Abstract
Background
Oxidized low-density lipoprotein (oxLDL) uptake by macrophages plays an important role in foam cell formation. It has been suggested the presence of heterogeneous subsets of macrophage, such as M1 and M2, in human atherosclerotic lesions. To evaluate which types of macrophages contribute to atherogenesis, we performed cDNA microarray analysis to determine oxLDL-induced transcriptional alterations of each subset of macrophages.
Results
Human monocyte-derived macrophages were polarized toward the M1 or M2 subset, followed by treatment with oxLDL. Then gene expression levels during oxLDL treatment in each subset of macrophages were evaluated by cDNA microarray analysis and quantitative real-time RT-PCR. In terms of high-ranking upregulated genes and functional ontologies, the alterations during oxLDL treatment in M2 macrophages were similar to those in nonpolarized macrophages (M0). Molecular network analysis showed that most of the molecules in the oxLDL-induced highest scoring molecular network of M1 macrophages were directly or indirectly related to transforming growth factor (TGF)-β1. Hierarchical cluster analysis revealed commonly upregulated genes in all subset of macrophages, some of which contained antioxidant response elements (ARE) in their promoter regions. A cluster of genes that were specifically upregulated in M1 macrophages included those encoding molecules related to nuclear factor of kappa light polypeptide gene enhancer in B-cells (NF-κB) signaling pathway. Quantitative real-time RT-PCR showed that the gene expression of interleukin (IL)-8 after oxLDL treatment in M2 macrophages was markedly lower than those in M0 and M1 cells. HMOX1 gene expression levels were almost the same in all 3 subsets of macrophages even after oxLDL treatment.
Conclusions
The present study demonstrated transcriptional alterations in polarized macrophages during oxLDL treatment. The data suggested that oxLDL uptake may affect TGF-β1- and NF-κB-mediated functions of M1 macrophages, but not those of M0 or M2 macrophages. It is likely that M1 macrophages characteristically respond to oxLDL.
Background
Atherosclerosis is a major cause of cardiovascular disease, which is one of the leading morbidities worldwide [1]. Atherosclerosis has been suggested to be merely a lipid-storage disease; however, it is now recognized as an inflammatory condition of the vessel wall characterized by infiltration of macrophages and T cells [2]. Monocytes are recruited into the arterial intima and differentiate into macrophages. They take up oxidized low-density lipoprotein (oxLDL) via scavenger receptors, and then become foam cells that play a crucial role in the initiation of atherosclerotic lesions [3]. Foam cells have been shown to affect many atherogenic events, including recruitment of monocytes and neutrophils by producing chemokines, such as monocyte chemoattractant protein (MCP)-1 [4] and interleukin (IL)-8 [5], formation of necrotic cores in atherosclerotic plaques [3], and production of matrix metalloproteases (MMPs), which degrade the extracellular matrix comprising the fibrous cap of plaque [6]. Therefore, macrophages immunologically interact with surrounding inflammatory cells during the process of differentiation into foam cells in atherogenic processes.
Over the past several decades, a number of studies have demonstrated that macrophages do not represent a homogenous cell population. Stein et al. described an alternative subset of macrophages induced by IL-4, characterized by high mannose receptor (MR) expression [7]. Since then, it has been demonstrated that monocyte-derived macrophages can be polarized into two subsets in vitro. One subset consists of classically activated macrophages (M1 macrophages) polarized with lipopolysaccharide (LPS) and interferon (IFN)-γ, which are characterized by CD86 expression and production of proinflammatory cytokines, such as tumor necrosis factor (TNF)-α, IL-1, and IL-6. The other subset consists of alternatively activated macrophages (M2 macrophages) polarized with Th2 cytokines, such as IL-4 and/or IL-13, which are characterized by MR expression [8].
Recently, Bouhlel et al. confirmed the presence of M2 macrophages within human atherosclerotic lesions by identifying the expression of M2 markers, including IL-10 and MR in human carotid plaques [9]. They also reported that macrophages expressing M2 markers show a different distribution from foam cells. These results suggested the presence of heterogeneous subsets of macrophages in human atherosclerotic lesions. However, it remains unclear which type of macrophages differentiate into foam cells or how they contribute to atherogenesis.
The present study was performed to elucidate the contributions of M1 and M2 macrophages to atherogenesis during differentiation into foam cells. Martinez et al. investigated the polarization of human monocytes toward M1 or M2 macrophages using cDNA microarray analysis, and found distinct sets of genes specifically upregulated in either subset of macrophages [10]. Cho et al. also examined the transcriptional differences in human monocyte-derived macrophages during oxLDL uptake by cDNA microarray analysis [11]. However, there have been no previous studies of the whole transcriptional alterations in human M1 or M2 macrophages during oxLDL uptake. To investigate the roles of these macrophage subsets during differentiation into foam cells, we examined the transcriptional alterations of M1 or M2 macrophages during oxLDL treatment.
Methods
Materials
Lymphoprep was purchased from AXIS-SHILD (Rodelokka, Oslo, Norway). Dulbecco's Modified Eagle's Medium: Nutrient Mixture F-12 (DMEM/F12) was obtained from Invitrogen (Carlsbad, CA), and RPMI-1640 culture medium (endotoxin-free) was from Sigma-Aldrich (St. Louis, MO). Recombinant human macrophage-colony stimulating factor (M-CSF), IFN-γ, and IL-4 were obtained from R&D Systems (Minneapolis, MN). LPS from Escherichia coli (serotype O111:B4) was obtained from List Biological Laboratories Inc. (Campbell, CA). OxLDL (endotoxin level < 0.5 EU/ml), which was prepared with 3.5 μM CuSO4 in PBS at 37°C for 24 h, was purchased from Biomedical Technologies (Stoughton, MA). The average level of thiobarbituric acid-reactive substances (TBARS) in this study was 76.23 ± 7.89 nmol malondialdehyde equivalents/mg LDL protein (mean ± SD). Anti-CD14 antibody, anti-CD86 antibody, and anti-MR antibody were obtained from eBioscience (San Diego, CA). All procedures were performed under endotoxin-free conditions.
Cells
Peripheral blood mononuclear cells (PBMCs) were obtained from healthy volunteers with informed consent from buffy coats by density-gradient centrifugation using Lymphoprep. The purity of monocytes was > 95% as determined by flow cytometric analysis using anti-CD14 antibody (data not shown). The monocytes were suspended in DMEM/F12, and plated onto tissue culture dishes at a density of 1 × 106 cells/cm2 for 2 h at 37°C. The adherent cells were differentiated into macrophages by incubation with 100 ng/mL M-CSF in RPMI-1640 medium supplemented with 20% fetal calf serum (FCS) for 7 days (these cells were defined as M0 macrophages). Macrophage polarization was performed as described by Martinez et al. with slight modifications [10]. To obtain M1 or M2 macrophages, M0 macrophages were further incubated with 10 pg/mL LPS plus 20 ng/mL IFN-γ or 20 ng/mL IL-4 in RPMI-1640 with 5% FCS for 18 h, respectively. After polarization, media were removed, and each subset of macrophages was incubated for a further 6 h in the presence or absence of 100 μg/mL oxLDL. The study was approved by the Ethical Committee of Juntendo University.
Flow cytometric analysis
The M1 or M2 polarized macrophages were washed with PBS. After washing, cells were stained with PE-Cy5- or FITC-conjugated antibodies or with corresponding isotype controls for 20 min at 4°C. Then, flow cytometry was performed to determine the expression of cell surface antigens using FACSCalibur (BD Biosciences, Franklin Lakes, NJ), as described previously [12]. Data were analyzed using Cell Quest software (BD Biosciences).
Quantitative real-time RT-PCR
Total RNA was extracted and purified from macrophages using an RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA was synthesized from 50 ng/μL of total RNA using an ExScript RT-PCR Kit (Takara-Bio, Shiga, Japan). Primers were selected using Perfect Real-Time Primer Support System provided by Takara. Real-time RT-PCR was performed using SYBR Premix Ex Taq (Takara-Bio) and an ABI 7900HT Sequence Detector System (Applied Biosystems, Foster City, CA). The amplification program included an initial denaturation step at 95°C for 10 s, 40 cycles of denaturation at 95°C for 10 s, and annealing and extension at 60°C for 30 s. After amplification, dissociation curves were acquired to determine the specificity of PCR products. The relative cDNA concentrations were established using a standard curve plotted with sequential tenfold dilutions of cDNA synthesized from QPCR Human Reference Total RNA (Stratagene, La Jolla, CA). The data were normalized relative to peptidylprolyl isomerase A (PPIA) as an internal control.
cDNA microarray analysis
cDNA synthesis and aminoallyl labeling of RNA were performed using an amino-allyl RNA amplification kit (Sigma-Aldrich) according to the manufacturer's instructions. The Cy3- or Cy5-labeled aminoallyl RNA was concentrated using Microcon YM-30 (Millipore, Bedford, MA), mixed with hybridization buffer supplied with the kit, and denatured at 95°C for 2 min. The hybridization mixture was applied onto a "3D-Gene" human oligo chip 25 k (Toray Industries, Tokyo, Japan), and incubated according to the manufacturer's instructions. After washing and drying the DNA chip slides, the fluorescent signals were quantified by ScanArray Lite (PerkinElmer Life Sciences, Boston, MA) and analyzed using ScanArray Express software. After subtraction of the mean background level, the fluorescence intensity was normalized relative to the mean sample intensity in each chip. Any given gene was analyzed if its normalized intensity was more than 2-4. We defined genes showing a change in expression of > 2-fold during oxLDL treatment as significantly up- or downregulated (log2 ratios were greater than +1 or less than -1).
Ingenuity pathway analysis
Ingenuity Pathway Analysis (IPA) software (version 8.7; Ingenuity Systems, Redwood, CA) was utilized to determine the possible biological pathways and intermolecular networks between candidate genes. A detailed description of IPA software can be found on the Ingenuity Systems website http://www.ingenuity.com/. The significantly up- or downregulated genes were overlaid onto a global molecular network developed from information contained in the Ingenuity Knowledge Base.
Functional gene ontology analysis identified the biological functions that were most significant to molecules in the network. The network molecules associated with biological functions in the Knowledge Base were considered for the analysis. Right-tailed Fisher's exact test was used to calculate the P-values determining the probability that each biological function assigned to that network was due to chance alone. IPA generates significant biological networks that are particularly enriched with the genes of interest, called "focus genes." It calculates a network score that takes into account the number of focus genes and the size of the networks, indicating the likelihood of focus genes in a network being found together by chance. The higher the score, the lower is the probability of finding the observed Network Eligible Molecules in a given network by chance. Network analysis produces a graphical representation of the molecular relationships between the identified genes. Molecules are represented as nodes, and the biological relationship between two nodes is represented as a line. All relationships are supported by at least 1 reference from the literature, from a textbook, or from canonical information stored in the Knowledge Base.
Statistical analysis
The data were expressed as the means ± SD and were analyzed for significant differences by one-way or two-way analysis of variance (ANOVA) and, Bonferroni's post hoc test using GraphPad Prism (version 5.00; GraphPad Software, La Jolla, CA).
