Abstract
The actin cytoskeleton has been implicated in restricting diffusion of plasma membrane components. Here, simultaneous observations of quantum dot-labelled FcεRI motion and GFP-tagged actin dynamics provide direct evidence that actin filament bundles define micron-sized domains that confine mobile receptors. Dynamic reorganisation of actin structures occurs over seconds, making the location and dimensions of actin-defined domains time dependent. Multiple FcεRI often maintain extended close proximity without detectable correlated motion, suggesting that they are co-confined within membrane domains. FcεRI signalling is activated by cross-linking with multivalent antigen. We show that receptors become immobilised within seconds of cross-linking. Disruption of the actin cytoskeleton results in delayed immobilisation kinetics and increased diffusion of cross-linked clusters. These results implicate actin in membrane partitioning that not only restricts diffusion of membrane proteins, but also dynamically influences their long-range mobility, sequestration, and response to ligand binding.
Signal transduction from the external environment to the cell interior is typically mediated by ligand-bound transmembrane receptors embedded in a lipid bilayer. In many systems, receptor activation is associated with changes in receptor dynamics and membrane topography1-3. Among these are the multi-chain immune recognition receptor family members that include the B-cell receptor (BCR) of B-cells, the T-cell receptor (TCR) of T-cells, and the high affinity IgE receptor (FcεRI) of mast cells and basophils, which are crucial to the execution of key events in the immune response. Cross-linking of these transmembrane receptors induces receptor oligomerisation, protein and lipid kinase activation and Ca2+ mobilisation, leading in turn to cytoskeletal reorganisation, receptor trafficking and cell-specific responses including altered gene expression4-6. These signalling events have been well studied by biochemical techniques, but the precise mechanism by which oligomerisation initiates these events has remained elusive. Full understanding of these complex signalling cascades will require a more complete description of receptor movements in the membrane, including restrictions that might limit receptor diffusion and accessibility.
A rich literature on single particle tracking (SPT) methods to follow the lateral diffusion of transmembrane and membrane-associated proteins7-10 has revealed nanometer-scale “confinement zones” that restrict lateral diffusion and supports the general notion that plasma membrane organisation is more structured than originally postulated by the fluid mosaic model11. A membrane-skeleton fence (picket fence) model has been proposed to explain confined diffusion12,13 at the scale of nanometers and microseconds. In this model, a relatively static meshwork of actin-defined domains transiently retain diffusing membrane proteins and lipids, leading to apparent long time-scale diffusion rates that are much slower than those measured in artificial lipid bilayers. A recent comparison of high resolution electron microscopy (EM) images of cytoskeletal “ghosts” with high-speed SPT data demonstrates that nanometer-sized transient confinement zones measured by SPT correlate well to regions delimited by membrane-associated cortical cytoskeleton visualised by EM14. However, technical limitations preclude simultaneous SPT and direct observation of nanometer-scale actin structures in living cells, leaving the precise mechanism of restricted diffusion at the nanometer scale open to debate. Specifically, other structural features in membranes, such as lipid rafts or protein islands15-18, have also been suggested to affect lateral diffusion.
Here, we directly characterise transmembrane receptor diffusion with respect to actin by simultaneous fluorescence imaging of micron-scale features of the cortical actin cytoskeleton and FcεRI. Quantum dot (QD)-labelled IgE was used to tag FcεRI. IgE binds tightly to the α subunit of FcεRI, essentially becoming another subunit of the receptor19,20. Exploiting this tight binding enables labelling of the resting receptor with minimal disruption to normal cell physiology, providing a highly specific and relevant probe of receptor dynamics and membrane topography. Micron scale features of the actin-based cytoskeleton have been described in detail using EM of detergent-extracted platelets21. We sought to visualise large membrane-proximal actin bundles in RBL-2H3 cells using GFP-tagged actin with confocal and total internal reflection fluorescence (TIRF) microscopy. We provide direct evidence that actin cables near the membrane define regions that limit receptor diffusion. These structures do not provide a static meshwork of enclosed domains, but instead form a dynamic labyrinth defined by micron-scale actin barriers that reorganise over time scales of 1-10 seconds. In contrast to the putative transient confinement zones of the picket fence model, here we describe a phenomenon that occurs in time scales of seconds and over distances of microns. In addition to restriction of membrane protein diffusion, we provide evidence that actin-based membrane partitioning dynamically influences long-range mobility, sequestration and response to ligand binding.
RESULTS
QD-IgE is functionally monovalent
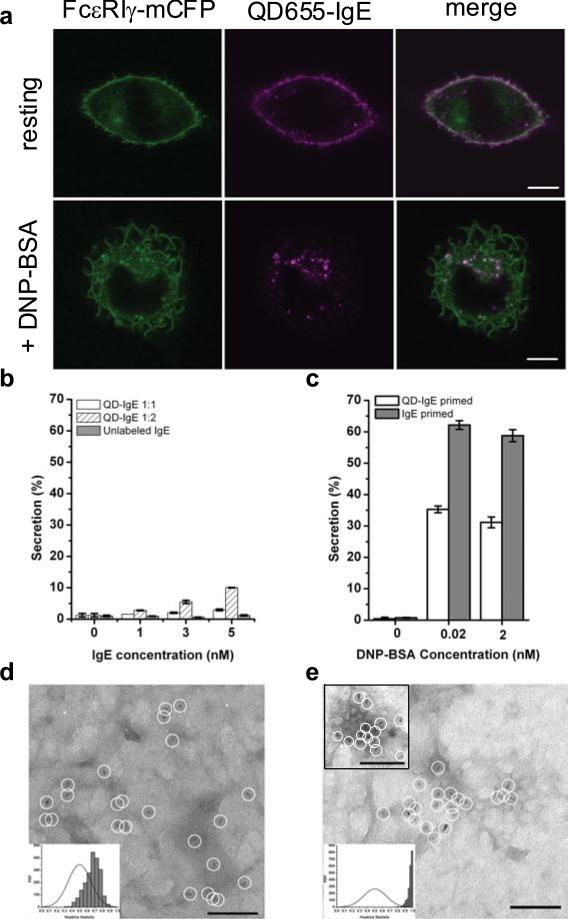
A monovalent QD-IgE probe was developed in order to study properties of the resting IgE receptor, FcεRI. The reaction conditions were carefully controlled to produce functionally monovalent complexes of QD with anti-DNP IgE (QD-IgE; see Materials and Methods). Several approaches were taken to evaluate valency (see Materials and Methods, Supplementary Information, Fig. S1). For functional evaluation, we determined that QD-IgE did not induce substantial activation of RBL-2H3 cells assessed by confocal microscopy as a lack of ruffle formation and QD-IgE internalisation (Fig. 1a; top panels) and by degranulation assay as a lack of β-hexosaminidase release (Fig. 1b). In addition, QD-IgE primed cells were capable of responding to stimulation with the multivalent antigen DNP-BSA. This was demonstrated visually as cell ruffling and QD-IgE internalisation (Fig. 1a, bottom panels and Supplementary Information, Video 1) and biochemically as significant β-hexosaminidase release (Fig. 1c). The somewhat blunted secretory response of QD-IgE primed cells (Fig. 1c) is likely due to steric limitations that the QD label imposes on the IgE-FcεRI aggregates22.
