Summary
The emerging discoveries on the link between polyadenylation and disease states underline the need to fully characterize genome-wide polyadenylation states. Here, we report comprehensive maps of global polyadenylation events in human and yeast generated using refinements to the Direct RNA Sequencing technology. This direct approach provides a quantitative view of genome-wide polyadenylation states in a strand-specific manner and requires only attomole RNA quantities. The polyadenylation profiles revealed an abundance of unannotated polyadenylation sites, alternative polyadenylation patterns, and regulatory element-associated polyA+ RNAs. We observed differences in sequence composition surrounding canonical and non-canonical human polyadenylation sites, suggesting novel non-coding RNA-specific polyadenylation mechanisms in humans. Furthermore, we observed the correlation level between sense and antisense transcripts to depend on gene expression levels, supporting the view that overlapping transcription from opposite strands may play a regulatory role. Our data provide a comprehensive view of the polyadenylation state and overlapping transcription.
Introduction
The known regulatory role of 3′-untranslated regions (3′UTRs) and poly-A tails in mRNA localization, stability and translation (reviewed by (Andreassi and Riccio, 2009)), and polyadenylation regulation defects leading to human diseases such as oculopharyngeal muscular dystrophy, thalassemias, thrombophilia and IPEX syndrome (Bennett et al., 2001; Brais et al., 1998; Gehring et al., 2001; Higgs et al., 1983; Lin et al., 1998; Orkin et al., 1985) underscores the need to fully characterize polyadenylation sites and mechanisms. Our knowledge in this area primarily originates from expressed sequence tag (EST) databases and predictions relying on polyadenylation-associated motif elements (Graber et al., 2002; Lutz, 2008; Tian et al., 2005). EST databases are valuable, but insufficient for in-depth mapping of polyadenylation sites due to data quality problems such as low numbers of full-length ESTs, chimeric sequences (due to cDNA template switching (Cocquet et al., 2006)), internal cDNA priming events leading to cloning of incomplete transcripts, and low quality sequences at the ends of ESTs (Zhang et al., 2005a; Zhang et al., 2005b). For applications requiring identification of polyadenylation site usage frequency changes across biological conditions, EST databases, motif searches and classical polyadenylation site mapping approaches (Slomovic et al., 2008) such as RACE, RT-PCR and nuclease sensitivity assays do not provide the required simplicity, sensitivity, depth and quantitative genome-wide view. Annotation of the 3′ ends of yeast genes were attempted previously with RNA-seq (Nagalakshmi et al., 2008) and microarray-based (David et al., 2006) approaches, but these studies did not have sufficient resolution to map individual cleavage sites for polyadenylation. Furthermore, despite much interest devoted to overlapping transcription, we still do not have a complete understanding of sense/antisense transcription (reviewed by (Faghihi and Wahlestedt, 2009)). To date, our knowledge in this area comes from methods relying on reverse transcription that suffers from spurious second-strand cDNA products (Gubler, 1987; Spiegelman et al., 1970), complicating analyses requiring unambiguous determination of RNA strand. While methods have recently been developed that preserve the RNA strand information through RNA-level modifications such as bisulfite treatment or RNA-level adapter ligation (He et al., 2008; Mamanova et al., 2010), these still rely on cDNA synthesis, ligation and amplification steps that may introduce artifacts and complicate the quantitation of various RNA species.
To avoid the known biases and artifacts introduced to RNA measurements during reverse transcription (Cocquet et al., 2006; Liu and Graber, 2006; Mamanova et al., 2010; Wu et al., 2008) or other sample manipulation steps, we recently developed the direct RNA sequencing (DRS) technology (Ozsolak et al., 2009). DRS sequences RNA molecules in a massively-parallel manner without its prior conversion to cDNA or the need for biasing ligation/amplification steps. Since this proof-of-concept study, we have improved and adapted DRS for use with the Helicos Genetic Analysis system. DRS produces alignable reads up to 55 nts (mean read length is 33–34 nts). Unlike other RNA analysis approaches which require multiple nucleic acid manipulation steps, DRS only requires polyadenylated and 3′ blocked RNA templates for sequencing.
We here applied DRS to generate a comprehensive and high resolution map of polyadenylation sites of human and yeast transcripts. Using multiple independent approaches, we validated our findings and demonstrated the usefulness of the approach to identify alternative polyadenylation events. We observed many unannotated polyadenylation sites and novel RNA species associated with open chromatin sites that may function to regulate gene expression. We also observed that sequence and motif contexts surrounding novel intergenic and genic sense/antisense polyadenylation sites away from 3′ ends of known genes exhibit significant differences than sequence and motif contexts surrounding polyadenylation sites near known gene 3′ ends. This observation suggests alternative mechanisms and/or purposes of RNA polyadenylation. In addition, we have examined overlapping transcription patterns of polyA+ transcripts. Between the steady-state quantities of sense and antisense transcripts, we observed a complex correlation pattern that depends on gene expression levels.
Results
Mapping Global 3′ Polyadenylation Sites with DRS
To determine polyadenylation locations, 200–300 picograms of human liver and yeast polyA+ RNAs, and 3ng human brain total RNA blocked at their 3′ ends were used per sequencing channel. Given that polyA+ RNA species already contain a natural poly-A tail, additional polyadenylation was not needed. After the capture of polyA+ RNA species on poly(dT)-coated flow cell surfaces by hybridization, a “fill” step with natural dTTP, and a “lock” step with fluorescently-labeled proprietary Virtual TerminatorTM (VT)-A,-C and -G nucleotides were performed. These steps correct for any misalignments that may be present in poly-A/T duplexes, and ensure that the sequencing starts in the template rather than the poly-A tail. After the completion of fill and lock steps, DRS was initiated. The 5′ ends of DRS reads signify cleavage locations. The resolution for identification of the polyadenylation cleavage nucleotide is dependent on fill and lock efficiency and the ability of the sequencing reaction to start immediately upstream of the polyA tails. We measured this efficiency using polyadenylated oligoribonucleotides, and determined the resolution to be ±2 nts (Figure S1A). To determine whether our results might have been negatively affected by potential internal priming events, we performed experiments to observe the sequencing behavior of templates containing internal polyA stretches with 3′ noncomplementary overhangs, and examined the fraction of polyadenylation regions containing downstream polyA-rich regions. We observed rare or no occurrence of internal priming events (Table S1 and Extended Experimental Procedures). Thus, the technology is capable of mapping the extensive 3′ end heterogeneity we and others (Iseli et al., 2002; Lopez et al., 2006; Muro et al., 2008) observed in the majority of yeast and human genes in a genome-wide manner and at nucleotide resolution (Figure 1A–D).