Results
Characteristics of M1 or M2 polarized macrophages
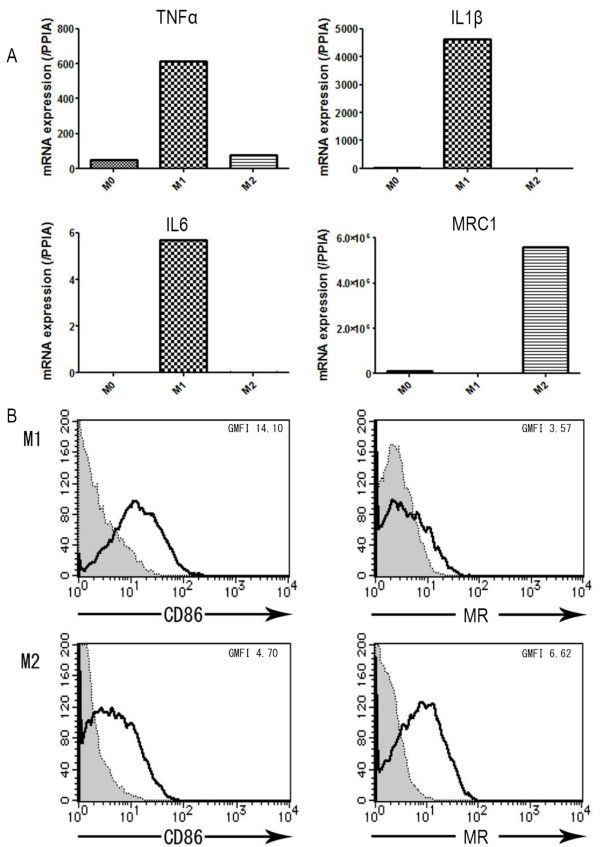
Human monocyte-derived macrophages cultured for 7 days in the presence of M-CSF can be polarized toward M1 macrophages by further treatment with 100 ng/mL LPS plus 20 ng/mL IFN-γ for 18 h [10]. However, under our experimental conditions, almost all cells were damaged by such high a concentration of LPS, as demonstrated by trypan blue staining (data not shown). Therefore, we differentiated M-CSF-treated monocytes into M1 macrophages by incubation with 10 pg/mL LPS plus 20 ng/mL IFN-γ. We confirmed the polarized cells as M1 and M2 macrophages by quantitative real-time RT-PCR and flow cytometric analysis (Figure 1). Consistent with the previous report of Martinez et al. [10], M1 macrophages showed higher levels of proinflammatory cytokine mRNA expression, such as TNF-α, IL-1β, and IL-6, than M0 or M2 macrophages. In contrast, M2 macrophages showed markedly elevated expression of MRC1, which encodes MR (Figure 1A). The level of CD86 expression on M1 macrophages was higher than that on M2 macrophages, while MR was expressed on M2 macrophages but not on M1 macrophages (Figure 1B).
Figure 1.
Expression of M1 and M2 macrophage polarized markers in human monocyte-derived macrophages. A. TNFα, IL1β, and IL6 gene expression as markers of M1, and MRC1 gene expression as a marker of M2 were analyzed by RT-PCR. Each panel shows data from one of 3 representative experiments. B. Surface expression of CD86 (M1 marker) and mannose receptor (MR) (M2 marker) were analyzed by flow cytometry. The level of CD86 expression was higher in M1 than in M2 macrophages, while the expression level of MR was higher in M2 than in M1 macrophages.
Transcriptional profile
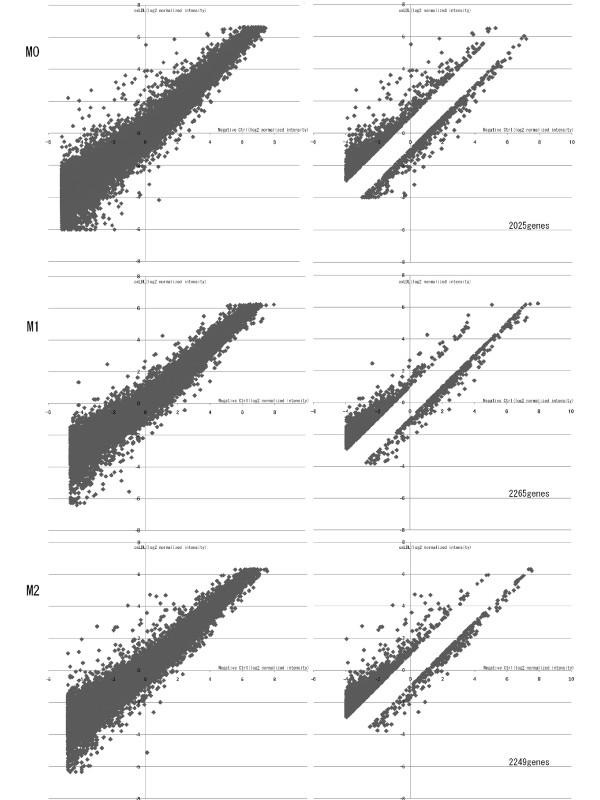
After confirming that the 2 subsets of macrophages were properly polarized to M1 or M2 macrophages, cDNA microarray analysis was performed to investigate the alterations during oxLDL treatment. Of the 25392 probe sets on "3D-Gene" human oligo chip 25 k, we eliminated 1125 probe sets as controls and backgrounds. Any given gene was eliminated if its normalized intensity was less than 2-4. We also eliminated non-altered genes that showed changes in expression level of less than 2-fold during oxLDL treatment. Finally, we identified 2025, 2265, and 2249 genes that were significantly up- or downregulated in M0, M1, and M2 macrophages, respectively (Figure 2). Among these genes, 1526, 1819, and 1880 genes were upregulated in M0, M1, and M2 macrophages by oxLDL treatment, respectively (All transcriptional profiles are shown in Additional file 1). Table 1 shows the top 30 genes that were most markedly upregulated by oxLDL. IL8, TRIM16, and ADM were commonly upregulated in all subsets of macrophages. Twenty-eight genes in the top 30 upregulated genes in M2 macrophages (93% of the top 30 genes) were also upregulated in M0, while 15 genes in the top 30 upregulated genes in M2 macrophages were upregulated in M1 cells.
Figure 2.
Genes showing significantly altered expression in oxLDL-treated macrophages on cDNA microarray analysis. The logarithmically transformed intensities of all the genes in oxLDL-treated macrophages were plotted against those in non-treated macrophages (Left panels). Genes were eliminated if the normalized intensity was less than 2-4, or if the change in alteration during oxLDL treatment was less than 2-fold (Right panels). Finally, 2025, 2265, and 2249 genes were identified as showing significantly regulated expression in M0, M1, and M2 macrophages, respectively.
Table 1.
Genes upregulated by oxLDL in polarized macrophages
| The top 30 genes upregulated by oxLDL in M0 | |||||
|---|---|---|---|---|---|
| Gene Symbol | Ref Seq ID | M0 | M1 | M2 | NCBI official full name |
| IL8 | NM_000584 | 6.200 | 3.200 | 5.177 | interleukin 8 |
| CCL1 | NM_002981 | 6.028 | 0.937 | - | chemokine (C-C motif) ligand 1 |
| SERPINB2 | NM_002575 | 5.485 | - | - | serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| IL1B | NM_000576 | 5.162 | 2.700 | 2.684 | interleukin 1, beta |
| PTGS2 | NM_000963 | 4.985 | - | - | prostaglandin-endoperoxide synthase 2 |
| TM4SF1 | NM_014220 | 4.836 | - | 1.238 | transmembrane 4 L six family member 1 |
| CCL19 | NM_006274 | 4.834 | 0.368 | - | chemokine (C-C motif) ligand 19 |
| POPDC3 | NM_022361 | 4.557 | 1.491 | - | popeye domain containing 3 |
| INHBA | NM_002192 | 4.476 | 2.335 | - | inhibin, beta A |
| CKB | NM_001823 | 4.378 | 0.920 | 2.107 | creatine kinase, brain |
| IFIT1 | NM_001001887 | 4.152 | 0.717 | 5.734 | interferon-induced protein with tetratricopeptide repeats 1 |
| CCL5 | NM_002985 | 4.075 | -0.218 | 1.616 | chemokine (C-C motif) ligand 5 |
| IFIT5 | NM_012420 | 4.039 | - | - | interferon-induced protein with tetratricopeptide repeats 5 |
| TNFAIP6 | NM_007115 | 4.033 | 0.019 | - | tumor necrosis factor, alpha-induced protein 6 |
| TRIM16 | NM_006470 | 3.990 | 2.992 | 3.453 | tripartite motif-containing 16 |
| ADM | NM_001124 | 3.973 | 4.807 | 3.202 | adrenomedullin |
| CCL4L2 | NM_207007 | 3.905 | 1.321 | 0.448 | chemokine (C-C motif) ligand 4-like 2 |
| ISG20 | NM_002201 | 3.839 | 0.952 | 4.442 | interferon stimulated exonuclease gene 20kDa |
| EREG | NM_001432 | 3.726 | 2.793 | - | epiregulin |
| CSF2 | NM_000758 | 3.644 | 1.605 | 0.724 | colony stimulating factor 2 (granulocyte-macrophage) |
| RHOF | NM_019034 | 3.638 | - | 3.020 | ras homolog gene family, member F (in filopodia) |
| GSTM3 | NM_000849 | 3.632 | 2.635 | 2.543 | glutathione S-transferase mu 3 (brain) |
| AKR1C3 | NM_003739 | 3.497 | 2.490 | 3.432 | aldo-keto reductase family 1, member C3 |
| CYP7B1 | NM_004820 | 3.482 | - | - | cytochrome P450, family 7, subfamily B, polypeptide 1 |
| CCL4 | NM_002984 | 3.466 | 0.742 | -0.123 | chemokine (C-C motif) ligand 4 |
| CCL24 | NM_002991 | 3.433 | 0.413 | - | chemokine (C-C motif) ligand 24 |
| NT5E | NM_002526 | 3.406 | - | - | 5'-nucleotidase, ecto (CD73) |
| IFI44L | NM_006820 | 3.325 | -0.074 | 4.474 | interferon-induced protein 44-like |
| COX11 | NM_004375 | 3.306 | 1.946 | 1.111 | COX11 cytochrome c oxidase assembly homolog (yeast) |
| NOL12 | NM_024313 | 3.281 | 0.937 | 1.313 | nucleolar protein 12 |
| The top 30 genes upregulated by oxLDL in M1 | |||||
| Gene Symbol | Ref Seq ID | M0 | M1 | M2 | NCBI official full name |
| ADM | NM_001124 | 3.973 | 4.807 | 3.202 | adrenomedullin |
| S1RT5 | NM_012241 | 3.039 | 3.486 | 2.137 | sirtuin 5 |
| CXCR5 | NM_032966 | 1.986 | 3.437 | 1.815 | chemokine (C-X-C motif) receptor 5 |
| KLHL21 | NM_014851 | 2.380 | 3.324 | - | kelch-like 21 (Drosophila) |
| IL8 | NM_000584 | 6.200 | 3.200 | 5.177 | interleukin 8 |
| DPYSL3 | NM_001387 | 3.064 | 3.192 | 3.497 | dihydropyrimidinase-like 3 |
| RAB43 | NM_198490 | 1.264 | 3.003 | 3.324 | RAB43, member RAS oncogene familyprovided |
| TRIM16 | NM_006470 | 3.990 | 2.992 | 3.453 | tripartite motif-containing 16 |
