Abstract
Background
Obesity causes insulin resistance in target tissues - skeletal muscle, adipose tissue, liver and the brain. Insulin resistance predisposes to type-2 diabetes (T2D) and cardiovascular disease (CVD). Adipose tissue inflammation is an essential characteristic of obesity and insulin resistance. Neuronatin (Nnat) expression has been found to be altered in a number of conditions related to inflammatory or metabolic disturbance, but its physiological roles and regulatory mechanisms in adipose tissue, brain, pancreatic islets and other tissues are not understood.
Results
We identified transcription factor binding sites (TFBS) conserved in the Nnat promoter, and transcription factors (TF) abundantly expressed in adipose tissue. These include transcription factors concerned with the control of: adipogenesis (Pparγ, Klf15, Irf1, Creb1, Egr2, Gata3); lipogenesis (Mlxipl, Srebp1c); inflammation (Jun, Stat3); insulin signalling and diabetes susceptibility (Foxo1, Tcf7l2). We also identified NeuroD1 the only documented TF that controls Nnat expression. We identified KEGG pathways significantly associated with Nnat expression, including positive correlations with inflammation and negative correlations with metabolic pathways (most prominently oxidative phosphorylation, glycolysis and gluconeogenesis, pyruvate metabolism) and protein turnover. 27 genes, including; Gstt1 and Sod3, concerned with oxidative stress; Sncg and Cxcl9 concerned with inflammation; Ebf1, Lgals12 and Fzd4 involved in adipogenesis; whose expression co-varies with Nnat were identified, and conserved transcription factor binding sites identified on their promoters. Functional networks relating to each of these genes were identified.
Conclusions
Our analysis shows that Nnat is an acute diet-responsive gene in white adipose tissue and hypothalamus; it may play an important role in metabolism, adipogenesis, and resolution of oxidative stress and inflammation in response to dietary excess.
Background
The WHO estimates there will be 2.3 billion overweight or obese adults world-wide by 2015; including 60% of adults and alarmingly 25% of children in developing countries http://www.who.int. This overweight and obesity epidemic is predicted to affect half the global population by 2025[1,2], largely because of increased urbanization, adaptation of the western diet, and sedentary lifestyle, particularly in China and India.
A serious and nearly ubiquitous complication of obesity is defective insulin signalling in target tissues - skeletal muscle, liver, adipose tissue and brain. Insulin resistance unites obesity with a major disease cluster - diabetes, fatty liver and heart disease - through compensatory hyperinsulinaemia leading to pancreatic β-cell failure, and complicating dyslipidaemia and hypertension, major risk factors for CVD. T2D affects 150 million people world-wide, and is estimated to double by 2025[1,2]. CVD is predicted to be the leading cause of death globally by 2020[3,4]. Dementia and cancer are also more common in the obese[4,5]. Therefore, understanding the mechanisms underlying obesity and insulin resistance is of great significance, as the need for new and more effective treatments grows.
Insulin resistance in obesity is causally associated with macrophage influx and low-grade, metabolic inflammation in adipose tissue. Macrophage-driven chronic inflammation in adipose tissue from the obese is considered to be triggered by endoplasmic reticulum (ER) and oxidative stress, together with hypoxia consequent on adiposity expansion and inadequate blood supply. Cellular stress leads to activation of stress kinesis and inflammatory effectors such as Jnk and Nfkb, with systemic release of cytokine and fatty acid mediators, which inhibit insulin signalling in target tissues[1-7].
While appetite, energy expenditure and metabolism are all critically regulated by hypothalamic brain circuits, white adipose tissue (WAT) is the major storage site for fat in the form of triglycerides, and therefore forms a repository balancing energy intake and expenditure. The overall adipose tissue burden of an individual is a combination of adiposity number (i.e. adipogenesis) and lipid load per cell (i.e. lipogenesis), and both processes may contribute to the adipose tissue mass in obesity.
Nnat is a paternally expressed imprinted gene first discovered in the developing brain and it is implicated in antenatal brain development and foetal growth[6-9]. In adult rodents and humans Nnat is highly expressed in neurons in many brain regions, including hypothalamic nuclei concerned with appetite regulation, and is concerned in nutrient sensing, and responds to feeding/fasting and leptin[10,11]. Nnat is also highly expressed in adult white adipose tissue and aortic endothelial cells, where expression is increased in obese and diabetic rodents[12,13]. Nnat is moreover expressed in pancreatic β-cell lines, where expression is glucose-dependent, and reduced in diabetic rodents[14-16]. By contrast Nnat expression in WAT is substantially reduced in lipodystrophic, in lean S6kko mice and in mice fed conjugated linoleic acid, which induces weight loss[16-18]. Recently a consistent association between single nucleotide polymorphisms in the human Nnat gene and severe obesity in both children and adults has been demonstrated[10]. This observation suggests that altered Nnat function in adipose tissue or the hypothalamic regions regulating energy homeostasis may be involved in the pathogenesis of human obesity.
Nnat has three exons, and resides within a large intron of Blcap[7,8], tissue specific enhancers lie well upstream of the CpG-rich Nnat promoter. Knowledge about the upstream regulation of Nnat is confined to the β-cell and control by NeuroD1, a transcription factor important for neuronal and endocrine cell differentiation and survival[14].
Knowledge concerning downstream effecter mechanisms through which Nnat operates, and its physiological functions are limited. Nnat is an amphipathic proteolipid. It has two isoforms, α of 81 and β of 54 amino acids, generated by alternative splicing of the middle exon, encoding a membrane-binding domain[13-15,19]. Both isoforms reside in the endoplasmic ER membranes, the β-isoform is implicated in ER stress and β-cell apoptosis[15], it is considered to regulate ER calcium ATPase through homology to other proteolipid. Nnat expression in vitro is associated with calcium-induced 3T3-L1 cell adipogenesis; glucose-stimulated, calcium-induced, insulin secretion from β-cells; activation of Pi3k, Erk, mTor and calcium signalling in medulloblastoma; Nfkb-regulated inflammation in aortic endothelial cells and protection against mitochondrial toxins and ionophores in PC12 cells and resolution of injury after ischemic/reperfusion of the kidney [12-14,19,20].
The physiological roles of Nnat in the brain, pancreas, adipose and other tissues are not understood. Through genome-wide gene expression analysis we sought to improve our understanding of the mechanisms of Nnat regulation and its biological functions. First we carried out an integrated bioinformatics analysis of the conserved Nnat promoter in seven species and derived a candidate list of potential Nnat transcription factors. Then we interrogated multi-tissue gene expression data to look for primary evidence on the applicability of potential Nnat transcriptional regulators. We then used Gene Set Enrichment Analysis[21] to understand which biological functions are most highly correlated with Nnat expression. Finally, we generated a mouse protein-protein interaction network in adipose tissue to probe the biological relationships among genes forming a co-expression network with Nnat.