Figure 1.
QD-IgE serves as a non-perturbing probe of FcεRI diffusion. (a) RBL-2H3 cells stably expressing FcεRI-gamma-mCFP were labelled with 1 nM QD655-IgE and then imaged (top panels) or cross-linked with 14 nM DNP-BSA for 10 min prior to imaging (bottom panels). (b,c) Degranulation assay plots showing the percentage of total β-hexosaminidase content secreted in response to 30 min incubation at 37°C with various IgE (b) or after priming with QD-IgE or IgE in response to various amounts of DNP-BSA (c). (d,e) Individual QD655-IgE-FcεRI are indicated by white circles in electron micrographs from membrane sheets prepared from RBL-2H3 cells labelled with QD655-IgE (d) and stimulated for 10 min with 14 nM DNP-BSA (e) before fixation. A right shift in the experimental data (grey bars) away from a random distribution (solid line) in the Hopkins plot indicates a slightly clustered distribution of QD655-IgE-FcεRI in the resting state (d, inset) and a highly clustered distribution after cross-linking (e,inset)3. The scale bars represent 10 μm in a and 100 nm in d and e.
Since QDs can serve as probes in both EM and fluorescence imaging23, we analysed the topography of QD-IgE-tagged receptors on native membrane sheets by TEM. A visual inspection of QD distribution on the sheets shows many singlets as well as small clusters of 20-40 nm in size (Fig. 1d), verified as statistically nonrandom by the Hopkins test (Fig. 1d; inset). These results are consistent with previous studies using immunogold3,24 and confirm that previously observed clustering of resting receptors cannot be trivially explained as an artifact of the multivalent gold probes used to label fixed cell membranes (described in13). Brief treatment of QD-IgE labelled cells with polyvalent antigen results in the formation of large clusters of receptors (Fig. 1e), as well as the appearance of QD-labelled receptors in clathrin coated pits (Fig. 1e, upper inset). These studies demonstrate that QD-IgE behaves comparably to unlabelled IgE and therefore serves as a reliable probe of IgE-FcεRI dynamics.
QD-IgE-FcεRI demonstrates characteristic membrane protein diffusion
Estimations of membrane protein diffusion are variable, depending on differences in acquisition rates, temperature and the method of measurement. For example, imaging at ambient temperatures is simpler, but the diffusion rate is slower than at physiological temperatures. The frequent use of cold (4°C) labelling conditions in studies of FcεRI dynamics25,26 is of particular concern because long duration cold exposure has been shown to decrease measured diffusion of other transmembrane proteins27. We compared the diffusion of QD-IgE-FcεRI at room temperature (22°C) and at 35°C and observed a faster diffusion coefficient (D) and larger restricted region of diffusion (L) at the higher temperature (Supplementary Information, Fig. S2). To avoid these potentially confounding effects, we labelled cells and acquired data at physiological temperatures (34-37°C). The median diffusion coefficient (D1-3), measured at video rate (33 frames/s), was 0.074 μm2/s (Table 1). This value is consistent with previous measurements performed at lower temperatures (D = 0.02-0.05 μm2/s25,26,28-30).
Table 1.
Median diffusion coefficients and actin overlap
| Imaging Condition | Cell Treatment | N | D1-3 (μm2/s)a | IQRc | Relative Actin Overlapd |
|---|---|---|---|---|---|
| Apical | resting | 236 | 0.074 | 0.065 | |
| Apical | Latrunculin B | 138 | 0.10 | 0.087 | |
| Apical | DNP-BSA | 647 | 0.010 | 0.038 | |
| Apical | Latrunculin B + DNP-BSA | 403 | 0.020 | 0.044 | |
| TIRF | resting | 303 | 0.077b | 0.074 | 1.43 |
| TIRF | Latrunculin B | 265 | 0.059b | 0.067 | 1.30 |
| TIRF | PMA | 102 | 0.075b | 0.078 | 1.32 |
D1-3 values were calculated as described in Materials and Methods. Values are reported as median.
Median D1-3 values from TIRF experiments were calculated after excluding “immobile” trajectories, defined as D1-3 ≤0.0009 μm2/s.
Interquartile Range (IQR), see Materials and Methods.
Ratio of actin crossings between simulated and real trajectories. A value > 1 indicates that simulated, freely-diffusing particles cross actin more frequently than real trajectories. The differences in behavior between real and simulated data are statistically significant (see text and Supplementary Information for details).
As documented in previous studies of other membrane proteins8,25,27,31-33, we observed four distinct modes of FcεRI diffusion in unstimulated cells: free, directed, restricted, and immobile (Supplementary Information, Fig. S3). Our observation of restricted and immobile receptors is consistent with the general notion that the plasma membrane contains structures that limit lateral diffusion, which are not present in artificial bilayers.
Receptors are co-confined in micron-scale membrane domains
Despite labelling conditions that resulted in only a few QD-IgE labelled receptors per cell, we often observed prolonged overlap between the emissions of two or three tracked receptors (Supplementary Information, Video 2). These observations raised the possibility that weak attractive interactions may exist between non-cross-linked receptors. However, when two QDs with the same emission spectra overlap, they cannot be easily distinguished until they separate. Thus, the analysis of close interactions between receptors is limited in protocols using only one colour of fluorophore.