| C1orf66 | NM_015997 | 0.382 | 2.970 | 1.953 | chromosome 1 open reading frame 66 |
| PIP4K2A | NM_005028 | 2.325 | 2.967 | 3.127 | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| AGAP3 | NM_031946 | 1.289 | 2.945 | 1.091 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| INPP4A | NM_001566 | 0.129 | 2.872 | 1.872 | inositol polyphosphate-4-phosphatase, type I, 107kDa |
| DAGLA | NM_006133 | 1.545 | 2.834 | 2.807 | diacylglycerol lipase, alpha |
| EREG | NM_001432 | 3.726 | 2.793 | - | epiregulin |
| AGAP11 | NM_133447 | 0.971 | 2.791 | 2.318 | ankyrin repeat and GTPase domain Arf GTPase activating protein 11 |
| ADAMTS10 | NM_030957 | 0.683 | 2.773 | 0.952 | ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
| KCP | NM_199349 | 1.269 | 2.746 | 1.861 | kielin/chordin-like protein |
| ZFAT | NM_020863 | 1.758 | 2.743 | 2.002 | zinc finger and AT hook domain containing |
| LZTR1 | NM_006767 | 1.092 | 2.714 | 3.134 | leucine-zipper-like transcription regulator 1 |
| MON1B | NM_014940 | 0.234 | 2.712 | -0.754 | MON1 homolog B (yeast) |
| IL1B | NM_000576 | 5.162 | 2.700 | 2.684 | interleukin 1, beta |
| STAM | NM_003473 | -0.222 | 2.697 | -0.150 | signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| MAFG | NM_032711 | 2.775 | 2.684 | 2.997 | v-maf musculoaponeurotic fibrosarcoma oncogene homologG (avian) |
| OSGIN1 | NM_182981 | 2.427 | 2.672 | 2.002 | oxidative stress induced growth inhibitor 1 |
| ZNF673 | NM_017776 | 0.606 | 2.669 | - | zinc finger family member 673 |
| IREB2 | NM_004136 | - | 2.648 | - | iron-responsive element binding protein 2 |
| GSTM3 | NM_000849 | 3.632 | 2.635 | 2.543 | glutathione S-transferase mu 3 (brain) |
| UBR2 | NM_015255 | 2.160 | 2.619 | 2.371 | ubiquitin protein ligase E3 component n-recognin 2 |
| SFT2D3 | NM_032740 | 1.146 | 2.577 | 1.609 | SFT2 domain containing 3 |
| FAM70A | NM_017938 | - | 2.577 | 2.614 | family with sequence similarity 70, member A |
| The top 30 genes upregulated by oxLDL in M2 | |||||
| Gene Symbol | Ref Seq ID | M0 | M1 | M2 | NCBI official full name |
| IFIT2 | NM_001547 | 2.559 | - | 5.893 | interferon-induced protein with tetratricopeptide repeats 2 |
| IFIT1 | NM_001001887 | 4.152 | 0.717 | 5.734 | interferon-induced protein with tetratricopeptide repeats 1 |
| IL8 | NM_000584 | 6.200 | 3.200 | 5.177 | interleukin 8 |
| MX2 | NM_002463 | 2.482 | 0.562 | 4.552 | myxovirus (influenza virus) resistance 2 (mouse) |
| IFI44L | NM_006820 | 3.325 | -0.074 | 4.474 | interferon-induced protein 44-like |
| ISG20 | NM_002201 | 3.839 | 0.952 | 4.442 | interferon stimulated exonuclease gene 20kDa |
| IDO1 | NM_002164 | - | 0.623 | 4.335 | indoleamine 2,3-dioxygenase 1 |
| PLSCR1 | NM_021105 | 2.002 | -0.179 | 4.119 | phospholipid scramblase 1 |
| IFIT3 | NM_001549 | 2.306 | -0.483 | 3.943 | interferon-induced protein with tetratricopeptide repeats 3 |
| SGPP2 | NM_152386 | 3.229 | - | 3.626 | sphingosine-1-phosphate phosphatase 2 |
| OR9I1 | NM_001005211 | 2.923 | 1.683 | 3.574 | olfactory receptor, family 9, subfamily I, member 1 |
| VNN1 | NM_004666 | 0.568 | 0.896 | 3.569 | vanin 1 |
| DPYSL3 | NM_001387 | 3.064 | 3.192 | 3.497 | dihydropyrimidinase-like 3 |
| TRIM16 | NM_006470 | 3.990 | 2.992 | 3.453 | tripartite motif-containing 16 |
| EPSTI1 | NM_033255 | 2.678 | -0.642 | 3.439 | epithelial stromal interaction 1 (breast) |
| AKR1C3 | NM_003739 | 3.497 | 2.490 | 3.432 | aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) |
| RAB43 | NM_198490 | 1.264 | 3.003 | 3.324 | RAB43, member RAS oncogene family |
| APOL6 | NM_030641 | 1.925 | 0.723 | 3.291 | apolipoprotein L, 6 |
| USP18 | NM_017414 | 1.686 | 0.052 | 3.288 | ubiquitin specific peptidase 18 |
| ADM | NM_001124 | 3.973 | 4.807 | 3.202 | adrenomedullin |
| RSAD2 | NM_080657 | 1.858 | 0.083 | 3.194 | radical S-adenosyl methionine domain containing 2 |
| LZTR1 | NM_006767 | 1.092 | 2.714 | 3.134 | leucine-zipper-like transcription regulator 1 |
| PIP4K2A | NM_005028 | 2.325 | 2.967 | 3.127 | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| CSF2RA | NM_172247 | 1.683 | 2.109 | 3.115 | colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| TNFRSF9 | NM_001561 | 2.899 | 1.702 | 3.100 | tumor necrosis factor receptor superfamily, member 9 |
| ARHGAP44 | NM_014859 | 1.136 | 1.919 | 3.053 | Rho GTPase activating protein 44 |
| RHOF | NM_019034 | 3.638 | - | 3.020 | ras homolog gene family, member F (in filopodia) |
| CWF19L1 | NM_018294 | 1.271 | 2.098 | 3.005 | CWF19-like 1, cell cycle control (S. pombe) |
| MAFG | NM_032711 | 2.775 | 2.684 | 2.997 | v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) |
| TNFRSF4 | NM_003327 | 1.815 | 1.376 | 2.974 | tumor necrosis factor receptor superfamily, member 4 |
Non-polarized (M0) and polarized M1 and M2 macrophages were treated with or without oxLDL for 6 h. The changes in gene expression were analyzed by cDNA microarray analysis as described in the Materials and methods section. The top 30 upregulated genes in each subset of macrophages are listed. The values denote fold changes (log2 ratio) of normalized intensities during oxLDL treatment. Letters in boldface indicate genes that were also upregulated by more than 2-fold in the two other subsets. "-" indicates genes that did not show significantly altered expression. Twenty-eight of the top 30 upregulated genes in M2 macrophages (93% of the top 30 genes) were also upregulated in M0, while 15 genes in the top 30 upregulated genes in M2 macrophages were upregulated in M1 cells.
Functional gene ontology
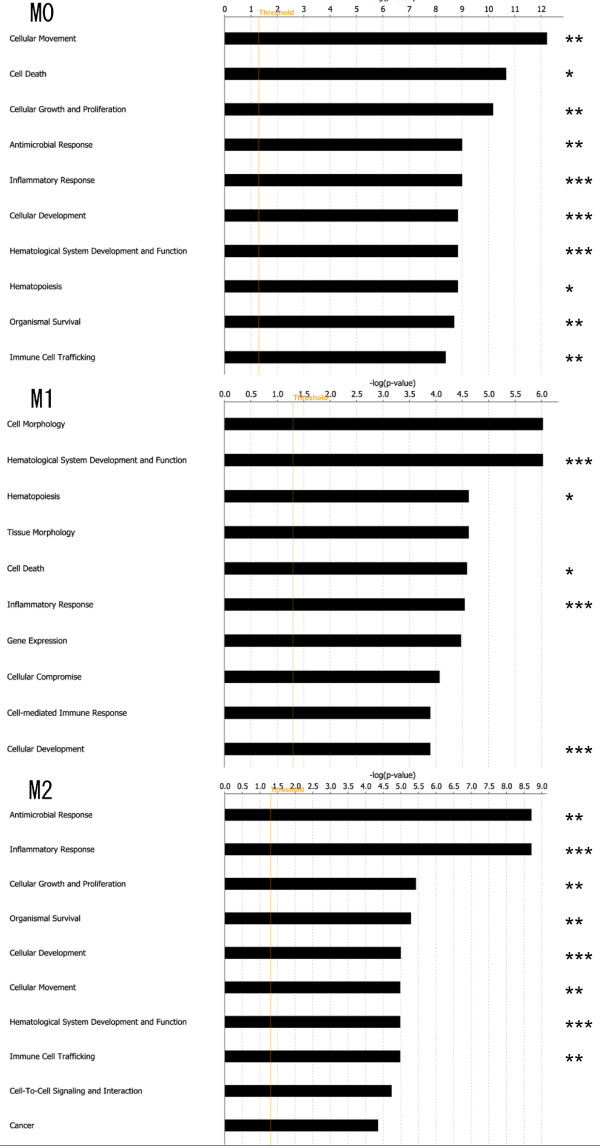
To identify oxLDL treatment-related biological functions of polarized macrophages, bioinformatics aspects of differentially expressed genes during oxLDL treatment were further analyzed using IPA software. The 1566, 1738, and 1749 genes of M0, M1, and M2 macrophages were identified by IPA software as functionally intentional genes, and categorized into 65, 84, and 80 groups according to functional gene ontology, respectively. Figure 3 shows the top 10 functional ontology categories which contain the molecules altered by oxLDL treatment. Eight of the top 10 ontology categories of M0 macrophages were also found in the top 10 of M2 macrophages, whereas only 3 ontology categories of M1 macrophages were found in the top 10 of M2.
Figure 3.
Gene ontology analysis. The genes showing significantly upregulated expression in oxLDL-treated macrophages were functionally categorized into groups according to gene ontology. The top 10 functional ontology categories in each subset of macrophages (M0, M1, and M2) are shown in order of P-value. Right-tailed Fisher's exact test was used to calculate the P-value determining the probability that each biological function assigned to that network was due to chance alone. Eight of the top 10 ontology categories were found in both M0 and M2. *Common in M0 and M1. **Common in M0 and M2. *** Common in M0, M1, and M2.