Methods
Microarray datasets and meta-analysis
In this study, we used four different microarray gene expression datasets from WAT of C57BL/6J mice, as described below, to complete a meta-analysis. The first dataset designated BAIR was obtained as part of the Biological Atlas of Insulin Resistance (BAIR, http://www.bair.org.uk) project, a multi-lab collaborative effort to define gene expression changes involved in insulin signalling and resistance. Male mice were weaned at 3 weeks of age and fed a control chow diet containing 5% w/w fat, 19% protein and 3.5% fibre (S&K Universal Ltd, Hull, UK). At 5 weeks of age, a group of mice was transferred to 40% w/w high-fat diet, of which saturated fat is 40%, containing 32% pig fat, 8% casein fat, 19% protein, 21% glucose (Special Diet Services, Witham, UK) ad libitum, whereas the control group remained on chow. Mice fat-fed for 2 days, 8 days, 3 weeks and 15 weeks, and with age-matched controls, were killed and epididymal WAT was obtained for microarray gene expression studies, employing Affymetrix mouse genome 4302 microarray. From these four time points a total of 40 Affymetrix gene expression raw CEL files were produced and RMA normalized by using the Affy package http://www.bioconductor.org; present/absent call information was also calculated based on Affymetrix MAS5 algorithm. This dataset has been deposited in EBI ArrayExpress Database with accession number E-BAIR-12 http://www.ebi.ac.uk/microarray-as/ae/.
The second dataset was obtained from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/, accession ID: GSE4671), which contains 28 Affymetrix mouse genome 4302 chips. Nine-week old male mice in this experiment were fed with either a control chow diet or a chow diet that also contained 0.5% conjugated linoleic acid (CLA). At each of seven time points (day 1, 2, 3, 4, 7, 10, 17), retroperitoneal WAT was obtained from two control mice and two test mice. Microarray data were processed using GCRMA algorithm.
The third dataset was also acquired from GEO with accession number GSE8831, in which 20 female and 15 male C57BL/6J mice, fed by ad libitum, varied in body weight and insulin sensitivity were studied. Fasting blood glucose and serum insulin concentrations were measured 2-4 days prior to collection of adipose tissue. RNA from perigonadal adipose tissue was extracted and processed for analysis on Affymetrix Mu74Av2 microarrays. RMA normalization was applied to the 35 raw CEL files, and MAS5 present/absent call information was calculated as well.
The fourth dataset consisted of 13 Affymetrix MG-U74av2 chips for epididymal WAT extracted from six-week old C57BL/6J male mice from the Diabetes Genome Anatomy Project DGAP http://www.diabetesgenome.org. There were four high-fat feeding mice (derives 55% calories from fat, 21% calories from protein, 24% calories from carbohydrates), nine low-fat feeding control mice (derives 14% calories from fat, 25% calories from protein and 61% calories from carbohydrates). Gene expression data was RMA normalized with present/absent call information.
In order to identify the gene co-vary with Nnat in WAT, we calculated Pearson correlation coefficients between the expression of Nnat and all other genes in each of the above four datasets separately, and chose genes showing the highest correlation with Nnat expression. Subsequently, rather than merely combining the p-values of the Pearson correlation coefficients, we used weighted unbiased Pearson statistical values, i.e. we normalized the statistical values to approximate a normal distribution with unit variance and zero mean. Some necessary filters were designed for quality control: (a) probesets with expression value lower than 100 or (b) absolute Pearson value less than 0.2 or (c) present/absent call less than 30% were excluded. Each gene was represented by the probesets which had the largest absolute Pearson correlation in each dataset in order to remove multiple annotated probesets for the same gene. In this way we obtained 2841, 3061, 2148 and 1201 unique genes within the four datasets respectively, giving 6120 unique genes in total.
The following formula was applied to calculate the combined Z-score
ωi denotes the number of samples in each dataset (i.e. 40, 28, 35, 13); Ti indicates the normalized Pearson association test statistic based on a t distribution. A two-tailed P-value can be derived by the normal distribution for each gene from this combined Zscore. Bonfferroni corrected FDR < 0.05 was applied, yielding the top 34 genes which are most strongly co-expressed with Nnat. For comparison we also applied Fisher's Z-transform to combine the transformed Pearson correlation coefficients in the meta-analysis, we got similar results. For the multi-species analysis, seven genes, Acp5, Tcta, Znrf2, Serpina3c, S3-12, Sgce and Lrg1, which are not conserved in rat, were excluded.
Promoter analysis
To delineate the Nnat promoter, we identified the conserved non-coding region of the Nnat locus in seven mammalian genera (Pan, Bos, Canis, Mus, Sus, Rattus and human). This corresponds to 1200 bp upstream and 800 bp downstream of the most utilized transcription start points (TSP) of Nnat, identified through the presence of CAGED transcripts in mice and human. For example, mouse Nnat has six transcripts, with two promoters having respectively 36 and 28 CAGE tags, while human NNAT has four transcripts with one promoter having 29 CAGE tags. For these analyses we used the Genome VISTA genome alignment tool http://www-gsd.lbl.gov/vista/ in conjunction with Genomatix Gene2promoter tool (Genomatix Software Inc., Ann Arbor, MI, USA). The Genomatix MatInspector programme with default parameter settings was used to detect common Transcription Factor Binding Sites (TFBS) between the seven promoter sequences of the seven genera. The occurrence of the TFBSs within all vertebrate genes was used as a control in MatInspector. Significant common TFBSs among the seven species were defined by having a p-value < 0.05 and presence in six or more species, compared with the control sets in the Genomatix database. We further explored the TF genes in each TFBSs family, examining expression data to determine whether they were expressed in human or mouse adipose, hypothalamus and pancreas islet (using BioGPS database http://biogps.gnf.org and our own BAIR data as reference sources). Promoter analysis was also applied to these 27 fully conserved Nnat co-expression genes (plus Nnat) all together, focusing on human, mouse and rat only. Again, their expression status in adipose, hypothalamus and pancreatic islets were included as well.
Gene set enrichment analysis
In order to inspect which biological pathways are significantly correlated with Nnat expression in adipose tissue, we employed Gene Set Enrichment Analysis, using the GSEA package http://www.broadinstitute.org/gsea/ for each of the above four datasets separately. We chose Pearson correlation and permuted 1000 times to detect statistical significance. Expression data were pre-filtered by using the present/absent call and background correction, similarly to the pre-process in meta-analysis above. In the GSEA database there were 200 KEGG pathway sets and 639 canonical pathway sets. We first focused on those approximately 100 KEGG gene sets which have more than 15 genes and fewer than 500 genes; the results returned were significant in the above four datasets as well as an additional dataset which included 45 sleep mice model with hypothalamus tissue (GEO: GSE6514). We then worked on those canonical pathway sets which passed the same filter only on BAIR fat-fed mice and human fat dataset about 20 lean and 19 obese person[22].
Protein-protein interaction network analysis
By using BioNetBuilder, a cytoscape plugin package http://err.bio.nyu.edu/cytoscape/bionetbuilder/ with supported databases DIP, BIND, Prolinks, KEGG, HPRD, The BioGrid and GO, we thus generated a mouse protein-protein interaction network which had 11777 unique nodes (genes) and 107813 unique edges (links). For the 27 fully conserved Nnat co-expression genes, we searched each of their direct correlation genes (first neighbours) in the PPI network, again filtered by expression value > 200 in adipose tissue by BioGPS.
Results
Nnat promoter analysis
We sought to investigate mechanisms of transcriptional regulation of Nnat and of co-varying genes in adipose tissue. The neuronatin gene is located on human chromosome 20 and mouse chromosome 2. To define the Nnat promoter we described the conserved non-coding region in seven mammalian species, including mouse and human, corresponding to 1200 bp upstream and 800 bp downstream of the most utilized transcription start points (TSP) of Nnat identified through the presence of CAGED transcripts in mice and humans. We used Genomatix software to determine the presence of binding sites for TF families in the conserved Nnat promoter, across seven mammalian species. We then filtered these potential regulators based on their expression in mouse WAT, and compared their expression also in hypothalamus and pancreatic β-cells in which Nnat is known to be expressed. Table 1 shows the gene expression behaviour for members of TFBS families scoring as significant for presence in the Nnat promoter (p-value < 0.05), and present in at least six of the Nnat promoter sequences; expression values were filtered on the basis of BioGPS expression bigger than 200, which therefore focuses attention on the most abundantly expressed TFs.
