Abstract
Background
Although much is known about molecular and chromosomal characteristics that distinguish glioma histological subtypes, DNA methylation patterns of gliomas and their association with other tumor features such as mutation of isocitrate dehydrogenase (IDH) genes have only recently begun to be investigated.
Methods
DNA methylation of glioblastomas, astrocytomas, oligodendrogliomas, oligoastrocytomas, ependymomas, and pilocytic astrocytomas (n = 131) from the Brain Tumor Research Center at the University of California San Francisco, as well as nontumor brain tissues (n = 7), was assessed with the Illumina GoldenGate methylation array. Methylation data were subjected to recursively partitioned mixture modeling (RPMM) to derive methylation classes. Differential DNA methylation between tumor and nontumor was also assessed. The association between methylation class and IDH mutation (IDH1 and IDH2) was tested using univariate and multivariable analysis for tumors (n = 95) with available substrate for sequencing. Survival of glioma patients carrying mutant IDH (n = 57) was compared with patients carrying wild-type IDH (n = 38) using a multivariable Cox proportional hazards model and Kaplan–Meier analysis. All statistical tests were two-sided.
Results
We observed a statistically significant association between RPMM methylation class and glioma histological subtype (P < 2.2 × 10−16). Compared with nontumor brain tissues, across glioma tumor histological subtypes, the differential methylation ratios of CpG loci were statistically significantly different (permutation P < .0001). Methylation class was strongly associated with IDH mutation in gliomas (P = 3.0 × 10−16). Compared with glioma patients whose tumors harbored wild-type IDH, patients whose tumors harbored mutant IDH showed statistically significantly improved survival (hazard ratio of death = 0.27, 95% confidence interval = 0.10 to 0.72).
Conclusion
The homogeneity of methylation classes for gliomas with IDH mutation, despite their histological diversity, suggests that IDH mutation is associated with a distinct DNA methylation phenotype and an altered metabolic profile in glioma.
CONTEXT AND CAVEATS
Prior knowledge
Human gliomas often have mutations in the isocitrate dehydrogenase genes (IDH1 and IDH2). IDH mutation is associated with improved survival in glioma patients. Epigenetic alterations like DNA methylation at CpG dinucleotides play an important role in gene regulation. Integration of genetic and epigenetic data is important for a better understanding of glioma development.
Study design
DNA methylation profile of CpG loci and methylation class of 131 glioma and seven non-glioma brain tissues were determined. The association between IDH mutation and methylation class was analyzed. Survival analysis of patients carrying IDH mutation vs wild-type IDH was also performed.
Contribution
CpG loci showed differential methylation between glioma and non-glioma tissues. Statistically significant associations were found between DNA methylation class and histological subtypes and between DNA methylation class and IDH mutation of gliomas. Patients carrying IDH mutation in gliomas showed improved survival compared with patients carrying IDH wild-type after adjustment for age and grade-specific tumor histology.
Implications
A distinct methylation pattern in glioma tissues is associated with IDH mutation.
Limitations
Mutation data were not available for all tissue samples, which may have limited the statistical power of the analyses.
From the Editors
Malignant glioma is the most common form of primary malignant brain tumor and the glioma histological subtypes include glioblastomas, grades 2 and 3 astrocytomas, grades 2 and 3 oligodendrogliomas, grades 2 and 3 oligoastrocytomas, ependymomas, and pilocytic astrocytomas (1). Presently, there are limited treatment options for glioma; glioblastoma, the most common glioma subtype, remains an incurable disease with a median survival of 15 months, even with radiation and temozolomide therapy (2).
A comprehensive appreciation of the integrated genomics and epigenomics of glioma is needed to better understand the multiple cellular pathways involved in their development, establish markers of resistance to traditional therapies, and contribute to the development of targeted therapies. Epigenetic alterations can alter gene expression, gene expression potential, or the regulation of gene function, and thereby contribute to gliomagenesis. Arguably, the most widely studied epigenetic mark is DNA methylation that occurs at cytosine residues in the context of CpG dinucleotides. Approximately half of human genes have concentrations of CpGs in their promoter regions and the methylation state of these and other gene-associated CpGs are widely regarded as critical indicators of gene regulation.
Since 2008, sequencing of gliomas has identified mutations in the isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) genes (3–5). The IDH1 and IDH2 enzymes convert isocitrate to alpha (α)-ketoglutarate producing NADPH and participate in cellular metabolic processes such as glucose sensing, lipid metabolism, and oxidative respiration [reviewed in (6)]. Mutations in IDH1 are consistently found in codon 132 for arginine (R132), and mutations in IDH2 consistently occur at the analogous amino acid R172 (3,7). Mutations in IDH1 and IDH2 (IDH when referring to both) are unlike most cancer-associated enzyme mutations because they confer neomorphic enzyme activity rather than inactivating, or constitutively activating, the enzyme. The mutant form of IDH enzymes convert α-ketoglutarate to 2-hydroxyglutarate in an NADPH-dependent manner, and via an unknown mechanism contribute to the pathophysiology of gliomas and leukemias (5,7,8). IDH mutations occur in approximately 80% of grades 2–3 gliomas and secondary glioblastomas, but less than 10% of primary glioblastomas (4,5). In gliomas, IDH mutation has been associated with genetic alterations in other genes including tumor suppressors and oncogenes (5). IDH mutation also has been associated with younger age and improved survival in glioma patients (5,9).
The somatic genetic signature of any individual tumor is critical to assessing its clinical and etiologic character. Similarly, the profile of somatic epigenetic alterations is central to forming a complete understanding of the pattern of disrupted cellular functioning responsible for the deadly behavior of gliomas. Major advances in the clinical role of epigenetics in gliomas include the findings that promoter methylation silencing of the O-6-methylguanine-DNA methyltransferase (MGMT) gene is associated with response to temozolomide treatment (10). Epigenetic silencing of MGMT gene is found in approximately 80% of gliomas with mutant IDH1 compared with approximately 60% of gliomas with wild-type IDH1 (9). Other common alterations in gliomas are mutations in tumor protein p53 (TP53) (11) and amplification of the epidermal growth factor receptor (EGFR) oncogene (12). Better definitions of the somatic nature of gliomas should integrate both their genetic and epigenetic alterations. In this study, we assessed CpG methylation patterns, IDH mutation, TP53 mutation, and EGFR amplification in histologically diverse gliomas to define epigenetic subgroups of potential clinical and etiologic relevance.
