Abstract
The echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4-ALK) fusion gene resulting from an inversion within chromosome 2p occurs in approximately 5% of non-small cell lung cancer and is mutually exclusive with Ras and EGFR mutations. In this study, we have used a potent and selective ALK small molecule inhibitor, NPV-TAE684, to assess the oncogenic role of EML4-ALK in non-small cell lung cancer (NSCLC). We show here that TAE684 inhibits proliferation and induces cell cycle arrest, apoptosis, and tumor regression in two NSCLC models that harbor EML4-ALK fusions. TAE684 inhibits EML4-ALK activation and its downstream signaling including ERK, AKT, and STAT3. We used microarray analysis to carry out targeted pathway studies of gene expression changes in H2228 NSCLC xenograft model after TAE684 treatment and identified a gene signature of EML4-ALK inhibition. The gene signature represents 1210 known human genes, and the top biologic processes represented by these genes are cell cycle, DNA synthesis, cell proliferation, and cell death. We also compared the effect of TAE684 with PF2341066, a c-Met and ALK small molecule inhibitor currently in clinical trial in cancers harboring ALK fusions, and demonstrated that TAE684 is a much more potent inhibitor of EML4-ALK. Our data demonstrate that EML4-ALK plays an important role in the pathogenesis of a subset of NSCLC and provides insight into the mechanism of EML4-ALK inhibition by a small molecule inhibitor.
Introduction
Anaplastic lymphoma kinase (ALK) is implicated in the oncogenesis of both hematopoietic and nonhematopoietic malignancies, and there is a strong rationale for ALK inhibition as a targeted therapy against cancers harboring oncogenic alterations of ALK [1]. ALK was first identified as a fusion protein containing the N-terminal portion of nucleophosmin (NPM) and the intracellular domain of ALK, which results from the t(2;5)(p23;q35) chromosomal rearrangement present in 60% to 70% of anaplastic large cell lymphomas (ALCLs) [2]. The oligomerization domain in NPM leads to dimerization and constitutive activation of NPM-ALK, which has been shown to transform various hematopoietic cell types in vitro and support tumor formation in vivo [3–6]. A causative role of NPM-ALK in pathogenesis of ALCL has been established [7–9]. NPM-ALK mediates oncogenesis, at least in part, through activation of PI3K/Akt, Jak/STAT, and Raf/MEK/ ERK signaling pathways, resulting in increased cell proliferation and resistance to apoptosis [10,11].
Recently, by screening a retroviral complementary DNA expression library generated from a non-small cell lung cancer (NSCLC) patient tumor sample, a novel ALK fusion protein EML4-ALK was identified [12] as a result of a small inversion within the short arm of chromosome 2. EML4-ALK is present in 3% to 7% of NSCLC and is mutually exclusive with K-Ras and EGFR mutations [12–16]. To date, at least seven EML4-ALK variants have been identified, based on the number of exons in EML4 fused to ALK [12–14,17,18]. All EML4-ALK fusions contain a coiled-coil domain within EML4 that mediates constitutive dimerization and activation of EML4-ALK. Overexpression of EML4-ALK in mouse 3T3 fibroblasts resulted in the formation of transformed foci in culture and subcutaneous tumors in nude mice [12]. Furthermore, transgenic mice that express EML4-ALK specifically in lung alveolar epithelial cells developed adenocarcinoma nodules in both lungs within a few weeks after birth, and treatment of these mice with an ALK small molecule inhibitor (SMI) resulted in rapid disappearance of the tumors [19]. These data suggest that EML4-ALK plays a pivotal role in the pathogenesis of NSCLC.
In this study, we used a potent and selective ALK SMI TAE684 [9,20] and two human NSCLC models [13,14,20] that harbor EML4-ALK fusion proteins to investigate further the oncogenic role of ALK fusions in NSCLC. Our results demonstrated that TAE684 inhibits cell proliferation, induces cell cycle arrest and apoptosis, and regresses established xenograft tumors of NSCLC. We show that EML4-ALK shares similar downstream signaling pathways with NPM-ALK, including Akt, ERK, and STAT3, which are inhibited by TAE684 treatment. We identified a gene signature of EML4-ALK inhibition by TAE684 in the NSCLC model that could be used as potential pharmacodynamic (PD) biomarkers to monitor the efficacy of treatment by ALK SMIs. In addition, we compared the efficacy of PF2341066, a c-met and ALK SMI in clinical development [21–23], with TAE684 in NSCLC models and demonstrated that PF2341066 is not as potent compared with TAE684 in inhibiting EML4-ALK oncogenic functions in vitro and in vivo.
Materials and Methods
Antibodies
Antibodies against human ALK, phospho-ALK (Tyr1604), Akt, phospho-Akt (Ser 473), ERK, phospho-ERK (Thr202/Tyr204), STAT3, and phospho-STATA3 (Tyr705) were obtained from Cell Signaling (Danvers, MA).
Cell Lines and Culture
Human NSCLC cell lines H2228 and H3122 were obtained from ATCC (Manassas, VA) and National Cancer Institute (Bethesda, MD), respectively. Cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. The cells have been tested for EML4-ALK fusions by reverse transcription-polymerase chain reaction regularly while maintained in culture.
Kinase Inhibitor
TAE684 and PF2341066 were synthesized following published procedures [9,21]. The structures of the compounds were confirmed by H nuclear magnetic resonance and the purity was determined by high-performance liquid chromatography at a wavelength of 254 nm as 100% pure.
Proliferation and Apoptosis Assays
Cells were seeded at 5000 cells per well in 96-well plates and treated with TAE684 at various doses (dose range, 1–1000 nM) for 24 to 72 hours. Cell proliferation was measured using CellTiter-Glo Luminescent Cell Viability Assay, and apoptosis was measured using Caspase3/7-Glo assay (Promega, Madison, WI) following the manufacturer's instructions.
Cell Cycle Analysis and Annexin V Staining
H2228 and H3122 cells were treated with 50 or 200 nM TAE684 for 24 hours and then synchronized [24] with hydroxyurea (HU). Cells were arrested in HU for 20 hours and released, and the cell cycle distribution was determined by flow cytometry. For cell cycle analysis, cells were harvested, fixed in 70% ethanol at 4°C overnight, washed in PBS, and treated with RNase A (0.1 mg/ml; Sigma, St Louis, MO) and propidium iodide (50 µg/ml; Sigma) for 30 minutes at 37°C. Samples were analyzed on FACScalibur Flow Cytometer (BD Bioscience, San Jose, CA). Cell apoptosis was determined using the annexin V-PE Apoptosis Detection Kit (BD Biosciences, San Jose, CA 95131) according to the manufacturer's instruction. Cell cycle distribution and percent of apoptotic cells were analyzed by FlowJo Data Analysis Software.