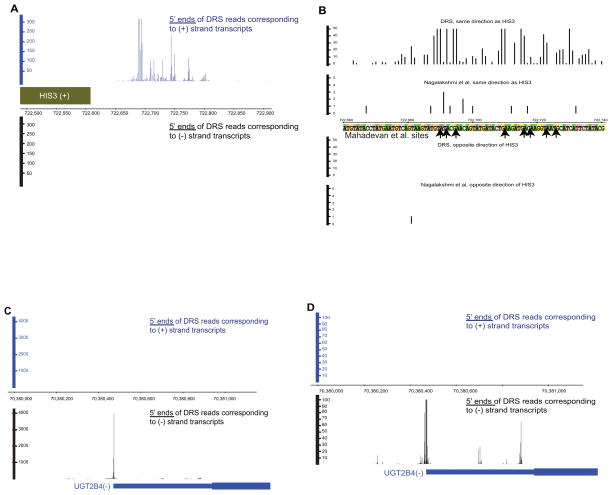
Figure 1. Polyadenylation site detection in yeast (A,B) and human (C,D).
(A) The blue and black panels show the DRS reads emanating from transcripts in the + and − direction, respectively. The major peaks in the blue panel correspond to the 13 polyadenylation sites at locations 722690, 722692, 722695, 722710, 722716, 722718, 722723, 722726, 722746, 722750, 722752, 722775, and 722777 previously identified for HIS3 (Mahadevan et al., 1997) using 3′ RACE-PCR. (B) Zoomed-in view of panel A. Y axis was reduced from 0–300 scale to 0–50. X-axis was reduced from 722,500–722,900 scale to 722,660–722–740. All “end tags” identified by Nagalakshmi et al. in this region are also shown (y axis for these tags is on the scale of 0–5). Arrows mark the sites identified by Mahadevan et al. in the region shown. (C, D) Overview (B) and a zoomed-in view (C) of reads mapping to UGT2B4 3′ annotated ends. Multiple potential polyadenylation sites are evident in panel C (see also Figure S2 and Table S1).
Genome-wide 3′ Polyadenylation State in Yeast
We obtained 7,036,730 DRS reads uniquely aligned to the yeast genome, each read representing a polyadenylation site of an independent transcript, to deduce the yeast polyadenylation landscape (Table S2). To verify our findings, we compared the polyadenylation sites identified here to the sites identified previously for 11 yeast genes using classical approaches, observing high overlap (Figures 1B). Because of its higher resolution, DRS found the frequently used cleavage locations reported previously and other generally lower-frequency cleavage positions (Figures 1A & S1B). In addition, DRS data agreed well with the polyadenylation sites mapped previously for ten genes and seven snoRNAs using PCR amplification of 3′ transcript ends in a manner that preserves the variability in the 3′ ends, followed by high-throughput DNA sequencing of the RT-PCR products (Ozsolak et al., 2009). Furthermore, we validated four previously unannotated intergenic and genic polyadenylation locations using cloning and RACE approaches (Figure S1C, Table S3). We also compared DRS reads to the 60,218 3′ end tags, which constitute ~0.2% of RNA-seq reads, are analogous to DRS reads and mark yeast polyadenylation sites (Nagalakshmi et al., 2008), observing 53,849 (89.4%) of end tags to be within 5 nts of DRS read start locations. The difference observed in the remaining ~10% may be due to differences in the resolution of both methods, different yeast strains and RNA preparation approaches used in both studies.
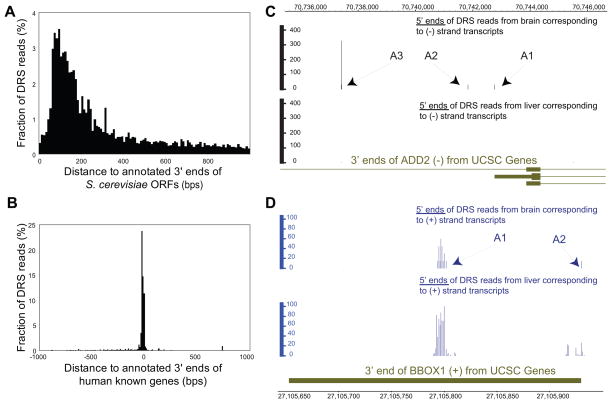
The median length of the 3′ UTRs of 5759 yeast open reading frames (ORFs) was 166 nts (Figure 2A, Table 1). With the number of reads and depth we generated for this study, we observed that 72.1% of the yeast genes exhibited polyadenylation locations separated by at least 50 nts, and frequently more, and thus have multiple polyadenylation sites. The higher levels observed here relative to the 10–15% level reported previously (Nagalakshmi et al., 2008) may be due to the higher resolution of the approach presented here and the higher number of transcripts analyzed. Similar to previous reports (Nagalakshmi et al., 2008), we observed 14% of genes to be orientated in tail-to-tail orientation and have overlapping 3′ ends (see below). 14% of yeast DRS reads mapped to regions within the yeast ORFs either in exons or introns (Table 1). Intronic polyadenylation sites are possibly due to a dynamic interplay between splicing and polyadenylation (Tian et al., 2007), and may represent transcripts encoding shorter proteins.
Figure 2. Characteristics of polyadenylation sites in yeast and human.
Y-axes in panels A and B indicate the fraction of DRS reads aligning at x-distances (in 10 bps bins) relative to the annotated 3′ ends of yeast ORFs (A) and the annotated 3′ ends of human UCSC genes (B). ADD2 (C) and BBOX1 (D) polyadenylation sites in human liver and brain. The polyadenylation sites identified (indicated as A1, A2 and A3) for both genes agree well with previous findings (Costessi et al., 2006; Rigault et al., 2006) (see also Figure S3 and Table S3).
Table 1. Distribution of yeast and human liver reads across genomic regions.