Patients, Materials, and Methods
Patients and Tissue Samples
Fresh frozen tumor tissues of patients (n = 131) diagnosed with glioma between 1990 and 2003 were obtained from the University of California San Francisco (UCSF) Brain Tumor Research Center Tissue Bank. Tumors were previously reviewed by UCSF neuropathologists to assign histological subtypes and grades according to the World Health Organization classification for patients operated on at the UCSF Medical Center (1). Tumor samples were defined as secondary glioblastoma if the patients had previous histological diagnosis of a lower-grade glioma. Nontumor brain tissue samples were obtained from cancer-free patients (n = 7) who underwent temporal lobe resection for treatment of epilepsy at the UCSF Medical Center. Patient ages were documented at the time of initial diagnosis. Other demographic and survival data were obtained from UCSF patient records and the California Cancer Registry. The Institutional Review Board approval certification was obtained from the UCSF Committee on Human Research, and subjects provided written informed consent for tissue collection.
Cell Lines, Cell Culture, and Reagents
A431 cells (a human epidermoid cancer cell line that is known to have EGFR amplification and overexpression) and HT29 cells (a human colon adenocarcinoma cell line without EGFR amplification) were obtained from American Type Culture Collection (Manassas, VA). Cell lines were maintained in Dulbecco’s modified Eagle medium and RPMI 1640 medium (both from Invitrogen, Carlsbad, CA), respectively, with 10% fetal bovine serum (Hyclone, Logan, UT) at 37°C in 5% CO2. When cultures reached 80% confluency, cells were harvested for DNA extraction.
DNA Extraction, Bisulfite Modification, and Methylation Analysis
Genomic DNA from 131 glioma tissue samples and seven nontumor brain tissue samples was isolated from approximately 25 mg wet weight of each frozen tissue sample using QIAamp DNA mini kit (Qiagen, Inc, Valencia, CA) according to the manufacturer’s instructions. DNA was eluted twice in a total of 100 μL of elution buffer. The same DNA extraction method was applied to A431 and HT29 cell lines that served as EGFR amplification controls.
For DNA methylation analysis, 1 μg of genomic DNA was first subjected to bisulfite modification using the EZ DNA Methylation Kit (Zymo Research Corporation, Orange, CA) according to the manufacturer’s instructions. Bisulfite modification converts unmethylated cytosine residues to uracil and preserves methylated cytosine residues as cytosines.
GoldenGate DNA methylation bead arrays (Illumina, Inc, San Diego, CA) were used to interrogate methylation of 1505 CpG loci associated with 803 cancer-related genes according to the manufacturer’s instructions. GoldenGate methylation arrays were used to analyze bisulfite-modified DNA from 131 glioma and seven nontumor samples for methylation and processed at the UCSF Institute for Human Genetics, Genomics Core Facility. The GoldenGate array methylation data were deposited in the Gene Expression Omnibus and are publicly available (accession GSE20395). The Cancer Genome Atlas (TCGA), a public data portal, was used to obtain GoldenGate methylation array data for validation of methylation classes. Quantitative methylation-specific polymerase chain reaction (PCR) (QMSP) was used to confirm methylation data from the GoldenGate array. Candidate genes were selected based on previous studies (13–16) that reported aberrant methylation in astrocytic glioma and included MGMT, Ras association domain family member 1 (RASSF1), PYD and CARD domain containing (PYCARD), homeobox A9 (HOXA9), paternally expressed 3 (PEG3), and slit homolog 2 (SLIT2). CpGenome Universal Methylated DNA (Millipore, Billerica, MA) was bisulfite modified and used as a positive control for QMSP analysis. QMSP was performed using Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA). The reaction plate was prepared using the Beckman Coulter automated liquid handler-Biomex 3000 (Beckman Coulter, Fullerton, CA). Each reaction contained 10.0 μL 2× Power SYBR Green PCR Master Mix (Applied Biosystems), 100–400 nM of forward and reverse primers (Supplementary Table 1, available online) and 25 ng of DNA template in a total reaction volume of 20 μL. For the amplification of RASSF1, 2%–3% dimethyl sulfoxide was added to the reaction mix. PCR conditions are modified by different primer concentrations, and dimethyl sulfoxide was added to ensure that primer dimers and nonspecific amplification products were not included in the threshold cycle (Ct) calculation. To confirm specificity of amplicons from QMSP, we performed dissociation curve analysis. The PCR conditions were 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. SYBR Green (the commonly used DNA binding dye) fluorescence data were collected only during the extension reaction at 72°C. Ct values were calculated by the 7900HT system software, and average relative quantification (RQ) values were obtained for each sample using actin, beta (ACTB) amplification as the referent, where RQ = (target gene/ACTB)/(Universal methylation calibrator/ACTB). Spearman rank correlation coefficients (rho) and P values were calculated to assess the correlation between GoldenGate array data and QMSP results.
Mutation Analysis
IDH Mutation.
The region spanning R132 codon of IDH1 and the region spanning R172 codon of IDH2 were amplified by PCR with primers designed with Primer 3 software (v.0.4.0) with the exception of the forward sequencing primer, which was selected from Balss et al. (4). PCR reaction mixtures contained 10–25 ng DNA, 1× buffer, 0.2 mM dNTP mix, 0.2 μM forward and reverse primers, 0.04 U of HotStarTaq, and 1 mM MgCl2 (Qiagen, Inc) in a 25 μL volume. The PCR conditions were 95°C for 10 minutes, 40 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute. The resulting products were analyzed on a 1.5% agarose gel. DNA was purified using the QIAquick PCR Purification Kit (Qiagen, Inc) and sent to Rhode Island Genomics and Sequencing Center at the University of Rhode Island, where it was sequenced in both directions using the BigDyeTerminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Sequences were analyzed with Applied Biosystems Sequence Scanner Software v1.0. All primers for IDH1 and IDH2 mutation analysis are listed in Supplementary Table 1 (available online).
TP53 Mutation.
For TP53 mutation analysis, PCR–single-strand conformation polymorphism technique was used, and DNA sequencing was done as previously described (8). Primers for PCR amplification of fragments of exons 5–8 of TP53 are listed in Supplementary Table 1 (available online). PCR reaction mixtures contained 50 ng DNA, 20 μmol/L dNTP, 10 mmol/L Tris–HCl (pH 9.0), 1.5 mmol/L MgCl2, 0.1% Triton X-100, 10 pmol of forward and reverse primers, 1 U Taq (Perkin-Elmer Cetus, Norwalk, CT), and 0.2 μCi [33P]dCTP (DuPont New England Nuclear, Boston, MA) in a 30 μL volume. DNA with TP53 mutation confirmed by sequencing was included as positive control. The PCR reaction was carried out using 35 cycles 94°C for 30 seconds, annealed for 30 seconds at 58°C for exons 5 and 8, and 60°C for exons 6 and 7 (primers listed in Supplementary Table 1, available online) and 72°C for 1 minute. Three microliters of PCR product was mixed with 2 μL of 0.1 N NaOH and then mixed with 5 μL of gel loading buffer solution (United States Biochemical Corp, Cleveland, OH) and heated at 94°C for 4 minutes. DNA was analyzed on 6% nondenatured polyacrylamide gel, supplemented with 10% glycerol. Electrophoresis was performed at room temperature for 20 hours and exposed to autoradiography films for 16 hours for detection of bands. Direct sequencing of PCR fragments for both DNA strands was done on all tumor DNAs that showed aberrant migration patterns on single-strand conformation polymorphism gel to determine the corresponding DNA sequences using dsDNA cycle sequencing system (Life Technologies, Gaithersburg, MD), as described in Wiencke et al. (17).
