Abstract
Poly(ethylene glycol) (PEG) chains were used to decorate microparticles with long adhesion ligands to emulate the efficacy of selectin-mediated leukocyte homing mechanisms. Ligands for P-selectin, an endothelial cell inflammatory marker, were coupled to PEG spacers of two sizes (MW 3400 Da and 10000 Da) to investigate the effects on adhesion kinetics to P-selectin substrates. Under shear flow 80 nm PEG spacers improved P-selectin-antibody adhesion frequency by up to 4.5-fold and bond lifetimes by 7-fold compared to microparticles bearing chemisorbed antibody. Presentation of the glycosulfopeptide P-selectin ligands (2-GSP-6) and its non-sulfated low affinity form (2-GP-6) by long PEG spacers lead to improved lifetimes of stressed bonds formed with P-selectin in shear flow and the rolling fluxes. Thus, structural features far removed from the binding pocket of a receptor that increase molecular contour length may enhance affinity in mechanically stressed environments such as those existing within the confines of the blood vessel. Such features may be useful for improving the performance of vascular-targeted micro- and nanoparticles used for drug, gene, and image contrast delivery. Ligand presentation on molecularly extended stalks may also serve to enhance any particle-surface interaction that takes place in laminar shear flow.
Introduction
Specialized blood cells of the immune system deliver what are essentially packets of protein and DNA to sites within the vasculature with impressive specificity, despite the stringent constraints created by blood hemodynamics. Synthetic drug and gene delivery platforms on the nano- to micron length scale face similar challenges for vascular targeting. However, despite years of intensive development, nano- and micro-scaled particulate delivery platforms still have yet to match the apparent ease with which leukocytes bind to the vessel wall and sense appropriate egress points from the vasculature. Mimicry of the targeting mechanisms used by leukocytes could, if successfully incorporated into a microparticle delivery system, impart the ability to bind under stringent flow conditions while carrying a substantial payload of drugs or biologic therapeutics. Model systems based on this concept have already demonstrated targeting capabilities in vivo.1, 2
Leukocytes express adhesion receptors called selectins that support tethering to the blood vessel wall as a first step leading to emigration into tissue. Selectins and their ligands share the general characteristic of being unusually long, with total bond lengths estimated to range from 60 to 90 nm unstressed, depending on the specific ligand-receptor combination.3–5 In addition to being long enough to reach past the glycocalyx, the selectin-ligand complex's length and flexibility may also boost the sampling volume of its binding pocket during an encounter with a surface-anchored ligand. Consequently, factors underlying the leukocyte’s robust ability to roll on blood vessel wall—such as the length of the adhesive crossbridges and their favorable formation rates and biomechanics—if appropriated into the design of a drug and gene delivery platform have the potential to improve targeting to endothelium.1, 6–8
Our objective was to examine the effect of lengthening bond structures on two dimensional (2-D) adhesion kinetics in shear flow. We used a microparticle as a surrogate cell and drug delivery platform to which was attached varying lengths of poly(ethylene glycol) (PEG) chains that had been functionalized with ligands to an endothelial cell inflammatory marker, P-selectin. By using bifunctional PEG linkages, functionalized microparticles could accommodate either a P-selectin monoclonal antibody (HuEP5C7.g2, HuEP).9, 10 or a glycosulfopeptide modeled on P-selectin Glycoprotein Ligand-1 (PSGL-1) as a targeting moiety.11 A laminar flow apparatus was used to create physiologic microparticle-substrate contact times, appropriate surface separation distances, and relevant distractive forces on the adhesive bond complex. Our results suggested that molecular spacers that increased total adhesive crossbridge length increased bond lifetimes and formation probability in shear flow by significant margins. Such design considerations may be broadly applicable to a variety of drug, gene, and even image contrast delivery systems that must bind under the stringent flow conditions present in the blood circulation.
Experimental Section
Antibodies and glycopeptides
HuEP5C7.g2, a humanized murine mEP-5c7 IgG2 P-and E- selectin blocking monoclonal antibody (HuEP), was developed as previously described 9. Biotin N-hydroxisuccinimide (NHS) ester (Sigma, St. Louis, MO) in dimethyl sulfoxide (DMSO) was used to biotinylate HuEP. The reaction was carried out following published protocols.12 Any unbound biotin and byproducts were removed by dialysis against PBS using a Spectra/Por Microdialyzer with a Spectra/Por 7 10kDa MWCO membrane (Spectrum, Gardania, CA). The amount of biotin bound to the antibody was determined using avidin-HABA (2-(4’-hydroxyazonbenzene) benzoic acid, Pierce) displacement method.13
A synthesized P-selectin glycoprotein ligand fragment of PSGL-1 known as 2-glycosulfopeptide-6 [2-(GSP)-6] was used as a targeting ligand to P-selectin. A non-sulfated version of the peptide, 2-GP-6, was used as lower affinity variant comparison. Both peptides are modeled after the NH2-terminal P-selectin binding region of PSGL-1 as previously described. 11
P-selectin substrate
Recombinant P-selectin (R&D Scientific, Flanders, NJ) was adsorbed onto polystyrene culture dishes (85 mm, Corning, Corning, NY) at a site density of 100 sites/µm2, unless otherwise noted, in phosphate buffered saline (PBS, Biowhittaker, Walkerville, MD), and were blocked against non-specific adhesion with 1% w/v of casein protein in PBS (Pierce, Rockford, IL). Site densities were determined using Europium assay as described previously.14
PEG microparticles
The PEG microparticles were functionalized through the following immobilization chemistry sequence: amino microbeads + NHS-PEG-biotin + streptavidin linker + biotinylated antibody/peptide. This sequence of assembly limited pre-clustering of the streptavidin-biotin-antibody conjugates. Polybead amino 6.0 micron microspheres (Polyscience, Warrington, PA) were used. The microparticles were washed in DMSO twice and re-suspended in DMSO at a final concentration of 1 × 107 microparticles per milliliter. Poly(ethylene glycol) (PEG) bi-functionalized with biotin on one end and NHS on the other end with MW = 3400 Da and MW = 10000 Da (Nektar, San Carlos, CA). was dissolved in DMSO at a concentration of 100 µg/µL. The biotin-NHS-PEG was added at a concentration of 100 µg per milliliter of microparticles for 2 hours to graft the PEG onto the microparticles (PEGylation). The microparticles were centrifuged to remove any unbound PEG and then blocked against non-specific interactions by adding 1 milliliter of 0.05% Tween 20 in PBS overnight. After removing the Tween 20 solution, streptavidin (Sigma) at a concentration of 2 mg for every 1 × 107 microparticle was added to the PEGylated microparticles. After 1 hour of incubation, the microparticles were washed by repeated centrifugation to remove any unbound streptavidin. The microparticles were then incubated at 4°C with 5 µg biotinylated HuEP antibody for every 1 × 107 microparticle. The biotinylated HuEP antibody was bound through a biotin-streptavidin bridge as previously described.10 The microparticles were stored in PBS buffer at a concentration of 5 × 105 microparticles/mL. Glycosulfopeptide microparticles were prepared two ways. Both 2-GSP-6 and its non-sulfated analog, 2-GP-6, were biotinylated and immobilized onto strepavidin-functionalized microparticles following the procedure used for PEG linking of HuEP to the microparticle surface. For the experiments in which 2-GSP-6 and 2-GP-6 were directly conjugated to the microparticle surface, the insertion of the PEG chain was omitted.
