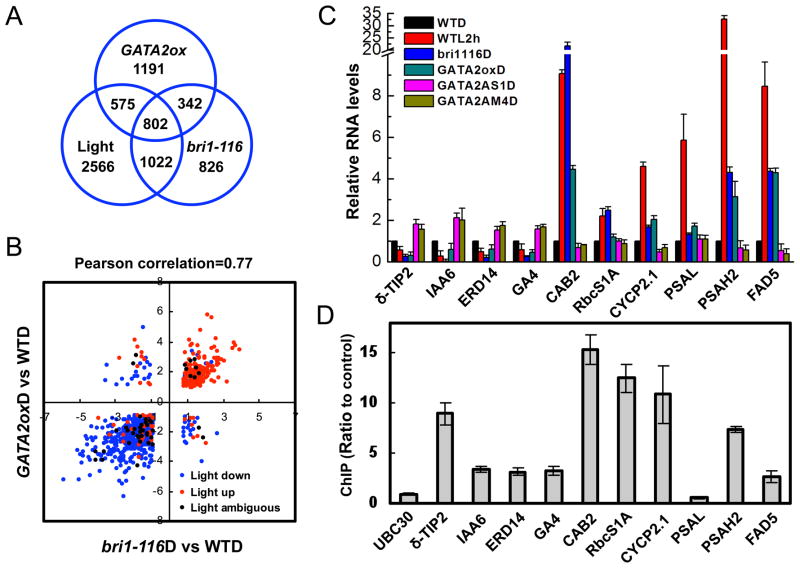
To further understand the function of GATA2 in the light- and BR-signaling pathways, we compared the transcriptomic changes caused by GATA2 overexpression, bri1 mutation, and light treatment. Four-day old dark-grown seedlings of GATA2-ox, bri1-116, and wild type were analyzed by microarray using the ATH1 array (Affymetrix). The results showed that expression of 2910 genes were altered in GATA2-ox plants, with 1743 genes repressed and 1167 activated by GATA2 overexpression (>2 fold and p<0.05, Table S1a). In the bri1-116 mutant, 2992 genes were differentially expressed compared to wild type, and about 38% (1144 of the 2992) of them were also affected in GATA2-ox (Figure 3A, Table S1b). More striking overlap was observed for the 120 most-repressed genes in GATA2-ox: 103 (86%) of them were also repressed in bri1-116 (Table S1c). Overall about 93% (1055) of the 1144 co-regulated genes were affected in the same way by GATA2-ox and bri1-116 (Figure 3B, Table S1d). Such similar genomic effects of GATA2-ox and bri1 mutation are consistent with elevated GATA2 expression in bri1 contributing to its altered gene expression and de-etiolation phenotype.
Figure 3. GATA2 directly regulates genes that are responsive to both BR and light.
(A) Venn diagrams of the number of genes differentially expressed in the dark-grown GATA2-ox vs WT, genes affected in the bri1-116 mutant, and genes affected in at least one of the light-treatment microarray experiments (see also Table S1). The numbers in the overlapping areas indicate the number of shared genes. (B) Scatter plot of log2 fold change values of GATA2-ox vs WT and bri1-116 vs WT for 802 genes differentially expressed in dark-grown GATA2-ox vs WT, bri1-116 vs WT, and light-grown vs dark-grown wild type seedlings. Effects of light treatment on the expression are denoted by color as shown (see also Table S1). (C) Quantitative RT-PCR analysis of a number of known light-responsive genes in GATA2-ox, GATA-AS-1, GATA-AM-4, or bri1-116 plants grown in the dark for 5 days, compared to wild type plants grown in the dark and then untreated or treated with white light for 2 hours. (D) Chromatin immunoprecipitation followed by real time PCR (ChIP-qPCR) assays of GATA2 binding to promoters of genes in panel C, performed using 35S::GATA2-YFP transgenic and wild type control seedlings grown in light for 2 weeks and an anti-GFP antibody. GATA2 binding was measured by qPCR as the ratio between GATA2-YFP and control sample. The UBC30 gene was used as a negative control. Error bars indicate SD. (See also Table S4).
When the gene expression changes of GATA2-ox were searched against an Arabidopsis microarray database that includes 1450 treatments (Zhang et al., 2010), the top nine best matches were microarray experiments that compared seedlings grown under various light conditions to those grown in the dark. The percent overlaps with the light datasets ranged from 27% to 48% (Table S2). The Pearson correlation coefficients of pair-wise comparison between the GATA2-ox vs WT data and various light vs dark data range from 0.57 to 0.75 (Table S2), suggesting that GATA2 overexpression causes a similar genomic response as light exposure. About 47% (1378) of the genes affected in GATA2-ox were affected by at least one of the light conditions (Table S3). Among these, 802 genes were affected by bri1 mutation (Figure 3A, Table S1e). About eighty seven percent of these shared genes were up or down regulated similarly by GATA2-ox, the bri1-116 mutation, and light treatments (Figure 3B, Table S1e). Such similar effects of GATA2 overexpression, bri1 mutation, and light on large numbers of genes strongly support an important role for GATA2 in mediating the antagonistic effects of BR and light on gene expression and photomorphogenesis.