The numbers indicate percentages of uniquely aligned yeast and human DRS reads (Table S2) as provided by the SeqSolve software (Integromics, S.L., Spain). The categories shown are not exclusive, and each proportion was computed independently. Hence proportions are not expected to add up to 100%. The relatively high percentage of reads in the category of antisense yeast reads within 1000 nts of 3′ ORF ends is due to ~2000 yeast ORFs whose 3′ ends are close to each other. CDS: Coding sequence, UTR: Untranslated region, ORF: Open reading frame, Transcripts: Within annotated gene boundaries (see also Figure S1 and Table S2).
| Human | 5′ UTR | 3′ UTR | CDS | Introns | Transcripts | ±200nts of 5′ ends | ±200nts of 3′ ends | ±10nts of 3′ ends |
|---|---|---|---|---|---|---|---|---|
| Sense | 6.46 | 79.38 | 1.02 | 8.8 | 83.94 | 0.59 | 71.32 | 55.7 |
| Antisense | 0.18 | 2.1 | 0.23 | 5.86 | 7.98 | 0.1 | 2.96 | 1.12 |
| Yeast | CDS | Introns | Transcripts | ±1000 nts of 3′ ends of ORFs |
|---|---|---|---|---|
| Sense | 4.68 | 0.19 | 4.86 | 91.36 |
| Antisense | 9.16 | 0.04 | 9.19 | 53.04 |
10.6% of yeast DRS reads did not map downstream of annotated yeast 3′ ends or within the ORFs. To examine the degree of association of yeast polyA+ transcripts with regulatory regions, we took advantage of the regulatory protein binding sites defined recently by DNAse I hypersensitive site (DHS) mapping (Hesselberth et al., 2009). We observed a significant enrichment of divergent transcripts (e.g. transcribed away from DHSs) in regions that are in proximity to intergenic DHSs (p-value = 8.041e-07, non-parametrical two-sample Kolmogorov-Smirnov test, Figure S2).
Genome-wide 3′ Polyadenylation State in Humans
11,882,580 uniquely mapping reads were obtained from human liver polyA+ RNA, of which 1,322,970 were derived from mitochondria and 2,570 reads from rRNA. This is consistent with the observations that human mitochondrial transcripts and a fraction of rRNAs are polyadenylated, perhaps for the purposes of degradation (Nagaike et al., 2005; Slomovic et al., 2010). 56.1% of DRS reads overlapped with 19,871 of 28,858 polyadenylation sites previously annotated using EST databases and motif searches (Zhang et al., 2005a). The differences observed may be due to the single tissue examined here while EST database searches include data from multiple tissue types. 55.7% of liver DRS reads emerged from within 10 nts of annotated 3′ ends of UCSC Genes (Figures 2B & S3A, Table 1). The remaining 44.3% of the reads represent either novel RNAs or alternative polyadenylation sites of known mRNAs. While estimation of the extend of non-coding transcription based on this data is difficult because the full-structure of transcripts represented by the DRS reads are not known, at the very least 9% of reads are located in intergenic regions that are at least 5 kilobases away from known genes and thus likely to represent novel RNAs. 37% of intergenic reads in humans are within 5 kb of known transcripts, 42% within 10 kb. Thus, a considerable fraction of intergenic reads are in proximity to known genes (van Bakel et al., 2010). Additional 14.7% of reads fall within introns on either strand. Polyadenylation events near the 3′ ends of known genes tend to happen more frequently in 3′ UTR regions rather than the region immediately downstream of the 3′ ends of genes (Table 1 and Figure S3A). This may be caused by degradation intermediates of prematurely terminated transcripts, or the 3′ end annotations generated from EST databases favoring more downstream polyadenylation locations over upstream ones due to concerns such as incomplete cDNA clones and sequences, and thus, underrepresenting the diversity of polyadenylation sites.
To exemplify DRS’ ability to identify alternative polyadenylation events, we profiled human brain total RNA. ADD2 mRNAs were found to have one major and additional minor polyadenylation sites in brain but none in liver (Figure 2C), as reported previously (Costessi et al., 2006). In addition, in concordance with previous results (Rigault et al., 2006), we observed two polyadenylation sites for BBOX1 and a higher quantity of the “short” versus the “long” transcript in both tissues (Figure 2D).
Sense/Antisense PolyA+ Transcripts in Yeast and Human
DRS can not only pinpoint the sites of sense and antisense transcription, but also provide quantification of such transcripts without biases introduced by steps such as ligation, amplification and other manipulations. Of the 5769 annotated yeast ORFs, at least 3492 (60.5%) had an antisense transcript as evidenced by at least 10 antisense reads within the annotated ORFs (Figure S3B&C). These antisense reads compose 9.2% of the total DRS reads. When we considered the ambiguity in yeast 3′ end annotations and included regions 200 nts downstream of the 3′ annotation, the fraction of antisense reads increased to 41.2% and the ORFs with antisense transcripts increased to 4641 (80.4%), in part due to the genes with overlapping 3′ ends.
In the human liver RNA, at least 19,680/65,260 (30.2%) of all annotated transcripts were found to have antisense transcription as defined by at least 10 antisense reads either in exons or introns (Figure S3D&E). While prevalent, the antisense transcription is still a minority in terms of transcript abundance: ~8% of all reads that overlap an annotated transcript are antisense to it. This number is similar to 11% reported previously (He et al., 2008). Importantly, these numbers were obtained from polyA+ RNA and do not represent the extent of polyA- antisense transcription (Dutrow et al., 2008; Kiyosawa et al., 2005).
Quantification of Sense/Antisense polyA+ Transcriptome
We then explored the correlation between the quantities of sense and antisense transcripts. This analysis was attempted to observe the relationship between sense and antisense transcripts encoded by the same genomic region, given the presence of certain biological constraints such as transcription in both directions in a locus and pathways degrading complementary RNA species, like microRNA or similar pathways in human. The distribution of the sense and antisense counts for yeast and human did not represent the normal distribution (Shapiro-Wilk test, p<0.0001), even after converting the values into log space. Thus, we used the non-parametric Spearman correlation for this analysis based on the raw (non-log converted) values of sense and antisense expression levels of annotated genes. We separated the annotated genes into four quartiles based on their sense expression levels (Table 2). We did find a weak, but significant (see below) negative correlation between the levels of sense and antisense polyadenylated RNAs in the top quartile (Q1). The correlation became progressively more positive as the levels of the sense transcripts decreased, as exemplified by the positive correlation for the bottom 4th and 3rd quartile of expression for the yeast and human samples, respectively. Since the expression levels of transcripts that do not overlap in the genome could also correlate and the negative correlations obtained for the high expressors could be influenced by the extreme values, we introduced a permutation test where pairing of sense-antisense values for each gene was reassigned: for each annotated gene the sense value was kept the same, the antisense value was randomly chosen from another gene, and the Spearman correlation was calculated. This test shows that all (even the lowest) correlations found between the real sense and antisense reads counts are indeed highly significant (p-value <0.001). Similar trends were observed when converging genes in yeast were omitted from the analyses (Table S4).