PEG and antibody concentration on the microparticle surface
The site density of the PEG chains on the microparticles was determined by time-resolved fluorescence detection of Europium (Eu)-labeled streptavidin (Perkin Elmer Wallac, Turku, Finland) 14 on a SPECTRAmax Gemini XS dual-scanning microplate spectrofluorometer (Molecular Devices, Sunnyville, CA). The microparticle concentration and surface area were determined using a Coulter Multisizer IIe counter (Beckman Coulter, Hialeah, FL). The amount of PEG on the microparticle surface was calculated from the europium fluorescence results and the surface area of the microparticles in solution. The concentration of the targeting antibody was measured as previously described.10
Flow chamber assay
The adhesion experiments of the PEGylated microparticles under shear stress were carried out using a parallel plate flow chamber (Glycotech; Gaithersburg, MD). The flow chamber was positioned on an inverted phase-contrast microscope (Diaphot-TMD; Nikon, Garden City, NY) equipped with a Photron FastCam Camera R2 model 1K (Photron USA Inc., San Diego, CA) allowing high temporal resolution video recordings. The microparticles were drawn through the flow chamber at wall shear stresses ranging from of 0.1 – 1 dyne/cm2 (0.01 – 0.1 Pa) by a Harvard Apparatus PHD2000 Syringe Pump (Instech Labs Inc., Plymouth Meeting, PA). Fluid was drawn from the stock solution through the flow chamber into a 10 mL glass “gas-tight” syringe (Hamilton, Reno, NV). The microparticle bond lifetimes and distances between adhesion events were recorded using a 20× objective and a time resolution of 125 frames per second (fps). Standard time resolutions of 30 fps were recorded with a VICON VC2410 camera (VICON, Hauppauge, NY) in all other assays unless indicated.
Microparticle rolling dynamics under shear stress
Under shear stress, targeting microparticles moving through the flow chamber translocated across the surface in a series of closely spaced transient interactions mediated by ligand-receptor bonds forming and breaking. The immobilized P-selectin interaction events were recorded 4× standard video rates to identify the distinct motions of microparticle rolling. The micro-motions of rolling at low ligand density can be divided into two states: a binding event (the pause time) where the microparticle is stationary and the time or distance between binding events (the step distance).15 The step distance is the distance traveled between the binding events caused by bond formation. The transient arrests characteristic of selectin binding events (pauses) were defined as any decrease in velocity 3 standard deviations away from the defined hydrodynamic velocity of a non-interacting particle of similar size traveling near the wall under shear. At 3 standard deviations, the instantaneous velocity is outside the 99% confidence interval for velocity variations of a non-interacting microparticle due to Brownian motion perpendicular to the direction of flow that must be accounted for to robustly identify adhesive interactions.16 For a 6 µm diameter microparticle with a separation distance of 100 nm, velocity ranged from 1.6 µm/s at a wall shear stress of 0.1 dyne/cm2 to 160 µm/s at a wall shear stress of 1 dyne/cm2. 17
Video imaging and data analysis
Video images of the rolling interactions were digitally recorded in .avi video format via the FastCam Motion Capture software (Photron, San Diego, CA). The interaction data: microparticle accumulation, microparticle detachment, rolling velocities, paused times, and skip distance between adhesion events was determined using a computer tracking algorithm coded in JAVA for the ImageJ software (NIH, Washington D.C.), which uses a cross-correlation interpolation that allows for subpixel resolution of changes in position to identify the particle in consecutive image frames.18
Statistical analysis
All differences were evaluated by one-tailed student’s t-test. P < 0.05 was considered statistically significant. All error bars represent standard deviations. For the non-linear fitting, chi squared distribution was used to evaluate the statistical significance of observed values to theoretical calculations. The coefficient of determination, R2, was used to evaluate linearity.
Results
In this study, we sought to determine the effect of varying adhesion receptor length on the 2-D kinetics of P-selectin targeted microparticles under physiological shear forces. To this end microparticle constructs were created with an adhesion receptor separated from the microparticle surface with different lengths of a spacer molecule. One set of microparticles had the adhesion receptor directly conjugated to their surfaces, another set had a short PEG3400 chain inserted between the adhesion receptor and the surface, and the final set had a long PEG10000 chain inserted between the adhesion receptor and the microparticle surface. The three linkage strategies created an estimated 80 nm difference in maximum receptor length.
Antibody functionalized PEG chains grafted to microparticles formed a polymer brush conformation
The dominant parameters of PEG coated microparticles are the polymer chain length and the surface grafting density. Both parameters directly determine the PEG layer conformation and thickness. Based on a Europium fluorescence assay, the PEG10000 chain site density was estimated to be ~5986 mol/µm2, or 1 PEG10000 chain for every 167 nm2. Similarly measured, the surface density of PEG3400 was ~20424 mol/µm2, or 1 PEG3400 chain for every 49 nm2. PEG chains functionalized with the HuEP antibody were grafted along with non-functionalized (neutral) PEG chains of the same molecular weight (PEG-NHS). Titration experiments using quantitative flow cytometry were performed to ensure that the site density of HuEP mAb was at ~3000 sites/µm2 for both PEG3400 and PEG10000 microparticles. Due to PEG’s hydrophilic and flexible nature, isolated chains are generally characterized by randomized mushroom conformations scaled to the Flory radius, where near full extension occurs infrequently.
The distance from the surface where the end monomer chain has the largest probability of residence in the mushroom conformation can be estimated by the Flory radius: Rf = aN0.64, where N is the number of monomer chains and a is the monomer length, estimated for the ethylene glycol monomers (- CH2 – CH2 – O -)n to be 3.5 angstroms Å.19, 20 Thus, PEG with a molecular weight of 3400 has an Rf = 5.66 nm and PEG with a molecular weight of 10000 has a Rf = 11.30 nm. However, as the surface density of PEG increases, the PEG chains are forced to form a brush conformation to minimize chain overlap. A brush conformation exists when the space between grafted chains has a Flory radius overlap of 50% or greater. With a PEG10000 Flory radius overlap of 62.7% and a PEG3400 Flory radius overlap of 73.8%, the PEG chains when grafted to the microparticle surface are in a packed brush conformation.13, 21, 22
From self-consistent field (SCF) calculations23 the average thickness of the grafted PEG brushes was predicted to be 18.9 nm for PEG3400 and 55.8 nm for PEG10000 at the measured chain density, in both cases ~ 70% of the fully extended length. In order to achieve similar levels of coverage and fractional extension of the PEG brushes for each size of polymer, the surface density of neutral PEG chains was necessarily different.