GATA2 directly regulates genes that respond to light and BR deficiency
Quantitative RT-PCR assays confirmed that the expression levels of light-repressed genes, such as TIP2 and IAA6, were repressed in GATA2-ox and bri1-116 plants but increased in the GATA-AS and GATA-AM plants, whereas the levels of light-induced genes, such as CAB2, PSAH2, were increased in GATA2-ox and bri1-116 plants but reduced in the GATA-AS and GATA-AM plants (Figure 3C). Chromatin immunoprecipitation (ChIP) assays for GATA sequence-containing regions of promoters demonstrated that GATA2 binds strongly to the promoters of TIP2, CAB2, CYCP2.1, RBCS1A, and PSAH2, and binds weakly to IAA6, ERD14, GA4, and FAD5, which are responsive to light treatment and affected in bri1 and the GATA2 transgenic plants. In contrast, GATA2 does not bind to PSAL, which is a light-responsive gene not affected by bri1 or GATA-AS (Figure 3D and Table S4). Furthermore, ChIP assays showed GATA2 binding to additional seven genes strongly repressed and three genes strongly activated in GATA2-ox, but not to the control gene UBC30 or two LHCB genes that were not affected in GATA2-ox (Table S4). These results demonstrate that GATA2 directly activates some of the light-induced and BR-repressed genes and inhibits light-repressed and BR-induced genes.
Light induces accumulation of GATA2 protein, which directly feedback inhibits its own transcription
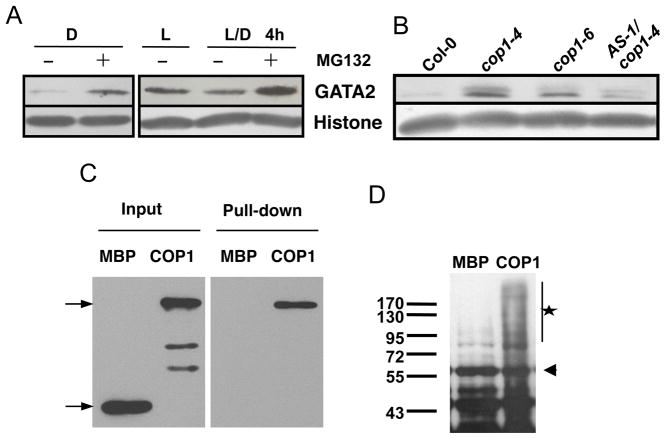
As a positive regulator of photomorphogenesis, GATA2 is expected to be activated by light. However, GATA2 and GATA4 are expressed at a higher level in dark-grown plants than in light-grown plants (Manfield et al., 2007). Quantitative RT-PCR analysis showed that the transcript levels of GATA2 and GATA4 rapidly decreased upon light treatment of dark-grown seedlings (Figure 4A). Interestingly, immunoblot analysis demonstrated that light treatment increased the GATA2 protein accumulation (Figure 4B). Opposite responses at protein and RNA levels are often caused by feedback inhibition of transcription by the protein product of the gene. Indeed, the levels of the endogenous GATA2 and GATA4 RNAs were reduced in the GATA2-ox transgenic plants, which overexpress the GATA2 RNA from the transgene (Figure 4C). Overexpression of GATA2 also led to reduced expression of a GATA2-GUS reporter gene in tobacco leaf cell (Figure S3). Chromatin immunoprecipitation assays further showed that the GATA2 protein directly binds to its own promoter in vivo (Figure 4D). These results demonstrate that light induces GATA2 protein accumulation at a posttranscriptional level, and light-activated GATA2 protein feedback inhibits the transcription of GATA2 and GATA4.
Figure 4. Light regulates GATA2 accumulation at the post-translational level.
(A) Light represses GATA2 and GATA4 transcription levels. Dark-grown Arabidopsis seedlings were treated with white light for indicated time and RNA levels of GATA2 and GATA4 were measured by real-time qRT-PCR. Error bars indicate standard deviation. (B) Light promotes GATA2 protein accumulation. Immunoblot analysis of GATA2 protein in 5-day-old dark-grown GATA2-ox L6 line seedlings treated with white light for the indicated time. (C) qRT-PCR analysis of the levels of RNA expressed from the endogenous GATA2 and GATA4 genes (endo) or total GATA2 RNA level in wild type (WT) and the GATA2-ox transgenic seedlings (L3 and L6). UBC30 was used as internal control. (D) ChIP-qPCR analysis of GATA2 binding to its own promoter. The upper panel shows a diagram of the promoter (open box), 5′UTR (black line) and the first exon (black box) of the GATA2 gene. Black circles indicate positions of putative GATA motifs. Lines marked a to f show GATA2-binding (solid) and non-binding (dashed) regions analyzed by qPCR. The lower panel shows ChIP-qPCR data. Error bars indicate SD.
GATA2 is degraded in the dark by the proteasome in a COP1-dependent manner
Several light-signaling transcription factors, such as HY5 and HFR1, are targeted for proteasomal degradation by the COP1 ubiquitin ligase, and light signaling stabilizes these transcription factors by inactivating COP1 (Osterlund et al., 2000; von Arnim et al., 1997). We investigated whether a similar COP1-dependent process is involved in light regulation of GATA2 accumulation. Treatment of dark-grown seedlings with MG132, an inhibitor of the proteasome, caused GATA2 protein accumulation, indicating that GATA2 is degraded by the proteasome in the dark. Upon transition from light to dark, GATA2 protein level decreased dramatically, and this decrease was blocked by MG132 treatment (Figure 5A), suggesting that light inhibits proteasomal degradation of GATA2. Immunoblotting data showed that the GATA2 protein level was increased in the cop1 mutants grown in the dark (Figure 5B), indicating that GATA2 degradation requires COP1. The accumulation of GATA2 obviously contributes to the de-etiolation phenotype of cop1, as suppressing GATA2 RNA levels in the cop1 mutant reduced the GATA2 protein level and increased the hypocotyl length (Figure 2G and Figure 5B). Furthermore, in vitro pull-down assays showed that COP1 can directly interact with GATA2 (Figure 5C). In vitro ubiquitination assay confirmed that COP1 can ubiquitinate GATA2 in vitro (Figure 5D). These results strongly support the possibility that the GATA2 protein is negatively regulated by COP1-dependent ubiquitination and proteasomal degradation, and inactivation of COP1 by light signaling leads to GATA2 accumulation.