Table 2. Spearman correlation coefficients between sense and antisense transcript levels.
Q1-4 indicates quartiles, with Q1 indicating the genes with highest sense expression values. For the human liver sample, we performed the analysis only for the top three quartiles since genes with zero expression level dominated the fourth quartile. The minimum and maximum correlation coefficients obtained after 1000 permutations were reported (thus p<0.001). Similar trends were observed for yeast after the removal of potentially overlapping transcripts (see also Table S4).
| Yeast | ||||
|---|---|---|---|---|
| Q1 | Q2 | Q3 | Q4 | |
| Actual Correlation | −0.11 | 0 | −0.01 | 0.36 |
| 1000 Permutations, minimum | −2.39E-05 | −5.55E-05 | −3.79E-05 | −6.78E-05 |
| 1000 Permutations, maximum | 9.05E-05 | 7.01E-05 | 8.47E-05 | 7.84E-05 |
| Human Liver | |||
|---|---|---|---|
| Q1 | Q2 | Q3 | |
| Actual Correlation | −0.11 | 0.02 | 0.12 |
| 1000 Permutations, minimum | −9.59E-05 | −3.40E-05 | −9.00E-05 |
| 1000 Permutations, maximum | 9.25E-05 | 9.80E-06 | 5.69E-07 |
Sequence Structure Surrounding Polyadenylation Sites
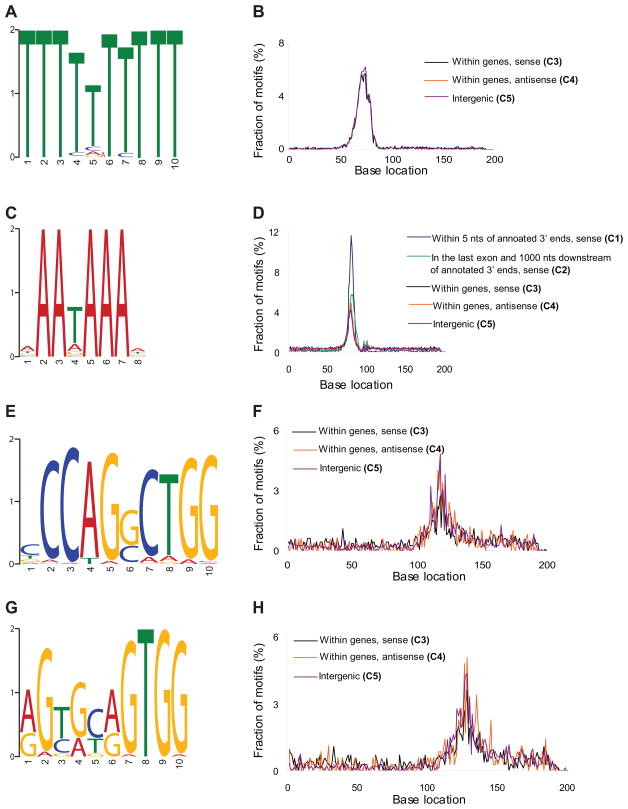
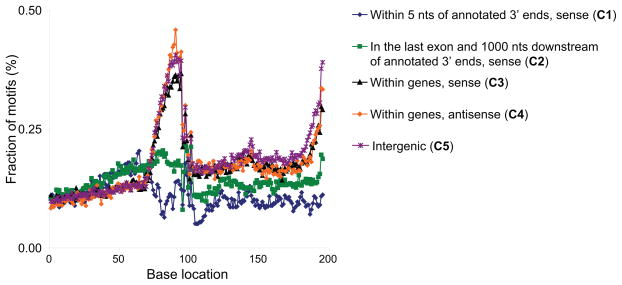
Having generated an in-depth view of polyadenylation cleavage locations, we examined the sequence patterns potentially governing transcription termination and polyadenylation. We first performed a de novo search for motifs near human polyadenylation locations and detected three novel motifs and the canonical signal (Figure 3). For this analysis, we used confident polyadenylation sites we defined using a clustering approach and supported by multiple reads (Figure S4, Table S5 and Extended Experimental Procedures). We identified a novel TTTTTTTTT motif (e=10−158, Figure 3A) and an AAWAAA motif closely resembling the canonical AWTAAA signal (e=10−112, Figure 3C) upstream of the polyadenylation sites (Zhao et al., 1999). We examined the distribution of these motifs across five polyadenylation site categories (C1-5) generated depending on site orientation (e.g. sense or antisense) and proximity relative to known 3′ ends of genes (Figure 3 and Experimental Procedures). Just like the canonical AWTAAA signal (Figure 3D, Table S6), TTTTTTTTT occurs in a highly position-specific manner, ~21 nt upstream of the polyadenylation site (Figure 3B), suggesting that these motifs are mechanistically important for polyadenylation. However, the TTTTTTTTT motif is largely present in the genic and intergenic regions (C3-5 in Figure 3), unlike the canonical motif which is largely present near the annotated 3′ ends of genes (C1-2).
Figure 3. Polyadenylation motif analyses.
Panels A, C, E and G indicate human motif elements identified. TTTTTTTTT (B), AWTAA (D), CCAGSCTGG (F) and RGYRYRGTGG (H) distance distribution are shown in respective panels. Human categories were defined as sites that are within 5 nucleotides of annotated 3′ ends of known human genes in sense orientation (category 1), in the last exon and 1 kb downstream of annotated 3′ ends of human known genes in sense orientation (category 2), located anywhere within the transcripts in sense orientation except in categories 1 and 2 (category 3), antisense to genes (category 4) and in intergenic regions (category 5). In distance plots, y-axis indicates the fraction of motifs (in percentages) at x-distances relative to the polyadenylation location (at base location 101) in each category. X-distances were calculated between the polyadenylation location identified with DRS and the first base immediately before the motif element. In panels B,F and H, only the categories 3,4 and 5 representing genic and intergenic sites were shown, because less than 10% (250–350) of these motifs were in categories 1 and 2, and not in sufficient numbers to be plotted in the graphs. Absolute numbers of motif counts for these latter three panels across all five human categories were provided in Figures S6A–C (see also Figure S4 and Table S5).