EGFR Amplification.
EGFR amplification was measured by a quantitative PCR method using the ABI 7900 Real-Time PCR system (Applied Biosystems) and SYBR Green I, which has been shown to be equivalent to TaqMan PCR assay for the assessment of gene copy number (18). Quality control measures for the real-time SYBR green assay included running both EGFR and control gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in triplicate. DNA from A431 and HT29 cell lines, with known copy number states for EGFR, served as positive and negative controls, respectively, for amplification.
Statistical Analysis
Data Assembly.
Methylation data were assembled with BeadStudio methylation software from Illumina. All GoldenGate methylation array data points are represented by fluorescent signals (Cy dyes) from both methylated (Cy5) and unmethylated (Cy3) alleles. The methylation level, designated as beta (β), is calculated as β = (max[Cy5, 0])/(|Cy3| + |Cy5| + 100), in which the average β value is derived from the approximately 30 replicate methylation measurements because each CpG probe set is present on the array and measured in each sample approximately 30 times. Raw average β values were analyzed without normalization as recommended by Illumina. At each CpG locus, for each tissue DNA sample, the detection P value was used to determine sample performance; all samples had detection P values less than 1 × 10−5 at more than 75% of CpG loci and passed performance criteria. There were eight CpG loci that had a median detection P value of greater than .05, and these eight CpGs were excluded from the analysis. All CpG loci on the X chromosome were excluded from analysis. The final dataset contained 1413 autosomal CpG loci associated with 773 genes. For each CpG locus, the differential methylation values (delta–beta [Δβ]) were calculated by subtracting the average β value of tumors from the mean β value of the seven nontumor brain samples. Subsequent analyses were carried out using the R software (19). All statistical tests were two-sided.
Unsupervised Clustering, Recursively Partitioned Mixture Modeling (RPMM), and Survival.
Hierarchical clustering of the DNA methylation data was performed using the R function hclust with Euclidean distance metric and Ward linkage. To discern and describe the relationships between CpG methylation data and patient and tumor covariates, a modified model-based form of unsupervised clustering known as RPMM was used as described in Houseman et al. (20) and as used in Christensen et al. (21). The analysis of associations between methylation class (categorical) and individual categorical covariates was performed using the Fisher exact test. To test for association between methylation class and continuous covariates, a permutation test was run with the Kruskal–Wallis test statistic, and a likelihood ratio test was used for comparing the association between methylation class and IDH mutation to a model including age and grade-specific histology. To test for associations between IDH mutation and grade-specific tumor histology, and IDH mutation and tumor grade, Fisher exact tests were used. To test for associations between IDH mutation and primary vs secondary glioblastoma, IDH mutation and TP53 mutation, and IDH mutation and EGFR amplification, χ2 tests were used. The assumption of proportionality for Cox proportional hazards modeling was verified by calculating Pearson correlation coefficients for the corresponding set of Schoenfeld residuals with a transformation of time based on the Kaplan–Meier estimate of the survival function (22) and graphically by plotting log(survival time) vs log(−log[survival as a function of time, t]).
Locus-by-Locus Analysis.
To examine differential methylation between tumor and nontumor tissues, gliomas were stratified by grade-specific histological subtypes, and individual CpG loci were compared between subtypes of glioma and nontumor samples using a Wilcoxon rank-sum test. Because this results in the simultaneous comparison of all CpG loci between glioma subtypes and nontumor sample types, false discovery rate estimation and Q values computed by the qvalue package in R (23) were used to adjust for multiple testing. Differentially methylated CpGs were counted as hyper- or hypomethylated if both the tumor vs nontumor Q less than .05 and the median methylation value |Δβ| greater than 0.2. An equivalent approach was used in the analysis of differential methylation for gliomas with mutant or wild-type IDH compared with nontumor tissues.
Pathway Analysis.
A canonical pathway analysis was conducted with the use of Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA). CpG gene-loci associated with the Illumina GoldenGate methylation array were used as reference, and loci from differential methylation analysis, as described later in the article, were investigated for pathways enrichment. The statistical significance of gene-locus enrichment within canonical pathways was measured with a Fisher exact test.
Results
Unsupervised Clustering and Modeling of Glioma and Nontumor DNA Methylation Data
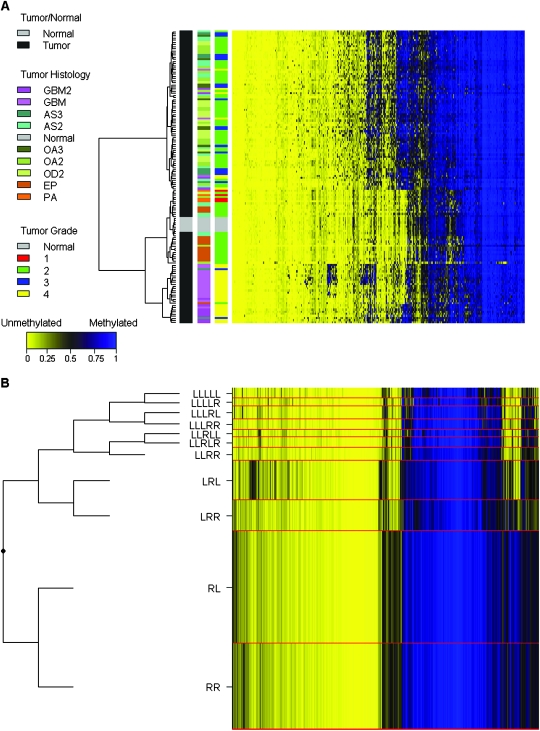
Histological grade and patient demographic data for the 131 gliomas and patient demographic data for the seven nontumor brain tissues are presented in Table 1. To characterize DNA methylation of gliomas and nontumor brain tissues, the bisulfite-modified DNA samples were hybridized to the GoldenGate DNA methylation array. Unsupervised clustering of DNA methylation data from 1413 autosomal CpG loci showed that nontumor brain tissues cluster with each other and are distinct from tumor tissues (Figure 1, A). Furthermore, we observed that oligodendrogliomas and astrocytomas generally clustered together and demonstrated a greater number of methylated loci relative to ependymomas, pilocytic astrocytomas, as well as nontumor brain tissues. Concomitantly, glioblastomas (also known as grade 4 astrocytoma), predominantly clustered together at the bottom of the heatmap (Figure 1, A) and displayed more hypermethylated CpG loci than ependymomas.