Figure 5. Light regulates GATA2 accumulation through a COP1 ubiquitin ligase-dependent process.
(A) Immunoblot analysis of GATA2 protein levels. Dark-grown (D) or light-grown (L or L/D) 5-day-old 35S:GATA2 transgenic seedlings (L3 line) were treated with mock solution (−) or 10 μM MG132 (+) for 4 hours in the dark (D and L/D) or light (L). Histone H3 was probed as a loading control. (B) Immunoblot assay of GATA2 protein level in 5-day old dark-grown wild type, the cop1 mutants, and cop1-4 crossed with the GATA-AS line. (C) In vitro pull-down assay showing the interaction between GATA2 and COP1. (D) In vitro ubiquitination assay showing ubiquitination of GST-GATA2 by MBP-COP1. The arrow points to the GST-GATA2 band and the star marks the ubiquitinated GST-GATA2 bands.
BZR1 binds to the GATA2 promoter in vivo
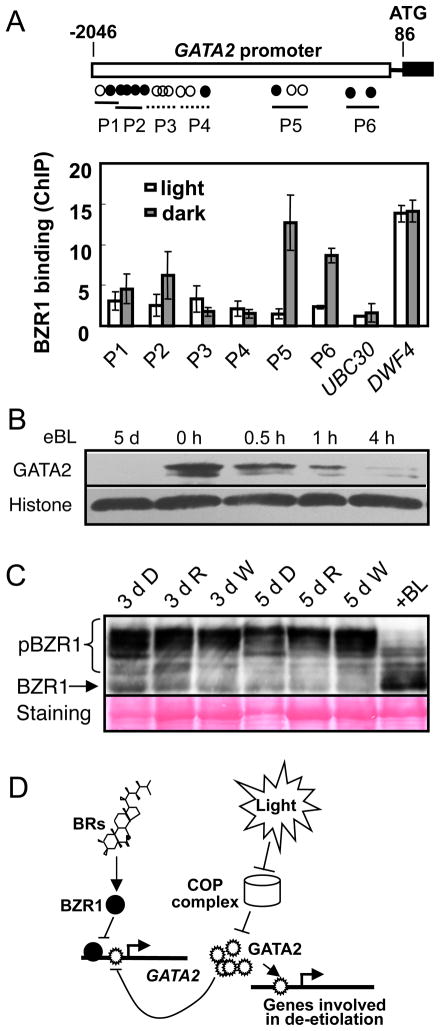
To test if BZR1 directly regulates GATA2 expression, we performed chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) assays using pBZR:BZR1-CFP transgenic plants and anti-GFP antibody, with 35S-GFP transgenic plants as a control. As shown in Figure 6A, BZR1 bound strongly to the GATA2 promoter in the dark-grown seedlings but only weakly in the light-grown seedlings, consistent with a prominent role of BR in repressing GATA2 expression in the dark (Figure 1A). In contrast, BZR1 bound to the DWF4 promoter strongly in both dark and light conditions. Such transcriptional regulation of GATA2 leads to altered levels of GATA2 protein, as BR treatment of the det2 mutant dramatically reduced the GATA2 protein levels (Figure 6B). In contrast, BR and the BR biosynthetic inhibitor brassinazole had little effect on the GATA2 protein level in the transgenic plants that constitutively express GATA2 from the 35S promoter (Figure S4), suggesting that BR represses GATA2 at the transcriptional but not posttranscriptional level.
Figure 6. BR represses GATA2 transcriptional level through BZR1 direct binding to its promoter.
(A) ChIP-qPCR assays of BZR1 binding to the GATA2 promoter. The pBZR1::BZR1-CFP and 35S::GFP transgenic Arabidopsis seedlings grown in dark or light for 5 days were used in ChIP using anti-GFP antibody. The upper panel shows a diagram depicting the putative promoter (open box), 5′UTR (black line) and the first exon (black box) of the GATA2 gene. Open and black circles indicate the positions of putative E-box and BRRE motifs, respectively. Thin lines marked P1 to P6 show BZR1-binding (solid) and nonbinding (dashed) regions analyzed by qPCR. The lower panel shows the quantitative PCR data for enrichment as ratio between BZR1-CFP and 35S-GFP normalized to the CNX5 control gene. Error bars indicate SD. (B) Immunoblot analysis shows BR repression of GATA2 accumulation. The det2 seedlings were grown in the dark on medium with 100 nM 24-epibrassinolide (eBL) for 5 days, or grown without eBL for 5 days and then treated with 10 μM eBL for 0 to 4 hours. The level of Histone H3 was used as a loading control. (C) Light does not have a significant effect on BZR1 phosphorylation status. Phosphorylated (pBZR1) and unphosphorylated (BZR1) BZR1 were analyzed by immunoblotting using an anti-BZR1 antibody in Arabidopsis seedlings grown in dark (D), under red (R) or white (W) light for three (3 d) or five days (5 d). Seedlings grown in white light for five days were treated with 100 nM brassinolide (+BL) for 30 min. The gel blot was stained with Ponceau S to show protein loading (the Rubisco major band is weaker in dark-grown samples). (D) A model for GATA2 function in BR- and light-regulation of photomorphogenesis. In the dark, the BR-activated BZR1 directly represses GATA2 transcription and COP1 promotes GATA2 ubiquitination and degradation, ensuring a low GATA2 level for etiolation/skotomorphogenesis. In the presence of light, COP1 is inactivated and the GATA2 protein accumulates to a high level to promote photomorphogenesis through binding to target genes. The GATA2 protein also feedback inhibits its own transcription by directly binding to its promoter, potentially desensitizing the system upon light-induced accumulation of GATA2 protein. When BR levels are low, reduced BZR1 activity leads to overexpression of GATA2, which promotes photomorphogenesis.