Xenograft Experiments
All studies were conducted in accordance with the Guidance for the Care and Use of Laboratory Animals (National Institutes of Health, Bethesda, MD) and approved by Institutional Animal Care and Used Committee. A total of 5 x 106 cells were implanted subcutaneously into the right flank of nude mice. When the tumor size reached 300 mm3 (H2228 and H3122) or 100 mm3 (A549), mice were randomized into different treatment groups (n = 10 for each group). TAE684 and PF2341066 were administered daily by oral gavage in formulations as described previously [9,21]. Tumor volume was measured twice weekly for 15 to 25 days. Statistical analyses were performed using two-way analysis of variance for comparison of tumor growth in different treatment groups.
For PD studies, mice bearing established tumors (mean size, 300 mm3) were treated with TAE684 at 15 mg/kg (H2228) or 30 mg/kg (H3122) for 0, 24, 48, and 72 hours. At each time point, tumors were excised, messenger RNA (mRNA) was extracted for microarray, and cell lysates were prepared for Western blot analysis. Tumor samples were fixed in formalin, and Ki-67 and cleaved caspase 3 (CC3) immunohistochemistry (IHC) was performed. For apoptosis analysis, tumor cells were separated from associated leukocytes by sorting out CD45-positive cells, stained with annexin V, and followed by flow cytometry.
Microarray Analysis
Messenger RNA expression profiling of H2228 xenograft tumors treated with TAE684 for 0, 24, 48, and 72 hours was carried out on Affymetrix GeneChip Human Genome U133 Plus 2.0 Array as per the manufacturer's protocol. Expression summary values for all probe sets were calculated using the RMA algorithm as implemented in the rma package from Bioconductor [25]. Statistical analyses of differentially expressed genes were performed using linear models and empirical Bayes moderated statistics as implemented in the limma package from Bioconductor. To obtain the biologic processes that are overrepresented by the differentially expressed genes, hypergeometric tests for association of Gene Ontology (GO) biologic process categories and genes were performed using the GOstats and Category packages, and P values for high-level generic GO slim terms were reported. The list of genes involved in cell cycle and apoptosis pathways was compiled from related canonical pathway gene sets from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/index.jsp). Hierarchical clustering of the expression profile was performed using the Pearson correlation as the similarity measure and complete linkage as the agglomeration method. The list of potential biomarkers was generated using Ingenuity Pathways Analysis (Ingenuity Systems, http://www.ingenuity.com).
Results
TAE684 Induces Apoptosis and Cell Cycle Arrest of H2228 NSCLC Cells
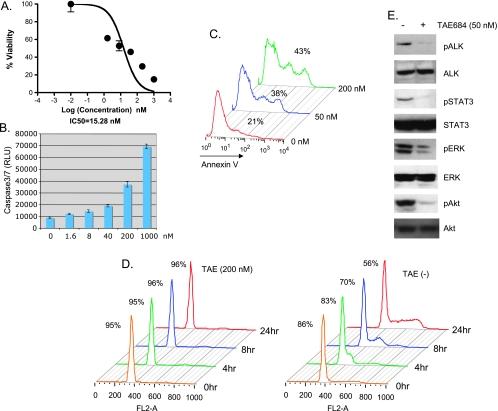
To evaluate the role of EML4-ALK in NSCLC, we first tested the effect of TAE684, a selective ALK SMI [9,20] (Supplemental Data, Figure W1) on NSCLC cell line H2228 that expresses EML4-ALK variant 3, containing exons 1 to 6 of EML4 [13]. TAE684 reduced viability of H2228 cells in a dose-dependent manner (Figure 1A), with an IC50 (50% inhibition) of 15 nM. This decrease in cell viability is caused in part by TAE684-induced apoptosis as demonstrated by the increased activation of caspase 3/7 (Figure 1B) and annexin V staining (Figure 1C). Seventy-two hours after TAE684 treatment, annexin V-positive cells increased from 21% to 38% (50 nM TAE684) and 43% (200 nM TAE684; Figure 1C). To test the impact of TAE684 on cell cycle progression, TAE684-treated H2228 cells were stained with propidium iodide (PI) and analyzed for cell cycle distribution. In H2228 cells treated with TAE684 for 24 hours, 96% cells were arrested in G1 phase compared with 56% of cells in vehicle-treated control (Figure 1D). Collectively, these results suggest that TAE684 inhibits the growth of H2228 NSCLC cells by both induction of apoptosis and inhibition of cell cycle progression, although TAE684-induced G1 arrest seems to be the major mechanism that reduces H2228 growth.
Figure 1.
TAE684 inhibits cell proliferation, induces apoptosis and cell cycle arrest, and inactivates EML4-ALK signaling of H2228 cells. (A) TAE684 reduces H2228 cell survival. Cells were treated with indicated concentrations of TAE684 and cell viability was measured by CellTiterGlo 72 hours after treatment. (B, C) TAE684 induces apoptosis. Cells were treated with indicated concentrations of TAE684. Caspase 3/7 activation was measured by Cap3/7Glo 48 hours after treatment (B). Cells were treated with 50 or 200 nM TAE684; apoptosis was measured at indicated time by annexin V stain 72 hours after treatment (C). (D) TAE684 induces G1 arrest. Cells were treated with 200 nM TAE684 for 24 hours, arrested at G1/S transition with HU, and released into the S phase. Cells were harvested at indicated time, stained with PI, and analyzed for cell cycle distribution. Percentage of cells in G1 at various time points is shown. (E) TAE684 inhibits ALK signaling. Cells were treated with 50 nM TAE684 for 6 hours, and Western blot analysis was performed to determine phosphorylation of ALK, Akt, ERK, and STAT3.
In addition, TAE684 inhibited ALK activation and downstream signaling. As shown in Figure 1E, 50 nM TAE684 inhibited phosphorylation of ALK, Akt, STAT3, and ERK. These results suggest that EML4-ALK activates ERK, PI3K/Akt, and STAT signaling in H2228 cells, similar to NPM-ALK in ALCL cells.