Table 1.
Expression of conserved TFBS in Nnat promoter
| TFs | TFBSs | p-value | WAT | hypo | panc | TFs | TFBSs | p-value | WAT | hypo | panc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pparγ | V$PPAR | 0 | 11959 | 253 | 357 | Hsf2 | V$HEAT | 0.0202 | 271 | 543 | 279 |
| Zfp161 | V$ZF5F | 2E-05 | 1309 | 1537 | 523 | Tef | V$PARF | 0.0209 | 4845 | 5297 | 846 |
| Rreb1 | V$RREB | 0.0005 | 3530 | 1344 | 117 | Myb | V$MYBL | 0.0217 | 730 | 51 | 81 |
| Tead4 | V$TEAF | 0.0005 | 271 | 239 | 367 | Meox2 | V$HBOX | 0.0221 | 555 | 37 | 47 |
| Hes2 | V$HESF | 0.0016 | 468 | 304 | 498 | Stat3 | V$STAT | 0.0236 | 3754 | 1438 | 617 |
| Gtf2i | V$DICE | 0.002 | 5309 | 7006 | 398 | Myt1l | V$MYT1 | 0.0247 | 327 | 1039 | 395 |
| Zfp384 | V$CIZF | 0.0023 | 1966 | 989 | 1073 | Neurod1 | V$NEUR | 0.0262 | 119 | 323 | 224 |
| Myog | V$MYOD | 0.0024 | 523 | 524 | 638 | Neurod2 | V$NEUR | 0.0262 | 209 | 216 | 237 |
| Glis2 | V$GLIF | 0.0026 | 308 | 365 | 390 | Hand1 | V$HAND | 0.0283 | 557 | 519 | 662 |
| Mef2d | V$MEF2 | 0.0027 | 2451 | 4184 | 668 | Bcl6 | V$BCL6 | 0.0301 | 4569 | 1672 | 82 |
| Tcf7l2 | V$LEFF | 0.0033 | 4252 | 1922 | 329 | Gata3 | V$GATA | 0.0323 | 432 | 54 | 72 |
| Ctcf | V$CTCF | 0.004 | 2655 | 1586 | 890 | Pbx2 | V$PBXC | 0.0345 | 524 | 456 | 253 |
| Srf | V$SRFF | 0.0041 | 340 | 275 | 212 | Hoxd9 | V$ABDB | 0.0364 | 230 | 139 | 190 |
| Zfp219 | V$ZBPF | 0.0042 | 1282 | 1712 | 270 | Hoxc8 | V$HOXF | 0.0371 | 1804 | 203 | 364 |
| Hif1a | V$HIFF | 0.0042 | 542 | 564 | 194 | Evi1 | V$EVI1 | 0.0412 | 274 | 240 | 335 |
| Mlxipl | V$EBOX | 0.006 | 304 | 27 | 32 | Creb1 | V$CREB | 0.0415 | 2387 | 1157 | 532 |
| Nfat5 | V$NFAT | 0.0069 | 1154 | 842 | 203 | Jun | V$CREB | 0.0415 | 2503 | 1955 | 1735 |
| Egr2 | V$EGRF | 0.0078 | 408 | 141 | 289 | Klf15 | V$KLFS | 0.0419 | 7360 | 2193 | 2438 |
| Tfdp1 | V$E2FF | 0.0125 | 1993 | 1575 | 77 | Klf4 | V$KLFS | 0.0419 | 4878 | 593 | 348 |
| Pou6f1 | V$BRN5 | 0.0146 | 632 | 973 | 446 | Klf6 | V$KLFS | 0.0419 | 3219 | 583 | 895 |
| Sp1 | V$SP1F | 0.0147 | 322 | 200 | 306 | Foxo1 | V$FKHD | 0.0424 | 400 | 129 | 106 |
| Dmrt2 | V$DMRT | 0.0157 | 275 | 28 | 57 | Sox4 | V$SORY | 0.043 | 4829 | 1217 | 74 |
| Nfyc | V$CAAT | 0.0161 | 2207 | 914 | 247 | Pou2f1 | V$OCT1 | 0.0443 | 225 | 229 | 238 |
| Irf1 | V$IRFF | 0.0166 | 5486 | 259 | 196 | Elf1 | V$ETSF | 0.0464 | 2419 | 78 | 134 |
| Ahr | V$AHRR | 0.0171 | 483 | 135 | 62 | Nr2f2 | V$NR2F | 0.0472 | 6090 | 8397 | 259 |
| Nfia | V$NF1F | 0.0171 | 3505 | 2335 | 505 | Rxrb | V$RXRF | 0.0479 | 1715 | 1309 | 1944 |
| Lhx4 | V$LHXF | 0.0177 | 269 | 242 | 225 | Msx1 | V$HOMF | 0.0479 | 392 | 360 | 435 |
Conserved TFBS found in Nnat promoter region across seven mammalian species and their gene expression in three tissues of mice.
The TFBS for the V$NEUR family (including NeuroD1 and NeuroD2) was found to be highly significant in six out of seven promoter sequences, but it was not found in the rat promoter sequences in our defined region. However, V$NEUR does exist farther upstream from the rat Nnat transcription start site (within 2.5kb). NeuroD1, the only confirmed transcriptional regulator of Nnat expression[14] is not one of the most highly expressed TFs in adipose, though its expression value is similar across all three tissues shown in Table 1, it is highly expressed in cerebellum.
The binding sites for the key adipose tissue regulator Pparγ are the most significant in the seven Nnat promoter sequences. Of the TFs with conserved binding sites it is also by far the most highly expressed in WAT, and has a crucial role in adipocyte metabolism. Other important TFs with conserved binding sites in the Nnat promoter and which are highly expressed in WAT include several genes which have crucial roles in the control of: adipogenesis (Klf15, Irf1, Creb1, Egr2, Gata3); lipogenesis (Mlxipl, Srebp1c); inflammation (Jun, Stat3); insulin signalling and diabetes susceptibility (Foxo1, Tcf7l2). Interestingly, the Rreb1 has a binding site in all seven Nnat promoters, and is expressed at high levels in adipose and hypothalamus. Rreb1 can enhance the transcription-activating function of NeuroD1, a property that requires both the physical interaction of Rreb1 with NeuroD1, and binding of Rreb1 to DNA[23]. Notable by their absence are binding sites for the C/EBP family of TFs, clearly suggesting that this family (several members of which are required for adipocyte differentiation) are not concerned with Nnat regulation, and may be down-stream of Nnat.