Molecular network analysis
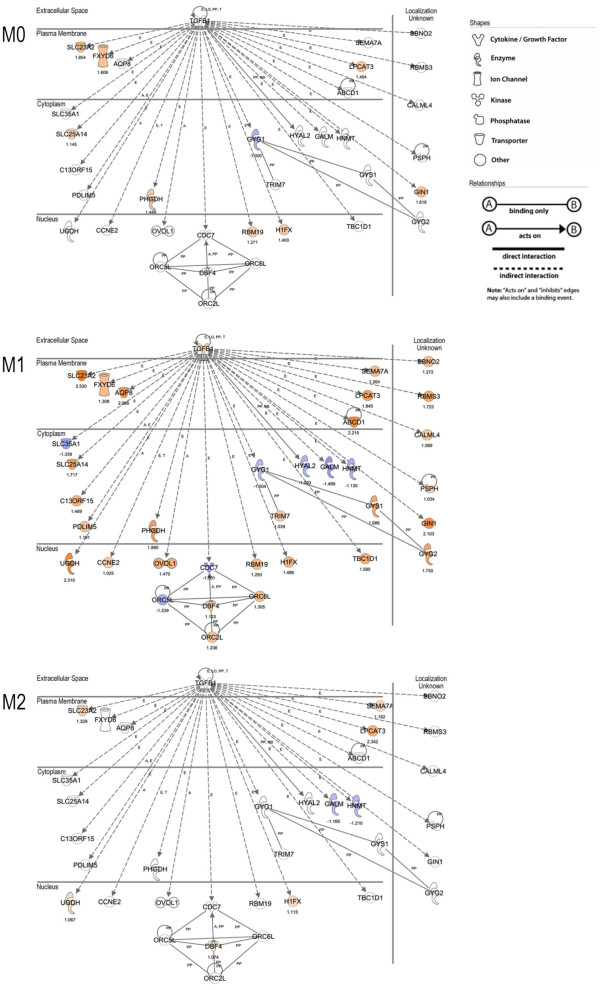
We performed molecular network analysis using IPA software to elucidate the molecular relationships when each subset of macrophages was treated with oxLDL. The top 5 highly scoring networks of each subset are shown in Table 2. Among these networks, the highest scoring network was found in M1 macrophages as Network #1, including molecules related to "carbohydrate metabolism," "DNA replication, recombination and repair," and "embryonic development." Interestingly, most of the molecules in network #1 were related to transforming growth factor (TGF)-β1 directly or indirectly (Figure 4).
Table 2.
Molecular network analysis using IPA
| Top 5 Networks in M0 | ||||
|---|---|---|---|---|
| ID | Molecules in Network | Score | Focus Molecules | Top Functions |
| #1 | BARX2, CD226, CRABP1, CTH, Cytokeratin, FOLR2, FOS, GCLM, GCNT1, HHEX, HTATIP2, IGDCC3, KCNJ2, KRT8, KRT18, KRT20, KRT6A, LPCAT3, MAF, MAFB, MAFF, MAFG, MT1A, musculoaponeurotic fibrosarcoma oncogene, NFE2L3, NRF1, PAX6, PLAGL1, PVR, S100P, TGFBI, TH1 Cytokine, TLE1, TMSB4, TMSB4X | 33 | 31 | Cellular Assembly and Organization, Cellular Function and Maintenance, Hair and Skin Development and Function |
| #2 | Alpha tubulin, APEX2, ARRDC3, CALB2, Calbindin, CSF1, CXORF21, DHCR24, DUB, EHMT2, FAM105A, FAM107A, GSC, HDAC6, HOOK2, LMO2, LRRC58, MIR124, NTRK3, oxidoreductase, POU5F1, PPP1R12C, REST, RYK, STAU1, TRPM2, TSPAN14, Tubulin, USP12, USP28, USP34, USP41, USP48, USP49, ZBED3 | 28 | 29 | Auditory and Vestibular System Development and Function, Genetic Disorder, Metabolic Disease |
| #3 | ANAPC1, ANAPC2, ANAPC5, APC, ASNS, BIRC3, CD3EAP, CDC20, CHD2, CKAP2, Cpt, CPT2, Cyclin A, DTYMK, E3 RING, EIF5, GLCE, GPC1, HEXIM1, HGF, IMPDH2, Integrin alpha V beta 3, ISG20, JMJD1C, MYL12A, NAMPT, POLR1A, SLC40A1, SPATC1, SPDEF, TMEM158, TOPBP1, UBR2, Vegf, VHL | 28 | 29 | Protein Degradation, Protein Synthesis, Cardiovascular System Development and Function |
| #4 | ALDH2, ASXL1, C14ORF1, CBX4, CCDC106, CDK5RAP2, Cyclooxygenase, DIO3, DUSP8, FHL1, JDP2, LSM2, MIP1, MYBPC3, P38 MAPK, PCGF2, PHYHIP, PLEKHN1, PORCN, SEMA7A, SENP2, Stat1 dimer, STK36, SUFU, Tnf receptor, TNFRSF9, TNFRSF11A, TNFSF9, TRAF, TRAF1, TRAF5, TRAF2-TRAF5, TRAIP, TREM1, WNT4 | 27 | 28 | Cellular Assembly and Organization, Cellular Function and Maintenance, Skeletal and Muscular System Development and Function |
| #5 | ANKRD29, APEX1, BAG5, CKB, CKMT1B, CNOT3, CNOT6, CNOT8, Creatine Kinase, DNAJA4, DNAJB6, DNAJC15, DNMBP, Fibrinogen, Hsp70, Hsp90, Hsp22/Hsp40/Hsp90, HSPA2, HSPA1A, ICAM1, IFT52, IP6K2, MIR1, Nos, Pka catalytic subunit, PLSCR1, SH3KBP1, SLC25A30, SPRY2, SRXN1, TIMP3, TNKS1BP1, TNPO2, TRIM2, XPNPEP3 | 26 | 27 | Cellular Assembly and Organization, Cellular Development, Cellular Growth and Proliferation |
| Top 5 Networks in M1 | ||||
| ID | Molecules in Network | Score | Focus Molecules | Top Functions |
| #1 | ABCD1, AQP8, C13ORF15, CALML4, CCNE2, CDC7, DBF4, FXYD6, GALM, GIN1, GYG1, GYG2, GYS1, H1FX, HNMT, HYAL2, LPCAT3, ORC2L, ORC5L, ORC6L, OVOL1, PDLIM5, PHGDH, PSPH, RBM19, RBMS3, SBNO2, SEMA7A, SLC23A2, SLC25A14, SLC35A1, TBC1D1, TGFB1, TRIM7, UGDH | 41 | 35 |
Carbohydrate Metabolism, DNA Replication, Recombination, and Repair, Embryonic Development |
| #2 | ATAD3B, CAPZB, CHTF18, CPD, CPT2, FXC1, FXYD1, GAMT, GPC1, GTF3C2, GTF3C4, HMG CoA synthase, HMGCS1, HNRNPA0, IQGAP3, IREB2, LMNB2, LSM2, LSM6, LSM7, MINA, Mir125b (mouse), MYC, Ndpk, NME2, NME6, NME7, PDS5B, RAD21, RANBP6, REC8, RFX2, RNGTT, RRM2B, TOR2A | 34 | 32 | Cellular Assembly and Organization, Genetic Disorder, Metabolic Disease |
| #3 | ACTN4, APEX2, ATRX, DNA-directed DNA polymerase, Erm, HOOK2, ICAM1, IGSF8, LAS1L, LIG3, LIG4, LRSAM1, MARCH3, MYOZ1, NEIL1, NOM1, PNKP, POLE2, POLG2, POLM, RNF2, RNF6, RNF10, RNF25, RNF166, RNF185, SLC9A1, TRIM37, TRP, TRPC4, UBE2D2, UBE2E3, UBE2 H (includes EG:7328), VIL1, ZNRF1 | 34 | 32 | DNA Replication, Recombination, and Repair, Gene Expression, Cell-mediated Immune Response |
| #4 | ARL15, BRD1, BTBD2, C14ORF1, C1ORF103, CCDC106, CCDC90B, CD3EAP, CDKN3, DNA-directed RNA polymerase, EGLN1, EHMT2, FEZ1, FXR1, GTF3A, histone-lysine N-methyltransferase, KBTBD7, KIF11, KIF2C, KLK10, LMO2, MELK, POLR1A, POLR3 D, POLR3E, POLR3F, RIT1, SETDB1, STAB2, THAP8 (includes EG:199745), TLE1, TMSB4X, UNC119, Vegf, ZNF24 (includes EG:7572) | 30 | 32 | Cell Cycle, Cellular Assembly and Organization, DNA Replication, Recombination, and Repair |
| #5 | ABCC1, ABCD3, ACIN1, ATP5B, ATPase, CRISPLD2 (includes EG:83716), DTX1, DUSP1, E2F1, FABP1, FAM177A1, GRAP, IL34, Immunoglobulin, INHBA, INHBC, LIF, MAFF, MARK4, MIR124, MYH9, NAA15, NOTCH1, POMC, RDH10, RSF1, RYK, SKIV2L, SLC22A18AS, STAB1, SYNGR2, Trypsin, WRN (includes EG:7486), WWP1, ZNF790 | 30 | 31 | Cellular Development, Nervous System Development and Function, Hematological System Development and Function |
| Top 5 Networks in M2 | ||||
| ID | Molecules in Network | Score | Focus Molecules | Top Functions |
| #1 | AKR1C3, ARL15, C1ORF103, CETN3, DCP1A, DCTPP1, DDX6, DDX20, DNA-directed RNA polymerase, DUB, ETV3, FILIP1L, FXYD6, GANAB, GSTM3 (includes EG:2947), GTF3C4, GXYLT1, JMJD1C, NUP155, POLR1A, POLR1E, POLR3F, PRMT1, RHOF, RIF1, SDCCAG8, SNRPA1, TARS, TRAF6, UNC119, USP20, USP41, USP45, VHL, ZFP161 | 36 | 33 | Infection Mechanism, Infectious Disease, Cell Cycle |
| #2 | ATN1, BANP, C8ORF4, CBFA2T2, CLK2, CLOCK, CRY1, DBP, EIF2B1, Fascin, H1FX, HIST1H2AE (includes EG:3012), Histone H1, HNRNPL, Importin beta, KHDRBS3, KIAA0913, LBR, LTBP4, LTC4 S, MAST2, OGG1, Pkc(s), RNF19A, SAFB, SDCBP2, SLC20A2, SRPK1, STK36, SUFU, TIMELESS, TM4SF1, TPM2, XPC, ZNF652 | 32 | 31 | Behavior, Nervous System Development and Function, Lipid Metabolism |
| #3 | ATF7IP, ATPase, BACE1, CAPN3, CASP2, CASP7, CASP10, Caspase, CTRL, ENTPD2, hydrolase, ICAM1, IDE, ITM2C, KIAA1632, KYNU, LGMN, MAGED1, MEOX2, NAIP, NANP, NGFR, NUP160, PAPPA, peptidase, PLEKHF2 (includes EG:79666), RECQL5, RTN3, SENP5 (includes EG:205564), SPPL2B, SPTBN1, THTPA, UBAC2, UBR5, XAF1 | 32 | 31 | Protein Degradation, Protein Synthesis, Post-Translational Modification |
| #4 | API5, ARRDC3, ASPH, Calbindin, CKB, CNNM4, COX11, COX15, COX10 (includes EG:1352), CPT1B, Creatine Kinase, Cytochrome c oxidase, ENO3, FICD, FOXP1, GPRASP2, HTR2B, HTT, JAKMIP1, KLF16, NDUFS1, NEFH, PHB (includes EG:5245), PPP1R16B, RCOR2, REST, RGS14, RNF34, SASH1, SCN4B, SNN, STRN, Thymidine Kinase, VAPA, WAC | 32 | 31 | Gene Expression, Neurological Disease, Cellular Compromise |
| #5 | ALDH7A1, BPI, Cbp, Ciap, CLIP2, CST7, DBT, DUSP2, G0S2, GFPT2, KLF3, KRT19, lymphotoxin-alpha1-beta2, MICA, NFkB (complex), NKIRAS1, PELI3, POU2F2, RBCK1, REL/RELA/RELB, RNF216, S100P, S100Z, SH3RF1, SIAH2, SLC2A6, SNIP1, SUMO4, TNIP2, TRIM32, TRIM69, TXNRD1, WTAP, ZMYND11, ZNF71 | 30 | 30 | Carbohydrate Metabolism, Lipid Metabolism, Small Molecule Biochemistry |
Molecular network analysis was performed using IPA software. The top 5 groups of molecules in the network are listed in order of network score, which IPA calculates as shown below. We focused on the highest scoring network #1 in M1 macrophages, which had a score of 41 and included 35 molecules matched with the Ingenuity Knowledge Base.