TAE684 Induces Regression of Established H2228 Xenograft Tumors
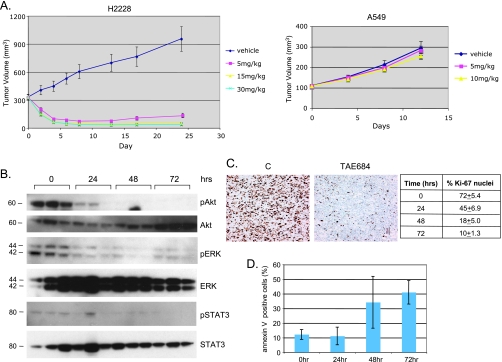
Previous study has shown that TAE684 induces regression of established lymphomas expressing NPM-ALK fusions [9]; we reasoned that if EML4-ALK is the oncogenic driver in NSCLC, TAE684 should have a similar effect on these tumors. To test this hypothesis, we established the H2228 xenograft model. When the tumor size reached an average of 300 mm3, mice were randomized into control and three treatment groups, and TAE684 was administered by oral gavage at 5, 10, and 30 mg/kg per day. After 7 days of treatment, tumors in the TAE684 treatment group at all dose levels showed almost complete regression, whereas tumors in the control group continues to grow (Figure 2A). TAE684 had no effect on xenograft tumor growth of A549, an NSCLC cell line that does not express ALK fusions, but contains K-Ras mutation and expresses wild-type EGFR [26] (Figure 2A) and it did not affect the body weight of treated mice (not shown). These results suggest that TAE684 specifically inhibits EML4-ALK in H2228 tumors.
Figure 2.
TAE684 induces regression of established H2228 xenograft tumors. (A) TAE684 inhibits H2228 but not A549 xenograft tumor growth. A total of 5 x 106 H2228 or A549 cells were implanted subcutaneously (sc) into nude mice. When the average tumor size reached 300 mm3 (H2228) or 100 mm3 (A549), mice were randomized into different treatment groups. TAE684 was administered per os daily at the indicated doses or vehicle (10% 1-methyl-2-pyrrolidinone/90% PEG30). n = 10 for each group. P= 7.6 x 10-32 (control vs TAE684 5 mg/kg), P = 8 x 10-11 (TAE684 5 vs 15 mg/kg). (B) TAE684 inhibits EML4-ALK signaling in H2228 xenograft tumors. Established H2228 xenograft tumors were treated with 15 mg/kg TAE684, cell lysates were prepared at the indicated time, and Western blot analysis was performed to measure phosphorylated Akt, ERK, and STAT3. (C, D) TAE684 inhibits proliferation and induces apoptosis of H2228 tumor cells. Established H2228 xenogaft tumors were treated with 15 mg/kg TAE684, and Ki-67 IHC was performed at the indicated time to measure cell proliferation. For apoptosis analysis, tumors were collected at the indicated time after dosing, and tumor cells were stained with annexin V (n = 3 for each time point).
To understand the mechanisms involved in TAE684 inhibition of H2228 tumor growth, we performed a pharmacodynamic study. Mice bearing established H2228 xenograft tumors were treated with either TAE684 (15 mg/kg) or vehicle for 3 days. Immunoblot analysis of protein extracted from tumor revealed a reduction in the phosphorylation levels of ALK downstream targets Akt, ERK, and STAT3, 24 hours after dosing (Figure 2B). There was a time-dependent decrease in Ki-67-positive cells with only 10% positive cells at 72 hours after dosing (Figure 2C), suggesting that TAE684 strongly inhibits tumor cell proliferation. TAE684 also induces tumor cell apoptosis as determined by annexin V stain, with 40% of tumor cells undergoing apoptosis 72 hours after dosing (Figure 2D). These results suggest that TAE684 inhibits NSCLC tumor growth by inhibition of EML4-ALK signaling, which in turn leads to decreased proliferation and increased apoptosis of tumor cells.
Effect of TAE684 on H3122 NSCLC Cells In Vitro and In Vivo
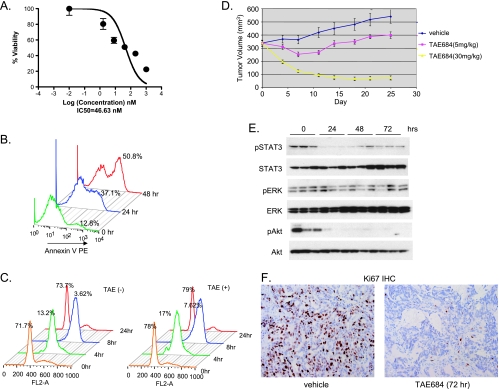
To further assess the oncogenic role of EML4-ALK in NSCLC, we tested the effect of TAE684 on another NSCLC model H3122, which harbors EML4-ALK variant 1 containing exons 1 to 13 of EML4 [14]. TAE684 reduces H3122 cell viability in a dose-dependent manner, with an IC50 of 47 nM (Figure 3A), which is higher than the 15-nM IC50 (Figure 1A) observed in H2228 cell. The reduced cell viability by TAE684 is likely due to the rapid induction of apoptosis; 50% of cells were stained annexin V-positive 48 hours after TAE684 treatment (Figure 3B). TAE684 does not seem to affect cell cycle progression in this cell line (Figure 3C), suggesting that induction of apoptosis plays a more important role in TAE684 inhibition of H3122 cell growth.
Figure 3.
Effect of TAE684 on H3122 NSCLC cells in vitro and in vivo. (A) TAE684 reduces H3122 viability. Cells were treated with indicated concentrations of TAE684. Cell viability was measured by CellTiterGlo 72 hours after treatment. (B) TAE684 induces apoptosis. Cells were treated with 200 nM TAE684, and apoptosis was measured at the indicated time after dosing by annexin V stain. (C) TAE684 does not affect cell cycle progression. Cells were treated with 200 nM TAE684 for 24 hours, arrested at G1/S transition with HU, and released into the S phase. Cells were harvested at indicated time, stained with PI, and analyzed for cell cycle distribution. Percentage of cells in G1 at various time points is shown. (D) TAE684 inhibits established H3122 xenograft tumor growth. A total of 5 x 106 cells were implanted sc into nude mice. When the average tumor size reached 300 mm3, mice were randomized into different treatment groups. TAE684 was administered per os daily at the indicated doses or vehicle (10% 1-methyl-2-pyrrolidinone/90% PEG30). n = 10 for each treatment group. P = 10-7 (control vs TAE684 5 mg/kg), P= 5.6 x 10-30 (TAE684 5 vs 30 mg/kg). (E) TAE684 inhibits EML4-ALK signaling in H3122 xenograft tumors. Established H3122 xenogaft tumors were treated with 30 mg/kg TAE684, cell lysates were prepared at the indicated time, and Western blot analysis was performed to measure phosphorylated Akt, ERK, and STAT3. (F) TAE684 inhibits proliferation of H3122 tumor cells. Established H3122 xenogaft tumors were treated with 30 mg/kg TAE684, and Ki-67 and CC3 IHC was performed at the indicated time to measure cell proliferation and apoptosis, respectively (n = 5 for each time point).