Light does not have a strong effect on BR signaling
Our observation of differential BZR1 binding to the GATA2 promoter in the dark and light suggests that light affects BZR1 activity. Because BZR1’s nuclear localization and DNA binding activity are tightly controlled by BR-regulated phosphorylation (Gendron and Wang, 2007), light could alter BZR1’s phosphorylation status if light has an effect on BR level or BR signal transduction. We therefore performed immunoblotting experiments to test whether light affects BZR1 accumulation and phosphorylation (Figure 6C). The results show that plants grown in the dark and under red light or white light conditions contain similar levels of phosphorylated and unphosphorylated BZR1, whereas treatment with BR caused dramatic dephosphorylation of BZR1 (Figure 6C). These results indicate that light does not have a significant effect on BR level or BR signaling upstream of BZR1.
DISCUSSION
Interactions between light and endogenous hormones are critical for plant development. It has been long recognized that BR plays a major role in light regulated plant development. The underlying molecular mechanism has remained unclear. This study identifies members of the GATA factor family (GATA2 and GATA4) as key transcription factors that integrate the BR and light-signaling pathways for coordinated regulation of gene expression and photomorphogenesis (Figure 6D). We show that GATA2 directly binds to light-responsive promoters in vivo and controls the expression of large numbers of genes that respond to both light and BR signaling. GATA2 is inhibited by BR signaling at the transcriptional level through BZR1 binding to its promoter and is activated by light at the protein level through inhibiting COP1-dependent proteolysis.
GATA2 also binds to its own promoter to feedback inhibit its own transcription. Such feedback mechanism could serve an important desensitizing mechanism during transition from dark to light, but would also lead to de-repression of GATA2 transcription and incomplete switch to skotomorphogenesis when GATA2 protein degradation is accelerated in the dark. As such repression of GATA2 expression by BR becomes essential for maintaining complete skotomorphogenesis in the dark. BR deficiency causes overexpression of GATA2, which contributes to de-etiolation in the dark. This study demonstrates a mode of BR-light crosstalk, in which BR signaling inhibits light responses through transcriptional repression of key components of the light-signaling pathway.
GATA2 is a key component for light-responsive gene expression
Analyses of light-regulated promoters have shown an essential role of the GATA element in light-regulated gene expression (Chattopadhyay et al., 1998b; Jeong and Shih, 2003; Terzaghi and Cashmore, 1995). It has been shown that combinations of different LREs, rather than individual elements, confer proper light-responsiveness to a promoter (Puente et al., 1996). The GATA element functions together with the G-box or GT1 motifs to confer normal response to a wide spectrum of light signals involving multiple photoreceptors and the COP/DET/FUS complex (Chattopadhyay et al., 1998a). These results indicated a role of the GATA element as an essential partner with other LREs in light-regulated gene expression. Previous studies have only identified the G-box-binding factors, including PIFs, HY5, and CIB1. This study identifies GATA2 and GATA4 as the missing transcription factors that act through the GATA element.
Our results provide strong genetic and molecular evidence for the role of GATA2 in light regulation of gene expression and photomorphogenesis. First, overexpression of GATA2 causes a typical de-etiolation phenotype and a transcriptomic change that resembles those caused by light exposure, whereas suppression of GATA2 by RNAi or antisense had an opposite effect on hypocotyl elongation and gene expression. Although the long hypocotyl phenotypes of the GATA-AM and –AS plants are relatively weak, this is likely due to incomplete suppression of GATA2 expression and/or redundant function of other homologous GATA factors. Second, ChIP assays showed that in vivo GATA2 binds to many light-responsive promoters at regions containing GATA motifs, providing direct evidence for GATA2 regulation of light-responsive genes. Finally, GATA2 protein is stabilized by light signaling, most likely through a COP1-dependent mechanism similar to the regulation of the light-signaling transcription factors HY5 and HFR1. GATA2 accumulates in the cop1 mutants and can interact with COP1 and be ubiquitinated by COP1 in vitro, though direct in vivo interaction is yet to be demonstrated. Therefore, GATA2 meets the criteria for a primary light signaling transcription factor.
GATA factors are a class of highly conserved transcription factors with a type IV zinc finger followed by a basic region, which are known to recognize the consensus sequence WGATAR (where W is T or A and R is G or A) (Lowry and Atchley, 2000). GATA factors are found in all eukaryotes from fungi to plants and metazoans. In fungi, GATA factors are involved in a number of different processes, ranging from nitrogen utilization, mating-type switch, and light responses (Scazzocchio, 2000). In Neurospora crassa, the White Collar-1 (WC1) and White Collar-2 (WC2) loci encoding “plant-like” GATA factors are required for light and circadian responses (Ballario and Macino, 1997). In addition to the GATA DNA binding domain, WC1 also contains a light-oxygen-voltage (LOV) domain and functions as a photoreceptor (Cheng et al., 2003). It seems that a function of GATA factors in light responses has been conserved during evolution from fungi to higher plants.