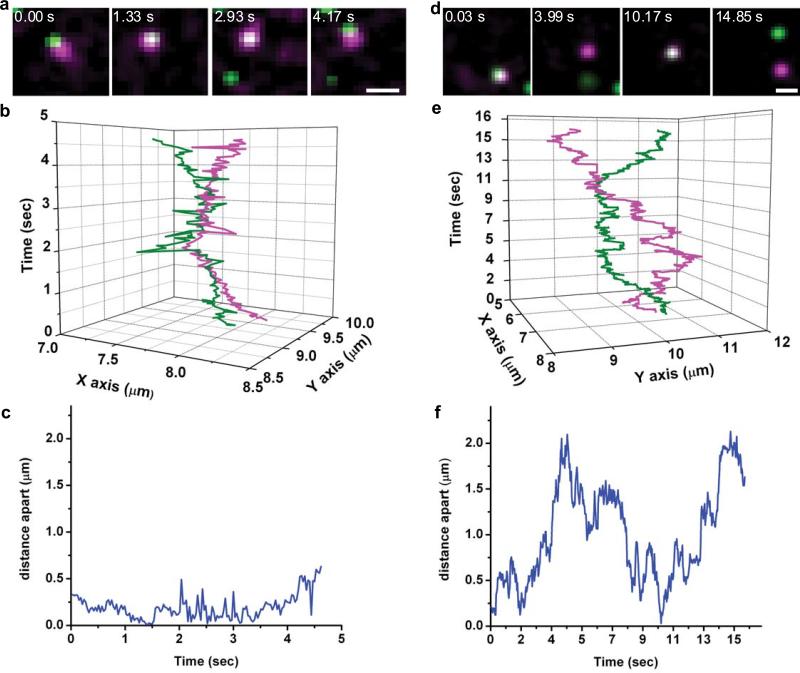
We overcame this limitation by labelling cells with a mixture of QD585-IgE and QD655-IgE and simultaneously tracking both QDs. This allowed us to independently localise each molecule even when their emissions overlapped. In the example shown in Figure 2a-c, two QD-labelled receptors repeatedly approach each other to within our localisation limits (see Materials and Methods) while never moving farther than 500 nm apart before coming back together (Fig. 2c). Despite their close proximity, the trajectories of the two molecules do not appear to be correlated (Fig. 2b). In the second example (Fig. 2d-f), a maximum separation of 2 μm occurs before the two molecules come back together (Fig. 2f). The observation that QD-IgE-FcεRI complexes can move microns apart before seeming to reverse direction to rejoin each other cannot be explained by attractive forces between the observed receptors.
Figure 2.
FcεRI are co-confined. (a) Sample images from a time series showing two QD-IgE-FcεRI complexes co-confined within a ~500 nm domain. (b) XY versus time plot of trajectories of QD655-IgE-FcεRI (magenta) and QD585-IgE-FcεRI (green) from time series shown in a. (c) Plot of interparticle distance versus time for particles shown in a. (d) Sample images from a time series showing two QD-IgE-FcεRI complexes co-confined within a ~2 μm domain. (e) XY versus time plot of trajectories of QD655-IgE-FcεRI (magenta) and QD585-IgE-FcεRI (green) from time series shown in d. (f) Plot of interparticle distance versus time for particles shown in d. Images in a and d have been Gaussian filtered. Images were acquired on the apical surface at 33 frames/s. The scale bars represent 1 μm in a and d.
To determine if protein-protein interactions (i.e., transient dimerisation) could explain the prolonged colocalisation of QD-IgE-FcεRI complexes, we developed mathematical analyses to quantify coordinated movement between receptors at separation distances less than 500 nm (see Supplementary Methods). In order to capture these potentially highly dynamic events, we used TIRF microscopy with a faster acquisition rate of 100 frames/s. The analysis involves measuring the magnitude of a receptor's displacement vector (jump magnitude) and the extent of correlated motion between nearby receptors (uncorrelated jump distance). If two receptors form a transient dimer, their diffusion is expected to be slowed13, producing a decrease in their jump magnitude. Additionally, if two receptors are forming a transient dimer, they would move together during the lifetime of this dimer; i.e., the motion of the two interacting receptors would be correlated, producing a decrease in the uncorrelated jump distance. By plotting the jump magnitude and the uncorrelated jump distance as a function of distance between receptor pairs, we can detect the presence of transient dimers, provided their lifetime is longer than the acquisition time for each image (10 ms).
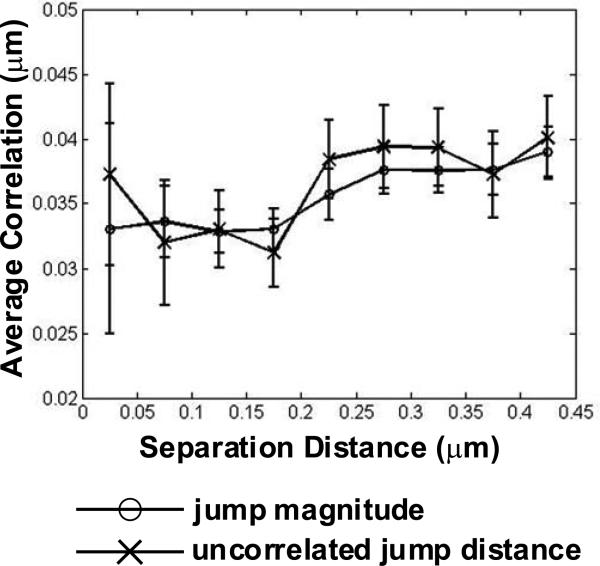
To model the observed behavior of prolonged colocalisation, we performed simulations of diffusing particles which formed transient dimers for 200 ms when two particles approached within 25 nm (see Supplementary Information, Fig. S4). This produced a dramatic decrease in the uncorrelated jump distance as receptors came close enough to interact (Supplementary Information, Fig. S4). In contrast to the simulations, analysis of real trajectories produced only gradual decreases in the jump magnitude (Fig. 3, open circles) and the uncorrelated jump distance (Fig. 3, x-marks). The weak dependence of these values on separation distance is consistent with measuring a slower diffusion constant when particles are confined in a small region (as described in reference 9). Together, these analyses do not support transient dimer formation as a mechanism for the prolonged colocalisation we observed and instead suggest co-confinement by membrane structure as the most likely explanation.
Figure 3.
Motion of QD-IgE-FcεRI is consistent with co-confinement; not attraction. Mean distance of uncorrelated jump distance (x-marks) and jump magnitude (open circles) plotted as a function of distance between QD-IgE-FcεRI complexes. Data are presented as mean ± s.e.m. of 1,128 instances of QD-IgE-FcεRI complexes approaching within 0.5 μm.
Dynamic actin structures restrict FcεRI diffusion
To determine the role of the actin cytoskeletal network in the lateral diffusion and observed co-confinement of QD-IgE-FcεRI complexes, RBL-2H3 cells were stably transfected with GFP-actin. Two-colour time series of QD-IgE-FcεRI motion with respect to the actin cytoskeleton were acquired. Due to its superior signal-to-noise ratio, TIRF microscopy was primarily used to track QD-IgE-FcεRI and GFP-actin movements on the adherent cell surface at 33 frames/s.