To test the effect of TAE684 on tumor growth in vivo, established H3122 xenograft tumors were treated with TAE684 at 5 and 30 mg/kg per day. Figure 3D shows that, at 30 mg/kg, TAE684 induces tumor regression, whereas at 5 mg/kg, it causes tumor growth stasis. These results are consistence with that of H2228 model; however, a higher dose of TAE684 was required to achieve tumor regression given the decreased potency in vitro.
We performed a pharmacodynamic study to examine the immediate molecular effects of short-term TAE684 treatment on the established H3122 tumors. Immunoblot analysis of protein extracts from xenograft tumors revealed a reduction in phosphorylation levels of EML4-ALK downstream signaling target STAT3 and Akt, but there was little change in phosphorylated ERK (Figure 3E). Ki-67 IHC showed that treatment of tumors with TAE684 resulted in a time-dependent reduction in Ki-67-positive nuclei, from 50% in vehicle treated tumors to 7% 72 hours after administration of TAE684 (Figure 3F). Furthermore, TAE684 induces rapid apoptosis of tumor cells, as demonstrated by cleaved caspase 3 (CC3) IHC (data not shown).
Taken together, these data showed that TAE684 is able to inactivate EML4-ALK signaling, reduce cell survival in vitro, and inhibit xenograft tumor growth in vivo. These results provide further evidence that the EML4-ALK plays a pivotal role in the oncogenesis of NSCLC.
TAE684 Is a More Potent Inhibitor of EML4-ALK than PF2341066
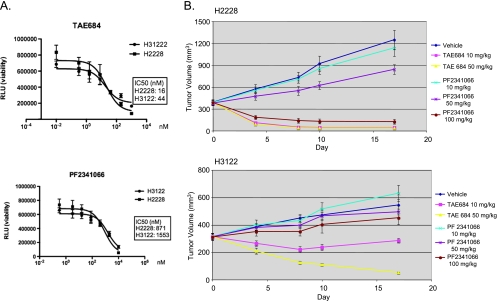
PF2341066, developed as c-Met SMI [21,22], also inhibits ALK kinase activity, with IC50 of 4 and 24 nM in in vitro kinase assays for c-met and ALK, respectively. It has been shown that PF2341066 inhibits ALCLs proliferation in vitro and xenograft tumor growth in vivo [22]. A recent phase 1 clinical trial demonstrated that PF2341066 exhibits activity in patients whose tumor harbor ALK fusion proteins [23]. However, there are few preclinical data for this compound in NSCLC models and how it compares with other ALK SMIs. We therefore compared TAE684 with PF2341066 in the two NSCLC models that contain EML4-ALK fusions. As shown in Figure 4A, although PF2341066 is able to reduce survival of H2228 and H3122 cells, it is much less potent compared with TAE684. The IC50 for PF2341066 is 871 and 1551 nM for H2228 and H3122, respectively, compared with 16 (H2228) and 44 nM (H3122) for TAE684.
Figure 4.
PF2341066 is not as potent as TAE684 in the inhibition of EML4-ALK fusions in NSCLC. (A) Effect on H2228 and H3122 in vitro growth. Cells were treated with indicated concentrations of TAE684 (upper panel) or PF2341066 (lower panel), and cell viability was measured by CellTiterGlo 72 hours after treatment. (B) Effect on H2228 and H3122 xenograft tumor growth. A total of 5 x 106 cells were implanted sc into nude mice. When the average tumor size reached 300 mm3, mice were randomized into different treatment groups. TAE684 and PF2341066 were administered per os daily at the indicated doses or vehicles (TAE684: 10% 1-methyl-2-pyrrolidinone/90% PEG30, PF2341066: water). n = 10 for each treatment group. Pvalues for H2228 model are as follows: P= .38 (PF 10 mg/kg vs vehicle), P= .0002 (PF 50 vs 10 mg/kg), P= 2 x 10-18 (PF 100 vs 50 mg/kg), P= 8 x 10-5 (TAE 10 mg/kg vs P F 100 mg/kg). H3122 model: P= .3 (PF 10 mg/kg vs vehicle), P= .22 (PF 50 mg/kg vs vehicle), P= .005 (PF 100 mg/kg vs vehicle), P= 10-6 (TAE 10 mg/kg vs PF 100 mg/kg), P= 1.7 x 10-11 (TAE 50 vs 10 mg/kg).
In xenograft models, TAE684 at 10 mg/kg resulted in complete regression of H2228 tumors within a week, whereas PF2341066 at the same dose has no effect on the tumor growth (Figure 4B, upper panel). The amount of 100 mg/kg of PF2341066 was needed for tumor regression in this model (n = 10, P = 2 x 10-18). However, even at this dose level, it took longer to achieve complete regression compared with TAE684 (Figure 4B, upper panel; P = 8.3 x 10-5, TAE684 10 mg/kg vs PF2341066 100 mg/kg). In the H3122 model, treatment with TAE684 at either 10 or 50 mg/kg resulted in tumor regression, whereas treatment with PF2341066 had a marginal effect on tumor growth at the same dose levels (Figure 4B, lower panel). Even at 100 mg/kg, PF2341066 only moderately inhibited tumor growth (Figure 4B, lower panel; P = .005). No significant body weight loss was observed in all treatment groups. These results suggest that PF2341066 is not as a potent inhibitor of EML4-ALK compared with TAE684.