Increased PEG spacer lengths enhance microparticle adhesive flux under flow conditions
We first addressed the effect that PEG linker length had upon the adhesion avidity of HuEP microparticles to immobilized P-selectin. The HuEP mAb receptor has been previously shown to be able to mediate microparticle binding under flow conditions both in vivo and in vitro.2, 10 HuEP is an unusual antibody in this regard as the majority of mAbs examined to date are ineffective at mediating microparticle or cell adhesion under flow.10, 24
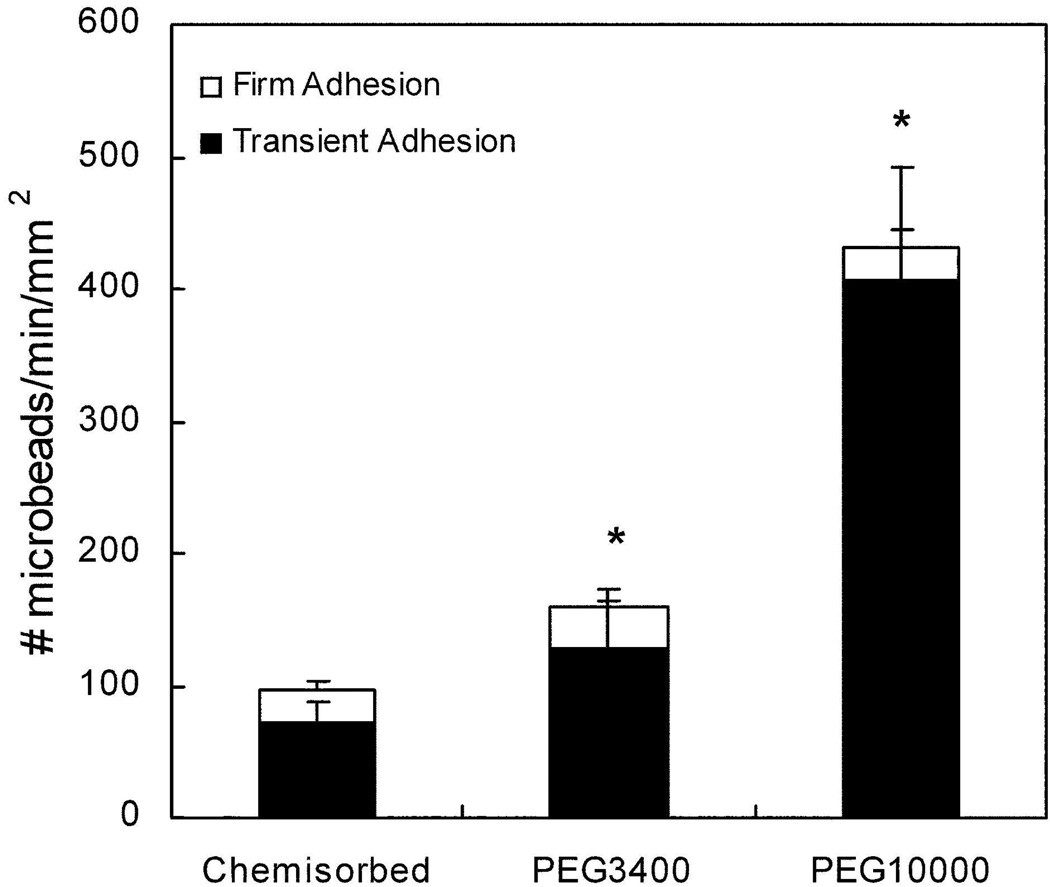
HuEP functionalized microparticles were perfused over P-selectin substrates and the flux of rolling interactions quantified. The majority of HuEP microparticles rolled on P-selectin (+80%), irrespective of the mode of HuEP immobilization. Microparticle rolling consists of a series of transient stops and repeated rebinding events leading to a salation motion on the P-selectin similar to that observed with PSGL-1 coated microparticles and leukocytes interacting with low density P-selectin substrates (Fig. 1).10, 25, 26 The number of rolling HuEP microparticles was defined as the adhesive flux. PEG3400 spacers increased the total HuEP-microparticle adhesive flux by 1.5-fold relative to chemisorbed HuEP microparticles at a wall shear stress of 1 dyne/cm2. The PEG10000 linkage further improved the adhesive flux of HuEP microparticles by 4-fold relative to chemisorbed HuEP microparticles.
Figure 1. Adhesive flux measurements on P-selectin for microparticles presenting HuEP immobilized by either chemisorption or by PEG linkages demonstrating PEG length enhances binding under flow conditions.
The flow rate was 1.0 dynes/cm2 wall shear stress and the P- selectin surface site density was set at 100 sites/µm2. Fluxes shown are the result of 2–4 independent measurements. Microparticles were counted as exhibiting transient interactions if their translocation velocity was 3 standard deviations less than the averaged threshold free-stream velocity. Firmly bound microparticles were defined as any microparticles remaining stationary for greater than 60 seconds. * p < 0.025 against the chemisorbed control and against the PEG3400 and PEG10000 microparticles (p = 0.050 for PEG3400. p = 0.0045 for PEG10000).
Adhesive dynamics during microparticle rolling
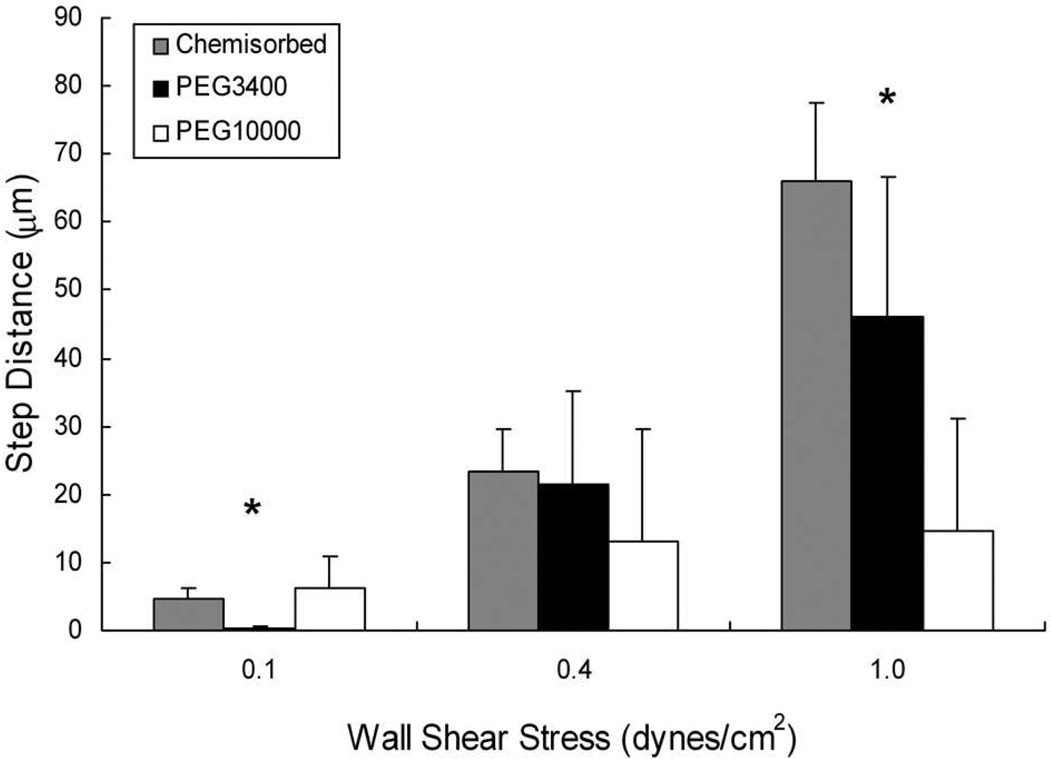
The adhesive flux, composed of microparticles rolling on the surface, is a combined measure of frequency of binding events and their duration.15, 27 To assess the targeting avidity of the system, we characterized the micro-motions of individual beads by measuring the distance between and duration of the transient adhesive events that are characteristic of rolling on low selectin densities.16 The distance between repeated microparticle binding events (also known as tethering) is hypothesized to correlate with the intrinsic selectin binding probability and is referred to as the 'step distance’.15, 28 At a wall shear stress of 1 dyne/cm2, the step distances of HuEP-PEG10000 microparticles rolling on P-selectin decreased 3-fold relative to those with PEG3400 linkers and 4-fold relative to microparticles presenting chemisorbed HuEP, indicative of a higher bond formation rate (Fig. 2). The effect of the PEG spacer length was significant, with the ~ 3-fold difference in brush conformation length between PEG10000 and PEG3400 leading to a 3-fold decrease in the step size at a wall shear stress of 1.0 dynes/cm2 (Fig. 2). The differences between HuEP chemisorbed, PEG3400, and PEG10000 microparticles were insignificant at lower wall shear stresses of 0.4 dyn/cm2, possibly due to the longer contact times and less stringent binding conditions at lower flow rates.