Identification of biological pathways associated with Nnat expression
In order to determine which biological pathways are significantly correlated with Nnat expression in WAT and hypothalamus, we employed Gene Set Enrichment Analysis. Initially we focused on KEGG pathways to investigate those processes that are most highly associated with Nnat expression in WAT and hypothalamus. In WAT, there are five KEGG pathways significantly correlated with Nnat expression (FDR < 0.05) in at least two of the four mouse WAT datasets (Table 2), these five pathways are all significant in BAIR fat-fed data, and include oxidative phosphorylation, ribosome, proteasome, cholera infection and neuroactive ligand receptor interaction, these last two indicating perturbation of ion transport and neuronal function. Furthermore, oxidative phosphorylation, ribosome and cholera infection are significant in the mouse hypothalamus dataset as well. Additional file 1, Figure S1 illustrates the enrichment plots and positively and negatively correlated genes in these three common pathways. Additional file 1, Table S1 lists the core enriched genes that correlate with Nnat in these three common pathways in both WAT and hypothalamus. GSEA on the canonical pathways (Additional file 1, Table S2) reveals that Nnat expression in mice on a high-fat diet and obese models displayed highly positive correlations with inflammatory pathways (complement and coagulation, cytokine and integrin signalling), and strongly negative correlations with metabolic pathways (most prominently oxidative phosphorylation, pyruvate metabolism, glycolysis and gluconeogenesis) and protein turnover (including proteasome, ribosome and mRNA processing). In human adipose tissue NNAT mRNA expression is decreased (Additional file 1, Table S3). In consequence NNAT expression displayed strong positive correlation with genes involved in protein turnover and fatty acid metabolism/degradation, and negatively correlated with genes involved in adhesion, complement and coagulation and cytokine. Additional file 1, Table S4 lists the common pathways correlated with Nnat expression between BAIR fat-fed mice and human obese WAT, and these are concerned with cytokine signalling and cell-cell interaction. Additional file 2, Table S5 contains the complete list of genes associated with Nnat in each KEGG pathway in mouse models.
Table 2.
Pathways associated with Nnat
| KEGG Pathway | Size | p-value | FDR | Correlation | Tissue |
|---|---|---|---|---|---|
| OXIDATIVE_PHOSPHORYLATION | 92 | < 0.001 | < 0.001 | Negative | WAT |
| RIBOSOME | 52 | < 0.001 | < 0.001 | Negative | WAT |
| PROTEASOME | 22 | < 0.001 | < 0.001 | Negative | WAT |
| CHOLERA_INFECTION | 33 | < 0.001 | < 0.01 | Negative | WAT |
| NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | 36 | < 0.001 | < 0.01 | Positive | WAT |
| OXIDATIVE_PHOSPHORYLATION | 93 | < 0.001 | 0.011 | Negative | Hypo |
| RIBOSOME | 52 | < 0.001 | 0.006 | Negative | Hypo |
| CHOLERA_INFECTION | 32 | < 0.001 | 0.031 | Positive | Hypo |
| ALANINE_AND_ASPARTATE_METABOLISM | 25 | < 0.002 | 0.029 | Positive | Hypo |
| ARGININE_AND_PROLINE_METABOLISM | 18 | < 0.001 | 0.026 | Positive | Hypo |
| TRYPTOPHAN_METABOLISM | 31 | < 0.001 | 0.025 | Positive | Hypo |
| SELENOAMINO_ACID_METABOLISM | 21 | 0.004 | 0.029 | Positive | Hypo |
| THYROID_CANCER | 21 | 0.003 | 0.025 | Positive | Hypo |
| BUTANOATE_METABOLISM | 29 | 0.004 | 0.036 | Positive | Hypo |
| GALACTOSE_METABOLISM | 16 | 0.006 | 0.04 | Positive | Hypo |
| GLYCOLYSIS_AND_GLUCONEOGENESIS | 30 | 0.004 | 0.05 | Positive | Hypo |
| PATHOGENIC_ESCHERICHIA_COLI_INFECTION_EHEC | 24 | 0.008 | 0.049 | Positive | Hypo |
Significant pathways associated with Nnat analyzed by GSEA.
Identification of genes that co-vary with Nnat
The genes whose expression co-varies with Nnat are likely to share regulatory mechanisms and, perhaps, biological functions. We therefore identified genes whose expression is highly correlated with Nnat in a meta-analysis of WAT gene expression data. Table 3 shows the correlation of gene expression behaviour with that of Nnat, in four microarray datasets; entries with a null value indicate that this gene is missing in the specific dataset; their fold change information in BAIR fat-fed mice and human with obesity are shown in Table 4. Additional file 1, Figure S2 gives the hierarchical clustering for these 28 genes in BAIR fat-fed dataset, here genes with expression most closely associated with Nnat include Aqp1, Sncg, Sulf2 and Cxcl9. Additional file 1, Figure S3 illustrates the expressions of the top four most correlated genes (Gstt1, Ccdc80, Hfe Sod3) with Nnat in BAIR and GSE6571 datasets. Among these genes is only one transcription factor, Ebf1 which has previously been suggested to have a regulatory role in adipogenesis[24]. Another of the Nnat-correlated genes, Lgals12, is a member of the galectin family of beta-galactoside-binding proteins, and is a major regulator of adipose tissue development[25].
Table 3.
Co-expression genes with Nnat in WAT
| Genes | GSE8831 | GSE4671 | BAIR | DGAP | meta.z | p-value | FDR |
|---|---|---|---|---|---|---|---|
| GSTT1 | 0.33 | 0.92 | 0.87 | 0.71 | 6.16 | 7.50E-10 | 4.59E-06 |
| CCDC80 | 0.36 | 0.93 | 0.86 | 5.82 | 5.85E-09 | 3.58E-05 | |
| HFE | 0.71 | 0.93 | 0.71 | 5.81 | 6.37E-09 | 3.90E-05 | |
| SOD3 | 0.64 | 0.84 | 0.79 | 0.51 | 5.55 | 2.93E-08 | 0.0002 |
| NPDC1 | 0.67 | 0.92 | 0.62 | 0.7 | 5.44 | 5.47E-08 | 0.0003 |
| AQP1 | 0.68 | 0.71 | 0.8 | 0.47 | 5.37 | 7.73E-08 | 0.0005 |
| EBF1 | 0.62 | 0.84 | 0.81 | 5.3 | 1.15E-07 | 0.0007 | |
| SNCG | 0.81 | 0.58 | 0.72 | 5.28 | 1.30E-07 | 0.0008 | |
| LGALS12 | 0.95 | 0.83 | 5.15 | 2.57E-07 | 0.0016 | ||
| SLC6A13 | 0.73 | 0.8 | 0.7 | 0.4 | 5.1 | 3.43E-07 | 0.0021 |
| HEBP1 | 0.73 | 0.84 | 0.69 | 5.09 | 3.53E-07 | 0.0022 | |
| NPR3 | 0.66 | 0.71 | 0.77 | 0.54 | 5.07 | 4.04E-07 | 0.0025 |
| RAD50 | 0.63 | 0.35 | 0.85 | 5.06 | 4.16E-07 | 0.0025 | |
| H6PD | 0.54 | 0.85 | 0.77 | 0.53 | 5.04 | 4.57E-07 | 0.0028 |
| HTRA3 | 0.48 | 0.95 | 0.67 | 5.01 | 5.76E-07 | 0.0035 | |
| ART3 | 0.63 | 0.81 | 0.77 | 4.95 | 7.28E-07 | 0.0045 | |
| CAV1 | 0.67 | 0.73 | 0.78 | 4.95 | 7.46E-07 | 0.0046 | |
| AOC3 | 0.27 | 0.95 | 0.76 | 0.25 | 4.94 | 7.73E-07 | 0.0047 |
| ANXA6 | 0.65 | 0.73 | 0.73 | 0.57 | 4.84 | 1.29E-06 | 0.0079 |
| CXCL9 | 0.33 | 0.53 | 0.88 | 4.71 | 2.51E-06 | 0.0154 | |
| JUP | 0.55 | 0.79 | 0.62 | 0.89 | 4.7 | 2.60E-06 | 0.0159 |
| BLCAP | 0.5 | 0.9 | 0.61 | 0.76 | 4.65 | 3.30E-06 | 0.0202 |
| FZD4 | 0.49 | 0.73 | 0.79 | 0.5 | 4.63 | 3.62E-06 | 0.0221 |
| SULF2 | 0.88 | 0.86 | 4.63 | 3.68E-06 | 0.0226 | ||
| CD151 | 0.83 | 0.87 | 0.46 | 4.62 | 3.79E-06 | 0.0232 | |
| MAP1LC3A | 0.68 | 0.78 | 0.59 | 0.67 | 4.56 | 5.05E-06 | 0.0309 |
| EPN2 | 0.54 | 0.91 | 0.53 | 0.71 | 4.48 | 7.42E-06 | 0.0454 |
High significant co-expression genes with Nnat in WAT based on gene expression meta-analysis. This table shows the Pearson correlation coefficient value of each gene in each of the four microarray datasets, blank value means that the relevant gene is not expressed (not passed the QC or even not included into the microarray chip). Top combined meta-zscore, meta-pvalue and FDR test are shown by FDR < 0.05.