Identification of a Gene Signature of EML4-ALK Inhibition by TAE684 in H2228 Cells
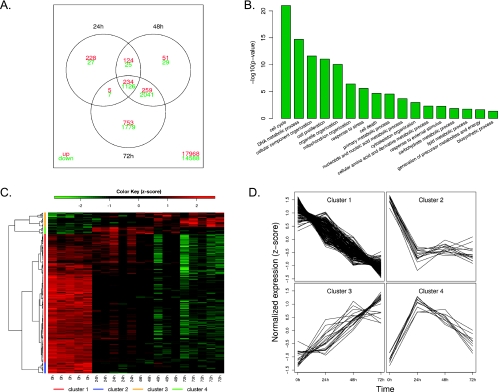
To investigate further the mechanisms involved in TAE684 inhibition of EML4-ALK, we performed mRNA profiling of H2228 cells after TAE684 treatment. Analysis of the microarray data revealed dramatic changes in the mRNA expression profile of H2228 xenografts on treatments with TAE684. The number of differentially expressed genes (compared with the pretreatment group, FDR < 0.1, fold change > 1.5) increases during the drug treatment with 1776, 3889, and 6204 genes at 24, 48, and 72 hours after treatment, respectively (Figure 5A). Among these genes, 234 are commonly upregulated and 1126 are commonly downregulated at all three time points. The top biologic processes (the number of genes in each process are listed in Table W1) represented by these genes include cell cycle, DNA metabolic process, and cell proliferation (Figure 5B), consistent with the known role of ALK fusion proteins in promoting cell cycle progression.
Figure 5.
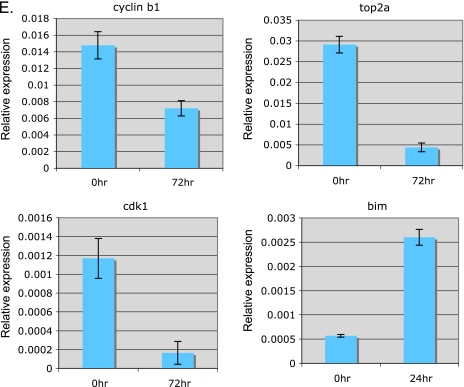
Microarray analysis gene expression profile after TAE684 treatment of H2228 xenograft tumors. (A) Venn diagram showing the overlap between the differentially expressed genes at three time points. (B) Top biologic processes represented by the differentially expressed genes. P values were from hypergeometric tests. (C) Hierarchical clustering of the expression profile of the genes involved in cell cycle and apoptosis pathways revealed four major groups. Each row represents one gene, whose expression values were scaled to means of zero and SDs of 1 (z-score transformation), and color-coded according to the color key. (D) Expression profiles of the four major clusters. Each line in the plots represents one gene. The normalized expression value of a gene at each time point is represented by the mean of z-scores from the five replicates. (E) Expression of representative genes by quantitative polymerase chain reaction. Quantitative polymerase chain reaction was performed on several selected genes using RNA samples from untreated or 24 hours (bim) or 72 hours (cyclin B1, CDK1, and TOP2A) after TAE684 treatment. Relative expression levels are shown.
We then focused our attention on genes known to be involved in cell cycle or apoptosis pathways. There are 210 genes (the complete list of the genes and changes in their expression levels after TAE684 treatment are in Table W2) in these pathways that are differentially expressed at least at one time point compared with the pretreatment group. Unsupervised hierarchical clustering of the expression profile of these genes suggested that there are four major groups (Figure 5, C and D).
Genes that are downregulated after TAE684 treatment are in clusters 1 and 2 (Figure 5D). Cluster 1 contains 168 genes that were downregulated over time, and cluster 2 has 14 genes that were rapidly downregulated 24 hours after dosing and then leveled off. These two clusters include ALK downstream signaling molecules AKT1, MEK, and ERK, as well as MAP kinases involved in stress response and apoptosis (JNK2 and MEKK7). The genes that exhibit strongest inhibition by TAE684 are those involved in cell cycle progression. TAE684 treatment resulted in more than a 10-fold decrease in mRNA levels of several cyclins and cyclin-dependent kinases. TAE684 also strongly downregulated the expression of topoisomerase IIα (TOP2A, >50-fold) and pituitary tumor transforming gene 1 (PTTG1, 3-fold), two proteins involved in chromosome condensation [27] and chromatid separation [28,29], respectively. Genes that are upregulated by TAE684 treatment are in clusters 3 (consistently upregulated over time) and 4 (transiently upregulated), representing a total of 28 genes. Bim, a known apoptosis enhancer protein, and p27/CDKN1B, a tumor suppressor protein that inhibits cell cycle progression are among the upregulated genes after TAE684 treatment. We confirmed the microarray results by performing quantitative polymerase chain reaction for several representative genes. Figure 5E shows that cyclin B1, TOP2A, and CDK1 mRNA levels decrease with TAE684 treatment, whereas the expression level of Bim increases, consistent with the microarray data.
To identify potential PD biomarkers for ALK inhibitor treatment, we analyzed the 193 genes that are consistently upregulated or down-regulated and are related to cell cycle and apoptosis for their known presence in human blood according to the Ingenuity Pathways Analysis tool. Twenty-seven genes that are downregulated (2- to 53-fold) on TAE684 treatment and are detectable in whole blood or plasma according to published literatures [30–34] are listed in Table 1. The expression of these genes could potentially be used to monitor PD properties of ALK SMIs.
Table 1.
Potential PD Biomarkers for ALK SMI Treatment in NSCLC.