Table 1.
Patient demographic and tumor characteristics*
| Characteristic | Nontumor brain tissue (n = 7) | Tumor histology and grade of glioma tissues (n = 131) |
||||||||
| Primary glioblastoma (n = 20) | Secondary glioblastoma (n = 12) | Grade 3 astrocytoma (n = 9) | Grade 2 astrocytoma (n = 20) | Grade 3 oligoastrocytoma (n = 9) | Grade 2 oligoastrocytoma (n = 22) | Grade 2 oligodendroglioma (n = 20) | Ependymoma (n = 15) | Pilocytic astrocytoma (n = 4) | ||
| Age at diagnosis, y | ||||||||||
| Median | 33 | 55 | 34.5 | 40 | 40 | 40 | 33 | 35.5 | 41 | 28.5 |
| Range | 23–42 | 21–78 | 18–49 | 23–57 | 21–64 | 26–52 | 19–48 | 20–59 | 19–70 | 22–39 |
| Sex, No. (%) | ||||||||||
| Female | 3 (43) | 7 (35) | 4 (33) | 6 (67) | 10 (50) | 4 (44) | 9 (41) | 10 (50) | 4 (27) | 2 (50) |
| Male | 4 (57) | 13 (65) | 8 (67) | 3 (33) | 10 (50) | 5 (56) | 13 (59) | 10 (50) | 11 (73) | 2 (50) |
| Race, No. (%) | ||||||||||
| White | — | 18 (90) | 11 (92) | 5 (56) | 17 (85) | 7 (78) | 16 (73) | 18 (90) | 12 (80) | 4 (100) |
| Hispanic | — | 1 (5) | 1 (8) | 2 (22) | 1 (5) | 0 | 1 (4) | 1 (5) | 2 (13) | 0 |
| American Indian | — | 0 | 0 | 0 | 1 (5) | 0 | 0 | 0 | 0 | 0 |
| Asian | — | 0 | 0 | 1 (11) | 1 (5) | 0 | 2 (9) | 1 (5) | 0 | 0 |
| Unknown | 7 (100) | 1 (5) | 0 | 1 (11) | 0 | 2 (22) | 3 (14) | 0 | 1 (7) | 0 |
| Survival, d | ||||||||||
| Median | NA | 759 | 1244 | 1933 | 1584 | 3007 | 2937 | 2532 | 2498 | 2789 |
| Range | NA | 108–2477 | 466–4973 | 516–4494 | 305–4043 | 603–6459 | 612–5843 | 4–5988 | 478–5983 | 948–3279 |
Nontumor brain tissues (n = 7) were obtained from cancer-free patients who underwent temporal lobe resection for treatment of epilepsy at the University of California San Francisco Medical Center. Glioma tissues (n = 131) were obtained between 1990 and 2003 from the University of California San Francisco Brain Tumor Research Center Tissue Bank.
Figure 1.
Association between glioma histological subtypes and DNA methylation pattern. A) The average methylation beta (β) values of both gliomas (n = 131) and nontumor tissue samples (n = 7) were subjected to unsupervised hierarchical clustering based on Euclidean distance metric and Ward linkage and are shown in the heatmap. Each row represents a sample and each column represents a CpG locus (all 1413 autosomal loci). The scale bar at the bottom shows the range of β values (0–1). Tissue histology and grade are defined in color keys next to the heatmap, on the left. GBM2 = secondary glioblastoma multiforme; GBM = primary glioblastoma multiforme; AS3 = grade 3 astrocytoma; AS2 = grade 2 astrocytoma; OA3 = grade 3 oligoastrocytoma; OA2 = grade 2 oligoastrocytoma; OD2 = grade 2 oligodendroglioma; EP = ependymoma; PA = pilocytic astrocytoma. B) Recursively partitioned mixture model (RPMM) of glioma and nontumor brain tissue samples (n = 138). Methylation profile classes are stacked in rows separated by red lines and class height corresponds to the number of samples in each class. Class methylation at each CpG locus (columns) is the mean methylation for all samples in a class. To the left of the RPMM is the clustering dendrogram. In the heatmap and RPMM, blue designates methylated CpG loci (average β = 1), and yellow designates unmethylated CpG loci (average β = 0).
To further investigate the DNA methylation patterns of gliomas and nontumor brain tissue, we implemented an agnostic approach by applying a modified model-based form of unsupervised clustering known as RPMM (20). RPMM allows for precise inference regarding the potential covariates associated with intrinsic similarities and differences in CpG methylation by generating distinct classes of DNA methylation for the modeled samples based on the DNA methylation array data. We applied RPMM clustering to all 131 tumors, which generated 11 methylation classes (Figure 1, B). Methylation classes contain samples with DNA methylation patterns that are most similar to each other, and samples with different DNA methylation patterns are distinguished by their membership in a different methylation class. Methylation class was statistically significantly associated with both tumor histological subtype (P < 2.2 × 10−16) and grade (P < 2.2 × 10−16) (Supplementary Table 2, available online).
Methylation Array and Methylation Class Validation
Methylation data from GoldenGate arrays have been extensively validated by our group and others using a variety of methods (24–28). The methylation array data presented in this study were validated by correlating CpG methylation array data to QMSP data for genes commonly methylated in gliomas—MGMT, RASSF1, PYCARD, HOXA9, PEG3, and SLIT2 (Supplementary Table 3, available online). To determine the validity of association between histology and methylation class, we utilized publicly available GoldenGate methylation array data for 71 glioblastoma samples from TCGA. Using the RPMM classification (Figure 1, B), we predicted the methylation class for each glioblastoma sample of TCGA and confirmed that 70 (99%) of the 71 TCGA glioblastoma samples were classified in RPMM methylation classes that contained glioblastoma samples (Supplementary Table 2, available online). The identification numbers and the predicted RPMM methylation classes of TCGA tumors are listed in Supplementary Table 4 (available online).