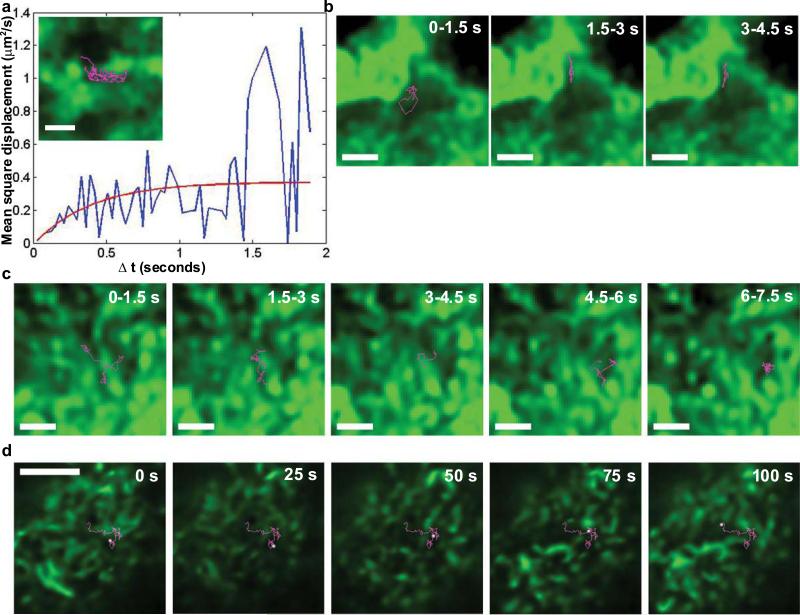
In initial experiments, cells were treated with the protein kinase C agonist, phorbol myristate acetate (PMA), to induce actin filament bundles resembling stress fibers34. QD-IgE-FcεRI diffusion was largely limited to actin-poor regions of the membrane, often remaining confined within a single actin-defined compartment for several seconds of tracking (Fig. 4a,b and Supplementary Information, Video 3). The size of these compartments (L) can be estimated based upon the plateau value on the Y axis of the mean square displacement (MSD) plot. Figure 4a illustrates a typical MSD plot, generated from the receptor trajectory trace shown in Figure 4a (inset), and fit by MSD = offset + (L2/3)(1-exp(-Δt / τ)) (ref 35), where D=L2/12τ. From the fit we find that L = 1.1 μm, which correlates very well with the actual, observed actin-defined region in Figure 4a.
Figure 4.
Actin defines regions of FcεRI motion in the membrane. (a) MSD plot of the trajectory shown in inset, demonstrating diffusion restricted to an area consistent with that delineated by the actin structures. (a, inset) QD-IgE-FcεRI trajectory (magenta) overlayed with the deconvolved GFP-actin image (green) from the adherent surface of a PMA-treated RBL-2H3 cell. (b,c) Image series of deconvolved GFP-actin (green) and 1.5 s long segments of a single QD655-IgE-FcεRI trajectory (magenta) on the adherent surface of a PMA treated (b) or untreated (c) RBL-2H3 cell. (d) Full 100 s QD-IgE-FcεRI trajectory (magenta) with position at 25 s intervals highlighted (white) overlayed with the deconvolved GFP-actin image (green) on the apical membrane of an RBL-2H3 cell. Panels a-c were acquired with TIRF microscopy at 33 frames/s. Panel d is from a 1 frame/s confocal time series with 1 μm slice thickness. The scale bars represent 1 μm in a-c and 5 μm in d.
Imaging of untreated RBL-GFP-actin cells revealed continuous re-organisation of the actin network, resulting in a nearly complete change in the pattern of GFP-actin fluorescence on a 10 s time scale (Supplementary Information, Video 4). Nevertheless, diffusion of QD-IgE-FcεRI still remained largely limited to actin-poor regions (Fig. 4c). Mathematical analysis of receptor/actin crossing frequency was performed to verify that receptors are avoiding actin structures. Comparing the real data trajectories with simulated, freely diffusing particles found a significant difference in actin/trajectory overlap, confirming that the real trajectories cannot be modelled by free diffusion unaffected by actin structures (Table 1 and Supplementary Information).
The data in Figure 4a-c above were acquired from the relatively flat, adherent cell surface in TIRF mode, which images structures within ~200 nm of the coverslip. We performed multiple control experiments to ensure that interactions between the plasma membrane and the coverslip were not responsible for the observed behavior of QD-IgE-FcεRI complexes (Supplementary Information, Fig. S5). The observation of similar behavior on the apical cell surface provides perhaps the most compelling support of the TIRF data for evidence of actin-restricted FcεRI diffusion. Confocal microscopy was used to eliminate the cytoplasmic GFP-actin signal and image the actin structure in a 1 μm slice at the apical cell surface. In confocal mode, the frame rate had to be reduced to 1 frame/s in order to image single QDs. Despite the slower frame rate, we were able to follow the trajectories of QD-labelled receptors over a period of 100 s while simultaneously imaging GFP-actin. Figure 4d shows that receptors on the apical surface were restricted from crossing actin-rich regions. As seen at the adherent cell surface, dynamic actin reorganisation accompanies receptor movement into previously restricted areas (Fig. 4d and Supplementary Information, Video 5).
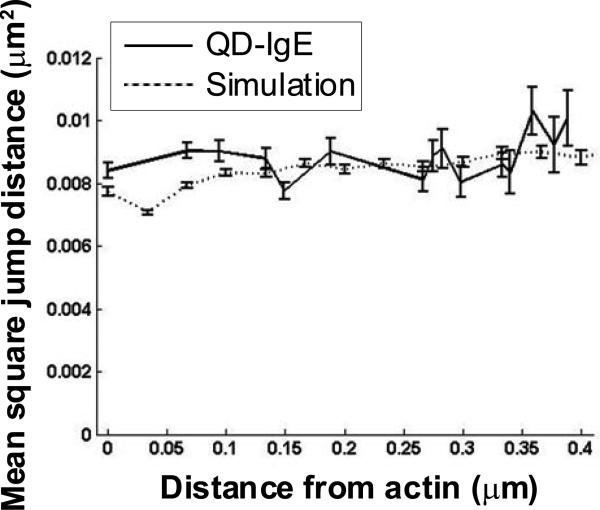
To determine if FcεRI alters its diffusion properties when encountering actin structures or is simply deflected by the physical barrier, we calculated the mean square single jump length of FcεRI as a function of distance from actin (Fig. 5). Although the data analysis in Figure 5 suggests a slight reduction in mean jump distance as the receptor approaches the actin (solid line), simulations (dashed line) show that this can be explained by a combination of geometric effects and finite detector integration time9. Thus, there is no indication of a real change in diffusion as close as 100 nm from underlying actin structures. These results favor the interpretation that membrane-associated actin structures or proteins bound to actin act on FcεRI as a physical barrier that deflects the receptor.
Figure 5.
Effect of actin proximity on FcεRI trajectories. All QD-IgE trajectories found by TIRF-SPT at 33 frames/s in PMA-treated cells were used to calculate single time step mean square jump distances as a function of distance from an actin structure (solid line). For comparison, simulations of particles diffusing near a reflecting boundary with constant membrane viscosity and finite localization accuracy, modelled after the real data, were used to calculate mean square jump distances as function of distance from the reflecting boundary (dashed line).