The function of Arabidopsis GATA factors in light response was not uncovered in previous genetic analysis, and suppression of GATA2 and GATA4 only partially suppressed cop1 and det2 mutants; these are most likely because of genetic redundancy. The Arabidopsis genome contains 29 genes that encode GATA factors (Reyes et al., 2004). Some members of the GATA family have been shown to play a role in regulating flower development (Zhao et al., 2004), chlorophyll synthesis, and carbon/nitrogen metabolism (Bi et al., 2005; Mara and Irish, 2008). In vitro DNA binding assays have shown binding of GATA1 to the GATA elements of the GAPB promoter that are essential for light responsive expression (Jeong and Shih, 2003). GATA2 shares 76% sequence identity with GATA1 in the DNA binding domain and is likely to have similar DNA binding specificity for GATA elements. Our ChIP experiment shows that GATA2 binds to promoter regions containing GATA sequence.
Additional GATA family members may be involved in light responses, as their expression levels are regulated by light. Higher expression in the light-grown than dark-grown seedlings has been observed for GATA6, GATA7, GATA21, GATA22, and GATA23 (Manfield et al., 2007). None of these genes are affected in the GATA2-ox plants based on our microarray data, suggesting their light regulation is independent of GATA2. In fact GATA21 and GATA22 are induced by red light in a PIF3-dependent manner (Monte et al., 2004). In contrast, four other genes, GATA2, GATA4, GATA9, and GATA12, showed stronger expression in the dark-grown than light-grown seedlings (Manfield et al., 2007), and they are all repressed in the GATA2-ox plants. Based on similarity in sequence, gene structure, and expression profiles, these four GATA genes have been predicted to share common ancestry, with GATA2 and GATA4 arisen from a recent chromosomal duplication (Reyes et al., 2004). GATA2 and GATA4 are coexpressed with each other and share common coexpressed genes, which include PHYA and light-signaling transcription factors PIF3, PIF1/PIL5, and HFR1 (Manfield et al., 2007), consistent with their role in light signaling. In contrast, GATA9 and GATA12 do not show significant coexpression with any of the genes known to be involved in light signaling. It has been suggested that GATA9 and GATA12 have diverged from GATA2 and GATA4 in expression and possibly in function as well (Manfield et al., 2007). Based on our expression microarray data, only GATA2 and GATA4, but not GATA9 and GATA12, are overexpressed in the bri1 mutant more than two fold and repressed in the bri1 bzr1-1D double mutant, indicating that BR regulates the transcription of GATA2 and GATA4 but not their close homologs GATA9 and GATA12. Further genetic analysis of double or multiple loss-of-function mutants will be required to understand whether other GATA factors also play a role in photomorphogenesis.
The relationship between GATA2 and other light-signaling transcription factors is key for understanding light-responsive gene expression. Several lines of evidence suggest that GATA2 functions together with the G-box-binding factors. First, GATA and G-box elements are found together in many light responsive promoters, and their dual presence is essential for normal light responsiveness in a synthetic promoter (Chattopadhyay et al., 1998b). Second, GATA2 shows strong coexpression with PIF3, PIF1/PIL5, SPT, and HFR1 (Manfield et al., 2007), many of which bind to the G-box. Third, GATA2 is stabilized by light at the posttranslational level, likely through the same COP1-dependent mechanism that regulates HY5 and HFR1. It is also worth noting that a higher percentage of the genes up regulated than down regulated in GATA2-ox are HY5 targets (Lee et al., 2007) (27% of 1167 up-regulated genes vs 21% of 1743 down-regulated genes), which is consistent with our hypothesis that GATA and G-box elements together confer light activated expression by recruiting GATA2 and HY5. Whether GATA2 directly interacts with other light-signaling transcription factors and how they orchestrate dynamic light-regulated gene expression are yet to be analyzed in future studies.
GATA2 is a key junction for the antagonism between BR and light signaling pathways
Genetic studies have long demonstrated a critical role of BR in skotomorphogenesis (Li et al., 1996; Szekeres et al., 1996). The antagonizing relationship between BR and light has been analyzed at the genetic and physiological levels. Mutations that reduce BR level enhanced the light responses (Neff et al., 1999), and a rice phyB mutant showed enhanced BR responses (Jeong et al., 2007). The antagonism at the level of gene expression was recognized in the initial studies of the BR deficient mutants (Chory et al., 1991) (Li et al., 1996; Szekeres et al., 1996), and confirmed by our microarray data showing similar transcriptomic changes caused by the bri1 mutation and light exposure. The similar effects of BR deficiency and light on seedling development and expression of large numbers of genes suggested three possible mechanisms of interaction between the BR and light-signaling pathways: (1) light reduces BR level or BR sensitivity, (2) BR regulates light-signaling components to inhibit light signaling, or (3) BR and light signaling pathways regulate common target genes through separate transcription factors independently controlled by each pathway. This study provides evidence for the second mechanism of BR-light crosstalk, and recent genomic analysis of BZR1 target genes supported the presence of also the third mechanism(Sun et al., 2010).
A previous study proposed that light inhibits BR biosynthesis by repressing a small G protein that binds to and activate a BR-biosynthetic enzyme (Kang et al., 2001). However, subsequent direct BR measurement failed to detect significant difference in BR levels between light-grown and dark-grown plants, but showed light reducing the level of gibberellin, another hormone that also promotes cell elongation (Symons and Reid, 2003). Our observations of no obvious effect of light on the phosphorylation status and accumulation of BZR1 or on BZR1 binding to the DWF4 promoter are consistent with the lack of change of BR level by light. Our results further suggest that light does not inhibit BR signaling upstream of BZR1. However, stronger BZR1 binding to the GATA2 promoter was observed in the dark-grown than light-grown seedlings. It is possible that light has an effect on the availability of BZR1 binding site or BZR1-interacting proteins at the GATA2 promoter. In contrast to the lack of strong effect of light on BR signaling, BR obviously has a strong effect on light signaling by repressing GATA2 expression.