We also detected a novel palindromic sequence, CCAGSCTGG (e=10−33, Figure 3E) downstream of the polyadenylation sites and manifests a strong position-specific pattern (Figure 3F). Further analysis using less stringent motif scans led to the identification of RGYRYRGTGG (Figure 3G) that co-occur (p value=~0) with the CCAGSCTGG motif at a frequency of ~45%, and localizes ~31 nt downstream from the polyadenylation location (Figure 3H). Notably, we found that CCAGSCTGG and RGYRYRGTGG also strongly co-occur with the TTTTTTTTT motif (p-value=~0) in the intergenic and genic regions (C3-5), while these motifs does not co-occur and anti-correlate with the canonical AWTAAA localization (p-value=~0). The pervasive presence of the TTTTTTTTT motif in the novel genic and intergenic polyadenylation sites, its similarity to the AWTAA signal with respect to its positional preference, its anti-correlation to AWTAAA localization and its co-occurrence with the CCAGSCTGG and RGYRYRGTGG motifs are intriguing and may point to uncharacterized polyadenylation mechanism(s) in humans. We applied similar approaches to yeast, detecting no additional motifs beyond the previously characterized positioning (PE, AAWAAA) and upstream efficiency (EE, TAYRTA) elements (Zhao et al., 1999). The general positioning of the upstream PE motif (Figure S5A) were closer to the cleavage site than the localization of the EE motif (Figure S5B), as expected (Zhao et al., 1999).
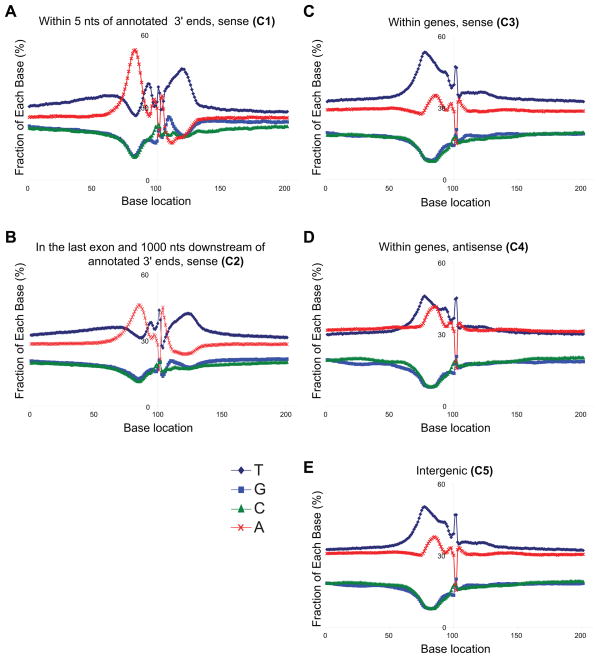
We then examined the nucleotide composition around the polyadenylation cleavage locations in each group. We observed a difference in the profiles of nucleotide frequency distributions surrounding human cleavage sites in regions near 3′ known gene ends (C1-2) and in genic and intergenic regions (C3-5, Figure 4). As expected, the categories 1 and 2 had the T-rich downstream sequence element (DSE) 20–30 bases downstream of the polyadenylation sites and A-rich sequences upstream (Zhao et al., 1999). On the other hand, the nucleotide profiles around the sites in the categories 3–5 were different and similar to the yeast sites (Figure S5C–F) with the pronounced upstream T-rich sequences, in line with the TTTTTTTTT motif identified in the upstream regions above (Figure 4). Presence of a T-rich polyadenylation enhancer sequence element upstream of the AATAAA motif is common among viruses and has been previously found in a few human genes (Bhat and Wold, 1987; Moreira et al., 1995). However, the T-rich pattern observed here is immediately upstream of the sense/antisense genic and intergenic cleavage sites, and therefore represents a different and novel observation. This latter similarity at the yeast and human nucleotide profiles prompted us to examine yeast motif presence in humans. Interestingly, we observed an enrichment of the yeast EE motif immediately upstream of the human cleavage sites in categories 3–5, but not in categories 1 and 2 (Figure 5). The yeast EE motif however does not co-occur with the novel CCAGSCTGG, RGYRYRGTGG and TTTTTTTTT motifs identified above, and thus may be present in an independent subset of genic and intergenic sites. This latter finding may point to the existence of another, perhaps yeast-like polyadenylation sequence structure in a subset of human polyadenylation sites.
Figure 4. The nucleotide composition surrounding polyadenylation cleavage locations in humans.
Category descriptions were provided in Figure 3. Y axis indicates the nucleotide composition (in percentages) at x-locations relative to the cleavage positions (at base location 101). Dark blue (diamond), blue (rectangle), green (triangle), and red (cross) lines indicate T,G,C, and A nucleotides, respectively. Polyadenylation locations in C3-5 differ from those in C1-2, and exhibit elevated T and A content in 40–50 nts upstream of polyadenylation cleavage positions (see also Figure S5 and Table S6).
Figure 5. Distance distribution of yeast EE (TAYRTA) motif across human categories.
Y axis indicates the fraction of motifs (in percentages) at x-distances relative to the cleavage positions (at base location 101) in each category. X-distances were calculated between the cleavage location identified with DRS and the first base immediately before the motif element. Human category descriptions were provided in Figure 3 legend. The enrichment of the EE motif immediately upstream of the cleavage sites in human categories 3,4 and 5, but not in categories 1 and 2, is in parallel to the upstream human T-enrichment pattern shown in Figure 4 (see also Figure S6).
Discussion
This study presents genome-wide polyadenylation maps that incorporate the accuracy of a high-throughput sequencing-based methodology and true strand-specificity. Other sequencing-based polyadenylation mapping approaches have recently become available (Mangone et al., 2010; Yoon and Brem, 2010). Compared to these, the DRS-based approach is in quantitative nature, free of reverse transcription and ligation artifacts, and requires only minute RNA quantities. The nucleotide resolution of the approach is similar to other classical methods of polyadenylation site mapping. However, just like these other approaches, the DRS-based approach cannot truly differentiate cases where the template cleavage may occur right after an A-residue. Such sites may cause the resolution of the approach to elevate from its current level of ±2 nts. Because sequencing technologies available or in development today, including DRS, do not provide the full transcript sequence, it is not possible to know the sequence of the entire RNA molecule represented by each read by any sequencing technology. It is therefore possible that the reads found around the annotated transcriptional start and polyadenylation sites may partly represent short polyA+ RNAs previously found to be associated with the gene termini (Kapranov et al., 2007a; Kapranov et al., 2010; Project., 2009). A fraction of reads found around the annotated polyadenylation site of known messages may not represent the annotated form, but other isoforms or correspond to other overlapping transcripts that share the same polyadenylation region. Furthermore, polyadenylation sites observed downstream of annotated 3′ ends may represent alternative polyadenylation events or transcription termination products (Kim et al., 2004; Teixeira et al., 2004; West et al., 2004).