To further discern the influence of the actin cytoskeleton on FcεRI diffusion, we treated cells with 500 nM latrunculin B for 10 minutes prior to imaging. This dose of latrunculin disrupts cytoskeletal architecture (see Supplementary Information, Fig. S6) without causing significant damage to cells36. Latrunculin produced a small but significant (p=1.9×10-5 by K-S test) increase in FcεRI diffusion on the apical membrane (Fig. 6a, Table 1), consistent with previous observations of less confined motion13. Unexpectedly, latrunculin treatment decreased diffusion at the adherent surface (Table 1). While the mechanism is unclear, we note that non-actin components of the cortical cytoskeleton (possibly spectrin) are still visible in EM as a fine meshwork on the adherent surface of treated cells (see Supplementary Information, Fig. S7).
Figure 6.
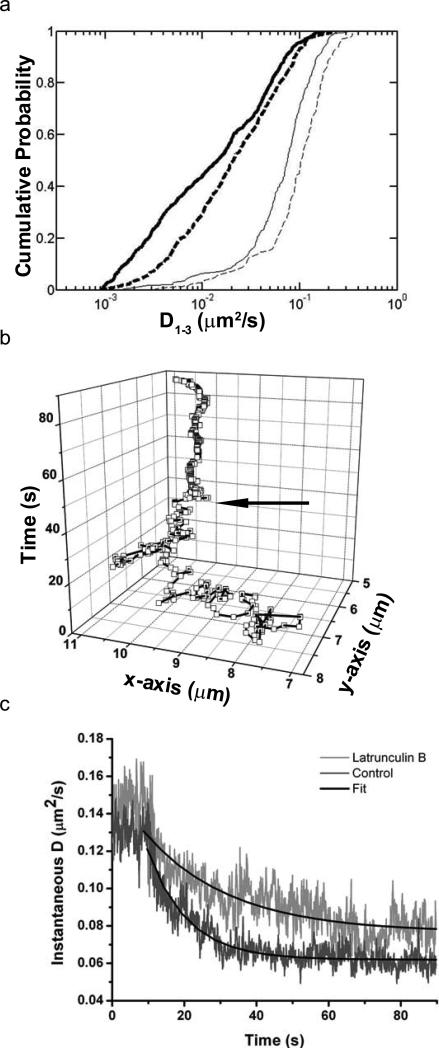
Actin facilitates cross-link-induced immobilisation of FcεRI. (a) Cumulative probability distribution plot of diffusion coefficients for QD-IgE in the absence (solid lines) and presence (dashed lines) of latrunculin B and before (thin lines) and after (thick lines) cross-linking with DNP-BSA. (b) XY versus time plot of a single QD-IgE-FcεRI. Arrow indicates time of DNP-BSA addition. (c) Kinetics of cross-link-induced immobilisation in the presence (light grey, n=12) and absence (dark grey, n=17) of latrunculin B. Black lines are fits to the exponential decay model: D(t) = Doe(-t/τ), where D is the instantaneous diffusion coefficient and τ is the decay constant.
Cytoskeletal disruption slows immobilisation kinetics
Activation of RBL-2H3 cells is well-known to induce substantial cytoskeletal remodelling and an increase in F-actin content36. There is also a dramatic decrease in FcεRI mobility within minutes of cross-linking37. To determine the kinetics of receptor immobilisation, we devised an assay to measure the mobility of QD-IgE-FcεRI complexes during cross-linking with 30 ms time resolution.
Upon addition of 14 nM DNP-BSA, FcεRI diffusion is rapidly decreased. The mobility transition is evident in trajectories of individual receptors, as seen in Figure 6b. Averaging the behaviour of many receptors over time yields the mean instantaneous diffusion coefficient plot (Fig. 6c). Note that diffusion reaches a baseline within seconds after cross-linking. An exponential decay model was used to fit the data after cross-linker addition, returning a characteristic decay time (τ) of 11.0 ± 0.19 s for untreated cells. In the presence of latrunculin, which disrupts both nanometer and micron-scale actin structures, τ increased to 30.0 ± 1.3 s (error indicates 95% confidence interval), suggesting a role for actin structure in the process of antigen-mediated receptor immobilisation. From these data, we were also able to calculate median diffusion constants both prior to and immediately after cross-linking. Latrunculin treatment significantly (p=1.8×10-13 by K-S test) increases the final diffusion rate after cross-linker addition (Fig. 6a and Table 1), confirming that the actin cytoskeleton plays an important role in cross-link-induced immobilisation of FcεRI.
DISCUSSION
Our results provide evidence for micron-scale restriction of membrane protein diffusion by dynamic actin structures and are particularly relevant to studies where membrane molecules are tracked on relatively long time scales (seconds). We frequently observed two or more QD-IgE-FcεRI complexes remaining in close proximity for several seconds (Fig. 2 and Supplementary Information, Video 2). However, close inspection of this behaviour revealed an absence of detectable correlated motion between individual receptors (Fig. 3). We suggest that this behaviour represents co-confinement of multiple receptors within the same membrane domain and support the idea that the micron-scale domains of co-confinement are defined by barriers of filamentous actin. These shifting actin barriers sub-divide the membrane, producing transiently co-confined sub-populations of membrane proteins. Actin could restrict diffusion through various mechanisms, including acting as a physical barrier (actin itself or associated proteins), directly binding FcεRI, or interacting with lipids and increasing membrane viscosity. Our analysis of FcεRI diffusion with respect to actin bundles (Fig. 5) favors the physical barrier explanation.
In addition to the micron-scale actin-mediated restriction we have observed, we recognise that there is a finer level of membrane organisation at work that we cannot directly visualise. This level of organisation has been described as a meshwork composed not only of actin, but also a wide variety of cell-type specific intermediate filament proteins (eg., spectrin38,39). This nano-scale organisation likely underlies the small “confinement zones” apparent in higher frame rate SPT measurements7-10 and may play a role in forming the smaller clusters (~50 nm) of resting FcεRI observed in electron micrographs (Fig. 1d). We speculate that the finer cytoskeletal meshwork may be involved in producing a heterogeneous distribution of membrane components on a nanometer scale. The literature offers several representations of this heterogeneity, which exists within the boundaries of large-scale actin-defined regions described in this work, including the protein islands model15,18, the lipid raft model40,41 and the membrane skeleton model12,13 (see Supplementary Information, Fig. S8).
The published models, while differing in their descriptions of the roles played by specific proteins in the cytoskeletal meshwork, are universally dependent upon an intact cortical cytoskeleton13,18,41. Each of these putative membrane features are reported to be highly dynamic. In the membrane skeleton model, tracked membrane components reside in individual domains on millisecond time scales13. Lipid rafts are also reported to be very dynamic40,41. In our data, the lack of correlated motion at small separation distances (Fig. 3) implies that receptors only briefly remain in the same nanometer-scale domain. These data suggest that the small clusters of resting FcεRI observed by EM (Fig. 1d) are highly dynamic, with exchange between neighbouring small domains occurring on time scales much more rapid than the diffusion of the domains themselves.