| Symbol | Entrez Gene Name | Family | Fold Change | FDR Adjusted P value |
| AKT1 | v-akt murine thymoma viral oncogene homolog 1 | Kinase | -3.23 | 2.48E-10 |
| ATF1 | Activating transcription factor 1 | Transcription regulator | -9.9 | 1.23E-10 |
| CCNA2 | Cyclin A2 | Other | -41.76 | 1.8E-13 |
| CCNB1 | Cyclin B1 | Other | -181.97 | 1.74E-17 |
| CCNB2 | Cyclin B2 | Other | -70.13 | 1.49E-17 |
| CCNE2 | Cyclin E2 | Other | -48.24 | 2.76E-16 |
| CDC2 | Cell division cycle 2, G1 to S and G2 to M | Kinase | -32.91 | 1.49E-17 |
| CDK2 | Cyclin-dependent kinase 2 | Kinase | -4.15 | 4.53E-12 |
| CHEK1 | CHK1 checkpoint homolog (Schizosaccharomyces pombe) | Kinase | -17.77 | 2.85E-11 |
| EGFR | Epidermal growth factor receptor | Kinase | -5.6 | 1.66E-10 |
| H2AFX | H2A histone family, member X | Other | -7.56 | 3.6E-15 |
| HMGB2 | High-mobility group box 2 | Transcription regulator | -8.3 | 2.88E-14 |
| HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Enzyme | -3.16 | 5.04E-09 |
| IGFBP3 | Insulin-like growth factor binding protein 3 | Other | -5.71 | .00000303 |
| IL8 | Interleukin 8 | Cytokine | -8.4 | 1.97E-13 |
| IL1R1 | Interleukin 1 receptor, type I | Transmembrane receptor | -5.2 | 1.16E-11 |
| IL1RAP | Interleukin 1 receptor accessory protein | Transmembrane receptor | -18.67 | .000848 |
| IRS1 | Insulin receptor substrate 1 | Other | -6.84 | 1.84E-11 |
| KITLG | KIT ligand | Growth factor | -4.77 | 1.03E-11 |
| LMNB1 | Lamin B1 | Other | -21.41 | 3.38E-15 |
| MAP2K1 | Mitogen-activated protein kinase kinase 1 | Kinase | -3.95 | 1.61E-11 |
| MAP2K2 | Mitogen-activated protein kinase kinase 2 | Kinase | -3.15 | 4.98E-08 |
| MYC | v-myc myelocytomatosis viral oncogene homolog | Transcription regulator | -5.59 | 5.3E-11 |
| PCNA | Proliferating cell nuclear antigen | Other | -5.98 | 4E-13 |
| PLK1 | Polo-like kinase 1 (Drosophila) | Kinase | -20.76 | 3.1E-14 |
| SHC1 | SHC (Src homolog 2 domain containing) transforming protein 1 | Other | -4.15 | 5.17E-12 |
| TOP2A | Topoisomerase (DNA) II alpha 170 kDa | Enzyme | -156.1 | 1.74E-20 |
Discussion
In this study, we have assessed the effect of a selective and potent ALK SMI TAE684 [9,20] on two NSCLC cell lines that contain EML4-ALK fusion proteins [13,14,20] in vitro and in vivo. Previous studies have shown that TAE684 exhibits more than 100-fold selectivity over insulin receptor in cell-based assays [9], and that screening of more than 600 cancer cell lines showed that only a few cancer cell lines that contain either ALK fusions or amplification/mutations are sensitive to TAE684 [20]. Our results show that TAE684 inhibits proliferation and induces cell cycle arrest, apoptosis, and tumor regression of NSCLC cell lines containing EML4-ALK fusions, confirming a pivotal role of EML4-ALK in NSCLC.
H2228, harboring EML4-ALK variant 3 [13], is slightly more sensitive to TAE684 inhibition than H3122 that expresses EML4-ALK variant 1 [14,20]. The in vitro IC50 on cell viability is 15 and 46 nM, and the dose required for tumor regression is 5 and 30 mg/kg for H2228 and H3122, respectively. Our results are consistent with previously published results by McDermott et al. [20], in that both H2228 and H31222 are extremely sensitive to TAE684. The results published by Koivunen et al. [14] showed that, whereas H3122 is sensitive to TAE684 inhibition, H2228 is not. It is well known that the same cell line, such as H2228, may evolve into distinct populations owing to different cell culture conditions and/or techniques, thus accounting for the differential sensitivity to TAE684.
Furthermore, TAE684 rapidly induces cell cycle arrest in H2228 (Figure 1), but it has no effect on cell cycle progression in H3122 (Figure 3C). Nevertheless, TAE684 has a greater impact on inducing apoptosis in H3122, with more than 50% cells undergoing apoptosis 48 hours after treatment, compared with 25% in H2228. The slightly higher concentration required to achieve EC50 (50% efficacy; 50 nM) in apoptosis assays compared with the IC50 (15 nM) to measure the metabolic activity in H2228 cell could be explained by the fact that TAE684 affects both cell cycle progression and apoptosis.
Consistent with these results, TAE684 inhibits different EML4-ALK downstream signaling molecules in the two cell lines. Whereas TAE684 inhibits phosphorylation of ERK, STAT3, as well as Akt in H2228, it affects only STAT3 and Akt but not ERK in H3122. These results suggest that ALK SMI may have different modes of action on various EML4-ALK fusion proteins.
PF2341066, an SMI originally developed for c-Met [21] but also inhibits ALK kinase activity [22], has been reported to exhibit clinical activity in cancer patients whose tumors harbor ALK fusion proteins [23]. However, there are few published data on the activity of this compound in NSCLC models containing EML4-ALK fusions. We therefore performed side-by-side comparison of TAE684 and PF2341066 in these models. Our results showed that both H2228 and H3122 are partially resistant to PF2341066 in the in vitro cell viability assay, with IC50 of 871 and 1553 nM, respectively, compared with IC50 of 15 and 46 nM for TAE684. In vivo, at least 100 mg/kg of PF2341066 is required to induce tumor regression in the H2228 model, whereas TAE684 at 10 mg/kg is more efficacious in the same model. In the H3122 model, PF2341066 only had a cytostatic effect even at 100 mg/kg, whereas TAE684 at 30 mg/kg induced tumor regression. These results suggest that PF2341066 is not as potent as TAE684 in inhibiting EML4-ALK. So far, PF2341066 was reported to achieve mostly partial responses or stable diseases but not complete response in clinical trials [23]. It is conceivable that a more potent and selective ALK SMI could achieve better responses in patients whose cancers harbor ALK fusion proteins.
To begin to understand the mechanisms involved in the inhibition of EML4-ALK by SMI, we conducted a pharmacodynamic study combined with gene profiling in a H2228 xenograft model treated with TAE684. We identified several biologic processes in which the gene expression is modulated by TAE684 treatment. On the top of the list are genes involved in cell cycle. Among the genes that are rapidly and persistently downregulated by TAE684 (cluster 1, Figure 5C) are CDC2, CDC7, and CDK4, involved in promoting the G1- to S-phase transition, and the prereplication complex machinery such as MCMs (minichromosome maintenance proteins) whose expression peaks at the G1-S boundary [35]. This change in gene expression profile is consistent with the observation that treatment of H2228 cells with TAE684 induces G1 arrest. In addition to the G1-S phase of the cell cycle, TAE684 modulates the expression of genes involved in chromosome condensation (TOP2A), chromatid separation (PTTG), and spindle checkpoint functions (BUBs, budding uninhibited by benzimidazole) [36], suggesting that TAE684 affects multiple aspects of the cell cycle. TAE684 seems to promote apoptosis by upregulating the expression of proapoptotic proteins such as Bim and by downregulating genes in Akt/JNK signaling pathways including Akt1, IRAK, and MAK9.