Our results show that GATA2 plays a key role in BR regulation of photomorphogenesis. GATA2 accumulates in the det2 mutant and GATA2 knockdown partially suppresses the photomorphogenic phenotypes of dark-grown det2 and bin2, indicating that de-etiolation in the BR mutants are at least partly due to the increased levels of GATA2. About one third of the genes affected in bri1 are affected similarly by GATA2-ox, suggesting that the elevated GATA2 level contributes to a major portion of bri1’s effect on genome expression and that BZR1 repression of GATA2 is a major mechanism for BR inhibition of light responses. By inhibiting transcription and promoting protein accumulation of GATA2, respectively, BR and light antagonistically regulate the level of GATA2 activity and consequently the expression of its downstream target genes. Thus, GATA2 represents a key junction of crosstalk between BR and light signaling pathways.
The mechanism of BR-light crosstalk through GATA2 is distinct from those for light crosstalk with GA and cytokinin. In addition to light repression of GA level, GA also affects the activity or accumulation of the light-signaling transcription factors PIF/PIL and HY5 (Alabadi et al., 2008; de Lucas et al., 2008; Feng et al., 2008). The DELLA proteins of the GA signaling pathway directly interact with and inhibit members of the PIF/PIL family, which are negative regulators of photomorphogenesis (de Lucas et al., 2008; Feng et al., 2008). GA also promotes degradation of HY5, possibly through a COP1-dependent process (Alabadi et al., 2008). In contrast, cytokinin, which promotes photomorphogenesis, induces HY5 protein accumulation (Vandenbussche et al., 2007). Whether other hormones also regulate GATA2 to modulate light responses remains to be tested by future studies.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The wild type, various mutants, and transgenic Arabidopsis thaliana plants were in the Columbia ecotype. Seeds were sterilized by incubation in freshly prepared 10% bleach plus 0.01% Triton X-100 for 15 min and then washed 3–4 times with sterilized water. The surface-sterilized seeds were treated in 4 °C for 2 days and at 22 °C under white light for 8 hours to induce uniform germination. For phenotype analyses, seedlings were grown on 0.8% phytoagar plates containing half-strength Murashige-Skoog (MS) nutrient and 1% sucrose. White light (about 100 μmol/m2/s) was provided by fluorescence light source in a growth room at 22 °C. Growth under red, far-red, and blue light was carried out in a LED light chamber (E-30LEDL3, Percival) at 22 °C. Seedlings were photographed next to a size reference (ruler) and their hypocotyl lengths measured using the Image J software. Seeds were harvested from plants grown in a greenhouse supplemented to 16-hour light/day and a temperature range of 18–28 °C.
Vector construction and transformation
A 1152-bp genomic fragment containing full-length GATA2 open reading frame was amplified by PCR and then cloned into the BamHI and KpnI sites of the pSN1301 binary vector to place GATA2 under the control of the CaMV 35S promoter.
The GATA2 antisense construct was made by inserting the GATA2 full-length cDNA fragment in reverse orientation into the pSN1301 plasmid. The artificial microRNA constructs were made using the vectors and methods previously reported (Schwab et al., 2006) (See Supplemental Information for details). The 35S::GATA2-YFP fusion construct was generated by inserting a full-length GATA2 cDNA without stop codon fused to the N terminus of the pEZR-LNY vector.
The GATA2-ox, GATA2-AS, GATA2-AM and 35S::GATA2-YFP binary constructs were transformed into the Agrobacterium tumefaciens strain GV3101 and then introduced into Arabidopsis thaliana Columbia wild-type plants via a floral dip method. About 20 T1 transgenic lines with single T-DNA insertion were selected for further analysis. Homozygous T3 or T4 transgenic seedlings were used for phenotype and molecular characterization.
Protein expression and antibody preparation
The full length GATA2 cDNA was cloned into the pGEX-4T-1 vector to express GST-GATA2 protein in E. coli Rosetta cells (Novagen). The recombinant fusion protein was purified using glutathione-agarose beads (GE Healthcare) and used to immunize rabbit. The anti-GATA2 antibody was purified from the immune serum using immobilized GST-GATA2 (Aminolink® Immobilization Kit, Pierce Biotechnology). The anti-Histone H3 antibody for loading control was from Millipore (Catalogue No. 07-690)
Total RNA isolation and quantitative RT-PCR analysis
Total RNA was extracted from Arabidopsis seedlings using the Trizol RNA extraction kit (Invitrogen, USA). The first-strand cDNA was synthesized by using M-MLV reverse transcriptase (Promega, USA) and used as RT-PCR templates. Quantitative real-time PCR analyses were carried out on Mx3000P (Stratagene, USA) by using the SYBR® Green reagent (TOYOBO, JAPAN) according to the manufacturer’s instructions. The RT-PCR was repeated at least three times using samples harvested separately. The UBC30 gene was used as internal reference. See Supplemental Information for primer sequences used for RT-PCR.