The immobilisation of IgE-FcεRI complexes in response to treatment with cross-linking agent has been previously studied by fluorescence recovery after photobleaching (FRAP)28,29,37,42 and by SPT25. Unique features of our study include labelling and tracking at physiological temperatures and acquisition of real-time kinetics. A previous attempt to measure the time course of cross-linking-induced immobilisation by FRAP was limited to events occurring at least 90 s after addition of cross-linking agent, at which time immobilisation had already reached a maximum37. We employed our real-time assay to evaluate the kinetics of cross-linking from the precise moment of cross-linker addition. Our results demonstrate that diffusion of QD-IgE-FcεRI decreases within seconds of adding cross-linking agent (Fig. 6b) and that the rate of immobilisation is slowed by actin disruption with latrunculin (Fig. 6a, Table 1). Combining our kinetic data with the SPT and EM results reveals that small, dynamic clusters of FcεRI transition to large, stable clusters by an actin-dependent mechanism within seconds of cross-linking. Disruption of actin by latrunculin also increased the apical diffusion coefficient of both resting and cross-linked receptors by two fold (Fig. 6a, Table 1). Interestingly, treatment with latrunculin is reported to prolong FcεRIγ phosphorylation and to increase degranulation36, suggesting that the more mobile clusters may have an increased signalling lifetime. Our observation that cross-linked receptors are not as immobile in the presence of latrunculin suggests that immobilisation is an important regulatory mechanism of FcεRI signalling.
Our direct and simultaneous observations of FcεRI motion and actin dynamics demonstrate that IgE receptors diffuse within micron-sized membrane domains defined by actin bundles and that confinement is dynamic over length scales of microns and time scales of seconds. Restricted motion has been observed for a wide variety of membrane proteins, including the erbB family33, glycine receptors43, and G-protein coupled receptors27. The ubiquitous nature of restricted diffusion, coupled with the observations here, suggest that actin-mediated restriction of membrane protein diffusion plays an important regulatory role in a wide variety of signalling pathways.
MATERIALS AND METHODS
Reagents
Mouse monoclonal anti-DNP IgE was prepared as described44. Monovalent biotin-IgE was prepared through modification of the FluoReporter® Mini-Biotin-XX Protein Labeling Kit protocol (Invitrogen, Carlsbad, CA): a biotin-succinimidyl ester to IgE ratio of 4:1 and a 15 min reaction time was used to generate biotin-IgE with an average biotin:IgE ratio of 0.83:1 as determined with the FluoReporter® Biotin Quantitation Assay Kit (Invitrogen, Carlsbad, CA). Biotin-IgE was combined in 1:1 stoichiometry with Qdot® 655 or Qdot® 585 streptavidin conjugate (Invitrogen, Carlsbad, CA) in PBS + 1% BSA to generate 20-60 nM stock solutions of monovalent QD-IgE. Stock solutions were stored at 4°C and used within four weeks. To facilitate comparisons between unlabelled IgE and QD-IgE, all concentrations are reported in nM, with 5 nM IgE or 14 nM DNP-BSA being equivalent to 1 μg/ml. Latrunculin B was from Sigma-Aldrich (St. Louis, MO). PMA was from Invitrogen (Carlsbad, CA). DNP-BSA was from Molecular Probes (Eugene, OR).
Cell culture
Rat basophilic leukemia (RBL-2H3) cells were grown as adherent monolayers in minimum essential medium with 10% fetal calf serum (Invitrogen, Carlsbad, CA) as described24. For microscopy, cell monolayers were cultured in 8-well Lab-Tek chambers (Nunc, Rochester, NY) 24 hours before experiments.
Degranulation assay
Cell monolayers were grown in 24-well tissue culture plates for 24 hours. Typically, FcεRI were primed by adding 5 nM anti-DNP-IgE to cultures overnight. After washing away excess IgE, cells were activated with 0.014-140 nM DNP-BSA. Release of β-hexosaminidase was measured as described22.
Cell treatment for SPT experiments
Cells in Lab-Tek chambers were labelled with 50 or 100 pM QD-IgE in Hanks’ balanced salt solution (HBSS) for 10 min at 37°C, then washed with HBSS prior to imaging. This yielded an average of ten QD-IgE-FcεRI complexes per cell. For TIRF imaging, washing was omitted. All imaging was performed at 34-36°C. PMA-treated samples were pre-incubated for 30 min and imaged in the presence of 50 nM PMA. Latrunculin B-treated samples were pre-incubated for 10 min and imaged in the presence of 500 nM latrunculin B. Where indicated, cells were stimulated with 14 nM DNP-BSA.
Cell treatment for immobilisation
Cells were labelled with 500 pM QD-IgE for 10 min at 37°C as above, then incubated at 37°C for 30 min with 140 nM unlabelled IgE. Cells were washed and imaged by wide field microscopy at 35°C in 200 μl HBSS for ~10 seconds, followed by addition of DNP-BSA to a final concentration of 14 nM. Where described, cells were treated with 500 nM latrunculin B during the last 10 min of IgE priming and latrunculin B was maintained in the medium during activation. The presence of 500 nM latrunculin B did not affect the rate or amount of IgE binding (data not shown).
Electron Microscopy
RBL-2H3 cell monolayers on glass coverslips were labelled with 30 nM QD655-IgE for 30 min at 37°C. Cells were fixed for seven min in 0.5% PFA and membrane sheets prepared as described in24. Electron micrographs were acquired using a transmission electron microscope (Hitachi H600). Probe identification and Hopkins statistical analysis were performed as described in3.
Fluorescence Microscopy
Confocal imaging was performed on a Zeiss LSM 510 META system equipped with a 63× 1.4 N.A. oil objective. Single 1 μm z-sections were acquired at 1 frame/s. GFP and QDs were excited simultaneously using 488 nm excitation and emission collected using a 545 nm dichroic mirror with 505 nm LP filter and the 625-689 nm range of the META detector, respectively.