Our results show that most yeast and human transcripts have yet un-characterized polyadenylation sites. This dataset, along with additional biological replicates and data from different cell types and states, will allow empirical annotation of such sites and provide the substrate for biological experimentation examining changes in these sites. The enrichment of reads in yeast intergenic functional transcription factor binding sites and DHSs suggests that these potential regulatory regions may indeed encode for RNAs. The presence of RNAs from a subset of potential mammalian enhancers (eRNAs) and open chromatin regions has recently been described (De Santa et al., 2010; Kim et al., 2010; van Bakel et al., 2010). Unlike the report by Kim et al., which found eRNAs to lack polyA tails, our results indicate the potential existence of polyA-tail containing RNAs associated with regulatory elements in yeast. We speculate that these regulatory region-associated reads may represent a recently-described class of polyadenylated non-coding RNAs that regulate gene expression (Bumgarner et al., 2009; Orom et al., 2010). They may also represent divergent transcription events from unannotated promoters (Neil et al., 2009; Seila et al., 2008; Xu et al., 2009). Alternatively, given the likely association of RNA polymerase II with the transcriptional factors binding to these regions, these RNAs may emerge from transcriptional noise events postulated to occur (Struhl, 2007). The lack of comprehensive transcription factor binding site and enhancer maps in humans prevented us from examining such RNAs in our human studies. However, the relatively high fraction of intergenic DRS reads obtained in the human samples suggest that at least a fraction of these reads may emerge from enhancers. Further studies are needed to delineate the functions, if any, of these RNAs and how they may be contributing to regulatory function.
Our observation of novel polyadenylation patterns including novel co-occurring motifs (CCAGSCTGG, RGYRYRGTGG, and TTTTTTTTT), and enrichment of T-rich and yeast EE motif sequences near sites corresponding to non-coding transcript categories (antisense, sense genic, and intergenic) compared to sites in proximity to the 3′ ends of known genes suggest interesting possibilities for human polyadenylation. Particularly, the anti-correlation we observed between the localizations of the three novel motifs above and the canonical AWTAAA suggests alternative and yet to be characterized mechanisms of transcription termination, cleavage and polyadenylation. Given that RNAs in these regions are likely to be non-coding, perhaps alternative modes of polyadenylation exist for non-coding RNAs. These three novel motifs are present in a relatively small fraction of polyadenylation sites and cleavage events (Table S6C). This may partly be explained by the relatively low fraction of polyadenylated non-coding RNAs relative to mRNAs of protein-coding genes in terms of mass. Combined with the recent observation that even very low abundance non-coding RNAs, as low as 4 copies per cell, can regulate target genes (Wang et al., 2008), these new motifs may be specific to such a subset of non-coding RNAs. Further in-depth de novo motif analyses in these novel regions and the identification of the components of this potential alternative polyadenylation machinery would open a number of conceptual and experimental possibilities. First, we may learn more about the RNAs they process, which may include various species (Buratowski, 2008) such as promoter-associated RNAs (Core et al., 2008; Neil et al., 2009; Seila et al., 2008), cryptic unstable RNAs (Preker et al., 2008; Wyers et al., 2005), long intergenic non-coding RNAs (Guttman et al., 2009) and polyadenylated RNAs resulting from degradation events (Slomovic et al., 2010). Second, we may get a more mechanistic understanding of polyadenylation and its connection with other cellular processes. For instance, the CCAGSCTGG palindromic motif identified here is a candidate binding site for human topoisomerase II (topo-II) (Spitzner and Muller, 1988). Topo-II is part of the RNA polymerase II holoenzyme, and relaxes the superhelical tension that accumulates during transcription elongation (Mondal and Parvin, 2001). Perhaps the presence of such a motif downstream of polyadenylation sites is to ensure that transcriptional superhelical tension does not extend beyond the boundaries of the transcripts and thus do not disturb downstream regions.
In line with previous studies (He et al., 2008; Kapranov et al., 2007b), we observed that antisense transcription is prevalent in the yeast and human genomes, and that the quantities of steady-state levels of sense and antisense transcripts occupying the same genomic space can negatively correlate with each other. Our results indicate a complex picture where the highly expressed genes in the top quartile tend to negatively correlate with the expression of antisense transcripts. On the other hand, the genes in the bottom quartile show a positive correlation between the sense and antisense transcription. While both results are significant, the former effect is relatively small and similar to what has been detected previously (Chen et al., 2005), while the latter effect is the strongest (at least in yeast) and is similar to the results obtained in S. pombe (Dutrow et al., 2008), and mouse (Katayama et al., 2005), where positive correlation was found. In view of these results, it is perhaps not surprising that the correlation of sense and antisense transcripts has remained a controversial issue as often both were found to be positively correlated (Kapranov et al., 2007b). The relatively low negative correlation values most likely reflect the fact the overlapping positioning in the genome is only one of many ways to regulate stable levels of polydenylated RNAs species. It is however tempting to speculate that in highly expressed genes, the physical interference of converging RNA polymerase complexes could exert a dominant effect, while this possibility may be less of a factor in the genes with lower transcriptional activity. In the latter cases, other factors, such as chromatin accessibility that could permit transcription from both strands could be a larger determining factor. To what extent the observed negative correlation is due to sense/antisense transcripts occupying the same genomic space and/or other transcriptional control mechanisms needs further exploration.
This study represents the first step for the adaptation of the direct RNA sequencing technology to decipher the genome and its functions. Future studies will focus on the functional characterization of novel polyA+ regulatory region-associated RNAs, antisense transcripts and polyadenylation sites identified in this study, and the adaptation of DRS for other existing and novel RNA applications.
Experimental Procedures
Sample Preparation for DRS
Yeast (Saccharomyces cerevisiae) and human liver polyA+ RNAs were obtained from Clontech, CA. Human brain total RNA was from Ambion. The 3′ blocking reaction was performed with poly(A) tailing kit (Ambion, TX) and 3′deoxyATP (Jena Biosciences, Germany), incubating the reaction mixture at 37ºC for 30 minutes. The blocked RNA was hybridized to flow cell surfaces for sequencing with DRS without additional cleaning steps (Ozsolak et al., 2009).