Microarray Data Analysis
Arabidopsis seedlings (Columbia, GATA2-ox, bri1-116) were grown on 1/2 MS medium in the dark for 4.5 days, and the seedlings were frozen in liquid nitrogen in complete darkness, and then the bri1-116 seedlings were selected from the segregating population. Ten micrograms (10 μg) of total RNA from the seedlings was used to prepare probes for hybridization, and each probe was hybridized independently to one chip according to the protocol of the ATH1 array manufacturer (Affymetrix). Three independent biological repeats were conducted. The data were analyzed using Genespring software ver. 7. Data that were flagged as absent, using the Affymetrix mismatch probes, in two or more of the repeats for each genotype were removed. Genes that passed this filter for any one of the genetic backgrounds were used for further analysis. P-value <0.05 and fold change >2 (for GATA2-ox) or fold change >1.8 (for bri1-116) were used to identify genes differentially expressed in GATA2-ox or bri1-116 compared to wild type control seedlings.
To determine what experimental conditions causes similar gene expression changes as GATA2-ox, we carried out expression fingerprint searching by comparing the differential gene expression pattern between GATA2-ox treatment and all available 1450 treatment/control microarray comparisons (T/Cs) in the Gene Expression Browser (GEB) database (http://www.expressionbrowser.com/) (Zhang et al., 2010). We inputted the pairs of GATA2-ox significant (2-fold and P<0.05 as cutoff) gene IDs and their log2 ratios, and compared them to each T/C of GEB with the following procedure: (1) Select the significant genes from the T/C using 2-fold and P<0.05 as cutoff. (2) Compute the overlapping genes between GATA2-ox and the T/C. The chi-square test was used for filtering out non-significant overlaps (P<0.01 as cutoff). (3) Compute the Pearson’s correlation coefficient using the paired log2 ratios of GATA2-ox and the T/C for the overlapping genes. The significance of correlation P value was also computed to reject non-significant correlations (p<0.01 as cutoff). As a result, all hits were significant in both number of overlapping genes and expression changes (Pearson correlation). Finally, the hit list was ordered by the Pearson correlation coefficient.
Chromatin immunoprecipitation
Chromatin immunoprecipitation experiments were performed following the protocol described previously (He et al., 2005), using 2-week-old light-grown wild type and 35S::GATA2-YFP transgenic Arabidopsis seedlings or 5-day-old dark- and light-grown 35S-GFP and pBZR::BZR1-CFP seedlings. An affinity-purified anti-GFP polyclonal antibody was used to immunoprecipitate the BZR1 or GATA2 protein-DNA complex, and the precipitated DNA was analyzed by real-time PCR using the SYBR® Green reagent (TOYOBO, JAPAN). Results were presented as the ratio of the amount of DNA immunoprecipitated from BZR1-CFP or GATA2-YFP samples to that of the control samples (35S-GFP or wide type). The UBC30 and PP2A genes were used as the negative controls. The ChIP experiments were performed 3 times, from which the means and standard deviations were calculated. The primer sequences for ChIP-qPCR are in Supplemental Information.
Protein purification and pull down assay
The GST-GATA2 protein was expressed using the pGEX-4T-1 vector in E. coli Rosetta cells (Novagen). The recombinant fusion protein was purified using glutathione-agarose beads (GE Healthcare). For pull-down assay, COP1 fused to maltose binding protein (MBP) was purified using amylose resin (NEB). Glutathione beads containing GST-GATA2 was incubated with MBP, MBP-COP1. The mixture was rotated in a cold room for 1 hr and the beads were washed 5 times with wash buffer (20 mM Tris-HCl [pH8.0], 200 mM NaCl). The proteins were eluted from the beads by boiling in equal volume of 2×SDS buffer and loaded onto a SDS-PAGE gel. Gel blots were analyzed using an anti-MBP antibody (NEB).
In vitro ubiquitination assay
The MBP-COP1 and GST-GATA2 proteins expressed in E. coli were affinity purified for in vitro ubiquitination assays. To improve the E3 activity of MBP-COP1, the purified MBP-COP1 and MBP control proteins on maltose beads were incubated with Arabidopsis cell extract for 30 min. After incubation, the cell extract was removed and the beads were washed. To perform the in vitro ubiquitination assay, crude extract containing recombinant wheat E1 (GI: 136632), human E2 (UBCh5b), His-UBI (UBQ14), purified GST-GATA2 and purified MBP-COP1 (or MBP control) were incubated at 30 °C with agitation in an Eppendorf Thermomixer for 1.5 hr. The proteins were immunoblotted after SDS-PAGE and GST-GATA2 was detected using an anti-GST antibody.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Institute of General Medical Sciences (R01GM066258), National Science Foundation of China (90817009), and Ministry of Agriculture of China (2008ZX08009-003). We thank Dr. Xing Wang Deng for providing COP1 clones and cop1 mutants, and Dr. Winslow Briggs for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFFERENCE
- Alabadi D, Gallego-Bartolome J, Orlando L, Garcia-Carcel L, Rubio V, Martinez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, et al. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 2008;53:324–335. doi: 10.1111/j.1365-313X.2007.03346.x. [DOI] [PubMed] [Google Scholar]
- Arguello-Astorga G, Herrera-Estrella L. Evolution of Light-Regulated Plant Promoters. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:525–555. doi: 10.1146/annurev.arplant.49.1.525. [DOI] [PubMed] [Google Scholar]
- Ballario P, Macino G. White collar proteins: PASsing the light signal in Neurospora crassa. Trends Microbiol. 1997;5:458–462. doi: 10.1016/S0966-842X(97)01144-X. [DOI] [PubMed] [Google Scholar]