Wide field imaging for SPT was performed using an Olympus IX71 inverted microscope equipped with a 60× 1.3 N.A. water, 100× 1.4 N.A. oil, or 150× 1.45 N.A. oil TIRF objective. Wide field excitation was provided by a mercury lamp with either a 436 nm BP or 543 nm BP excitation filter. Objective-based total internal reflection fluorescence (TIRF) microscopy was performed using excitation from a 472 nm continuous wave laser (CrystaLaser, Reno NV), which was expanded and entered the microscope through a laser side port of the microscope filter turret. Fluorescence emission was filtered with a 473 nm long pass filter (LP01-473RU, Semrock, Rochester, NY). Emission was collected by an electron multiplying CCD camera (Andor iXon 887 for widefield; Andor Luca for TIRF). In both cases, an image splitter (Cairn Research OptoSplit II, Kent, U.K.) was used to simultaneously image two spectrally distinct QDs or QD655 and GFP. Images were registered using a calibration image of multi–fluorophore fluorescent beads (0.2 μm Tetraspeck, Invitrogen, Carlsbad, CA) that have an emission spectrum covering the two spectral windows. For further details see Supplementary Information. QD emission was collected using the appropriate QD (20 nm bandpass) emission filters (Chroma, Rockingham, VT). GFP emission was collected using a 510/40 BP emission filter. The sample temperature (34-36 °C) was maintained by an objective heater (Bioscience Tools, San Diego, CA).
Single Particle Tracking
Images were acquired at either 33 or 100 fames/s for a total of 3,000 frames. Analysis of the acquired image series was performed as described previously45 and similar to43 to obtain trajectories, which were used to generate mean square displacement (MSD) plot46 (see Supplementary Methods for more detail). Diffusion coefficients were found by fitting the first three points of the MSD plots to MSD = offset + 4D1-3Δt, (ref33). Localisation accuracy for each individual trajectory is described by the offset from the origin of the resulting fit. Localisation accuracy of the system was determined by imaging QD-IgE-FcεRI in cells treated with 14 nM DNP-BSA for 5 minutes, then fixed in 2% paraformaldehyde for 20 minutes at room temperature. Imaging under various experimental conditions yielded localisation accuracies of 11.86 and 13.81 nm for QD655 and QD585, respectively imaged at 100 frames/s in TIRF mode; 9.32 and 10.37 nm for QD655 and QD585, respectively imaged at 33 frames/s in TIRF mode; and 50.00 nm for QD655 imaged on the apical surface at 33 frames/s in wide-field mode. Note that all interaction analysis was performed on data acquired under TIRF imaging conditions, where the localisation is most accurate. Instantaneous diffusion coefficients are average diffusion coefficients calculated over all tracked QDs and using all points within a sliding window of 10 frames (.3 s). The instantaneous diffusion coefficient is given by D=(1/N)Σt,τ,n((rt+τ-rt)2-offset)/4τ where r is a trajectory position, offset is found by a fit to the longest trajectory, and the sum is over all N valid trajectory coordinate pairs in the time window.
Statistical Analysis
Due to the wide range of values and nonparametric distribution of D1-3, values are reported as medians and interquartile range is provided as a measure of statistical dispersion. Statistical comparisons between data sets were made using the Kolmogorov-Smirnov test unless otherwise noted and significance was defined as p<0.05.
Image Processing
All image processing was performed using Matlab (The MathWorks, Inc., Natick, MA) in conjunction with the image processing library DIPImage (Delft University of Technology). For descriptions of specific analysis routines, see Supplementary Information.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01 GM49814, R01 AI051575 and P20 GM 67594, the Oxnard Foundation, ACS IRG 192, and by the Sandia LDRD and SURP programs. Sandia is a multi-program laboratory operated by Sandia Corporation, a Lockheed Martin Company, for the United States Department of Energy under contract DE-AC04-94AL85000. Nicholas Andrews was supported by NSF IGERT DGE-0549500 and the UNM-SOM MD/PhD Program. We thank Sheli Ryan for cell culture assistance, Jun Zhang for EM image analysis, and Jonas Anderson and Bernd Rieger for image processing assistance. The UNM Cancer Center Fluorescence Microscopy Facility received support from NIH grants S10 RR14668, S10 RR19287, S10 RR016918, P20 RR11830 and P30 CA118100 and from NSF grant MCB9982161. Electron micrographs were generated in the University of New Mexico Electron Microscopy Facility, which received support from NIH grants P20 GM067594, S10 RRI5734 and RR022493.
Abbreviations
- BSA
bovine serum albumin
- COM
center of mass
- DNP
2,4-dinitrophenol
- EM
electron microscopy
- FcεRI
high affinity IgE receptor
- FRAP
fluorescence recovery after photobleaching
- MSD
mean square displacement
- QD
quantum dot
- SPT
single particle tracking
- TIRF
total internal reflection fluorescence
- TPA
time-resolved phosphorescence anisotropy
REFERENCES
- 1.Dustin ML, Cooper JA. The immunological synapse and the actin cytoskeleton: molecular hardware for T cell signaling. Nat. Immunol. 2000;1:23–9. doi: 10.1038/76877. [DOI] [PubMed] [Google Scholar]
- 2.Lidke DS, Lidke KA, Rieger B, Jovin TM, Arndt-Jovin DJ. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J. Cell. Biol. 2005;4:619–26. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, et al. Characterizing the topography of membrane receptors and signaling molecules from spatial patterns obtained using nanometer-scale electron-dense probes and electron microscopy. Micron. 2006;1:14–34. doi: 10.1016/j.micron.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Boniface JJ, et al. Initiation of signal transduction through the T cell receptor requires the multivalent engagement of peptide/MHC ligands [corrected]. Immunity. 1998;4:459–66. doi: 10.1016/s1074-7613(00)80629-9. [DOI] [PubMed] [Google Scholar]
- 5.Kraft S, Kinet JP. New developments in FcepsilonRI regulation, function and inhibition. Nat. Rev. Immunol. 2007;5:365–78. doi: 10.1038/nri2072. [DOI] [PubMed] [Google Scholar]
- 6.Thyagarajan R, Arunkumar N, Song W. Polyvalent antigens stabilize B cell antigen receptor surface signaling microdomains. J. Immunol. 2003;12:6099–106. doi: 10.4049/jimmunol.170.12.6099. [DOI] [PubMed] [Google Scholar]
- 7.Kusumi A, Ike H, Nakada C, Murase K, Fujiwara T. Single-molecule tracking of membrane molecules: plasma membrane compartmentalization and dynamic assembly of raft-philic signaling molecules. Semin. Immunol. 2005;1:3–21. doi: 10.1016/j.smim.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Murase K, et al. Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys J. 2004;6:4075–93. doi: 10.1529/biophysj.103.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritchie K, et al. Detection of non-Brownian diffusion in the cell membrane in single molecule tracking. Biophys. J. 2005;3:2266–77. doi: 10.1529/biophysj.104.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxton MJ, Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997:373–99. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 11.Singer S, Nicolson G. The fluid mosaic model of the structure of cell membranes. Science. 1972;4023:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson K, Sheets ED, Simson R. Revisiting the fluid mosaic model of membranes. Science. 1995;5216:1441–2. doi: 10.1126/science.7770769. [DOI] [PubMed] [Google Scholar]