Network Score = -log10(P-value)
Figure 4.
Molecular network analysis of the highest scoring network in Table 2. The molecular network in network #1 of M1 macrophages shown in Table 2 and corresponding data of M0 and M2 macrophages are shown. Nearly all the molecules in M1 were related to TGF-β1 directly or indirectly. Molecules are represented as nodes, and the biological relationships between pairs of nodes are represented as lines. The intensity of the node color indicates the degree of upregulation (red) or downregulation (blue). The numbers below the nodes denote fold changes (log2 ratio) of normalized intensities during oxLDL treatment. Nodes are displayed using various shapes representing the functional class of the gene product. Lines are displayed with various labels describing the nature of the relationship between the nodes; i.e., A for Activation, E for Expression, LO for Localization, PD for Protein - DNA binding, PP for Protein - Protein binding, RB for Regulation of Binding, and T for Transcription.
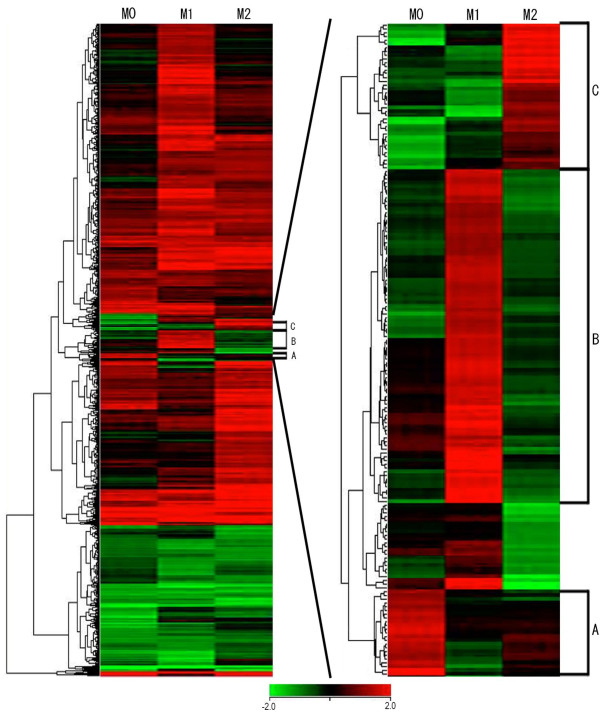
Hierarchical cluster analysis
Hierarchical cluster analysis allows us to visually comprehend differential patterns over multiple microarray datasets. To analyze the hierarchical clusters over subsets of macrophages, we constructed a heat map where red and green indicated up- and downregulation, respectively (Figure 5). We employed 3196 genes, expression levels of which were significantly altered by oxLDL treatment in at least one subset of macrophages. A total of 251 genes were commonly identified as upregulated genes in all subsets of macrophages, including TRIM16, HMOX1, TXNRD1, GCLM, and DUSP1, all of which contain an antioxidant response element (ARE) in their promoter regions, which serves as a binding site for nuclear factor erythroid 2-related factor 2 (Nrf2). Hierarchical cluster analysis identified 3 clusters the genes of which were upregulated in one subset but not in the other subsets (Figure 5). Cluster A included 17 annotated genes that were upregulated in M0, but not in the other subsets (Table 3). The genes in cluster A belonged to ontology categories including "cell-mediated immune response," "cellular movement," "hematological system development and function," and "immune cell trafficking." There were 72 annotated genes in cluster B, which were specifically upregulated in M1. These 72 genes were related to "gene expression" and "cellular development." They included NFKB2, encoding nuclear factor of kappa light polypeptide gene enhancer in B-cells (NF-κB), and PIK3R4, encoding phosphoinositide-3-kinase (PI3K), both of which are molecules related to the NF-κB signaling pathway. In cluster C, 28 annotated genes were identified as specifically upregulated in M2. These genes were associated with "carbohydrate metabolism," "lipid metabolism," and "small molecule biochemistry."
Figure 5.
Heat map constructed by hierarchical cluster analysis. Red and green in the heat maps indicate up- and downregulation during oxLDL treatment, respectively. The left map includes 3196 genes expression levels of which were significantly altered by oxLDL treatment in at least one subset of macrophages. Genes in three clusters, denoted as A to C, were specifically upregulated only in one subset. There were 17, 72, and 28 annotated genes in cluster A, B, and C, respectively (right map).
Table 3.
Genes included in each cluster
| Cluster A. | |||||
|---|---|---|---|---|---|
| Gene Symbol | Ref Seq ID | M0 | M1 | M2 | NCBI official full name |
| CHI3L2 | NM_004000 | 1.170712 | 0.008113 | -0.453072 | chitinase 3-like 2 |
| PARP8 | NM_024615 | 1.018767 | 0.147437 | -0.206628 | poly (ADP-ribose) polymerase family, member 8 |
| MAP4K4 | NM_145686 | 1.115230 | -0.302140 | -0.763189 | mitogen-activated protein kinase kinase kinase kinase 4 |
| CD44 | NM_001001391 | 1.312520 | -0.201936 | -0.048693 | CD44 molecule |
| CYP27B1 | NM_000785 | 1.111109 | -0.055705 | -0.015432 | cytochrome P450, family 27, subfamily B, polypeptide 1 |
| SLC1A2 | NM_004171 | 1.466166 | -0.088519 | 0.225126 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| CKMT1 | NM_020990 | 1.436353 | 0.264923 | 0.338353 | creatine kinase, mitochondrial 1B |
| MYOD1 | NM_002478 | 1.665245 | 0.211625 | 0.330158 | myogenic differentiation 1 |
| CD40 | NM_001250 | 1.319905 | -0.606033 | 0.379319 | CD40 molecule, TNF receptor superfamily member 5 |
| PRKAR2B | NM_002736 | 1.349370 | -0.472458 | 0.347752 | protein kinase, cAMP-dependent, regulatory, type II, beta |
| KIAA0913 | NM_015037 | 1.244477 | -0.318545 | 0.447287 | KIAA0913 |
| SNX10 | NM_013322 | 1.138265 | -0.351416 | 0.635676 | sorting nexin 10 |
| HHIP | NM_022475 | 1.177555 | -0.537186 | 0.506197 | hedgehog interacting protein |
| BIRC3 | NM_001165 | 1.499122 | -0.583781 | -0.065715 | baculoviral IAP repeat-containing 3 |
| ZBTB7C | NM_001039360 | 1.293925 | -1.001009 | 0.404020 | zinc finger and BTB domain containing 7C |
| PACRGL | NM_145048 | 2.026548 | -0.460488 | 0.187722 | PARK2 co-regulated-like |
| CCL23 | NM_005064 | 2.471357 | -0.192407 | -0.036793 | chemokine (C-C motif) ligand 23 |
| Cluster B. | |||||
| Gene Symbol | Ref Seq ID | M0 | M1 | M2 | NCBI official full name |
| NDN | NM_002487 | -0.169667 | 1.457695 | -0.617320 | necdin homolog (mouse) |
| FAM129C | NM_173544 | -0.110208 | 1.321559 | -0.841954 | family with sequence similarity 129, member C |
| NME6 | NM_005793 | -0.315804 | 1.345342 | -0.759802 | non-metastatic cells 6, protein expressed in (nucleoside-diphosphate kinase) |
| TMEM99 | NM_145274 | -0.390527 | 1.497197 | -0.676125 | transmembrane protein 99 |
| ATP2B1 | NM_001682 | -0.371182 | 1.535878 | -0.824423 | ATPase, Ca++ transporting, plasma membrane 1 |
| KIF1A | NM_004321 | -0.362399 | 1.333531 | -0.928733 | kinesin family member 1A |
| TMEM188 | NM_153261 | -0.375073 | 1.157188 | -0.955238 | transmembrane protein 188 |
| PLEKHG4 | NM_015432 | -0.414580 | 1.052587 | -0.706013 | pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| NOXA1 | NM_006647 | -0.322207 | 1.015456 | -0.592170 | NADPH oxidase activator 1 |
| CUX1 | NM_001913 | -0.236206 | 1.147118 | -0.551592 | cut-like homeobox 1 |
| SBNO2 | NM_014963 | -0.294540 | 1.271977 | -0.630553 | strawberry notch homolog 2 (Drosophila) |
| TIGD5 | NM_032862 | -0.376362 | 1.224225 | -0.526717 | tigger transposable element derived 5 |
| CTSL2 | NM_001333 | -0.428772 | 1.153800 | -0.359408 | cathepsin L2 |
| ZNF24 | NM_006965 | -0.385988 | 1.069071 | -0.360632 | zinc finger protein 24 |
| STK32C | NM_173575 | -0.634840 | 1.124350 | -0.544619 | serine/threonine kinase 32C |
| NUDT17 | NM_001012758 | -0.524910 | 1.111636 | -0.519731 | nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| CALML4 | NM_033429 | -0.433422 | 1.089113 | -0.492229 | calmodulin-like 4 |
| C7orf33 | NM_145304 | -0.435642 | 1.106760 | -0.512504 | chromosome 7 open reading frame 33 |
| FBXO22 | NM_012170 | -0.203452 | 1.309369 | -0.481216 | F-box protein 22 |
| LINGO1 | NM_032808 | -0.174931 | 1.312372 | -0.445152 | leucine rich repeat and Ig domain containing 1 |
| ZMYND19 | NM_138462 | -0.243665 | 1.351306 | -0.517691 | zinc finger, MYND-type containing 19 |
| CCR1 | NM_001295 | -0.165232 | 1.419892 | -0.480175 | chemokine (C-C motif) receptor 1 |
| ATRX | NM_000489 | -0.498685 | 1.285463 | -0.371568 | alpha thalassemia/mental retardation syndrome X-linked |
| FRMD4B | NM_015123 | -0.477381 | 1.252296 | -0.287931 | FERM domain containing 4B |
| CEP164 | NM_014956 | -0.419638 | 1.269277 | -0.283263 | centrosomal protein 164kDa |
| LANCL2 | NM_018697 | -0.393507 | 1.324253 | -0.242555 | LanC lantibiotic synthetase component C-like 2 (bacterial) |
| UBA5 | NM_024818 | -0.478021 | 1.390785 | -0.479541 | ubiquitin-like modifier activating enzyme 5 |
| GGCX | NM_000821 | -0.380201 | 1.330677 | -0.476370 | gamma-glutamyl carboxylase |
| FOXN3 | NM_005197 | -0.772837 | 1.196028 | -0.268613 | forkhead box N3 |
| TP53BP1 | NM_005657 | -0.797822 | 1.249895 | -0.460222 | tumor protein p53 binding protein 1 |
| ORC6 | NM_014321 | -0.633454 | 1.305326 | -0.322922 | origin recognition complex, subunit 6 |