Figure 2. Distances between successive transient bonding events (step size) is lowered by longer PEG spacers suggesting higher bond formation rates.
The average step distances between interactions for HuEP microparticles during low site density rolling with direct antibody chemisorption (grey), PEG3400 linkers (black), and PEG10000 linkers (white). P-selectin site density was set at 100 sites/µm2. The step distance is defined as the distance traveled between paused events. * p < 0.05 against each condition (p = 0.000002 for 0.1 dynes/cm2. p = 0.00019 for 1.0 dynes/cm2.)
We observed that at shear stresses of 0.3 dynes/cm2 and below, HuEP-PEG10000 microparticles displayed a decreased likelihood of a repeated interaction when compared to HuEP-PEG3400 microparticles. The entropically forced brush conformation of the PEG spacer may have limited the ability of the PEG10000 microparticle to sample its immediate environment by repelling fluctuations closer to the surface, thus reducing the probability of binding at low flow rates.29, 30 Increasing the contact force at higher flow rates may therefore be necessary to overcome repulsion forces between the bead and surface created by the neutral PEG chains on the microparticle surface.31
Effect of PEG spacer length on the HuEP-P-selectin bond strength of PEG grafted microparticles in shear flow
The adhesive dynamics of rolling microparticles were influenced significantly by the fluid forces on the particle and its binding geometry. When a microparticle touches down on a surface, it immediately experiences forces created by the bonding of its adhesion receptors, leading to arrest of the particle for a period of time that can be determined by measuring the duration of time it’s stationary.32 The dissociation of this bond can be described through first order reaction kinetics when the interactions are dominated by single bond events as is the case with low site density substrates. 32–34
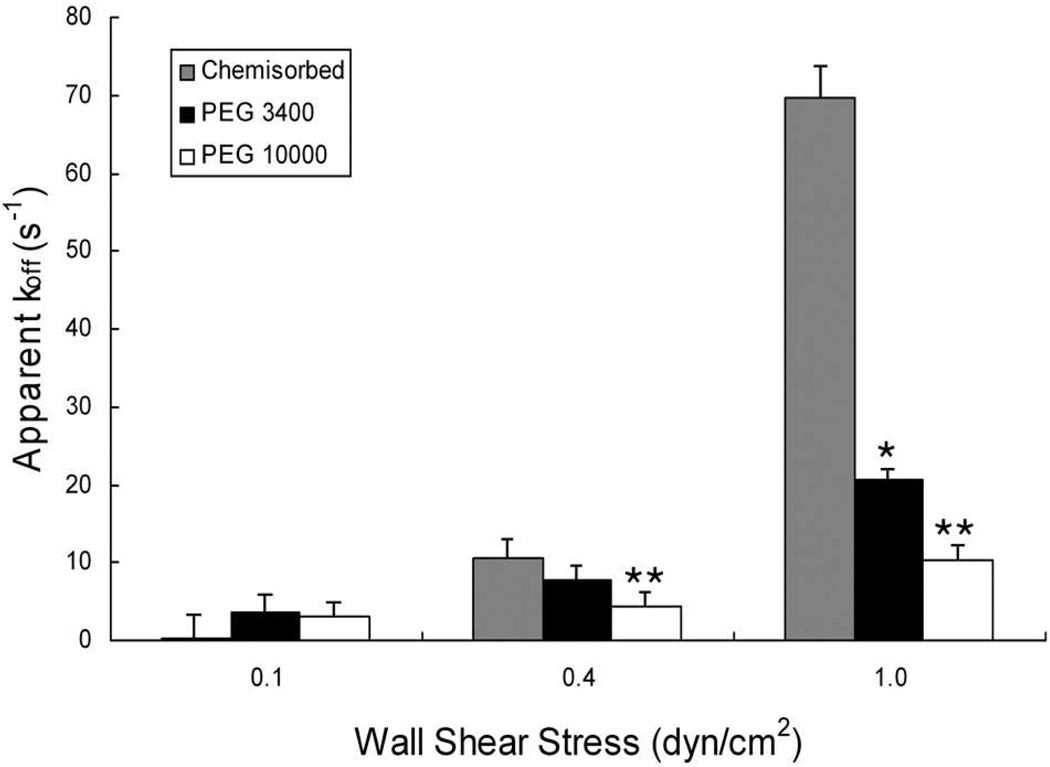
The dissociation constant (koff) of HuEP-P-selectin bonds made with different PEG lengths were measured by analysis of the lifetime of the microparticle's transient adhesive interactions (referred to often as ‘tether bonds’) during rolling on P-selectin.26, 35 The 2-fold decrease in apparent koff of HuEP-P-selectin bonds conferred by PEG10000 compared to PEG3400 at 1 dyne/cm2 resulted in 2-fold increase in average bond lifetime (Fig. 3). For the HuEP functionalized microparticles at a shear stress of 1 dyne/cm2, adding the long PEG insert improved the bond lifetime by 7-fold compared to that of chemisorbed HuEP. Within the range of shear stresses examined, the data indicated that with increasing receptor length, the rate of bond breakage decreased independent of the bond’s intrinsic force sensitivity (Table I). Wall shear stresses above 1 dyne/cm2 were not investigated in this study in order to focus on the dynamics of small numbers of bonds. Higher forces would necessitate multivalent binding events in order to create detectable adhesive interactions.
Figure 3. Bond dissociation constants (koff) are lowered by increasing the length of the HuEP-PEG spacer on the microparticle.
Distributions of the lifetimes of the transient adhesions were used to calculate the bond dissociation constants (koff) of chemisorbed, PEG3400, and PEG10000 linked HuEP microparticles interacting with the P-selectin surface at wall shear stresses of 0.1 dynes/cm2, 0.4 dynes/cm2, and 1.0 dynes/cm2. The relationship between koff and wall shear rate was determined for (grey) chemisorbed, (black) PEG3400 and (white) PEG10000 linked microparticles. The P- selectin surface site density was 100 sites/µm2. R2 > 0.94 for the bi-molecular reaction model equation: ln[C] = −koff t + constant. 26,37 Dissociation constant differences that achieve significance are indicated by asterisk (* p<0.01) for HuEP3400 koff versus chemisorbed HuEP and (** p<.01) for HuEP10000 koff versus chemisorbed HuEP koff.
Table I. Effect of PEG length on the selectin bonds force.