Data Analysis
Raw DRS reads were filtered using a suite of Helicos tools available at: http://open.helicosbio.com/mwiki/index.php/Releases and described at: http://open.helicosbio.com/helisphere_user_guide/. Alignments were conducted with indexDPgenomic available on the Helicos website (http://open.helicosbio.com/mwiki/index.php/Releases). For the genomic alignments, reads were aligned to the yeast SGD/sacCer2 and human NCBIv36 version of the genome supplemented with the complete ribosomal repeat unit (Gen. Bank. Accession U13369.1). Reads with a minimal length of 25 nts and alignment score of 4.3 and above were allowed. Aligned reads were further filtered for reads having a unique best alignment score. Total raw per base error rate was 4–5%, dominated by missing base errors (2–3%).
Downstream analysis was performed with the SeqSolve NGS software (Integromics, S.L., Spain). Annotated yeast or human transcriptome was defined as either the SGD Genes from Saccharomyces Genome Database track or UCSC Genes track on the UCSC Genome Browser. Counts within each annotation were derived from either the sense or antisense strand using the positions of the 5′ ends of reads aligned to the appropriate strand. Yeast median UTR length was calculated by taking the median of the distances between the annotated 3′ end locations of yeast ORFs and the reads that map in the sense orientation and within 1000 bps downstream of ORF 3′ ends.
For the sequence composition surrounding polyadenylation cleavage site analysis, the 5′ ends of reads representing the 3′ cleavage sites were grouped based on overlap with the genomic annotation as described in figures 3 and S5. Mitochondrial reads were not used for the sequence analysis. These categories for human were: 1) Sense cleavage locations that are within 5 bases of annotated 3′ ends; 2) Sense cleavage locations that are not in category #1 and are in the last exons or 1 kb downstream of the annotated 3′ ends; 3) Sense cleavage locations that are not in categories 1 & 2 and are within annotated genes; 4) Antisense cleavage locations that are within annotated genes; 5) Intergenic cleavage locations that are not in categories 1–4. The categories for yeast were: 1) Sense cleavage locations that are located within 200 bps downstream of the annotated 3′ ends of yeast ORFs; 2) Sense cleavage locations that are not within category 1 and are within bodies of ORFs; 3) Antisense cleavage locations that are not within category 1 and are within bodies of ORFs; 4) Intergenic cleavage locations that are not in categories 1, 2 and 3, and are at least of 1 kb away from the 3′ ends of yeast ORFs. Reads in each category were then collapsed based on their unique 5′ ends representing unique polyadenylation cleavage locations. Sequences 100 bases on each side of each collapsed locations were analyzed as described in the text.
Detection of novel motifs
To investigate the presence of new sequence motifs, upstream and downstream genomic sequences (50 bases) of novel polyadenylation sites (Figure S4) were scanned independently using MEME (Bailey et al., 2006). To reduce the occurrence of spurious motifs, motif searches were performed using a highly stringent E-value (10−25) threshold, based on a non-redundant set of 1000 sequences that were sampled uniformly from the complete set of upstream/downstream sequences. The threshold (10−25) was used because even when sites across each chromosome was separately analyzed (24 control experiments) to rule out dataset artifacts, the three human motifs were consistently detected. The various motif variants were manually inspected to select a single motif for display representation. For additional validations of the motifs, the up-/down-stream occurrences and co-occurrences were analyzed. Total occurrences of motifs in up-/downstream sequences were determined by searching for all short strings that matched (>90%) the position-specific scoring (log-odds) matrix profile of the motifs detected by MEME. To test the statistical significance of co-occurrence between two motifs, hypergeometric tests (Lee et al., 2007) were performed based on the total number of occurrences of the two motifs in the complete set of non-redundant sequences. Since only four motifs were compared (6 comparisons) to each other for co-occurrence analysis, and because the reported p-values are close to zero, the Bonferroni correction factor of 6 was not used.
Supplementary Material
Acknowledgments
We thank our Helicos and Integromics colleagues for technical assistance and discussions. This work was supported by the National Human Genome Research Institute grant R01-HG005230 to F.O. and P.M.M. B.J. is supported by NIH (GM079756) and American Cancer Society (RSG0905401), A.P.M. by NIH (MH60774), and S.F. by the Spanish Ministry of Science and Innovation - FEDER (CDTI loan IDI-20091293).
Footnotes
Accession Numbers
Sequencing datasets described in this study have been deposited at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), accession no SRA012232. The datasets are also available as wiggle files at the Helicos open access website (HeliSphere, http://open.helicosbio.com/) along with yeast and human polyadenylation sites defined in this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental information includes Extended Experimental Procedures, six figures, six tables and Supplemental References.