Table 4.
The fold change of co-expression genes
| Genes | BAIR.2d | BAIR.8d | BAIR.3w | BAIR.15w | Human |
|---|---|---|---|---|---|
| NNAT | 7.82 | 3.54 | 1.02 | 1.19 | 0.72 |
| GSTT1 | 2.5 | 1.43 | 1.1 | 0.89 | 0.86 |
| CCDC80 | 2.65 | 2.2 | 0.69 | 1.05 | 2.57 |
| HFE | 1.59 | 1.16 | 0.77 | 1.34 | 0.87 |
| SOD3 | 1.99 | 1.64 | 0.87 | 1.45 | 0.89 |
| NPDC1 | 1.28 | 1.14 | 0.92 | 0.96 | 1.88 |
| AQP1 | 1.64 | 1.32 | 1.27 | 1.01 | 1.52 |
| EBF1 | 2.07 | 1.54 | 0.89 | 1.28 | 0.68 |
| SNCG | 6.16 | 8.06 | 1.53 | 1.53 | 1.26 |
| LGALS12 | 3.12 | 1.47 | 0.89 | 0.93 | 1.03 |
| SLC6A13 | 2.09 | 1.74 | 0.54 | 1.89 | 0.77 |
| HEBP1 | 1.35 | 1.18 | 0.77 | 1 | 1.08 |
| NPR3 | 5.28 | 3.14 | 0.7 | 2.87 | 1.69 |
| RAD50 | 2.02 | 1.67 | 0.85 | 1.24 | 0.71 |
| H6PD | 1.82 | 1.38 | 1.05 | 0.95 | 1.16 |
| HTRA3 | 1.65 | 1.23 | 0.8 | 1.27 | 1.12 |
| ART3 | 2.03 | 1.41 | 0.92 | 0.88 | 1.33 |
| CAV1 | 1.48 | 1.18 | 0.88 | 1.14 | 0.93 |
| AOC3 | 1.91 | 1.11 | 0.75 | 1.37 | 0.83 |
| ANXA6 | 1.64 | 1.59 | 0.97 | 1.4 | 0.85 |
| CXCL9 | 3.48 | 2.3 | 1.26 | 1.25 | 1.22 |
| JUP | 1.54 | 1.07 | 0.89 | 1 | 1.06 |
| BLCAP | 1.58 | 1.11 | 1.03 | 1.1 | 0.92 |
| FZD4 | 2.24 | 1.42 | 0.88 | 1.9 | 1.11 |
| SULF2 | 1.59 | 1.31 | 1 | 1.11 | 2.08 |
| CD151 | 1.54 | 1.23 | 0.95 | 1.18 | 1.1 |
| MAP1LC3A | 1.32 | 0.95 | 1.03 | 1.32 | 0.93 |
| EPN2 | 1.14 | 1.03 | 1.18 | 1.17 | 1.24 |
This table shows the fold change (FC) of those co-expression genes of Nnat in adipose tissue in fat-fed mice during development of diet-induced obesity, and in human obese adipose tissue.
In order to investigate whether these 27 genes share conserved TFBS with Nnat as identified in Table 1, we used Genomatix tools to analyze the promoters of all 28 genes in human, mouse and rat separately. We found that nine TFBS families in Table 1, V$CREB, V$EVI1, V$GATA, V$KLFS, V$NR2F, V$RXRF, V$SORY, V$STAT, V$HAND, which are conserved in the promoter region of Nnat from seven species, are significant TFBS of all these 28 genes in human, mouse and rat, indicating these TFBSs are likely to be important in WAT. We also found that V$RREB and V$TEAF are highly significant TFBSs (p-value < 3.31e-10, p-value < 6.22e-8, p-value < 5.58e-7) in most of the 28 genes, while V$PPAR is highly significant (p-value < 5.7e-4, p-value < 4.14e-6, p-value < 5.7e-4) half in of the 28 genes (Table 5).
Table 5.
Common TFBS among Nnat and its co-vary genes
| TFBS | hs_pval | mm_pval | rat_pval | #hs | #mm | #rat |
|---|---|---|---|---|---|---|
| V$PPAR | 5.70E-04 | 4.14E-06 | 5.70E-04 | 12 | 15 | 12 |
| V$ZF5F | 1.62E-06 | 2.34E-04 | 0.0033 | 18 | 15 | 13 |
| V$RREB | 3.31E-10 | 6.22E-08 | 5.58E-07 | 26 | 24 | 23 |
| V$TEAF | 6.22E-08 | 6.22E-08 | 5.34E-09 | 24 | 24 | 25 |
| V$STAT | 9.89E-09 | 1.29E-07 | 8.77E-06 | 26 | 25 | 23 |
| V$HESF | 2.35E-09 | 4.30E-08 | 2.83E-05 | 27 | 26 | 23 |
| V$MYOD | 1.29E-08 | 1.69E-05 | 9.92E-05 | 27 | 24 | 23 |
| V$GLIF | 5.36E-09 | 3.90E-06 | 3.00E-07 | 28 | 26 | 27 |
| V$MEF2 | 0.0025 | 2.52E-05 | 0.0025 | 21 | 24 | 21 |
| V$LEFF | 0.0042 | 5.25E-05 | 2.80E-04 | 21 | 24 | 23 |
| V$CTCF | 9.37E-05 | 1.43E-05 | 1.59E-06 | 24 | 25 | 26 |
| V$SRFF | 1.50E-05 | 0.002 | 1.67E-06 | 25 | 22 | 26 |
| V$ZBPF | 1.23E-06 | 2.30E-08 | 1.23E-06 | 27 | 28 | 27 |
| V$HIFF | 2.45E-07 | 8.69E-12 | 2.45E-07 | 22 | 26 | 22 |
| V$EBOX | 8.80E-06 | 6.89E-05 | 1.34E-08 | 26 | 25 | 28 |
| V$NFAT | 5.79E-05 | 5.79E-05 | 0.0025 | 27 | 27 | 25 |
| V$EGRF | 1.84E-04 | 2.35E-06 | 1.84E-04 | 25 | 27 | 25 |
| V$SP1F | 8.94E-06 | 8.94E-06 | 8.94E-06 | 27 | 27 | 27 |
| V$CAAT | 2.19E-06 | 2.19E-06 | 1.00E-04 | 28 | 28 | 27 |
| V$CART | 0.0029 | 0.0029 | 0.0029 | 25 | 25 | 25 |
| V$IRFF | 6.53E-04 | 0.0034 | 6.53E-04 | 26 | 25 | 26 |
| V$NF1F | 4.37E-06 | 5.72E-08 | 4.37E-06 | 23 | 25 | 23 |
| V$AHRR | 6.01E-07 | 6.07E-08 | 1.42E-04 | 24 | 25 | 21 |
| V$HEAT | 0.0015 | 2.07E-04 | 2.07E-04 | 26 | 27 | 27 |
| V$MYBL | 0.0021 | 3.12E-04 | 7.05E-06 | 26 | 27 | 28 |
| V$STAT | 1.38E-05 | 1.38E-05 | 1.38E-05 | 28 | 28 | 28 |
| V$NEUR | 1.64E-04 | 2.64E-05 | 3.09E-06 | 24 | 25 | 26 |