- Bi YM, Zhang Y, Signorelli T, Zhao R, Zhu T, Rothstein S. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 2005;44:680–692. doi: 10.1111/j.1365-313X.2005.02568.x. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998a;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Puente P, Deng XW, Wei N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 1998b;15:69–77. doi: 10.1046/j.1365-313x.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Cheng P, He Q, Yang Y, Wang L, Liu Y. Functional conservation of light, oxygen, or voltage domains in light sensing. Proc Natl Acad Sci U S A. 2003;100:5938–5943. doi: 10.1073/pnas.1031791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA. Phenotypic and Genetic Analysis of det2, a New Mutant That Affects Light-Regulated Seedling Development in Arabidopsis. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Deng XW, Caspar T, Quail PH. cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Wang ZY. Multiple mechanisms modulate brassinosteroid signaling. Curr Opin Plant Biol. 2007;10:436–441. doi: 10.1016/j.pbi.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, MJ, Gendron, Sun YL, SS, Gampala, Gendron N, Sun CQ, Wang ZY. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DH, Lee S, Kim SL, Hwang I, An G. Regulation of brassinosteroid responses by phytochrome B in rice. Plant Cell Environ. 2007;30:590–599. doi: 10.1111/j.1365-3040.2007.01644.x. [DOI] [PubMed] [Google Scholar]
- Jeong MJ, Shih MC. Interaction of a GATA factor with cis-acting elements involved in light regulation of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis. Biochem Biophys Res Commun. 2003;300:555–562. doi: 10.1016/s0006-291x(02)02892-9. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- Kang JG, Yun J, Kim DH, Chung KS, Fujioka S, Kim JI, Dae HW, Yoshida S, Takatsuto S, Song PS, et al. Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell. 2001;105:625–636. doi: 10.1016/s0092-8674(01)00370-1. [DOI] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang J, Sun Y, Burlingame AL, Wang ZY. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Bio. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Wang ZY. Brassinosteroid Signal Transduction from Receptor Kinases to Transcription Factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- Lowry JA, Atchley WR. Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding domain. J Mol Evol. 2000;50:103–115. doi: 10.1007/s002399910012. [DOI] [PubMed] [Google Scholar]
- Luccioni LG, Oliverio KA, Yanovsky MJ, Boccalandro HE, Casal JJ. Brassinosteroid mutants uncover fine tuning of phytochrome signaling. Plant Physiol. 2002;128:173–181. [PMC free article] [PubMed] [Google Scholar]
- Ma L, Gao Y, Qu L, Chen Z, Li J, Zhao H, Deng XW. Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis. Plant Cell. 2002;14:2383–2398. doi: 10.1105/tpc.004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield IW, Devlin PF, Jen CH, Westhead DR, Gilmartin PM. Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the Arabidopsis GATA gene family. Plant Physiol. 2007;143:941–958. doi: 10.1104/pp.106.090761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mara CD, Irish VF. Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol. 2008;147:707–718. doi: 10.1104/pp.107.115634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci U S A. 2004;101:16091–16098. doi: 10.1073/pnas.0407107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, et al. BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Wei N, Deng XW. The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Physiol. 2000;124:1520–1524. doi: 10.1104/pp.124.4.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente P, Wei N, Deng XW. Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 1996;15:3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Reyes JC, Muro-Pastor MI, Florencio FJ. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004;134:1718–1732. doi: 10.1104/pp.103.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio C. The fungal GATA factors. Curr Opin Microbiol. 2000;3:126–131. doi: 10.1016/s1369-5274(00)00063-1. [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhou XY, Li L, Xue LJ, Yang X, Xue HW. Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis. Mol Plant. 2009;2:755–772. doi: 10.1093/mp/ssp039. [DOI] [PubMed] [Google Scholar]
- Sun Y, Fan X-Y, Cao D-M, He K, Tang W, Zhu J-Y, He J-X, Bai M-Y, Zhu S, Oh E, et al. Integration of Brassinosteroid Signal Transduction with the Transcription Network for Plant Growth Regulation in Arabidopsis. Dev Cell. 2010 doi: 10.1016/j.devcel.2010.10.010. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons GM, Reid JB. Hormone levels and response during de-etiolation in pea. Planta. 2003;216:422–431. doi: 10.1007/s00425-002-0860-z. [DOI] [PubMed] [Google Scholar]
- Symons GM, Smith JJ, Nomura T, Davies NW, Yokota T, Reid JB. The hormonal regulation of de-etiolation. Planta. 2008;227:1115–1125. doi: 10.1007/s00425-007-0685-x. [DOI] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Tang W, Deng Z, Wang ZY. Proteomics shed light on the brassinosteroid signaling mechanisms. Curr Opin Plant Biol. 2010;23:27–33. doi: 10.1016/j.pbi.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated Transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Vandenbussche F, Habricot Y, Condiff AS, Maldiney R, Van der Straeten D, Ahmad M. HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. 2007;49:428–441. doi: 10.1111/j.1365-313X.2006.02973.x. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Osterlund MT, Kwok SF, Deng XW. Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol. 1997;114:779–788. doi: 10.1104/pp.114.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science. 2001;294:154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW. The role of the COP/DET/FUS genes in light control of arabidopsis seedling development. Plant Physiol. 1996;112:871–878. doi: 10.1104/pp.112.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhang Y, Liu L, Yu L, Tsang S, Tan J, Yao W, Kang MS, An Y, Fan X. Gene Expression Browser: large-scale and cross-experiment microarray data integration, management, search & visualization. BMC Bioinformatics. 2010;11:433. doi: 10.1186/1471-2105-11-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Medrano L, Ohashi K, Fletcher JC, Yu H, Sakai H, Meyerowitz EM. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell. 2004;16:2586–2600. doi: 10.1105/tpc.104.024869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.