Ratios of Hypermethylated to Hypomethylated CpG Loci and Tumor Histology
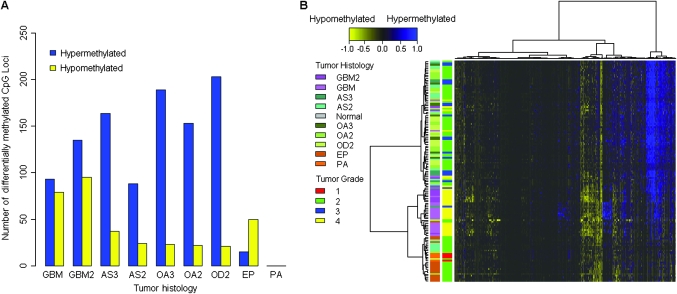
We examined the differential methylation (Δβ) between tumor and nontumor brain tissues and observed a striking pattern of the number of hyper- and hypomethylated CpG loci among different tumor subtypes (Figure 2, A). Glioblastomas showed a low ratio of hyper- to hypomethylated loci (ratio = 1.3) compared with the ratio for grades 2 and 3 astrocytomas, grades 2 and 3 oligoastrocytomas, and grade 2 oligodendrogliomas (ratios = 3.7, 7.6, and 9.7, respectively). Conversely, ependymomas showed increased hypomethylation (ratio = 0.3). The ratios of hyper- to hypomethylated CpG loci were statistically significantly different across glioma tumor histological subtypes (permutation P < .0001). Histology-related hyper- and hypomethylation patterns were also evident in unsupervised hierarchical clustering of Δβ methylation values for all 1413 autosomal CpG loci (Figure 2, B).
Figure 2.
Differential methylation and the ratio of hyper- to hypomethylated loci in gliomas. Differential methylation values (Δβ) were calculated by subtracting tumor average β value from the mean β value of the nontumor brain samples (n = 7) for each CpG locus. A) The number of statistically significantly differentially hyper- and hypomethylated loci (Q < .05 and |Δβ| > 0.2) are plotted by grade-specific glioma histology. GBM2 = secondary glioblastoma multiforme; GBM = primary glioblastoma multiforme; AS3 = grade 3 astrocytoma; AS2 = grade 2 astrocytoma; OA3 = grade 3 oligoastrocytoma; OA2 = grade 2 oligoastrocytoma; OD2 = grade 2 oligodendroglioma; EP = ependymoma; PA = pilocytic astrocytoma. B) Δβ values for all tumors (n = 131) were subjected to unsupervised hierarchical clustering based on Euclidean distance metric and Ward linkage. Each row represents a sample and each column represents a CpG locus (all 1413 autosomal loci). The scale bar at the top shows the range of Δβ values (−1 to 1). Tissue histology and grade are defined in color keys next to the heatmap on the left. In the heatmap, blue designates differentially hypermethylated CpG loci in tumors (Δβ = 1), and yellow designates differentially hypomethylated CpG loci in tumors (Δβ = −1).
We next assessed the cellular pathways associated with statistically significantly differentially hypomethylated and (separately) hypermethylated CpG loci that were common among glioblastomas, astrocytomas, oligoastrocytomas, and oligodendrogliomas. There were 18 CpG loci with statistically significant differential hypomethylation (Q < .05) and common among glioblastomas, astrocytomas, oligoastrocytomas, and oligodendrogliomas. An analysis of cellular pathways enriched among these 18 CpG loci, compared with all genes represented on the methylation array, revealed statistically significant enrichment of metabolism and biosynthesis pathways (Supplementary Table 5, available online). In addition, there were 35 statistically significantly differentially hypermethylated (Q < .05) CpG loci common among glioblastomas, astrocytomas, oligoastrocytomas, and oligodendrogliomas. An analysis of cellular pathways enriched among these 35 CpG loci showed that oxidative stress response and retinoic acid–mediated apoptosis signaling pathways were statistically significantly enriched (Supplementary Table 5, available online). For each grade-specific tumor histology, all statistically significant differentially hypomethylated and hypermethylated CpG loci are detailed in Supplementary Tables 6 and 7, respectively (available online).
Glioma Methylation Classes, IDH Mutation, and Survival
The analysis of differentially methylated CpG loci in cellular pathways suggested that metabolic pathways as a group were commonly hypomethylated in gliomas. We hypothesized that genetic mutations in the metabolic pathways were associated with the observed DNA methylation phenotype. To test this hypothesis, we sequenced a subset of 95 tumors with available DNA for IDH1 and IDH2 mutations. IDH2 mutation was detected in only two tumors, and IDH1 mutation was detected in 56 tumors (total IDH mutation prevalence = 60.0%). IDH mutations were more common in oligoastrocytoma, oligodendroglioma, or astrocytoma histological subtypes than in glioblastomas, pilocytic astrocytomas, or ependymomas (P = 6.4 × 10−9); in lower-grade than higher-grade tumors (P = .01); in tumors with TP53 mutation compared with wild-type TP53 (P = .06); and in younger patients (mean age = 36.6 vs 47.4 years, P = .0009) (Table 2). However, IDH mutation was not associated with EGFR amplification (P = .10) (Table 2). Additionally, tumors with IDH mutation showed statistically significantly higher MGMT methylation (P = 3.6 × 10−4) (Supplementary Figure 1, available online).
Table 2.
Patient age, grade-specific glioma histology, grade, TP53 mutation, and EGFR amplification stratified by IDH mutation status*
| Patient age and tumor characteristic |
IDH mutation† |
|
| No | Yes | |
| Age at diagnosis, y | P = 9.0 × 10−4‡ | |
| Median age (range) | 49 (17–78) | 35 (20–59) |
| Mean age (SD) | 47.4 (17.5) | 36.6 (8.7) |
| Tumor histology,§ No. (%) | P = 6.4 × 10−9║ | |
| Grade 2 astrocytoma | 5 (26) | 14 (74) |
| Grade 3 astrocytoma | 0 (0) | 4 (100) |
| Ependymoma | 14 (100) | 0 (0) |
| Primary glioblastoma | 15 (79) | 4 (21) |
| Secondary glioblastoma (P = .005)¶ | 1 (14) | 6 (86) |
| Grade 2 oligoastrocytoma | 2 (13) | 13 (87) |
| Grade 3 oligoastrocytoma | 0 (0) | 1 (100) |
| Grade 2 oligodendroglioma | 1 (6) | 15 (94) |
| Tumor grade, No. (%) | P = .01# | |
| 1 | — | — |
| 2 | 22 (34) | 42 (66) |
| 3 | 0 (0) | 5 (100) |
| 4 | 16 (62) | 10 (38) |
| TP53 mutation, No. (%) | P = .06** | |
| No | 27 (63) | 16 (37) |
| Yes | 5 (31) | 11 (69) |
| EGFR amplification, No. (%) | P = .10†† | |
| No | 28 (51) | 27 (49) |
| Yes | 5 (100) | 0 (0) |
Analysis of patient age and tumor characteristics vs isocitrate dehydrogenase (IDH) gene mutation status. TP53 = tumor protein 53; EGFR = epidermal growth factor receptor.
IDH gene mutation was assessed by sequencing tumor DNA.
Association between age and IDH mutation was assessed using two-sided Student t test.