We also performed gene profiling in H3122 xenograft tumors (data not shown). The gene signature in H3122 cell on TAE684 treatment is overlapping but also different from that of H2228. For example, cell cycle is not a top biologic process in H3122, but apoptosis is. This is consistent with our results that TAE684 reduces cell viability in H3122 by inducing apoptosis with no effect on cell cycle progression.
Among the 210 genes in Figure 5C, many can be detected in blood [30–34]. These include several cyclins (A2, B1, B2, D3, E1, and E2), CDC2, CDK2, as well as ALK downstream signaling molecules (Table 1). The changes in mRNA levels for most of these genes on TAE684 treatment are dramatic (e.g., 7-fold for cyclin B2 and 53-fold for TOP2A). TOP2A is frequently amplified in cancers including breast [36], colon [37], as well as prostate [38] and is a predictive marker to cytotoxic drugs such as anthracycline [39]. Cyclin B2 is one of the key genes required for progression through mitosis and is frequently over-expressed in cancer. The expression of cyclin B2 is used as a diagnostic marker for lung cancer [40], a prognostic marker for colorectal cancer [41], and a PD biomarker for the cyclin-dependent kinase inhibitor seliciclib [42]. These genes can therefore be potential PD biomarkers for monitoring ALK SMI in the treatment of NSCLC.
In conclusion, we have demonstrated that EML4-ALK fusion is an oncogenic driver in two NSCLC models that harbor this genetic alteration. The primary human NSCLC tumors are more heterogeneous compared with cell line models and thus may have less dramatic responses to ALK SMI. PF2341066, a moderately potent inhibitor of EML4-ALK as demonstrated here, exhibited clinical activity in multiple patients harboring ALK fusion proteins in their tumors [23], confirming the pivotal role of ALK fusions in oncogenesis. Therefore, a more potent and selective ALK SMI should be able to achieve superior clinical efficacy akin to the effect of Gleevec on BCR-Abl in CML and GIST.
Supplementary Material
Acknowledgments
The authors thank Allen Ebens and Deepak Sampath for reading the manuscript and providing insightful suggestions.
Abbreviations
- EML4-ALK
echinoderm microtubule-associated protein like-4-anaplastic lymphoma kinase
- NSCLC
non-small cell lung cancer
- ALCL
anaplastic large cell lymphoma
- TOP2A
topoisomerase II α
- PD
pharmacodynamics
Footnotes
There are no financial support and potential conflicts of interest.
This article refers to supplementary materials, which are designated by Figures W1, Tables W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Mossé YP, Wood A, Maris JM. Inhibition of ALK signaling for cancer therapy. Clin Cancer Res. 2009;15:5609–5614. doi: 10.1158/1078-0432.CCR-08-2762. [DOI] [PubMed] [Google Scholar]
- 2.Morris SW, Kirstein MN, Valentine MB, Dittmer K, Shapiro DN, Look AT, Saltman DL. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 3.Duyster J, Bai RY, Morris SW. Translocations involving anaplastic lymphoma kinase (ALK) Oncogene. 2001;20:5623–5637. doi: 10.1038/sj.onc.1204594. [DOI] [PubMed] [Google Scholar]
- 4.Turner SD, Alexander DR. What have we learnt from mouse models of NPM-ALK-induced lymphomagenesis? Leukemia. 2005;19:1128–1134. doi: 10.1038/sj.leu.2403797. [DOI] [PubMed] [Google Scholar]
- 5.Kuefer MU, Look AT, Pulford K, Behm FG, Pattengale PK, Mason DY, Morris SW. Retrovirus-mediated gene transfer of NPM-ALK causes lymphoid malignancy in mice. Blood. 1997;90:2901–2910. [PubMed] [Google Scholar]
- 6.Chiarle R,, Gong JZ, Guasparri I, Pesci A, Cai J, Liu J, Simmons WJ, Dhall G, Howes J, Piva R, et al. NPM-ALK transgenic mice spontaneously develop T-cell lymphomas and plasma cell tumors. Blood. 2003;101:1919–1927. doi: 10.1182/blood-2002-05-1343. [DOI] [PubMed] [Google Scholar]
- 7.Wan W, Albom MS, Lu L, Quail MR, Becknell NC, Weinberg LR, Reddy DR, Holskin BP, Angeles TS, Underiner TL, et al. Anaplastic lymphoma kinase activity is essential for the proliferation and survival of anaplastic large-cell lymphoma cells. Blood. 2006;107:1617–1623. doi: 10.1182/blood-2005-08-3254. [DOI] [PubMed] [Google Scholar]
- 8.Piva R, Chiarle R, Manazza AD, Taulli R, Simmons W, Ambrogio C, D'Escamard V, Pellegrino E, Ponzetto C, Palestro G, et al. Ablation of oncogenic ALK is a viable therapeutic approach for anaplastic large-cell lymphomas. Blood. 2006;107:689–697. doi: 10.1182/blood-2005-05-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galkin AV, Melnick JS, Kim S, Hood TL, Li N, Li L, Xia G, Steensma R, Chopiuk G, Jiang J, et al. Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc Natl Acad Sci USA. 2007;104:270–275. doi: 10.1073/pnas.0609412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amin HM, Lai R. Pathobiology of ALK+ anaplastic large-cell lymphoma. Blood. 2007;110:2259–2267. doi: 10.1182/blood-2007-04-060715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, Raghunath PN, Xue L, Majewski M, Carpentieri DF, Odum N, Morris S, Skorski T, Wasik MA. Multilevel dysregulation of STAT3 activation in anaplasticlymphoma kinase-positive T/null-cell lymphoma. J Immunol. 2002;168:466–474. doi: 10.4049/jimmunol.168.1.466. [DOI] [PubMed] [Google Scholar]