The effect of PEG molecular contour length on the force on the selectin mediated bonds. From calculated and experimentally confirmed results, the lever arm of chemisorbed and PEGylated microparticles was determined. The contour length of each component of the linkage is comprised of PEG3400 (~25 nm fully stressed), PEG10000 (~80 nm fully stressed), biotin/avidin linkage (~4 nm53), and the selectin/Ab linkage (~40 nm).
| HuEP Linkage | |||
|---|---|---|---|
| None | PEG3400 | PEG10000 | |
| Contour Length (nm) | ~ 40 | 65.3 ± 7.3 | 161.5 ± 5.9 |
| Lever Arm (nm) | 491.5 | 629.3 | 994.4 |
| koff at 1 dyne/cm2 (s−1) | 69.8 | 20.7 | 10.4 |
| Force on the Bond (pN) | 140.2 | 109.9 | 70.6 |
| Bond Compliance (Ang) | 0.94 ± 0.43 | 1.22 ± 0.07 | 1.03 ± 0.21 |
Intrinsic reactive bond compliance appeared unaffected by immobilization strategies
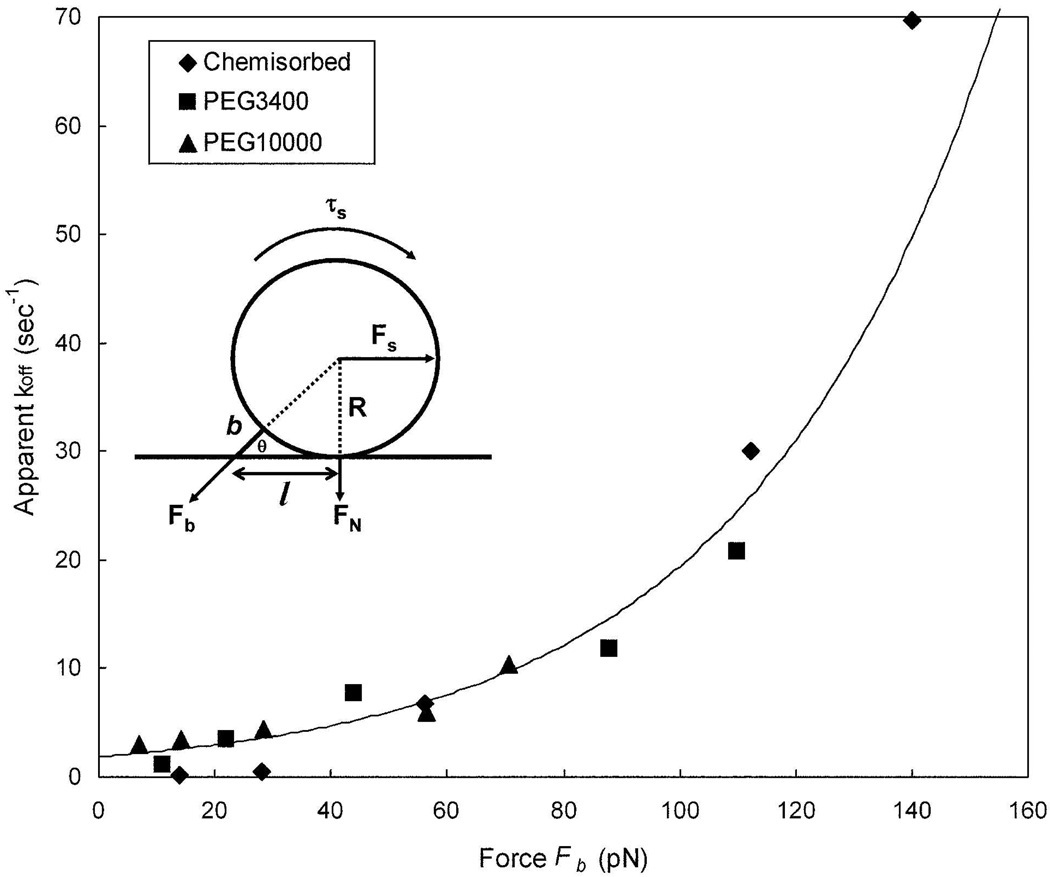
The force of the bond was calculated using the geometrical parameters of the system and fluid dynamic equations that describe the magnitude of the shear and torque acting on a bond17 (Fig. 4 insert), assuming static equilibrium.36, 37 From the change in measured koff as a function of force it is possible to estimate the reactive bond compliance represented by σ in the Bell equation and the unstressed dissociation constant (Fig. 4),
| (1) |
where kb is the Boltzmann's constant, and T is the absolute temperature.38 The Bell model predicts that the lifetime of a stressed bond will decline exponentially with increasing force. Nonlinear fitting using the Leverburg-Marquardt algorithm yielded an unstressed dissociation rate constant of for the HuEP-P-selectin bond. For HuEP linked via PEG10000 and PEG3400, the reactive bond compliance (σ) was 1.03 ± 0.21 Å and 1.22 ± 0.07 Å, respectively (Table 1). The reactive bond compliance of chemisorbed HuEP-P-selectin bonds was virtually identical to that of PEG-linked HuEP-P-selectin bonds within statistical error (0.94 ± 0.43 Å).10 The reactive bond compliance for the entire population was calculated to be 1.06 ± 0.14 Å, suggestive that the intrinsic bond mechanics of HuEP-P-selectin were indifferent to the mode of surface anchorage.
Figure 4. Off rate of the HuEP / P-selectin bond as a function of the shear force is independent of the size of the molecular spacer between HuEP and the microparticle surface.
Comparison between chemisorbed (diamond), PEG3400 linkers (square), and PEG10000 linkers (triangle). The bond lifetime, microbead bond geometry, and shear parameters were fit into the Bell model to calculate the reactive bond compliance from the relationship between force and bond lifetimes under wall shear stress. The non-linear curve fit line was evaluated from Levenberg-Marquardt algorithm. The averaged fitted unstressed dissociation constant was 1.8 s−1 with a bond compliance (σ) of 1.06 angstroms. Diagram of a microbead in static equilibrium and the estimation of force on a single bond (insert).
Despite the similar reactive bond compliance and zero-shear of the chemisorbed and PEG-linked HuEP-P-selectin bonds, microparticles with longer PEG spacers had longer lived adhesive events at higher flow rates (> 0.4 dynes/cm2). If the force-bond lifetime relationships (the reactive bond compliance, or σ) were significantly different, it would suggest that the linkage and immobilization processes were changing the intrinsic biomechanical properties of the bond. Since the differences in reactive bond compliance between the various immobilization strategies were insignificant, we therefore concluded that the microparticles with the longer PEG spacers formed longer-lived bonds--not necessarily more bonds. One caveat is that the disparity between the grafting density of PEG3400 and PEG10000 might lead to clusters of HuEP bound to the tetravalent streptavidin linkers leading to multiple ligand binding if P-selectin site densities were insufficiently sparse.