| ST7 | NM_018412 | -0.678132 | 1.309238 | -0.290689 | suppression of tumorigenicity 7 |
| OGFOD1 | NM_018233 | 0.061541 | 1.413933 | -0.438702 | 2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| UBE2B | NM_003337 | 0.146284 | 1.431435 | -0.295974 | ubiquitin-conjugating enzyme E2B (RAD6 homolog) |
| GALNTL4 | NM_198516 | 0.178950 | 1.470650 | -0.270045 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4 |
| HSPA14 | NM_016299 | 0.185892 | 1.419392 | -0.264205 | heat shock 70kDa protein 14 |
| PEX13 | NM_002618 | 0.137507 | 1.527385 | -0.274508 | peroxisomal biogenesis factor 13 |
| ATP2B2 | NM_001001331 | 0.084953 | 1.689567 | -0.394052 | ATPase, Ca++ transporting, plasma membrane 2 |
| TAAR2 | NM_014626 | 0.168770 | 1.695874 | -0.439255 | trace amine associated receptor 2 |
| HUS1 | NM_004507 | 0.098273 | 1.666199 | -0.226370 | HUS1 checkpoint homolog (S. pombe) |
| RIC8B | NM_018157 | 0.214093 | 1.650594 | -0.307883 | resistance to inhibitors of cholinesterase 8 homolog B (C. elegans) |
| DNAJC17 | NM_018163 | 0.172153 | 1.482705 | -0.492935 | DnaJ (Hsp40) homolog, subfamily C, member 17 |
| TMEM130 | NM_152913 | 0.206368 | 1.399311 | -0.480833 | transmembrane protein 130 |
| AP2A2 | NM_012305 | 0.285524 | 1.391232 | -0.458091 | adaptor-related protein complex 2, alpha 2 subunit |
| TTTY13 | NR_001537 | 0.381981 | 1.560081 | -0.451429 | testis-specific transcript, Y-linked 13 (non-protein coding) |
| AKT1S1 | NM_032375 | 0.385837 | 1.517956 | -0.371995 | AKT1 substrate 1 (proline-rich) |
| WNT3 | NM_030753 | 0.009319 | 1.560600 | -0.801987 | wingless-type MMTV integration site family, member 3 |
| ZNF443 | NM_005815 | 0.154517 | 1.457293 | -0.742146 | zinc finger protein 443 |
| TFCP2L1 | NM_014553 | 0.207710 | 1.525949 | -0.955561 | transcription factor CP2-like 1 |
| KIAA0355 | NM_014686 | 0.278629 | 1.724807 | -0.576470 | KIAA0355 |
| ASAP2 | NM_003887 | 0.524513 | 1.733462 | -0.459752 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| C14orf49 | NM_152592 | 0.578042 | 1.624871 | -0.365254 | chromosome 14 open reading frame 49 |
| NFKB2 | NM_002502 | 0.652839 | 1.882528 | -0.343188 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| ZNF586 | NM_017652 | 0.328381 | 1.848104 | -0.284595 | zinc finger protein 586 |
| RNMTL1 | NM_018146 | 0.277170 | 1.645924 | -0.133286 | RNA methyltransferase like 1 |
| FOSL2 | NM_005253 | 0.386974 | 1.569486 | -0.229451 | FOS-like antigen 2 |
| SATB1 | NM_002971 | 0.542138 | 1.252296 | -0.749110 | SATB homeobox 1 |
| ASB6 | NM_177999 | 0.666548 | 1.536652 | -0.584820 | ankyrin repeat and SOCS box-containing 6 |
| SLC5A3 | NM_006933 | 0.682674 | 1.388148 | -0.424733 | solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| C8orf84 | NM_153225 | 0.637255 | 1.709652 | -0.981619 | chromosome 8 open reading frame 84 |
| MON1B | NM_014940 | 0.234404 | 2.712191 | -0.753618 | MON1 homolog B (yeast) |
| PIGO | NM_152850 | 0.134053 | 2.574539 | -0.004491 | phosphatidylinositol glycan anchor biosynthesis, class O |
| STAM | NM_003473 | -0.221822 | 2.697280 | -0.150394 | signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| CCDC45 | NM_138363 | -0.046055 | 2.118025 | -0.312274 | coiled-coil domain containing 45 |
| GYS1 | NM_002103 | -0.699205 | 1.685994 | -0.629003 | glycogen synthase 1 (muscle) |
| RENBP | NM_002910 | -0.403657 | 1.957579 | -0.580833 | renin binding protein |
| BTD | NM_000060 | -0.348646 | 1.647239 | -0.685758 | biotinidase |
| FAM172A | NM_032042 | -0.409833 | 1.717425 | -0.665763 | family with sequence similarity 172, member A |
| PIK3R4 | NM_014602 | -0.188899 | 2.325311 | -0.729078 | phosphoinositide-3-kinase, regulatory subunit 4 |
| RSAD1 | NM_018346 | -0.251002 | 2.260460 | -0.748812 | radical S-adenosyl methionine domain containing 1 |
| ZNF626 | NM_145297 | -0.252970 | 1.842391 | -0.910235 | zinc finger protein 626 |
| CTH | NM_001902 | -1.060839 | 2.365953 | -1.038817 | cystathionase (cystathionine gamma-lyase) |
| Cluster C. | |||||
| Gene Symbol | Ref Seq ID | M0 | M1 | M2 | NCBI official full name |
| MXD3 | NM_031300 | -0.800223 | 0.116059 | 1.984245 | MAX dimerization protein 3 |
| CD14 | NM_000591 | -1.193454 | -0.266058 | 2.053786 | CD14 molecule |
| BARX1 | NM_021570 | -1.650218 | 0.236670 | 1.728783 | BARX homeobox 1 |
| ZNF331 | NM_018555 | -1.729498 | -0.028187 | 1.786943 | zinc finger protein 331 |
| GPR54 | NM_032551 | -0.085064 | -0.929503 | 1.669680 | KISS1 receptor |
| ANP32A | NM_006305 | -0.155917 | -0.929503 | 1.827632 | acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| C19orf54 | NM_198476 | -0.205921 | -0.803972 | 1.634746 | chromosome 19 open reading frame 54 |
| G0S2 | NM_015714 | -0.367904 | -0.858834 | 2.131860 | G0/G1switch 2 |
| SPATA3 | NM_139073 | -0.301818 | -0.432004 | 1.766064 | spermatogenesis associated 3 |
| FOXA2 | NM_153675 | -0.306075 | -0.533883 | 1.983144 | forkhead box A2 |
| PSORS1C2 | NM_014069 | -0.423428 | -0.713119 | 1.449567 | psoriasis susceptibility 1 candidate 2 |
| SNN | NM_003498 | -0.492216 | -1.239138 | 1.185086 | stannin |
| CDKN2B | NM_078487 | -0.404103 | -0.847300 | 1.024029 | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| ZNF257 | NM_033468 | -0.916132 | -1.388935 | 1.189490 | zinc finger protein 257 |
| ZBTB4 | NM_020899 | -0.296438 | -1.533325 | 0.937391 | zinc finger and BTB domain containing 4 |
| CASP2 | NM_032983 | -1.527530 | -0.751505 | 1.026536 | caspase 2, apoptosis-related cysteine peptidase |
| TCTEX1D1 | NM_152665 | -1.661006 | -0.348442 | 1.280638 | Tctex1 domain containing 1 |
| FILIP1L | NM_014890 | -0.967885 | -0.212315 | 1.018932 | filamin A interacting protein 1-like |
| USP52 | NM_014871 | -0.885665 | -0.310932 | 1.097774 | PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| SETD8 | NM_020382 | -1.020324 | -0.599401 | 0.697655 | SET domain containing (lysine methyltransferase) 8 |
| C13orf31 | NM_153218 | -1.442594 | -0.330041 | 0.688789 | chromosome 13 open reading frame 31 |
| POLD1 | NM_002691 | -1.311086 | -0.314006 | 0.812769 | polymerase (DNA directed), delta 1, catalytic subunit 125kDa |
| DNMT3A | NM_022552 | -1.207327 | -0.289399 | 0.489584 | DNA (cytosine-5-)-methyltransferase 3 alpha |
| TGFBI | NM_000358 | -1.324217 | -0.324120 | 0.308245 | transforming growth factor, beta-induced, 68kDa |
| ANP32C | NM_012403 | -1.493138 | -0.380761 | 0.494101 | acidic (leucine-rich) nuclear phosphoprotein 32 family, member C |
| ITGA2B | NM_000419 | -1.109218 | 0.018385 | 1.026536 | integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| RGS3 | NM_130795 | -1.022299 | -0.038087 | 0.656812 | regulator of G-protein signaling 3provided |
| MARCH1 | NM_017923 | -1.374411 | 0.233896 | 0.729622 | membrane-associated ring finger (C3HC4) 1provided |
| The top functional ontology categories | |||||
| Cluster A. | P-value | ||||
| Cell-mediated Immune Response | 9.46E-06-2.65E-02 | ||||
| Cellular Movement | 9.46E-06-3.76E-02 | ||||
| Hematological System Development and Function | 9.46E-06-3.57E-02 | ||||
| Immune Cell Trafficking | 9.46E-06-2.49E-02 | ||||
| Cardiovascular Disease | 2.64E-05-3.76E-02 | ||||
| Cluster B. | P-value | ||||
| Cell Cycle | 1.31E-04-3.28E-02 | ||||
| Cell Cycle | 1.31E-04-3.28E-02 | ||||
| DNA Replication, Recombination, and Repair | 1.31E-04-4.51E-02 | ||||
| Gene Expression | 2.16E-03-3.35E-02 | ||||
| Cellular Development | 3.51E-03-3.64E-02 | ||||
| Nervous System Development and Function | 3.51E-03-4.7E-02 | ||||
| Cluster C. | P-value | ||||
| Carbohydrate Metabolism | 1.12E-04-4.09E-02 | ||||
| Lipid Metabolism | 1.12E-04-4.09E-02 | ||||
| Small Molecule Biochemistry | 1.12E-04-4.09E-02 | ||||
| Cell Death | 3.76E-04-4.81E-02 | ||||
| Nervous System Development and Function | 7.79E-04-3.33E-02 | ||||
Three clusters were extracted from the heat map shown in Figure 5. There were 17, 72, and 28 annotated genes in clusters A, B, and C, respectively. The top functional ontology categories and the corresponding P-values in each subset are shown. NFKB2 and PIK3R4 were found in cluster B. The values denote fold changes (log2 ratio) of normalized intensities during oxLDL treatment.