- 13.Kusumi A, et al. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 2005:351–78. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 14.Morone N, et al. Three-dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J. Cell. Biol. 2006;6:851–862. doi: 10.1083/jcb.200606007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;6:937–50. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Draber P, Draberova L. Lipid rafts in mast cell signaling. Mol. Immunol. 2002;16-18:1247–52. doi: 10.1016/s0161-5890(02)00071-8. [DOI] [PubMed] [Google Scholar]
- 17.Frankel DJ, et al. Revealing the topography of cellular membrane domains by combined atomic force microscopy/fluorescence imaging. Biophys. J. 2006;7:2404–13. doi: 10.1529/biophysj.105.073692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 2006;50:18992–7. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garman SC, Wurzburg BA, Tarchevskaya SS, Kinet JP, Jardetzky TS. Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc epsilonRI alpha. Nature. 2000;6793:259–66. doi: 10.1038/35018500. [DOI] [PubMed] [Google Scholar]
- 20.Kulczycki A, Jr., Metzger H. The interaction of IgE with rat basophilic leukemia cells. II. Quantitative aspects of the binding reaction. J. Exp. Med. 1974;6:1676–95. doi: 10.1084/jem.140.6.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartwig JH, DeSisto M. The cytoskeleton of the resting human blood platelet: structure of the membrane skeleton and its attachment to actin filaments. J. Cell. Biol. 1991;3:407–25. doi: 10.1083/jcb.112.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortega E, Schweitzer-Stenner R, Pecht I. Possible orientational constraints determine secretory signals induced by aggregation of IgE receptors on mast cells. EMBO J. 1988;13:4101–9. doi: 10.1002/j.1460-2075.1988.tb03304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giepmans BN, Deerinck TJ, Smarr BL, Jones YZ, Ellisman MH. Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nat. Methods. 2005;10:743–9. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- 24.Wilson BS, Pfeiffer JR, Oliver JM. Observing FcepsilonRI signaling from the inside of the mast cell membrane. J. Cell. Biol. 2000;5:1131–42. doi: 10.1083/jcb.149.5.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barisas BG, et al. Compartmentalization of the Type I Fc epsilon receptor and MAFA on mast cell membranes. Biophys. Chem. 2007;1-3:209–17. doi: 10.1016/j.bpc.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 26.Feder TJ, Brust-Mascher I, Slattery JP, Baird B, Webb WW. Constrained diffusion or immobile fraction on cell surfaces: a new interpretation. Biophys. J. 1996;6:2767–73. doi: 10.1016/S0006-3495(96)79846-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki K, Ritchie K, Kajikawa E, Fujiwara T, Kusumi A. Rapid hop diffusion of a G-protein-coupled receptor in the plasma membrane as revealed by single-molecule techniques. Biophys. J. 2005;5:3659–80. doi: 10.1529/biophysj.104.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pyenta P, Schwille P, Webb WW, Holowka D, Baird B. Lateral diffusion of membrane lipid-anchored probes before and after aggregation of cell surface IgE-receptors. J. Phys. Chem. A. 2003:8310–8318. [Google Scholar]
- 29.Schlessinger J, Webb WW, Elson EL, Metzger H. Lateral motion and valence of Fc receptors on rat peritoneal mast cells. Nature. 1976;5586:550–2. doi: 10.1038/264550a0. [DOI] [PubMed] [Google Scholar]
- 30.Thomas JL, Feder TJ, Webb WW. Effects of protein concentration on IgE receptor mobility in rat basophilic leukemia cell plasma membranes. Biophys. J. 1992;5:1402–12. doi: 10.1016/S0006-3495(92)81946-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daumas F, et al. Confined diffusion without fences of a g-protein-coupled receptor as revealed by single particle tracking. Biophys. J. 2003;1:356–66. doi: 10.1016/S0006-3495(03)74856-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacquier V, Prummer M, Segura JM, Pick H, Vogel H. Visualizing odorant receptor trafficking in living cells down to the single-molecule level. Proc. Natl. Acad. Sci. U. S. A. 2006 doi: 10.1073/pnas.0603942103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kusumi A, Sako Y, Yamamoto M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 1993;5:2021–40. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfeiffer JR, Oliver JM. Tyrosine Kinase-Dependent Assembly of Actin Plaques Linking Fc-Epsilon-R1 Cross-Linking to Increased Cell Substrate Adhesion in Rbl-2h3 Tumor Mast-Cells. J. Immunol. 1994;1:270–279. [PubMed] [Google Scholar]
- 35.Destainville N, Salome L. Quantification and correction of systematic errors due to detector time-averaging in single-molecule tracking experiments. Biophys. J. 2006;2:L17–9. doi: 10.1529/biophysj.105.075176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frigeri L, Apgar JR. The role of actin microfilaments in the down-regulation of the degranulation response in RBL-2H3 mast cells. J. Immunol. 1999;4:2243–50. [PubMed] [Google Scholar]
- 37.Menon AK, Holowka D, Webb WW, Baird B. Cross-linking of receptor-bound IgE to aggregates larger than dimers leads to rapid immobilization. J. Cell. Biol. 1986;2:541–50. doi: 10.1083/jcb.102.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dahl SC, Geib RW, Fox MT, Edidin M, Branton D. Rapid capping in alpha-spectrin-deficient MEL cells from mice afflicted with hereditary hemolytic anemia. J. Cell. Biol. 1994;5:1057–65. doi: 10.1083/jcb.125.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Q, Edidin M. Lowering the barriers to random walks on the cell surface. Biophys. J. 2003;1:400–7. doi: 10.1016/S0006-3495(03)74860-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 2006;7:1597–8. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Viola A, Gupta N. Tether and trap: regulation of membrane-raft dynamics by actin-binding proteins. Nat. Rev. Immunol. 2007;11:889–96. doi: 10.1038/nri2193. [DOI] [PubMed] [Google Scholar]
- 42.Mao SY, Varin-Blank N, Edidin M, Metzger H. Immobilization and internalization of mutated IgE receptors in transfected cells. J. Immunol. 1991;3:958–66. [PubMed] [Google Scholar]
- 43.Dahan M, et al. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;5644:442–5. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 44.Liu FT, et al. Monoclonal dinitrophenyl-specific murine IgE antibody: preparation, isolation, and characterization. J. Immunol. 1980;6:2728–37. [PubMed] [Google Scholar]
- 45.Arndt-Jovin DJ, et al. In vivo cell imaging with semiconductor quantum dots and noble-metal nanodots. Proc. SPIE. 2006:60960P1–10. [Google Scholar]
- 46.Martin DS, Forstner MB, Kas JA. Apparent subdiffusion inherent to single particle tracking. Biophys. J. 2002;4:2109–17. doi: 10.1016/S0006-3495(02)73971-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.