References
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CL, Brunkow ME, Ramsdell F, O’Briant KC, Zhu Q, Fuleihan RL, Shigeoka AO, Ochs HD, Chance PF. A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA-->AAUGAA) leads to the IPEX syndrome. Immunogenetics. 2001;53:435–439. doi: 10.1007/s002510100358. [DOI] [PubMed] [Google Scholar]
- Bhat BM, Wold WS. A small deletion distant from a splice or polyadenylation site dramatically alters pre-mRNA processing in region E3 of adenovirus. J Virol. 1987;61:3938–3945. doi: 10.1128/jvi.61.12.3938-3945.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brais B, Bouchard JP, Xie YG, Rochefort DL, Chretien N, Tome FM, Lafreniere RG, Rommens JM, Uyama E, Nohira O, et al. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet. 1998;18:164–167. doi: 10.1038/ng0298-164. [DOI] [PubMed] [Google Scholar]
- Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci U S A. 2009;106:18321–18326. doi: 10.1073/pnas.0909641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Transcription. Gene expression--where to start? Science. 2008;322:1804–1805. doi: 10.1126/science.1168805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 2005;21:326–329. doi: 10.1016/j.tig.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Cocquet J, Chong A, Zhang G, Veitia RA. Reverse transcriptase template switching and false alternative transcripts. Genomics. 2006;88:127–131. doi: 10.1016/j.ygeno.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costessi L, Devescovi G, Baralle FE, Muro AF. Brain-specific promoter and polyadenylation sites of the beta-adducin pre-mRNA generate an unusually long 3′-UTR. Nucleic Acids Res. 2006;34:243–253. doi: 10.1093/nar/gkj425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutrow N, Nix DA, Holt D, Milash B, Dalley B, Westbroek E, Parnell TJ, Cairns BR. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat Genet. 2008;40:977–986. doi: 10.1038/ng.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Frede U, Neu-Yilik G, Hundsdoerfer P, Vetter B, Hentze MW, Kulozik AE. Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat Genet. 2001;28:389–392. doi: 10.1038/ng578. [DOI] [PubMed] [Google Scholar]
- Graber JH, McAllister GD, Smith TF. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 2002;30:1851–1858. doi: 10.1093/nar/30.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U. Second-strand cDNA synthesis: mRNA fragments as primers. Methods Enzymol. 1987;152:330–335. doi: 10.1016/0076-6879(87)52038-9. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs DR, Goodbourn SE, Lamb J, Clegg JB, Weatherall DJ, Proudfoot NJ. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983;306:398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Iseli C, Stevenson BJ, de Souza SJ, Samaia HB, Camargo AA, Buetow KH, Strausberg RL, Simpson AJ, Bucher P, Jongeneel CV. Long-range heterogeneity at the 3′ ends of human mRNAs. Genome Res. 2002;12:1068–1074. doi: 10.1101/gr.62002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007a;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Ozsolak F, Kim SW, Foissac S, Lipson D, Hart C, Roels S, Borel C, Antonarakis SE, Monaghan P, et al. New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism. Nature. 2010 doi: 10.1038/nature09190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007b;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa H, Mise N, Iwase S, Hayashizaki Y, Abe K. Disclosing hidden transcripts: mouse natural sense-antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res. 2005;15:463–474. doi: 10.1101/gr.3155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Li Z, Brower-Sinning R, John B. Regulatory circuit of human microRNA biogenesis. PLoS Comput Biol. 2007;3:e67. doi: 10.1371/journal.pcbi.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CL, Bristol LA, Jin L, Dykes-Hoberg M, Crawford T, Clawson L, Rothstein JD. Aberrant RNA processing in a neurodegenerative disease: the cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis. Neuron. 1998;20:589–602. doi: 10.1016/s0896-6273(00)80997-6. [DOI] [PubMed] [Google Scholar]
- Liu D, Graber JH. Quantitative comparison of EST libraries requires compensation for systematic biases in cDNA generation. BMC Bioinformatics. 2006;7:77. doi: 10.1186/1471-2105-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez F, Granjeaud S, Ara T, Ghattas B, Gautheret D. The disparate nature of “intergenic” polyadenylation sites. RNA. 2006;12:1794–1801. doi: 10.1261/rna.136206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- Mahadevan S, Raghunand TR, Panicker S, Struhl K. Characterisation of 3′ end formation of the yeast HIS3 mRNA. Gene. 1997;190:69–76. doi: 10.1016/s0378-1119(96)00708-1. [DOI] [PubMed] [Google Scholar]
- Mamanova L, Andrews RM, James KD, Sheridan EM, Ellis PD, Langford CF, Ost TW, Collins JE, Turner DJ. FRT-seq: amplification-free, strand-specific transcriptome sequencing. Nat Methods. 2010;7:130–132. doi: 10.1038/nmeth.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. The landscape of C. elegans 3′UTRs. Science. 2010;329:432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal N, Parvin JD. DNA topoisomerase IIalpha is required for RNA polymerase II transcription on chromatin templates. Nature. 2001;413:435–438. doi: 10.1038/35096590. [DOI] [PubMed] [Google Scholar]
- Moreira A, Wollerton M, Monks J, Proudfoot NJ. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 1995;14:3809–3819. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro EM, Herrington R, Janmohamed S, Frelin C, Andrade-Navarro MA, Iscove NN. Identification of gene 3′ ends by automated EST cluster analysis. Proc Natl Acad Sci U S A. 2008;105:20286–20290. doi: 10.1073/pnas.0807813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Cheng TC, Antonarakis SE, Kazazian HH., Jr Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human beta-globin gene. EMBO J. 1985;4:453–456. doi: 10.1002/j.1460-2075.1985.tb03650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Project., AETPCSHLET. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigault C, Le Borgne F, Demarquoy J. Genomic structure, alternative maturation and tissue expression of the human BBOX1 gene. Biochim Biophys Acta. 2006;1761:1469–1481. doi: 10.1016/j.bbalip.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Fremder E, Staals RH, Pruijn GJ, Schuster G. Addition of poly(A) and poly(A)-rich tails during RNA degradation in the cytoplasm of human cells. Proc Natl Acad Sci U S A. 2010;107:7407–7412. doi: 10.1073/pnas.0910621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Portnoy V, Schuster G. Detection and characterization of polyadenylated RNA in Eukarya, Bacteria, Archaea, and organelles. Methods Enzymol. 2008;447:501–520. doi: 10.1016/S0076-6879(08)02224-6. [DOI] [PubMed] [Google Scholar]
- Spiegelman S, Burny A, Das MR, Keydar J, Schlom J, Travnicek M, Watson K. DNA-directed DNA polymerase activity in oncogenic RNA viruses. Nature. 1970;227:1029–1031. doi: 10.1038/2271029a0. [DOI] [PubMed] [Google Scholar]
- Spitzner JR, Muller MT. A consensus sequence for cleavage by vertebrate DNA topoisomerase II. Nucleic Acids Res. 1988;16:5533–5556. doi: 10.1093/nar/16.12.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Teixeira A, Tahiri-Alaoui A, West S, Thomas B, Ramadass A, Martianov I, Dye M, James W, Proudfoot NJ, Akoulitchev A. Autocatalytic RNA cleavage in the human beta-globin pre-mRNA promotes transcription termination. Nature. 2004;432:526–530. doi: 10.1038/nature03032. [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Pan Z, Lee JY. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007;17:156–165. doi: 10.1101/gr.5532707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Proudfoot NJ. Human 5′ --> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Du J, Rozowsky J, Zhang Z, Urban AE, Euskirchen G, Weissman S, Gerstein M, Snyder M. Systematic analysis of transcribed loci in ENCODE regions using RACE sequencing reveals extensive transcription in the human genome. Genome Biol. 2008;9:R3. doi: 10.1186/gb-2008-9-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon OK, Brem RB. Noncanonical transcript forms in yeast and their regulation during environmental stress. RNA. 2010;16:1256–1267. doi: 10.1261/rna.2038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hu J, Recce M, Tian B. PolyA_DB: a database for mammalian mRNA polyadenylation. Nucleic Acids Res. 2005a;33:D116–120. doi: 10.1093/nar/gki055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005b;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.