| V$HAND | 2.30E-04 | 2.30E-04 | 2.30E-04 | 28 | 28 | 28 |
| V$BCL6 | 5.24E-06 | 0.0027 | 3.39E-05 | 24 | 20 | 23 |
| V$GATA | 6.49E-05 | 6.49E-05 | 6.49E-05 | 28 | 28 | 28 |
| V$EVI1 | 3.23E-04 | 3.23E-04 | 3.23E-04 | 28 | 28 | 28 |
| V$CREB | 3.64E-04 | 3.64E-04 | 3.64E-04 | 28 | 28 | 28 |
| V$KLFS | 6.56E-04 | 6.56E-04 | 6.56E-04 | 28 | 28 | 28 |
The common TFBS among the 28 (Nnat and its co-vary) genes in three species respectively. The last three columns give the number of promoter sequences among the 28 which share the same TFBS in their promoter sequences
We used the Genomatix Promoter Module Library and were able to identify highly conserved models of five transcription factor elements highly conserved across the Nnat promoter. Among these promoter sequences, we got 20 significant models which consist of elements from five transcription factor family. Figure 1 shows TFBPs for V$EVI1 - V$EGRF - V$E2FF - V$NRF1 - V$LEFF, on the Nnat promoter close together in a similar region for six species separately just upstream of the major TSP. V$EVI1 - V$EGRF - V$E2FF and -V$LEFF are in Table 1 and TF members of these families are expressed in WAT, hypothalamus and pancreas. V$EVI1 - V$EGRF and - V$LEFF in Table 5 indicate conservation in the promoters of genes that co-vary with Nnat. There are important transcription factors in these families, for example Egr2 and Tcf7l2, which are implicated in stress responses and T2D; E2f which is concerned with adipogenesis and Evi1, which activates Stat3, a transcription factor concerned with leptin signalling in the hypothalamus.
Figure 1.
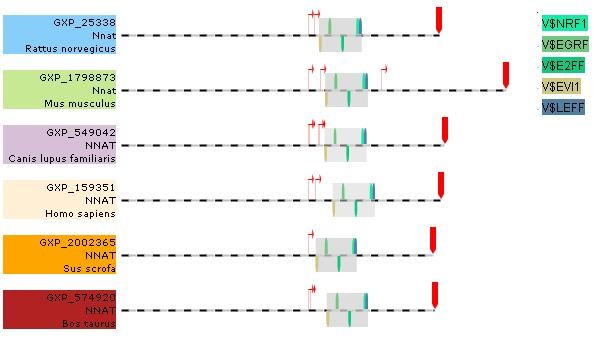
Transcription factor module in six Nnat promoter sequences. There are 20 significant five-element modules found in the six promoter sequences of Nnat for six species (Human, Mus, Can, Sus, Bos, Rat), sequence in 1200 bp upstream and 800 bp downstream, here shows TFBSs for V$EVI1 - V$EGRF - V$E2FF - V$NRF1 - V$LEFF, on the Nnat promoter closely and together in the similar region of the six species separately just upstream of the major TSP.
Functional networks formed by genes that co-vary with Nnat
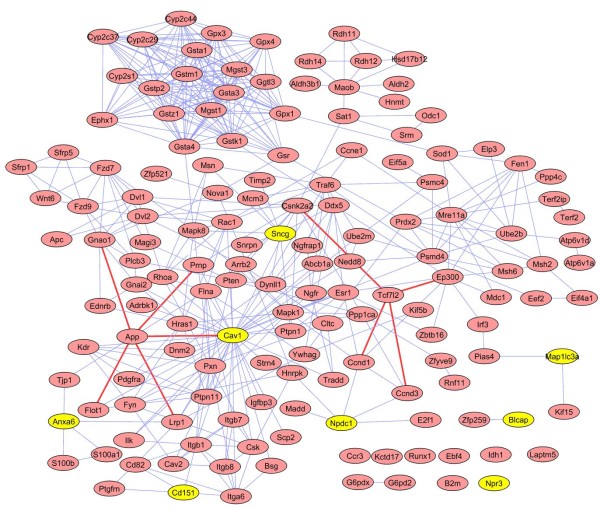
In the mouse PPI network, we sought Nnat, and the 27 genes that co-vary with it, and their respective first neighbours. 21 of the 27 Nnat co-expression genes can be mapped to this PPI network, which contains 375 first neighbours, leading to a sub-network with a total of 396 nodes and 3804 edges. Among these genes, 191 (48%) are expressed in hypothalamus (including 11 of the 21 Nnat co-varying genes) and 275 (69%) are expressed in WAT (including all 21 of the Nnat co-expression genes), 190 (48%) are expressed in pancreas (including 8 of the 21 Nnat co-varying genes, all expressed in hypothalamus as well). The functional groups formed by the PPI involving the 21 Nnat co-varying genes (examples given in parentheses with sentinel gene emboldened) correspond to the following major biological functions: (1) cellular stress, inflammation, adipocyte metabolism, metabolic syndrome and type 2 diabetes (Sncg, Erk1/2, Jnk, Hnf4a and Fabp4); (2) detoxification of aldehydes, retinol and products of oxygen free radical metabolism (Aoc3, Aldh1a3/9a1, Rdh11/12/13); (3) local natriuretic peptide system and growth (Npr3, Nppa); (4) glutathione and xenobiotic metabolism (Gstt1/3, Cyp1a1/1b1); (5) iron metabolism (Hfe and Tfrc); (6) Wnt signaling (Fzd6, Sfrp1 and Wtn11); (7) adipogenesis (Ebf1, Ebf4, Crebbp, Ep300 ); (8) protein turnover (Rad50, Eif4a1, Psmc4 and Ube2b); (9) integrins, neurodegeneration, and metabolic, inflammatory and growth-control signalling (Cav1, Ngfr, Tnfrsf1b, Hras1, Pten, App, Hnf4a and Esr). Figure 2 shows a sub-network with genes expressed in WAT, hypothalamus and pancreatic β-cells, which has 151 nodes and 447 links. Eight genes that co-vary with Nnat in the three tissues are shown in yellow, and comprise: Anxa6, Cav1, Cd151, Gstt1, Map1lc3a, Npdc1, Npr3, and Sncg. Nnat does not appear since it is not included in the mouse PPI database.
Figure 2.