Quantitative real-time RT-PCR analysis
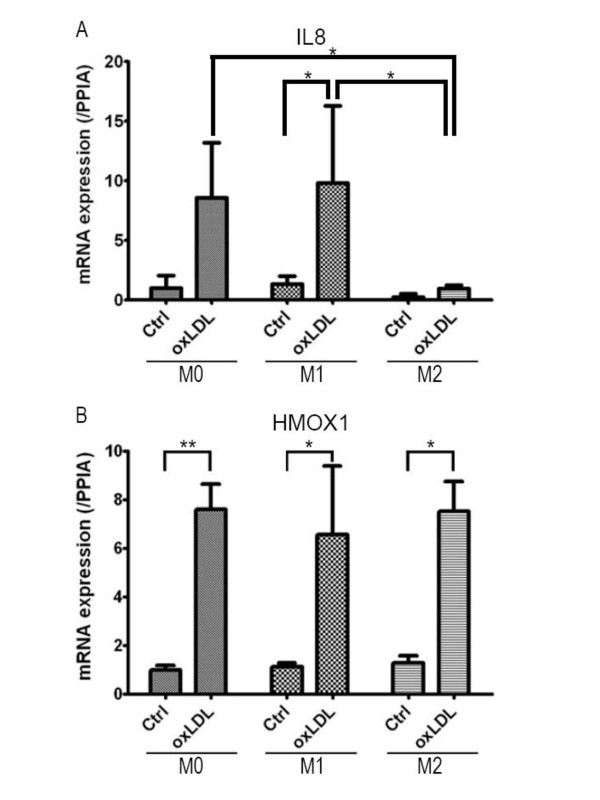
All data from cDNA microarray analysis in the present study were comparisons between cells with and without oxLDL treatment. To compare gene expression levels among subsets of macrophages, we performed quantitative real-time RT-PCR for two genes: IL8 listed in the top 30 genes that were commonly upregulated in all subsets of macrophages (Table 1) and HMOX1 encoding heme oxygenase (HO)-1 as a representative of genes containing an ARE.
Consistent with previous reports, the expression levels of IL8 were higher in non-stimulated M1 macrophages than in M2 [10], and oxLDL treatment induced higher levels of IL8 expression in M0 macrophages [13,14] (Figure 6A). Moreover, the expression level of IL8 was significantly upregulated by oxLDL treatment in M1 macrophages, whereas its expression level after oxLDL treatment in M2 was markedly lower than those in M0 and M1 macrophages (P < 0.05). It has been known for several decades that oxLDL treatment increases, while IL-4 treatment decreases IL-8 production in human monocyte-derived macrophages [13,15]. However, a recent report of microarray analysis indicated that oxLDL treatment induced no changes in human monocyte-derived macrophages [11]. This may have been due to various factors, such as individual variations or duration of oxLDL treatment [16].
Figure 6.
Quantitative real-time RT-PCR. A. IL8 mRNA expression levels. The expression level of IL8 was significantly upregulated by oxLDL treatment in M1 macrophages (P < 0.05). IL8 expression level after oxLDL treatment in M2 was markedly lower than those in M0 and M1 macrophages (P < 0.05). B. HMOX1 mRNA expression levels. OxLDL treatment significantly enhanced HMOX1 gene expression by 7.6-fold (**P < 0.0005), 5.8-fold (*P < 0.05), and 5.9-fold (*P < 0.05) in M0, M1, and M2 macrophages, respectively. HMOX1 gene expression levels were almost the same in all 3 subsets of macrophages even after oxLDL treatment. Each bar shows the mean ± SD of 3 experiments.
HO-1 is expressed in vascular endothelial cells and macrophages in the early stages of atherosclerotic lesions and in foam cells in the advanced stages, and is known for its antiinflammatory actions [17,18]. HMOX1 is known to be upregulated by oxidized phospholipids in murine macrophages polarized toward M1 or M2 [19]. Treatment with oxLDL yielded markedly higher levels of HMOX1 expression in all subsets of macrophages: i.e., 7.6-fold (P < 0.0005), 5.8-fold (P < 0.05), and 5.9-fold (P < 0.05) changes in M0, M1, and M2 macrophages compared to corresponding non-treated controls, respectively. HMOX1 gene expression levels were almost the same in all 3 subsets of macrophages even after oxLDL treatment (Figure 6B).
Discussion
In the present study, we demonstrated transcriptional alterations during oxLDL treatment, which has been suggested to be a model of the early stages of foam cell formation, in human polarized macrophages. Our study demonstrated that: 1) 93% of the top 30 genes upregulated by oxLDL treatment in M2 macrophages were also upregulated in M0; 2) the top 10 functional ontology categories in M2 macrophages were similar to those in M0; 3) almost all of the molecules in the highest scoring molecular network of M1 were related either directly or indirectly to TGF-β1; 4) there were commonly upregulated genes in all subset of macrophages, some of which contained ARE in their promoter regions; 5) hierarchical cluster analysis revealed a cluster specifically upregulated in M1, including genes encoding molecules related to the NF-κB signaling pathway; 6) in quantitative real-time RT-PCR, the level of IL8 gene expression after oxLDL treatment in M2 macrophages was markedly lower than those in M0 and M1 macrophages; and 7) HMOX1 gene expression levels were almost the same in all 3 subsets of macrophages even after oxLDL treatment.
The top genes expression of which was upregulated by oxLDL treatment in M2 but not M1 macrophages were highly correlated with the genes that were upregulated in M0 (Table 1). Moreover, the top altered ontology categories during oxLDL treatment in M2 macrophages were more similar to those of M0 than M1 (Figure 3). It has been reported that M-CSF-induced macrophages (M0 macrophages in the present study) have a similar transcriptional profile to M2 macrophages [10]. The transcriptional alteration during oxLDL treatment in M2 macrophages was also relatively similar to that in M0 macrophages but not M1 cells (Table 1 and Figure 3).
The data of hierarchical cluster analysis are shown in Figure 5. Commonly upregulated genes in all subsets of macrophages included some ARE-containing genes; e.g., TRIM16, HMOX1, TXNRD1, GCLM, and DUSP1. TRIM16, HMOX1, TXNRD1, GCLM, and DUSP1 encode tripartite motif-containing 16 (TRIM16), HO-1, thioredoxin reductase (Txnrd) 1, glutamate-cysteine ligase, modifier subunit (GCLM), and dual specificity phosphatase (DUSP) 1, respectively. These genes were upregulated during oxLDL treatment in all subsets of macrophages in the present study. ARE is a binding site for the transcription factor Nrf2 [20]. The Nrf2-ARE pathway plays a crucial role in protection against oxidative stress [21]. On exposure to oxidative stress, Nrf2 translocates to the nucleus, binds to the ARE [22], and activates the genes, including TRIM16, HMOX1, TXNRD1, GCLM, and DUSP1. These data were consistent with the recent report that oxidized phospholipids upregulated expression of ARE-containing genes in murine bone marrow-derived macrophages [19].
TGF-β has been suggested to have antiinflammatory properties [23], and it thought to be produced by alternatively activated macrophages [24]. Activation of M1 macrophages might be altered by M2-derived TGF-β. As TGF-β downregulates scavenger receptors, such as scavenger receptor type A (SR-A) I/II and CD36 [25], and upregulates ATP-binding cassette (ABC) transporters, ABCA1 and ABCG1 [26], TGF-β is also thought to have protective effects against the development of atherosclerosis. However, the contribution of TGF-β to the development of atherosclerosis is more complicated, taking account of clinical data. It is controversial whether TGF-β levels in blood from patients are positively or negatively correlated with cardiovascular disease [27]. In molecular network analysis, the molecules in the highest scoring network of M1 macrophages, but not M0 or M2, were related directly or indirectly to TGF-β1 (Figure 4). However, no molecules in the known TGF-β signal transduction pathway, including TGF-β receptors (TβRs) and SMADs, were altered by oxLDL treatment in this study. The results of cDNA microarray analysis (Figure 4) and real-time RT-PCR analysis (data not shown) indicated that oxLDL treatment slightly induced TGF-β1 gene expression in M1 macrophages. TGF-β generally plays an important role in maintaining normal vessel wall conditions, including the expression of contractile proteins in vascular smooth muscle cells (VSMCs) [28]. Under atherogenic conditions, however, TGF-β reduces extracellular matrix production from VSMCs and enhances leukocytes recruitment to atherosclerotic plaques, resulting in plaque rupture. Our results suggest that TGF-β-related molecules were affected by oxLDL stimulation, and that TGF-β promoted proinflammatory activities in M1 macrophages as in VSMCs. These findings suggest that oxLDL regulates the functions of M1 macrophages through an as yet unknown TGF-β-mediated cascade. It is therefore necessary to elucidate the detailed TGF-β-related functions regulated by oxLDL stimulation in various cells.
NF-κB is present in an inactive form bound to an inhibitor protein (I-κB) in the cytoplasm. On stimulation, NF-κB is released from I-κB, is translocated to the nucleus, and binds to the promoter DNA, followed by production of many types of inflammatory cytokine [29,30]. The NF-κB signaling pathway is known to be activated by oxLDL in a CD36-dependent manner [31]. Interestingly, cluster B included genes related to the NF-κB signaling pathway, such as NF-κB and PI3K (Figure 5 Table 3). The results of molecular network analysis indicated that oxLDL treatment induced upregulation of the growth factor receptor-mediated NF-κB signaling pathway in M1 but not M0 or M2 macrophages, while I-κB was upregulated in M0 and M2 but not M1 (Additional file 2). Thus, it seems that oxLDL stimulated the NF-κB signaling pathway specifically in M1.
There have been some reports partially conflicting with this study [10,11], probably due to differences in experimental conditions, such as oxLDL concentrations, TBARS levels, or duration of treatment. Further studies are required to determine whether M1 macrophages contribute to foam cell formation. In this study, we primarily measured mRNA levels, and all samples were obtained from healthy volunteers. Measurements of protein levels and data derived from atherosclerotic subjects should be included in the next study.
Conclusions
The present study demonstrated the effects of oxLDL on transcriptional alterations in polarized macrophages. The data suggested that oxLDL uptake may affect TGF-β1- and NF-κB-mediated functions of M1 macrophages, but not M0 or M2 macrophages. It is likely that M1 macrophages characteristically respond to oxLDL. Further studies are required to evaluate the roles of TGF-β1- and NF-κB-mediated macrophage functions in the early stages of foam cell formation.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KI, KS, and HD designed the study. KH participated at all stages and drafted the manuscript. CI conducted cDNA microarray analysis, HN performed flow cytometry analysis, and TK provided valuable help with cell cultures. KH, CI and HN analyzed the data together with KI, KS, and HD. KH, KI, and KS co-wrote the paper. All authors read and approved the final draft of the manuscript.