Tumors were previously reviewed by neuropathologists at the University of California San Francisco to assign histological subtypes and grades according to the World Health Organization classification.
Association between grade-specific histology and IDH mutation was assessed using two-sided Fisher exact test.
Association between primary vs secondary glioblastoma and IDH mutation was assessed using two-sided χ2 test.
Association between tumor grade and IDH mutation was assessed using two-sided Fisher exact test.
Association between TP53 mutation and IDH mutation was assessed using two-sided χ2 test.
Association between EGFR amplification and IDH mutation was assessed using two-sided χ2 test.
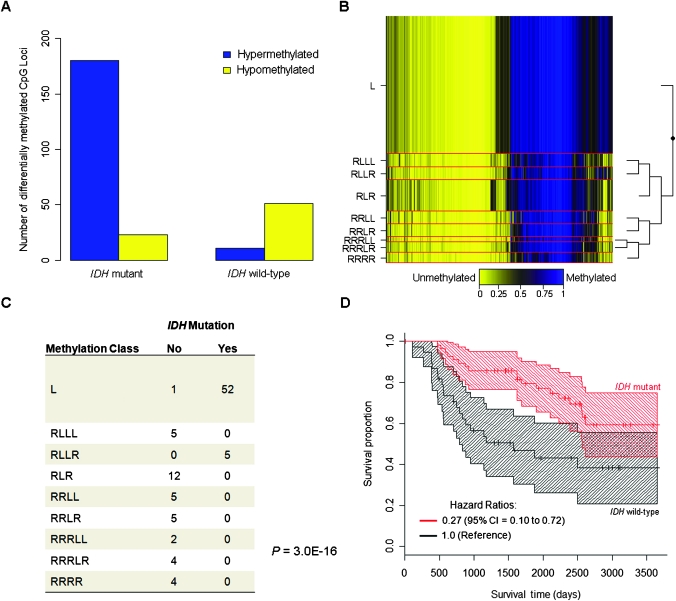
Next, we investigated the number of statistically significantly differentially methylated CpG loci between tumor and nontumor samples stratified by IDH mutation status. Tumors with IDH mutation revealed a striking contrast between the number of statistically significantly differentially hypermethylated loci, as well as the ratio of hyper- to hypomethylated loci in IDH mutant tumors vs IDH wild-type tumors (mutant = 7.8 vs wild-type = 0.22) (Figure 3, A). We used the statistically significantly differentially hypermethylated and hypomethylated CpG loci in IDH mutant tumors to conduct an enrichment analysis of cellular pathways. We found that cellular signaling pathways were hypermethylated, whereas metabolism and biosynthesis pathways that included starch and sucrose metabolism and pentose and glucuronate interconversion pathways were hypomethylated in IDH mutant tumors (Supplementary Table 8, available online).
Figure 3.
Association between IDH mutation and methylation phenotype in gliomas. A) The number of statistically significantly differentially hyper- and hypomethylated loci (Q < .05 and |Δβ| > 0.2), are plotted by tumor IDH mutation status. B) Recursively partitioned mixture model (RPMM) of glioma samples with both methylation and mutation data (n = 95). Methylation profile classes are stacked in rows separated by red lines, class height corresponds to the number of samples in each class. Class methylation at each CpG locus (columns) is the mean methylation for all samples in a class where blue designates methylated CpG loci (average β = 1), and yellow designates unmethylated CpG loci (average β = 0). To the right of the RPMM is the clustering dendrogram. C) Methylation-class-specific IDH mutation status (Fisher P = 3.0 × 10−16). D) Kaplan–Meier survival probability strata for IDH mutant (red, n = 57) and IDH wild-type (black, n = 38) tumors, tick marks are censored observations and banding patterns represent 95% confidence intervals.
Methylation profiling with RPMM of the 95 gliomas with both methylation data and IDH mutation status resulted in nine methylation classes (Figure 3, B). Methylation classes were statistically significantly associated with patient age (permutation P = 3.0 × 10−4), histology (P < 2.2 × 10−16), and grade (P = 6.0 × 10−9) (Supplementary Table 9, available online). IDH mutation was also strongly associated with methylation class (P = 3.0 × 10−16) (Figure 3, C), and this association remained statistically significant when controlling for age and histology (likelihood ratio P < .0001). Only two methylation classes had IDH mutant tumors (class L and class RLLR), and greater than 98% of the tumors (all but one) in these two classes had an IDH mutation (Figure 3, C). Furthermore, methylation classes L and RLLR were both more highly methylated than the other methylation classes (Figure 3, B).
Last, we examined the potential association between IDH mutation and patient survival among cases with available mutation data (n = 95) because previous studies reported increased survival among glioma patients with IDH mutation (3,5). In a multivariable Cox proportional hazards model controlling for age at diagnosis, sex, and grade-specific histology, we observed that patients whose tumors harbored IDH mutation showed statistically significantly better survival compared with patients (n = 38) whose tumors harbored wild-type IDH (hazard ratio of death = 0.27, 95% confidence interval = 0.10 to 0.72) (Figure 3, D, and Table 3).
Table 3.
Survival analysis using multivariable Cox proportional hazards model*
| Variable | HR† (95% CI) |
| Age | 1.03 (1.00 to 1.06) |
| Sex | |
| Female | 1.0 (referent) |
| Male | 0.73 (0.34 to 1.55) |
| IDH mutation‡ | |
| No | 1.0 (referent) |
| Yes | 0.27 (0.10 to 0.72) |
| Histology§ | |
| Grade 2 astrocytoma | 1.0 (referent) |
| Grade 3 astrocytoma | 1.79 (0.35 to 9.13) |
| Ependymoma | 0.25 (0.06 to 1.06) |
| Primary glioblastoma | 1.77 (0.60 to 5.22) |
| Secondary glioblastoma | 3.94 (1.20 to 12.9) |
| Grade 2 oligoastrocytoma | 2.8 (0.06 to 1.39) |
| Grade 3 oligoastrocytoma║ | — |
| Grade 2 oligodendroglioma | 0.75 (0.21 to 2.69) |
Cox proportional hazards model of survival included age, sex, IDH mutation, and grade-specific histology. HR = hazards ratio; CI = confidence interval; IDH = isocitrate dehydrogenase gene.
Adjusted hazard ratio values.
IDH gene mutation was assessed by sequencing tumor DNA.
Tumors were previously reviewed by neuropathologists at the University of California San Francisco to assign histological subtypes and grades according to the World Health Organization classification.
n = 1, HR = 1.4 × 10−7, SE = 4,910, confidence interval indeterminable.