Mouse Nnat co-expression gene tissue-specific PPI network. All Nnat co-expression genes in Table 3 and their first neighbours in mouse PPI network showing expressed in WAT, hypothalamus and pancreas, generated a tissue-specific network with 151 nodes and 447 links. The eight genes in yellow are Nnat co-varying genes in Table 3. Two obesity relative genes App and Tcf7l2 and their direct associated genes linked by red edges.
Discussion
This study explored the behaviour of the diet-responsive gene Nnat in multiple gene expression datasets from mouse adipose tissue, and subsequently analysed in silico the promoter of Nnat in seven mammalian species. Rosen and MacDougald[24] have pointed out that at least 100 TFs are expressed in adipocyte, and our analyses here have sought to focus attention on those which could play a part in the regulation of expression of Nnat and its co-varying genes. Based on our study we find 50 TFBSs conserved across seven species on the Nnat promoter, and expression of the TFs corresponding to these in adipose tissue, many of which have known roles in adipocyte processes including, adipogenesis (Klf15, Irf1, Creb1, Egr2, Gata3); lipogenesis (Mlxipl, Srebp1c); inflammation (Jun, Stat3); insulin signalling and diabetes susceptibility (Foxo1, Tcf7l2). These TFs are also well expressed in the hypothalamus and pancreas.
NeuroD1 is the only confirmed transcription factor for Nnat, whose expression regulates in the developing pancreas[14]. Binding sites for the TF family V$NEUR (which includes NeuroD1 and NeuroD2) are predicted in six of the seven mammalian Nnat proximal promoter regions examined here (all except rat), and are also present further upstream in the rat Nnat promoter region. Members of this family are expressed at reasonable levels in all three mouse tissues analysed here, so it remains possible that the V$NEUR family participates in Nnat regulation in several tissues. NeuroD1 is considered to be a weak activator of transcription, but is known to cooperate with other transcription factors, including Rreb1 and Sp1[23,26], in the stimulation of gene expression. Binding sites for both of these families are conserved in the Nnat promoter across mammalian species, as well as in the promoters for the Nnat co-varying genes analysed above. Indeed this property requires both the physical interaction of Rreb1 with NeuroD1, and binding of Rreb1 to DNA. Direct evidence for such a mechanism of regulation requires experimental validation. The expression of Nnat remains strong in the developing pituitary of NeuroD1 knockout mice[14], so it is clear that different TFs (or combinations thereof) are likely to be of varying importance in regulating Nnat expression in different tissues.
Pparγ is both necessary and sufficient for adipocyte differentiation of fibroblast/pre-adipocyte cell lines[24] and the presence, in all the Nnat promoters analysed here, of Pparγ binding sites is consistent with it playing a role in regulation of Nnat expression in WAT. Nnat expression is strongly reduced in WAT of a mouse model in which a dominant negative version of Pparγ is expressed[17]. Activation of Pparγ with Pparγ agonists improves insulin signalling and decreases inflammation. This is consistent with the view that Nnat protects against toxins and is concerned with resolution of tissue injury. Indeed our unpublished results in 3T3L1 cells, where we have over expressed Nnat indicate an anti-inflammatory function. However there is currently no evidence on whether Pparγ plays a direct role in the regulation of Nnat. Likewise, in mice expressing an inactive mutant version of Srebp1c, Nnat expression in WAT is severely reduced as well[17]. In both of these mouse models complex changes in WAT gene expression make it difficult to draw conclusions as to the regulation of particular genes by specific TFs. Pparγ is known to operate in a cellular network with other TFs of the EBF, C/EBP, KLF and IRF families in regulating adipogenesis[24,27-29]. Our bioinformatical analysis suggests that Pparγ could be a candidate for Nnat regulation, meanwhile the absence of V$SREB binding sites in the Nnat promoter does not support a direct role for Srebp1c.
The pattern of gene expression observed in the mouse WAT upon high fat feeding in experiments analysed here suggests a critical in oxidative stress; indeed sentinel indicators of this Gstt1, Ccdc80, Hfe and Sod3 all concerned with cellular management of free radicals are the genes most highly correlated with Nnat in the meta-analysis (Additional file 1, Figure S3). Genes concerned with adipogenesis (Ebf1, Fzd4 and Lgals12) and inflammation (Sncg, Cxcl9 and Aoc3) are also highly correlated in mouse WAT upon high fat feeding (Additional file 1, Figure S2). This is consistent with data indicating that Nnat expression favours adipogenesis and inflammation. Of the aforementioned adipogenic TFs, only Ebf1 expression correlates well with Nnat, but the conserved Nnat promoter does not possess Ebf1 binding sites (TF family V$NOLF). Thus it is unlikely that Ebf1 regulates Nnat expression; instead they may both be targets of the same TFs. By treating the Nnat co-varying genes as a set, and carrying out promoter analysis on this set, we discovered that many wide-acting TFs have binding sites in all of these genes. This does not itself indicate which, if any, of these TFs may regulate the Nnat co-varying genes, but does draw attention to the families V$HIFF (example: HIF1a) V$RREB (example: Rreb1) and V$TEAF (example: Tead4) as being the most highly over-represented TFBS in this group of genes.
Our gene set enrichment analysis showed that expression of Nnat is strongly negatively correlated with energy metabolism (most notably oxidative phosphorylation) and positively correlated with inflammation. Repression of oxidative phosphorylation gene expression is one of the major changes seen in human diabetes[21], and a metabolic/inflammatory network is thought to underlie metabolic syndrome in susceptible mice[30]. Although in the human data NNAT expression is not correlated with oxidative phosphorylation in the GSEA in a more general analysis of gene expression in this human data the oxidative phosphorylation gene set is enriched. We would expect that major changes in gene set regulation require the participation of wide-acting TFs and/or extensive networks of TFs and their cofactors.
We found 27 genes that show strong co-variation with Nnat. 21 of these can be mapped to the PPI network, and eight of these (Anxa6, Cav1, Cd151, Gstt1, Map1lc3a, Npdc1, Npr3, and Sncg) are expressed in adipose tissue, pancreas and hypothalamus. App and Tcf7l2, genes implicated in metabolic inflammation, Alzheimer's disease and diabetes are shown linked to their respective first neighbours (Figure 2). Sncg along with Nnat is a leptin responsive gene[11].
Conclusions
In conclusion we identify numerous TFBSs, conserved among mammals, on the Nnat promoter, and expression of TFs corresponding to these in adipose tissue, hypothalamus and pancreas. Many of these have known roles in adipocyte, as well as pancreatic and hypothalamic processes. We observe clustering of TFBSs for Egr1 and Tcf7l2, which are implicated in stress responses and T2D; E2f which is concerned with adipogenesis and Evi1, a developmental gene highly expressed in pancreatic islets. GSEA analysis suggests the roles for Nnat in oxidative stress, resolution of inflammation, energy metabolism, mainly oxidative phosphorylation, and protein turnover. Genes that co-vary with Nnat, such as Sncg, Gstt1, Hfe and Ebf1, share some of these TFBSs and form functional networks. The recent demonstration that SNPs in Nnat are found in obese humans suggest that altered Nnat function in adipose tissue or the hypothalamus may be involved in the pathogenesis of obesity via such mechanisms.
Authors' contributions
XL and JS conceived and designed this study, XL and PT performed the data analysis, XL, PT, DJW and JS wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
This PDF file includes supplementary figures and supplementary table 1-4.
SupplementaryTable5
Contributor Information
Xinzhong Li, Email: xinzhong.li@imperial.ac.uk.