Supplementary Material
All transcriptional profile. After eliminated controls and backgrounds from all the probe sets on "3D-Gene" human oligo chip 25 k, the remained 24267 probe sets are listed. The values denote fold changes (log2 ratio) of normalized intensities during oxLDL treatment. Pink and blue cells indicate the genes showing a change in expression of >2-fold during oxLDL treatment as significantly up- or downregulated (log2 ratios were greater than +1 or less than -1). Genes intensities of which were less than 2-4 are shown as blank cells.
Molecular network focusing on the NF-κB signaling pathway. This network consisted of the known NF-κB signaling pathway. The intensity of the node color indicates the degree of upregulation (red) or downregulation (blue). Nodes are displayed using various shapes represent the functional class of the gene product. Lines are displayed with various labels that describe the nature of the relationship between the nodes: A for Activation, M for Biochemical Modification, P for Phosphorylation/Dephosphorylation, PP for Protein - Protein binding, PR for Protein - RNA binding, RB for Regulation of Binding, T for Transcription, and TR for Translocation. The growth factor receptor-mediated NF-κB signaling pathway was upregulated in M1, but not in M0 or M2 macrophages.
Contributor Information
Kuniaki Hirose, Email: khirose@juntendo.ac.jp.
Kazuhisa Iwabuchi, Email: iwabuchi@juntendo.ac.jp.
Kazunori Shimada, Email: shimakaz@juntendo.ac.jp.
Takashi Kiyanagi, Email: kiyanagi@juntendo.ac.jp.
Chihiro Iwahara, Email: iwahara@juntendo.ac.jp.
Hitoshi Nakayama, Email: nhitoshi@juntendo.ac.jp.
Hiroyuki Daida, Email: daida@juntendo.ac.jp.
Acknowledgements and Funding
This study was supported in part by the "High-Tech Research Center" Project for Private Universities: matching fund subsidy, by a Grant-in-Aide (S0991013) for the Foundation of Strategic Research Projects in Private Universities from Ministry of Education, Culture, Sport, Science, and Technology, Japan, and by the Mizutani Foundation for Glycoscience (to K. I.).
References
- Rosamond W, Flegal K, Furie K, Go A, Greenlund K, Haase N, Hailpern SM, Ho M, Howard V, Kissela B, Kittner S, Lloyd-Jones D, McDermott M, Meigs J, Moy C, Nichol G, O'Donnell C, Roger V, Sorlie P, Steinberger J, Thom T, Wilson M, Hong Y. Heart disease and stroke statistics--2008 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2008;117:e25–146. doi: 10.1161/CIRCULATIONAHA.107.187998. [DOI] [PubMed] [Google Scholar]
- Rocha VZ, Libby P. Obesity, inflammation, and atherosclerosis. Nat Rev Cardiol. 2009;6:399–409. doi: 10.1038/nrcardio.2009.55. [DOI] [PubMed] [Google Scholar]
- Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- Yla-Herttuala S, Lipton BA, Rosenfeld ME, Sarkioja T, Yoshimura T, Leonard EJ, Witztum JL, Steinberg D. Expression of monocyte chemoattractant protein 1 in macrophage-rich areas of human and rabbit atherosclerotic lesions. Proc Natl Acad Sci USA. 1991;88:5252–5256. doi: 10.1073/pnas.88.12.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Tabas I, Winchester R, Ravalli S, Rabbani LE, Tall A. Interleukin 8 is induced by cholesterol loading of macrophages and expressed by macrophage foam cells in human atheroma. J Biol Chem. 1996;271:8837–8842. doi: 10.1074/jbc.271.15.8837. [DOI] [PubMed] [Google Scholar]
- Ardans JA, Economou AP, Martinson JM, Zhou M, Wahl LM. Oxidized low-density and high-density lipoproteins regulate the production of matrix metalloproteinase-1 and -9 by activated monocytes. J Leukoc Biol. 2002;71:1012–1018. [PubMed] [Google Scholar]
- Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- Bouhlel MA, Derudas B, Rigamonti E, Dievart R, Brozek J, Haulon S, Zawadzki C, Jude B, Torpier G, Marx N, Staels B, Chinetti-Gbaguidi G. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Me tab. 2007;6:137–143. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- Cho HJ, Shashkin P, Gleissner CA, Dunson D, Jain N, Lee JK, Miller Y, Ley K. Induction of dendritic cell-like phenotype in macrophages during foam cell formation. Physiol Genomics. 2007;29:149–160. doi: 10.1152/physiolgenomics.00051.2006. [DOI] [PubMed] [Google Scholar]
- Iwabuchi K, Nagaoka I. Lactosylceramide-enriched glycosphingolipid signaling domain mediates superoxide generation from human neutrophils. Blood. 2002;100:1454–1464. [PubMed] [Google Scholar]
- Terkeltaub R, Banka CL, Solan J, Santoro D, Brand K, Curtiss LK. Oxidized LDL induces monocytic cell expression of interleukin-8, a chemokine with T-lymphocyte chemotactic activity. Arterioscler Thromb. 1994;14:47–53. doi: 10.1161/01.atv.14.1.47. [DOI] [PubMed] [Google Scholar]
- Kiyanagi T, Iwabuchi K, Shimada K, Hirose K, Miyazaki T, Sumiyoshi K, Iwahara C, Nakayama H, Masuda H, Mokuno H, Sato S, Daida H. Involvement of Cholesterol-enriched Microdomains in Class A Scavenger Receptor-mediated Responses in Human Macrophages. Atherosclerosis. in press . [DOI] [PubMed]
- Standiford TJ, Strieter RM, Chensue SW, Westwick J, Kasahara K, Kunkel SL. IL-4 inhibits the expression of IL-8 from stimulated human monocytes. J Immunol. 1990;145:1435–1439. [PubMed] [Google Scholar]
- Martin-Fuentes P, Civeira F, Recalde D, Garcia-Otin AL, Jarauta E, Marzo I, Cenarro A. Individual variation of scavenger receptor expression in human macrophages with oxidized low-density lipoprotein is associated with a differential inflammatory response. J Immunol. 2007;179:3242–3248. doi: 10.4049/jimmunol.179.5.3242. [DOI] [PubMed] [Google Scholar]
- Otterbein LE, Bach FH, Alam J, Soares M, Tao Lu H, Wysk M, Davis RJ, Flavell RA, Choi AM. Carbon monoxide has anti-inflammatory effects involving the mitogen-activated protein kinase pathway. Nat Med. 2000;6:422–428. doi: 10.1038/74680. [DOI] [PubMed] [Google Scholar]
- Morita T. Heme oxygenase and atherosclerosis. Arterioscler Thromb Vasc Biol. 2005;25:1786–1795. doi: 10.1161/01.ATV.0000178169.95781.49. [DOI] [PubMed] [Google Scholar]
- Kadl A, Meher AK, Sharma PR, Lee MY, Doran AC, Johnstone SR, Elliott MR, Gruber F, Han J, Chen W, Kensler T, Ravichandran KS, Isakson BE, Wamhoff BR, Leitinger N. Identification of a novel macrophage phenotype that develops in response to atherogenic phospholipids via Nrf2. Circ Res. 2010;107:737–746. doi: 10.1161/CIRCRESAHA.109.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Ohta T, Yamamoto M. Unique function of the Nrf2-Keap1 pathway in the inducible expression of antioxidant and detoxifying enzymes. Methods Enzymol. 2004;378:273–286. doi: 10.1016/S0076-6879(04)78021-0. full_text. [DOI] [PubMed] [Google Scholar]
- Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, Annunziata N, Doetschman T. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CG, Homer RJ, Zhu Z, Lanone S, Wang X, Koteliansky V, Shipley JM, Gotwals P, Noble P, Chen Q, Senior RM, Elias JA. Interleukin-13 induces tissue fibrosis by selectively stimulating and activating transforming growth factor beta(1) J Exp Med. 2001;194:809–821. doi: 10.1084/jem.194.6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draude G, Lorenz RL. TGF-beta1 downregulates CD36 and scavenger receptor A but upregulates LOX-1 in human macrophages. Am J Physiol Heart Circ Physiol. 2000;278:H1042–1048. doi: 10.1152/ajpheart.2000.278.4.H1042. [DOI] [PubMed] [Google Scholar]
- Argmann CA, Van Den Diepstraten CH, Sawyez CG, Edwards JY, Hegele RA, Wolfe BM, Huff MW. Transforming growth factor-beta1 inhibits macrophage cholesteryl ester accumulation induced by native and oxidized VLDL remnants. Arterioscler Thromb Vasc Biol. 2001;21:2011–2018. doi: 10.1161/hq1201.099426. [DOI] [PubMed] [Google Scholar]
- Grainger DJ. TGF-beta and atherosclerosis in man. Cardiovasc Res. 2007;74:213–222. doi: 10.1016/j.cardiores.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Grainger DJ. Transforming growth factor beta and atherosclerosis: so far, so good for the protective cytokine hypothesis. Arterioscler Thromb Vasc Biol. 2004;24:399–404. doi: 10.1161/01.ATV.0000114567.76772.33. [DOI] [PubMed] [Google Scholar]
- Baeuerle PA, Henkel T. Function and activation of NF-kappa B in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- Lenardo MJ, Baltimore D. NF-kappa B: a pleiotropic mediator of inducible and tissue-specific gene control. Cell. 1989;58:227–229. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- Janabi M, Yamashita S, Hirano K, Sakai N, Hiraoka H, Matsumoto K, Zhang Z, Nozaki S, Matsuzawa Y. Oxidized LDL-induced NF-kappa B activation and subsequent expression of proinflammatory genes are defective in monocyte-derived macrophages from CD36-deficient patients. Arterioscler Thromb Vasc Biol. 2000;20:1953–1960. doi: 10.1161/01.atv.20.8.1953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All transcriptional profile. After eliminated controls and backgrounds from all the probe sets on "3D-Gene" human oligo chip 25 k, the remained 24267 probe sets are listed. The values denote fold changes (log2 ratio) of normalized intensities during oxLDL treatment. Pink and blue cells indicate the genes showing a change in expression of >2-fold during oxLDL treatment as significantly up- or downregulated (log2 ratios were greater than +1 or less than -1). Genes intensities of which were less than 2-4 are shown as blank cells.
Molecular network focusing on the NF-κB signaling pathway. This network consisted of the known NF-κB signaling pathway. The intensity of the node color indicates the degree of upregulation (red) or downregulation (blue). Nodes are displayed using various shapes represent the functional class of the gene product. Lines are displayed with various labels that describe the nature of the relationship between the nodes: A for Activation, M for Biochemical Modification, P for Phosphorylation/Dephosphorylation, PP for Protein - Protein binding, PR for Protein - RNA binding, RB for Regulation of Binding, T for Transcription, and TR for Translocation. The growth factor receptor-mediated NF-κB signaling pathway was upregulated in M1, but not in M0 or M2 macrophages.