Discussion
In this study, we demonstrate a distinct pattern of methylation across histological subtypes of glioma that is associated with genetic mutation in IDH gene loci. The two methylation classes associated with mutant IDH tumors had a homogeneous hypermethylation-rich character compared with the methylation classes for tumors with wild-type IDH. Additionally, the tumors with wild-type IDH belonged to several distinct methylation classes. The contrast between a homogenous hypermethylated profile and several heterogeneous hypomethylated profiles (associated with distinct histological types) strongly suggests that IDH mutation “drives” the observed hypermethylated phenotype, irrespective of tumor histology. In support of this, we note that IDH mutation is more robustly associated with methylation class compared with the classical glioma tumor genetic markers like TP53 mutation and EGFR amplification.
IDH mutations are heterozygous and allow the enzyme normally responsible for conversion of isocitrate to α-ketoglutarate to convert α-ketoglutarate to 2-hydroxyglutarate in an NADPH-dependent manner and results in accumulation of 2-hydroxyglutarate (7,8). Despite the observed hypermethylated profile of IDH mutant tumors, analysis of cellular pathways showed hypomethylation of several metabolic pathways, potentially to compensate for mutation-related metabolic stress. Because the methylation profile of IDH mutant tumors is generally homogenous, it is possible that the hypermethylation phenotype is either selected for, or driven by, the hypomethylation of compensatory metabolic pathways, thus directly linking and temporally situating these events. The level of α-ketoglutarate has been shown to be slightly lower in IDH1 mutant gliomas, though this decrease was not statistically significant (8). However, IDH1 localizes to the cytosol and peroxisomes, whereas IDH2 localizes to mitochondria; and because most IDH mutations in gliomas are in IDH1, pan-cellular α-ketoglutarate levels may not represent available cytosolic α-ketoglutarate levels. Furthermore, IDH1 R132 mutation has been shown to favor an active conformation of the enzyme, increase its affinity for NADPH, and favor reduction of α-ketoglutarate to 2-hydroxyglutarate over the conversion of isocitrate to α-ketoglutarate, which may reduce the availability of cytosolic α-ketoglutarate and NADPH (8). Hence, a potential mechanism responsible for the strong association between epigenetic profile and IDH mutation is related to potentially altered availability of α-ketoglutarate in these tumors. The Jumonji domain–containing histone demethylases require α-ketoglutarate as a substrate for their enzymatic activity (29), and altered activity of these histone demethylases could lead to aberrantly remodeled chromatin, potentially resulting in epigenetic alterations at the DNA level as well. However, studies that are beyond the scope of this article would be necessary to disentangle the complex networks of chromatin remodeling enzymes, their targets, and their responses to altered levels of enzymatic substrate. Alternatively (or perhaps in conjunction), lower concentrations of NADPH associated with mutant IDH1 (30) may result in a decreased capacity for reductive processes in defense against reactive oxygen species. Furthermore, α-ketoglutarate itself is a potent antioxidant (6) and its decreased availability in IDH mutant cells alone, or together with lower NADPH levels, could drive the selection of cells with compensatory metabolic gene expression profiles mediated by altered epigenetic patterns including chromatin configuration and DNA methylation. Consistent with the suggestion that gene expression profiles are altered in association with DNA methylation related to IDH mutation, an analysis of glioblastoma gene expression subtypes showed that IDH mutation occurred almost exclusively in proneural glioblastomas (31).
More broadly, and similar to the hypermethylation phenotype we describe here, hypermethylator phenotypes have previously been associated with other cancers. This phenotype was first described in colon cancer and is commonly referred to as CpG Island Methylator Phenotype (CIMP) (32). Specifically, colorectal cancers can be divided in CIMP-high, CIMP-low, and non-CIMP based on the methylation of five to eight specific gene promoters (33,34). Similar to IDH in glioma, CIMP status in colon tumors has been associated with specific mutations; CIMP-high with BRAF and CIMP-low and non-CIMP with KRAS (35). Recently, Noushmehr et al. (36) described a CIMP in glioblastomas, termed G-CIMP, which they found to be tightly associated with IDH1 mutation. In a number of lower-grade gliomas, Noushmehr et al. performed methylation profiling of eight markers of G-CIMP and confirmed that IDH1 mutation is associated with G-CIMP in low-grade tumors, which is consistent with our array-based findings. Furthermore, more than 83% of G-CIMP-positive glioblastomas with IDH1 mutation were of the proneural glioblastoma gene expression subtype (36), additional evidence supporting an association between distinct, IDH-related methylation in our data (from diverse glioma histological subtypes), and a specific gene expression phenotype. In addition, MGMT methylation is often investigated in glioma because it has been associated with increased sensitivity to alklyating agents such as temozolomide and can affect response to therapy (37). In fact, increased MGMT methylation can also distinguish CIMP-high and CIMP-low from non-CIMP in colon cancer (38). Our results, consistent with previous work (9), demonstrate an association between increased MGMT methylation and IDH mutation. Finally, some studies have reported CIMP-positive colon cancers to have a relatively better prognosis (39), and from both the work of Noushmehr et al. and ours, this appears to be consistent with the pattern of survival observed in CIMP gliomas.
The association between IDH mutation and a homogenous methylation profile across several histological subtypes suggests that genetic and epigenetic alterations are not independent. This observation also has profound implications for the development of new therapies for glioma. Although pharmacological inhibition of 2-hydroxyglutarate has been suggested as a possible approach to treating IDH mutant gliomas (40) such drugs do not yet exist. However, DNA methylation is a modifiable therapeutic target; DNA methyltransferase inhibitors and histone deacetylase inhibitors are in clinical trials and showing some promise for the treatment of hematopoietic malignancies (41–43). Our work suggests that a simple diagnostic test for DNA methylation (or mutation) can identify a class of tumors for which the modification of DNA methylation may have therapeutic efficacy. This class of tumors is not discernable by any of the classic histopathologic or tumor markers for glioma. The recognition that IDH mutation has value as a clinical prognostic marker and is associated with a broad DNA methylation phenotype suggests that glioma therapeutic protocols that reverse DNA methylation should be pursued.
Our study has a few limitations. Although we studied 131 histologically diverse tumors, we did not have IDH mutation, TP53 mutation, and EGFR amplification data on all subjects and had somewhat limited statistical power to explore the relationships between IDH mutation and these alterations. Future investigations that include larger numbers of histologically diverse samples and higher-resolution methylation array techniques, along with measurements of other somatic alterations (IDH mutation, mRNA expression, and copy number), will afford a more comprehensive understanding of the molecular and chromosomal characteristics that distinguish glioma subtypes. Understanding whether these glioma molecular and chromosomal subtypes are differentially associated with glioma risk loci (44) also will help to understand the etiology and possibly outcomes of this often catastrophic disease.