Peter A Thomason, Email: p.thomason@beatson.gla.ac.uk.
Dominic J Withers, Email: d.withers@imperial.ac.uk.
James Scott, Email: j.scott@imperial.ac.uk.
Acknowledgements
This work was supported by Wellcome Trust and BBSRC.
References
- Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414(6865):782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- Saltiel AR. New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell. 2001;104(4):517–529. doi: 10.1016/S0092-8674(01)00239-2. [DOI] [PubMed] [Google Scholar]
- Breslow JL. Cardiovascular disease burden increases, NIH funding decreases. Nat Med. 1997;3(6):600–601. doi: 10.1038/nm0697-600. [DOI] [PubMed] [Google Scholar]
- Yach D, Stuckler D, Brownell KD. Epidemiologic and economic consequences of the global epidemics of obesity and diabetes. Nat Med. 2006;12(1):62–66. doi: 10.1038/nm0106-62. [DOI] [PubMed] [Google Scholar]
- de la Monte SM, Wands JR. Alzheimer's disease is type 3 diabetes-evidence reviewed. J Diabetes Sci Technol. 2008;2(6):1101–1113. doi: 10.1177/193229680800200619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans HK, Weidman JR, Cowley DO, Jirtle RL. Comparative phylogenetic analysis of blcap/nnat reveals eutherian-specific imprinted gene. Mol Biol Evol. 2005;22(8):1740–1748. doi: 10.1093/molbev/msi165. [DOI] [PubMed] [Google Scholar]
- Evans HK, Wylie AA, Murphy SK, Jirtle RL. The neuronatin gene resides in a "micro-imprinted" domain on human chromosome 20q11.2. Genomics. 2001;77(1-2):99–104. doi: 10.1006/geno.2001.6612. [DOI] [PubMed] [Google Scholar]
- John RM, Aparicio SA, Ainscough JF, Arney KL, Khosla S, Hawker K, Hilton KJ, Barton SC, Surani MA. Imprinted expression of neuronatin from modified BAC transgenes reveals regulation by distinct and distant enhancers. Dev Biol. 2001;236(2):387–399. doi: 10.1006/dbio.2001.0327. [DOI] [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138(5):885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Vrang N, Meyre D, Froguel P, Jelsing J, Tang-Christensen M, Vatin V, Mikkelsen JD, Thirstrup K, Larsen LK, Cullberg KB. et al. The imprinted gene neuronatin is regulated by metabolic status and associated with obesity. Obesity (Silver Spring) 2010;18(7):1289–1296. doi: 10.1038/oby.2009.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung YC, Ma M, Piper S, Coll A, O'Rahilly S, Yeo GS. Novel leptin-regulated genes revealed by transcriptional profiling of the hypothalamic paraventricular nucleus. J Neurosci. 2008;28(47):12419–12426. doi: 10.1523/JNEUROSCI.3412-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mzhavia N, Yu S, Ikeda S, Chu TT, Goldberg I, Dansky HM. Neuronatin: a new inflammation gene expressed on the aortic endothelium of diabetic mice. Diabetes. 2008;57(10):2774–2783. doi: 10.2337/db07-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh YH, Kim WH, Moon C, Hong YH, Eun SY, Lim JH, Choi JS, Song J, Jung MH. Ectopic expression of Neuronatin potentiates adipogenesis through enhanced phosphorylation of cAMP-response element-binding protein in 3T3-L1 cells. Biochem Biophys Res Commun. 2005;337(2):481–489. doi: 10.1016/j.bbrc.2005.09.078. [DOI] [PubMed] [Google Scholar]
- Chu K, Tsai MJ. Neuronatin, a downstream target of BETA2/NeuroD1 in the pancreas, is involved in glucose-mediated insulin secretion. Diabetes. 2005;54(4):1064–1073. doi: 10.2337/diabetes.54.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe MK, Lee HJ, Suh YH, Han KL, Lim JH, Song J, Seong JK, Jung MH. Crucial roles of neuronatin in insulin secretion and high glucose-induced apoptosis in pancreatic beta-cells. Cell Signal. 2008;20(5):907–915. doi: 10.1016/j.cellsig.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F. et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326(5949):140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Huang LW, Snow KJ, Ablamunits V, Hasham MG, Young TH, Paulk AC, Richardson JE, Affourtit JP, Shalom-Barak T. et al. A mouse model of conditional lipodystrophy. Proc Natl Acad Sci USA. 2007;104(42):16627–16632. doi: 10.1073/pnas.0707797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRosa PC, Miner J, Xia Y, Zhou Y, Kachman S, Fromm ME. Trans-10, cis-12 conjugated linoleic acid causes inflammation and delipidation of white adipose tissue in mice: a microarray and histological analysis. Physiol Genomics. 2006;27(3):282–294. doi: 10.1152/physiolgenomics.00076.2006. [DOI] [PubMed] [Google Scholar]
- Siu IM, Bai R, Gallia GL, Edwards JB, Tyler BM, Eberhart CG, Riggins GJ. Coexpression of neuronatin splice forms promotes medulloblastoma growth. Neuro Oncol. 2008;10(5):716–724. doi: 10.1215/15228517-2008-038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel B, Metsuyanim S, Schmidt-Ott KM, Fridman E, Jacob-Hirsch J, Simon A, Pinthus J, Mor Y, Barasch J, Amariglio N. et al. Multiple imprinted and stemness genes provide a link between normal and tumor progenitor cells of the developing human kidney. Cancer Res. 2006;66(12):6040–6049. doi: 10.1158/0008-5472.CAN-05-4528. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Nair S, Lee YH, Rousseau E, Cam M, Tataranni PA, Baier LJ, Bogardus C, Permana PA. Increased expression of inflammation-related genes in cultured preadipocytes/stromal vascular cells from obese compared with non-obese Pima Indians. Diabetologia. 2005;48(9):1784–1788. doi: 10.1007/s00125-005-1868-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray SK, Nishitani J, Petry MW, Fessing MY, Leiter AB. Novel transcriptional potentiation of BETA2/NeuroD on the secretin gene promoter by the DNA-binding protein Finb/RREB-1. Mol Cell Biol. 2003;23(1):259–271. doi: 10.1128/MCB.23.1.259-271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7(12):885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Yang RY, Hsu DK, Yu L, Chen HY, Liu FT. Galectin-12 is required for adipogenic signaling and adipocyte differentiation. J Biol Chem. 2004;279(28):29761–29766. doi: 10.1074/jbc.M401303200. [DOI] [PubMed] [Google Scholar]
- Ray SK, Leiter AB. The basic helix-loop-helix transcription factor NeuroD1 facilitates interaction of Sp1 with the secretin gene enhancer. Mol Cell Biol. 2007;27(22):7839–7847. doi: 10.1128/MCB.00438-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J, Yan QW, Schones DE, Kamal M, Hsu CH, Zhang MQ, Crawford GE, Rosen ED. Interferon regulatory factors are transcriptional regulators of adipogenesis. Cell Metab. 2008;7(1):86–94. doi: 10.1016/j.cmet.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7(4):339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao D, Lazar MA. Peroxisome proliferator activated receptor gamma, CCAAT/enhancer-binding protein alpha, and cell cycle status regulate the commitment to adipocyte differentiation. J Biol Chem. 1997;272(34):21473–21478. doi: 10.1074/jbc.272.34.21473. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK. et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452(7186):429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This PDF file includes supplementary figures and supplementary table 1-4.
SupplementaryTable5