Adhesive flux benefits of PEG spacer length were directly applicable to a natural P-selectin binding glycopeptide
While it appeared that the increased PEG spacer length improved the mechanically stressed affinity of HuEP for P-selectin, it was important to determine if long PEG spacer effects could be generalized to other types of receptors with different affinities. Therefore, we examined glycopeptides based on the principle natural ligand for P-selectin that has a well-characterized binding pocket and that supports rolling of microparticles on P-selectin similar to that of leukocytes. The glycopeptide is based on the N-terminal 19 amino acids of PSGL-1 and has been generated with (2-GSP-6) and without the critical tyrosine sulfation (2-GP-6). The peptide, 2-GSP-6, supports rolling on P-selectin when attached to microparticles.11 Tyrosine sulfation on the peptide increases its affinity to P-selectin, by 40-fold relative to the non-sulfated variant, 2-GP-6.39 Consistent with a lower affinity, the 2-GP-6 which lacks sulfation supports much less robust microparticle rolling on P-selectin.27, 40
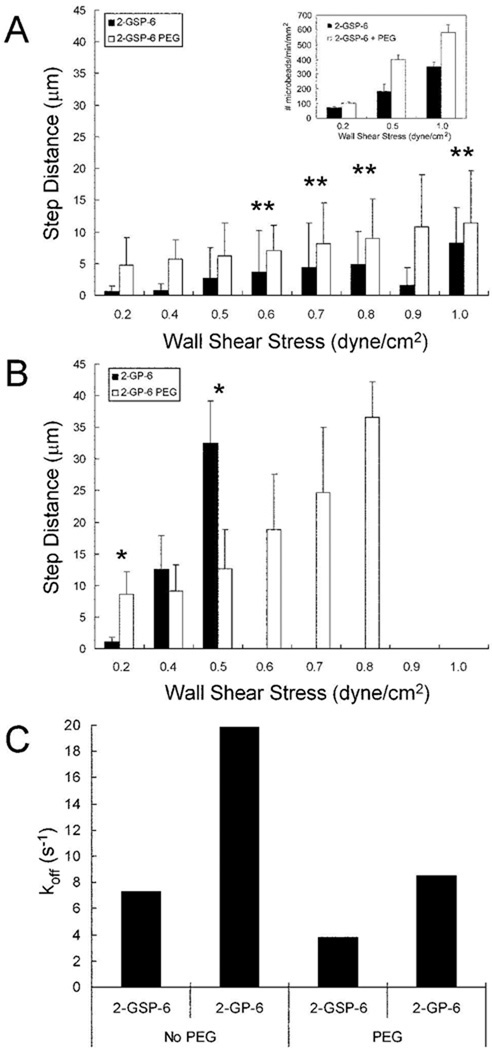
When 2-GSP-6 was linked to the microparticle surface with a PEG10000 insert, there was a 40% increase in rolling flux compared to chemisorbed 2-GSP-6 (Fig. 5A insert). Despite the increase in rolling flux, the step size of 2-GSP-6 conjugated microparticles was unaffected within statistical error (Fig. 5). The lack of sulfation on chemisorbed 2-GP-6 reduced microparticle rolling to within a narrow range of wall shear stresses compared to chemisorbed 2-GSP-6 microparticles (Fig 5B), with no binding or rebinding observed at wall shear stresses above 0.5 dyn/cm2. However, when 2-GP-6 was conjugated to the microparticle surface using a PEG10000 linker, the 2-GP-6 microparticles displayed a 2-fold improvement in binding frequency at 0.5 dyn/cm2 wall shear stress and were able to support rolling at flow rates up to 0.8 dyn/cm2 wall shear stress. For both 2-GSP-6 and 2-GP-6, the inclusion of long PEG chains lowered the apparent dissociation rate constant (koff) values under shear stress by 2-fold relative to chemisorbed immobilization (Fig. 5C). In the case of the glycopeptides, the PEG spacer appeared to both improve binding frequency and allow the bonds so formed to last longer after loading. Thus, long PEG tethers showed evidence of mechanically improving the affinity of a relatively poor binding molecule such as 2-GP-6 by both improving bond formation probability and the lifetimes of stressed adhesive bonds.
Figure 5. Long PEG spacers improved the microparticle binding performance of a low affinity P-selectin ligand.
The averaged step distances between interactions for microparticles over a range of shear rates is shown. (A) Comparison of chemisorbed 2-GSP-6 (black) and 2-GSP-6 with PEG10000 (white). Insert in panel (A) shows the adhesive flux of chemisorbed 2-GSP-6 microparticles (white) compared to 2-GSP-6+PEG10000 microparticles (black) at flow rates of 0.2, 0.5, and 1.0 dyn/cm2 wall shear stress. (B) Comparison of averaged step distances of chemisorbed 2-GP-6 (black) and 2-GP-6 with PEG10000 (white). Wall shear rates with zero step distance measurements indicates step distances larger than the microscope field of view (100+ µm), and were considered non-interacting. (C) The mean koff of 2-GSP-6 microparticles, 2-GP-6 microparticles, 2-GSP-6+PEG10000 microparticles, and 2-GSP-6+PEG10000 microparticles interacting with the P-selectin surface at a wall shear stress of 0.5 dynes/cm2. The P-selectin surface density for all experiments was set at 100 sites/µm2. ** p > 0.05 (p = 0.058 for 0.6 dynes/cm2. p = 0.350 for 0.7 dynes/cm2. p = 0.117 for 0.8 dynes/cm2. p = 0.365 for 1.0 dynes/cm2). * p < 0.05 (p = 0.000158 for 0.2 dynes/cm2. p = 0.0078 for 0.5 dynes/cm2). R2 > 0.96 for the bi-molecular reaction model equation.
Discussion
It has long been known that molecular spacers inserted between a recognition motif and other parts of a molecule can markedly improve its accessibility to ligand, as is commonly done with purification handles such as 6-His tags. Insertions of short spacers have also been shown to improve the binding avidity of large protein domains in the example of a growth-factor targeted gene delivery system.41 In the case of interactions between receptor decorated polymersomes and reactive surfaces, it has been shown that inserting a PEG spacer results in greater adhesion strength due to the larger reactive cap and consequent formation of multiple bonds.42–44 Several lines of experimentation therefore suggest that positioning a receptor's binding pocket away from the surface can improve accessibility. However, in none of the examples cited above has the effective affinity of the molecular interaction been shown to increase or be otherwise modulated by spacer length.
This study illustrated that lengthening selectin-targeted receptors with PEG spacers improved the two dimensional kinetics of the reaction in two ways: 1) by increased microparticle binding frequency and 2) by reducing the total stress on the bonds formed with the P-selectin and thereby lengthening adhesive lifetimes. The faster bond formation rates and the longer adhesive lifetimes suggested that the mechanically stressed affinity of P-selectin and its ligands were therefore increased. Consequently, the introduction of PEG spacers of sufficient length may generally improve two-dimensional reaction kinetics in shear flow environments and potentially lead to greater binding robustness of intravascular targeted drug, gene, and image contrast delivery systems.
Under static conditions, the addition of polymer chain spacers extends the effective range of binding interaction events, given time for binding on the order of seconds.20, 45 In a setting of low PEG chain density, the functionalized and mobile end of the polymer resides near the Flory radius. Excursions to its maximum contour length where binding with a ligand would be predicated to occur are exceedingly rare, consistent with random walk statistics. Under flow conditions, however, relative velocities between surfaces such as a cell and the blood vessel wall lead to contact times on the order of a millisecond or less. The combination of rare excursions to the maximum contour length of the PEG spacer and brief contact times at normal blood flow rates would be expected to result in relatively infrequent adhesion events unless surfaces were brought to within the Flory radius.
To overcome some of the barriers to cell and particle interactions with surfaces, both biologic and non-biologic, the leukocyte model of long adhesion bonds offers a valuable lesson for successful targeting. Hydrodynamic effects of erythrocytes in the blood are essential for leukocyte margination to the vessel wall, but it is unclear whether such forces are sufficient to bring cell membranes into molecular contact. Given the low diffusivity of cells near a surface, adhesion may ultimately be limited by whether the receptor and its ligand can span the fluid layer between cell and surface. For this reason, the relatively long reach of leukocyte homing receptors may therefore be critical for bridging cell surfaces in close apposition.