In summary, our work demonstrates a clear relationship between genetic and epigenetic events in human gliomas by associating IDH mutations with a homogenous methylation profile, and demonstrates that profiles of methylation differ by histological subtype of disease. Additionally, and consistent with previous work, we also showed that patients with IDH mutation have significantly improved survival. Advances in therapy for glioma may be realized by targeting DNA methylation. Much attention has recently been given to the utility of MGMT methylation in predicting response to therapy, and our data further suggest that other DNA methylation markers may improve clinical assessment, guide therapies, and potentially uncover novel therapeutic avenues altogether.
Funding
National Institute of Health (R01CA52689 to M.R.W.; P50CA097257 to M.R.W. and J.K.W; R01CA078609, R01CA121147, R01CA126939, and R01CA100679 to K.T.K.; R01ES06717 and R01CA126831 to J.K.W.; P30CA077598 to H.H.N.); Tobacco-Related Diseases Research Program (18CA-0127 to J.L.W.).
Supplementary Material
Footnotes
B. C. Christensen and A. A. Smith contributed equally to the work. M. R. Wrensch, K. T. Kelsey, and J. K. Wiencke are joint lead investigators.
The funders did not have any role in the study design, collection of data, interpretation of the results, preparation of the article, or the decision to submit the article for publication.
References
- 1.Kleihues P, Burger PC, Scheithauer BW. The new WHO classification of brain tumours. Brain Pathol. 1993;3(3):255–268. doi: 10.1111/j.1750-3639.1993.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 5.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102(13):932–941. doi: 10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 10.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 11.Mashiyama S, Murakami Y, Yoshimoto T, Sekiya T, Hayashi K. Detection of p53 gene mutations in human brain tumors by single-strand conformation polymorphism analysis of polymerase chain reaction products. Oncogene. 1991;6(8):1313–1318. [PubMed] [Google Scholar]
- 12.Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84(19):6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallol A, Krex D, Hesson L, Eng C, Maher ER, Latif F. Frequent epigenetic inactivation of the SLIT2 gene in gliomas. Oncogene. 2003;22(29):4611–4616. doi: 10.1038/sj.onc.1206687. [DOI] [PubMed] [Google Scholar]
- 14.Maegawa S, Itaba N, Otsuka S, et al. Coordinate downregulation of a novel imprinted transcript ITUP1 with PEG3 in glioma cell lines. DNA Res. 2004;11(1):37–49. doi: 10.1093/dnares/11.1.37. [DOI] [PubMed] [Google Scholar]
- 15.Stone AR, Bobo W, Brat DJ, Devi NS, Van Meir EG, Vertino PM. Aberrant methylation and down-regulation of TMS1/ASC in human glioblastoma. Am J Pathol. 2004;165(4):1151–1161. doi: 10.1016/S0002-9440(10)63376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, Zhang H, Gu J, et al. Methylation profiles of thirty four promoter-CpG islands and concordant methylation behaviours of sixteen genes that may contribute to carcinogenesis of astrocytoma. BMC Cancer. 2004;4:65. doi: 10.1186/1471-2407-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiencke J, Aldape K, McMillan A, et al. Molecular features of adult glioma associated with patient race/ethnicity, age, and a polymorphism in O6-methylguanine-DNA-methyltransferase. Cancer Epidemiol Biomarkers Prev. 2005;14(7):1774–1783. doi: 10.1158/1055-9965.EPI-05-0089. [DOI] [PubMed] [Google Scholar]
- 18.De Preter K, Speleman F, Combaret V, et al. Quantification of MYCN, DDX1, and NAG gene copy number in neuroblastoma using a real-time quantitative PCR assay. Mod Pathol. 2002;15(2):159–166. doi: 10.1038/modpathol.3880508. [DOI] [PubMed] [Google Scholar]
- 19.R Development CT. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2007. [Google Scholar]
- 20.Houseman EA, Christensen BC, Marsit CJ, et al. Model-based clustering of DNA methylation array data: a recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinformatics. 2008;9(365) doi: 10.1186/1471-2105-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen BC, Houseman EA, Godleski JJ, et al. Epigenetic profiles distinguish pleural mesothelioma from normal pleura and predict lung asbestos burden and clinical outcome. Cancer Res. 2009;69(1):227–234. doi: 10.1158/0008-5472.CAN-08-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grambsch P, Therneau T. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81(3):515–526. [Google Scholar]
- 23.Storey J, Taylor J, Siegmund D. Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: a unified approach. J Royal Stat Soc. 2004;Series B(66):187–205. [Google Scholar]
- 24.Bibikova M, Lin Z, Zhou L, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16(3):383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byun HM, Siegmund KD, Pan F, et al. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet. 2009;18(24):4808–4817. doi: 10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen BC, Houseman EA, Marsit CJ, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irizarry RA, Ladd-Acosta C, Carvalho B, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18(5):780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ladd-Acosta C, Pevsner J, Sabunciyan S, et al. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81(6):1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30(1):344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winkler BS, DeSantis N, Solomon F. Multiple NADPH-producing pathways control glutathione (GSH) content in retina. Exp Eye Res. 1986;43(5):829–847. doi: 10.1016/s0014-4835(86)80013-6. [DOI] [PubMed] [Google Scholar]
- 31.Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogino S, Nosho K, Kirkner G, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100(23):1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Issa J. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4(12):988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 35.Ogino S, Kawasaki T, Kirkner G, Loda M, Fuchs C. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8(5):582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chin L, Meyerson M, Aldape K, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogino S, Kawasaki T, Kirkner G, Suemoto Y, Meyerhardt J, Fuchs C. Molecular correlates with MGMT promoter methylation and silencing support CpG island methylator phenotype-low (CIMP-low) in colorectal cancer. Gut. 2007;56(11):1564–1571. doi: 10.1136/gut.2007.119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10(1):13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frezza C, Tennant DA, Gottlieb E. IDH1 mutations in gliomas: when an enzyme loses its grip. Cancer Cell. 2010;17(1):7–9. doi: 10.1016/j.ccr.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 41.Borthakur G, Huang X, Kantarjian H. Report of a phase 1/2 study of a combination of azacitidine and cytarabine in acute myelogenous leukemia and high-risk myelodysplastic syndromes. Leuk Lymphoma. 2010;51(1):73–78. doi: 10.3109/10428190903318329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santos FP, Kantarjian H, Garcia-Manero G, Issa JP, Ravandi F. Decitabine in the treatment of myelodysplastic syndromes. Expert Rev Anticancer Ther. 2010;10(1):9–22. doi: 10.1586/era.09.164. [DOI] [PubMed] [Google Scholar]
- 43.Mercurio C, Minucci S, Pelicci PG. Histone deacetylases and epigenetic therapies of hematological malignancies. Pharmacol Res. 62(1):18–34. doi: 10.1016/j.phrs.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.