- 12.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 13.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, Choi HG, Kim J, Chiang D, Thomas R, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perner S, Wagner PL, Demichelis F, Mehra R, Lafargue CJ, Moss BJ, Arbogast S, Soltermann A, Weder W, Giordano TJ, et al. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia. 2008;10:298–302. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC, Ho KK, Au JS, Chung LP, Pik Wong M. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 17.Choi YL, Takeuchi K, Soda M, Inamura K, Togashi Y, Hatano S, Enomoto M, Hamada T, Haruta H, Watanabe H, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, Takada S, Ueno T, Yamashita Y, Satoh Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 19.Soda M, Takada S, Takeuchi K, Choi YL, Enomoto M, Ueno T, Haruta H, Hamada T, Yamashita Y, Ishikawa Y, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci USA. 2008;105:19893–19897. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDermott U, Iafrate AJ, Gray NS, Shioda T, Classon M, Maheswaran S, Zhou W, Choi HG, Smith SL, Dowell L, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68:3389–3395. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
- 21.Zou HY, Li Q, Lee JH, Arango ME, McDonnell SR, Yamazaki S, Koudriakova TB, Alton G, Cui JJ, Kung PP, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007;67:4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
- 22.Christensen JG, Zou HY, Arango ME, Li Q, Lee JH, McDonnell SR, Yamazaki S, Alton GR, Mroczkowski B, Los G. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther. 2007;6:3314–3322. doi: 10.1158/1535-7163.MCT-07-0365. [DOI] [PubMed] [Google Scholar]
- 23.Kwak EL, Camidge DR, Clark J, Shapiro I, Maki RG, Ratain MJ, Solomon B, Bang Y, Ou S, Salgia R. Clinical activity observed in phase I dose escalation trial of an oral c-met and ALK inhibitor, PF-02341066. J Clin Oncol. 2009;27 (suppl; abstr 3509) [Google Scholar]
- 24.Scaife RM. Selective and irreversible cell cycle inhibition by diphenyleneiodonium. Mol Cancer Ther. 2005;4:876–884. doi: 10.1158/1535-7163.MCT-05-0009. [DOI] [PubMed] [Google Scholar]
- 25.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yauch RL, Januario T, Eberhard DA, Cavet G, Zhu W, Fu L, Pham TQ, Soriano R, Stinson J, Seshagiri S, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005;11:8686–8698. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- 27.Tan KB, Dorman TE, Falls KM, Chung TD, Mirabelli CK, Crooke ST, Mao J. Topoisomerase II alpha and topoisomerase II beta genes: characterization and mapping to human chromosomes 17 and 3, respectively. Cancer Res. 1992;52:231–234. [PubMed] [Google Scholar]
- 28.Pei L, Melmed S. Isolation and characterization of a pituitary tumor-transforming gene (PTTG) Mol Endocrinol. 1997;11:433–441. doi: 10.1210/mend.11.4.9911. [DOI] [PubMed] [Google Scholar]
- 29.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 30.Banerjee SK, Weston AP, Zoubine MN, Campbell DR, Cherian R. Expression of cdc2 and cyclin B1 in Helicobacter pylori-associated gastric MALT and MALT lymphoma: relationship to cell death, proliferation, and transformation. Am J Pathol. 2000;156:217–225. doi: 10.1016/S0002-9440(10)64722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naderi S, Blomhoff HK. Retinoic acid prevents phosphorylation of pRB in normal human B lymphocytes: regulation of cyclin E, cyclin A, and p21 (Cip1) Blood. 1999;94:1348–1358. [PubMed] [Google Scholar]
- 32.Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Post-translational methylation of high mobility group box 1 (HMGB1) causes its cytoplasmic localization in neutrophils. J Biol Chem. 2007;282:16336–16344. doi: 10.1074/jbc.M608467200. [DOI] [PubMed] [Google Scholar]
- 34.Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, et al. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 35.Kearsey SE, Labib K, Maiorano D. Cell cycle control of eukaryotic DNA replication. Curr Opin Genet Dev. 1996;6:208–214. doi: 10.1016/s0959-437x(96)80052-9. [DOI] [PubMed] [Google Scholar]
- 36.Kang J, Yang M, Li B, Qi W, Zhang C, Shokat KM, Tomchick DR, Machius M, Yu H. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol Cell. 2008;32:394–405. doi: 10.1016/j.molcel.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arriola E, Rodriguez-Pinilla SM, Lambros MB, Jones RL, James M, Savage K, Smith IE, Dowsett M, Reis-Filho JS. Topoisomerase II alpha amplification may predict benefit from adjuvant anthracyclines in HER2 positive early breast cancer. Breast Cancer Res Treat. 2007;106:181–189. doi: 10.1007/s10549-006-9492-5. [DOI] [PubMed] [Google Scholar]
- 38.Al-Kuraya K, Novotny H, Bavi P, Siraj AK, Uddin S, Ezzat A, Sanea NA, Al-Dayel F, Al-Mana H, Sheikh SS, et al. HER2, TOP2A, CCND1, EGFR and C-MYC oncogene amplification in colorectal cancer. J Clin Pathol. 2007;60:768–772. doi: 10.1136/jcp.2006.038281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy AJ, Hughes CA, Barrett C, Magee H, Loftus B, O'Leary JJ, Sheils O. Low-level TOP2A amplification in prostate cancer is associated with HER2 duplication, androgen resistance, and decreased survival. Cancer Res. 2007;67:2893–2898. doi: 10.1158/0008-5472.CAN-06-2962. [DOI] [PubMed] [Google Scholar]
- 40.Stav D, Bar I, Sandbank J. Usefulness of CDK5RAP3, CCNB2, and RAGE genes for the diagnosis of lung adenocarcinoma. Int J Biol Markers. 2007;22:108–113. doi: 10.1177/172460080702200204. [DOI] [PubMed] [Google Scholar]
- 41.Lyall MS, Dundas SR, Curran S, Murray GI. Profiling markers of prognosis in colorectal cancer. Clin Cancer Res. 2006;12:1184–1191. doi: 10.1158/1078-0432.CCR-05-1864. [DOI] [PubMed] [Google Scholar]
- 42.Whittaker SR, Te Poele RH, Chan F, Linardopoulos S, Walton MI, Garrett MD, Workman P. The cyclin-dependent kinase inhibitor seliciclib (R-roscovitine; CYC202) decreases the expression of mitotic control genes and prevents entry into mitosis. Cell Cycle. 2007;6:3114–3131. doi: 10.4161/cc.6.24.5142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.