Emulation of the model of an extended leukocyte adhesion receptor was achieved by insertion of ligand-functionalized PEG chains in a sea of neutral PEG, forcing the reactive pocket to reside at much nearer its maximum contour length than its Flory radius. PEG chain overlap at the site densities of neutral PEG examined favors an extended conformation that is many-fold longer than the Flory radius of an isolated ligand-bearing PEG chain. When surface roughness is accounted for, which is estimated to range from 50 to slightly over 100 nm depending on material, the combined receptor-ligand complexes plus PEG spacers appeared long enough to span the gap (see Table 1) of ~ 75 nm at which the microparticles reside under flow conditions when gravity and vertical Brownian motion are included.46–48 As microparticle binding frequencies were proportional to estimated contour length, we speculate that in both cases the ligands were capable of sampling space near the P-selectin receptors a significant fraction of the time and not simply too far from the surface to bind. Hence, differences in microparticle binding frequency may depend more on the variety of molecular configurations afforded by a longer PEG spacer.
To minimize multivalent interactions, we focused on examination of microparticle interactions at low site densities. By setting up the experimental conditions to favor single or small bond cluster interactions, it was possible to decouple intrinsic bond mechanics from the engineered effects of increasing receptor length. Nevertheless, multiple bonding events due to nanoscale clustering would be predicted to improve stressed lifetime and bond formation frequency and would likely be a desirable design feature for situations needful of fast bonding. Higher site densities, higher ligand densities, and higher flow rates would be important parameters to examine in the future to further test the hypothesis that receptor length might modulate microparticle and cell adhesion in physiologic blood flow.
In addition to the apparent enhancement in HuEP-P-selectin association rate by lengthening the PEG spacers, we also observed that sheared bonds lasted longer. The force dependence of HuEP-P-selectin bond lifetimes as modeled by the Bell equation38 suggested that the reactive compliance of the antibody-antigen bond, and therefore its intrinsic affinity, was unaffected by the PEG spacers and immobilization strategies. How bond length could influence bond lifetime can be explained in part by analogy to how extensible microvilli strengthen leukocyte rolling on P-selectin.26, 49, 50 In brief, the longer bond structure allows the bond to preferentially localize at the trailing edge of the moving leukocyte, or microparticle. As the bond stretches the moment arm is increased, leading to a reduction in the torque acting on the adhesive site.26, 50 The proposed mechanism for strengthening leukocyte rolling through microvillus extension has been mimicked on a micron length scale by creation of surface wrinkles on lipid-shelled microbubbles that improved binding in flow to P-selectin surfaces.51 We observed what appeared to be a similar effect on the nanometer length scale by insertion of a PEG spacer between the microparticle’s surface and the HuEP ligand.
While it appeared that the longer PEG spacers improved the mechanically stressed affinity of a bond, it was important to determine if it's effects could be generalized to other types of receptors with different affinities. The peptide, 2-GSP-6, is modeled after the NH2-terminal P-selectin binding region of PSGL-1 and supports rolling on P-selectin when attached to microparticles.11 Tyrosine sulfation on the peptide increases the affinity to P-selectin, thus the non-sulfated variant, 2-GP-6, has a 40-fold reduction in affinity.39 As expected, the 2-GP-6 supported much less robust microparticle rolling on P-selectin.52 But anchorage of the glycopeptides via PEG spacers lead to improved microparticle rolling flux for both the 2-GSP-6 and 2-GP-6 peptides, with the lower affinity 2-GP-6 microparticles displaying the greatest improvement in rolling. Thus, the insertion of PEG spacers that restore 2-GP-6 peptide to an approximation of its physiologic length showed evidence of mechanically improving the binding of a relatively low affinity molecule such as 2-GP-6.
Conclusion
In the case of leukocyte trafficking and homing of vascular-targeted microparticles, blood flow creates a highly dynamic two dimensional confinement zone that not only limits the time for bond formation, but also leads to rapid mechanical loading of any biomolecular complex. We observed that in such confinement zones, increases in the length of the adhesive bond’s contour length by insertion of PEG spacers significantly increased the mechanical affinity of sheared bonds formed with P-selectin. This is a novel observation that not only gives insight into the design principles of leukocyte homing receptors, but also may serve as an approach to improve vascular targeting of micron and sub-micron scaled drug and gene delivery moieties.
ACKNOWLEDGEMENTS
This work was supported by a National Institute of Health Grant NIH R01 HL54614 (M.B.L) and a Bioengineering Research Partnership Grant HL64381 (M.B.L.).
Contributor Information
Anthony Sang Won Ham, Email: aham@imquest.com.
Alexander L. Klibanov, Email: alk6n@virginia.edu.
Michael B. Lawrence, Email: mbl2a@virginia.edu.
REFERENCES
- 1.Sakhalkar HS, Dalal MK, Salem AK, Ansari R, Fu J, Kiani MF, Kurjiaka DT, Hanes J, Shakesheff KM, Goetz DJ. Proc. Natl. Acad. Sci. U.S.A. 2003;100(26):15895–15900. doi: 10.1073/pnas.2631433100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakhalkar HS, Hanes J, Fu J, Benavides U, Malgor R, Borruso CL, Kohn LD, Kurjiaka DT, Goetz DJ. FASEB J. 2005 doi: 10.1096/fj.04-2668fje. 04-2668fje. [DOI] [PubMed] [Google Scholar]
- 3.Mehta P, Cummings RD, McEver RP. J. Biol. Chem. 1998 December 4;49:32506–32513. doi: 10.1074/jbc.273.49.32506. [DOI] [PubMed] [Google Scholar]
- 4.Huang J, Chen J, Chesla SE, Yago T, Mehta P, McEver RP, Zhu C, Long M. J. Biol. Chem. 2004;279(43):44915–44923. doi: 10.1074/jbc.M407039200. [DOI] [PubMed] [Google Scholar]
- 5.Somers WS, Tang J, Shaw GD, Camphausen RT. Cell. 2000;103(3):467–479. doi: 10.1016/s0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- 6.Bendas G, Krause A, Bakowsky U, Vogel J, Rothe U. Int. J. Pharm. 1999;181(1):79–93. doi: 10.1016/s0378-5173(99)00002-2. [DOI] [PubMed] [Google Scholar]
- 7.Spragg DD, Alford DR, Greferath R, Larsen CE, Lee K-D, Gurtner GC, Cybulsky MI, Tosi PF, Nicolau C, Gimbrone MA., Jr Proc. Natl. Acad. Sci. U.S.A. 1997;94(16):8795–8800. doi: 10.1073/pnas.94.16.8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajitou A, Pasqualini R, Arap W. Trends Cardiovasc. Med. 2006;16(3):80–88. doi: 10.1016/j.tcm.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He X-Y, Xu Z, Melrose J, Mullowney A, Vasquez M, Queen C, Vexler V, Klingbeil C, Co MS, Berg EL. J. Immunol. 1998;160(2):1029–1035. [PubMed] [Google Scholar]
- 10.Ham ASW, Goetz DJ, Klibanov AL, Lawrence MB. Biotechnol. Bioeng. 2006 doi: 10.1002/bit.21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leppanen A, Mehta P, Ouyang Y-B, Ju T, Helin J, Moore KL, van Die I, Canfield WM, McEver RP, Cummings RD. J. Biol. Chem. 1999;274(35):24838–24848. doi: 10.1074/jbc.274.35.24838. [DOI] [PubMed] [Google Scholar]
- 12.Hermanson GT. Bioconjugate Techniques. San Diego, CA: Academic Press; 1996. [Google Scholar]
- 13.Takalkar AM, Klibanov AL, Rychak JJ, Lindner JR, Ley K. J. Controlled Release. 2004;96:473–482. doi: 10.1016/j.jconrel.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Suonpaa M, Markela E, Stahlberg T, Hemmila I. J. Immunol. Methods. 1992;149(2):247–253. doi: 10.1016/0022-1759(92)90256-s. [DOI] [PubMed] [Google Scholar]
- 15.Smith MJ, Smith BRE, Lawrence MB, Snapp KR. J. Biol. Chem. 2004;279(21):21984–21991. doi: 10.1074/jbc.M402731200. [DOI] [PubMed] [Google Scholar]
- 16.Paschall CD, Guilford WH, Lawrence MB. Biophys J. 2008;94(3):1034–1045. doi: 10.1529/biophysj.106.098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldman AJ, Cox RG, Brenner H. Chem. Eng. Sci. 1967;22:653–660. [Google Scholar]
- 18.Cheezum MK, Walker WF, Guilford WH. Biophys. J. 2001;81(4):2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Gennes PG. Adv. Colloid Interface Sci. 1987;27(3–4):189–209. [Google Scholar]
- 20.Jeppesen C, Wong JY, Kuhl TL, Israelachvili JN, Mullah N, Zalipsky S, Marques CM. Science. 2001 July 20;293:265–468. doi: 10.1126/science.293.5529.465. [DOI] [PubMed] [Google Scholar]
- 21.Borden MA, Zhang H, Gillies RJ, Dayton PA, Ferrara KW. Biomaterials. 2008;29(5):597–606. doi: 10.1016/j.biomaterials.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DH, Klibanov AL, Needham D. Langmuir. 2000;16(6):2808–2817. [Google Scholar]
- 23.Murat M, Grest GS. Macromolecules. 1989;22:4054–4059. [Google Scholar]
- 24.Chen S, Alon R, Fuhlbrigge RC, Springer TA. Proc. Natl. Acad. Sci. U.S.A. 1997;94(7):3172–3177. doi: 10.1073/pnas.94.7.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goetz DJ, Greif DM, Ding H, Camphausen RT, Howes S, Comess KM, Snapp KR, Kansas GS, Luscinskas FW. J. Cell Biol. 1997;137(2):509–519. doi: 10.1083/jcb.137.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park EYH, Smith MJ, Stropp ES, Snapp KR, DiVietro JA, Walker WF, Schmidtke DW, Diamond SL, Lawrence MB. Biophys. J. 2002 April;82:1835–1847. doi: 10.1016/S0006-3495(02)75534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yago T, Leppanen A, Qiu H, Marcus WD, Nollert MU, Zhu C, Cummings RD, McEver RP. J. Cell Biol. 2002;158(4):787–799. doi: 10.1083/jcb.200204041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramachandran V, Yago T, Epperson TK, Kobzdej MM, Nollert MU, Cummings RD, Zhu C, McEver RP. Proc. Natl. Acad. Sci. U S A. 2001;98(18):10166–10171. doi: 10.1073/pnas.171248098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Efremova NV, Bondurant B, O'Brien DF, Leckband DE. Biochemistry. 2000;39:3441–3451. doi: 10.1021/bi992095r. [DOI] [PubMed] [Google Scholar]
- 30.Wong JY, Park CK, Seitz M, Israelachvili J. Biophys. J. 1999;77:1458–1468. doi: 10.1016/S0006-3495(99)76993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y, Chien S, Weinbaum S. Biophys. J. 2001;80(3):1124–1140. doi: 10.1016/S0006-3495(01)76090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanley WD, Wirtz D, Konstantopoulos K. J Cell Sci. 2004;117(12):2503–2511. doi: 10.1242/jcs.01088. [DOI] [PubMed] [Google Scholar]
- 33.Tees DF, Waugh RE, Hammer DA. Biophys. J. 2001;80:668–682. doi: 10.1016/S0006-3495(01)76047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alon R, Hammer DA, Springer TA. Nature. 1995;374(6522):539–542. doi: 10.1038/374539a0. [DOI] [PubMed] [Google Scholar]
- 35.Zhu C, Long M, Chesla SE, Bongrand P. Ann. Biomed. 2002;30(3):305–314. doi: 10.1114/1.1467923. [DOI] [PubMed] [Google Scholar]
- 36.Smith MJ, Berg EL, Lawrence MB. Biophys. J. 1999;77:3371–3383. doi: 10.1016/S0006-3495(99)77169-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Nature. 2003;423(6936):190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 38.Bell GI. Science. 1978 May 12;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 39.Hicks AER, Leppanen A, Cummings RD, McEver RP, Hellewell PG, Norman KE. FASEB J. 2002;16(11):1461–1462. doi: 10.1096/fj.02-0075fje. [DOI] [PubMed] [Google Scholar]
- 40.Rodgers SD, Camphausen RT, Hammer DA. Biophys. J. 2001;81:2001–2009. doi: 10.1016/S0006-3495(01)75850-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaffer DV, Lauffenburger DA. J. Biol. Chem. 1998;273(43):28004–28009. doi: 10.1074/jbc.273.43.28004. [DOI] [PubMed] [Google Scholar]
- 42.Lin JJ, Silas J, Bermudez H, Milam VT, Bates FS, Hammer D. Langmuir. 2004;20:5493–5500. doi: 10.1021/la036417a. [DOI] [PubMed] [Google Scholar]
- 43.Burridge KA, Figa MA, Wong JY. Langmuir. 2004;20(23):10252–10259. doi: 10.1021/la0489099. [DOI] [PubMed] [Google Scholar]
- 44.Wong JY, Kuhl TL. Langmuir. 2008;24(4):1225–1231. doi: 10.1021/la702357a. [DOI] [PubMed] [Google Scholar]
- 45.Wong JY, Kuhl TL, Israelachvili JN, Mullah N, Zalipsky S. Science. 1997 February 7;275:820–8222. doi: 10.1126/science.275.5301.820. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt BJ. Microfluidic, Computational, and Recombinant Protein Design for the Analysis of the Molecular Properties of Dynamic Cell Capture Systems. Charlottesville: Doctorate, University of Virginia; 2009. [Google Scholar]
- 47.Schmidt BJ, Huang P, Breuer KS, Lawrence MB. Anal. Chem. 2008;80(4):944–950. doi: 10.1021/ac071529i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierres A, Benoliel AM, Zhu C, Bongrand P. Biophys. J. 2001;81(1):25–42. doi: 10.1016/S0006-3495(01)75677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pospieszalska MK, Zarbock A, Pickard JE, Ley K. Microcirculation. 2009;16(2):115–130. doi: 10.1080/10739680802462792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao J-Y, Ting-Beall HP, Hochmuth RM. Proc. Natl. Acad. Sci. U.S.A. 1998;95(12):6797–6802. doi: 10.1073/pnas.95.12.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rychak JJ, Lindner JR, Ley K, Klibanov AL. J. Controlled Release. 2006;114(3):288–299. doi: 10.1016/j.jconrel.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Leppanen A, White SP, Helin J, McEver RP, Cummings RD. J. Biol. Chem. 2000;275(50):39569–39578. doi: 10.1074/jbc.M005005200. [DOI] [PubMed] [Google Scholar]
- 53.Noppl-Simson DA, Needham D. Biophys. J. 1996;70(3):1391–1401. doi: 10.1016/S0006-3495(96)79697-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cozen-Roberts C, Lauffenburger DA, Quinn JA. Biophys. J. 1990;58:841–856. doi: 10.1016/S0006-3495(90)82430-9. [DOI] [PMC free article] [PubMed] [Google Scholar]