Abstract
Here we describe the cloning and initial characterization of a previously unidentified CRF-related neuropeptide, urocortin II (Ucn II). Searches of the public human genome database identified a region with significant sequence homology to the CRF neuropeptide family. By using homologous primers deduced from the human sequence, a mouse cDNA was isolated from whole brain poly(A)+ RNA that encodes a predicted 38-aa peptide, structurally related to the other known mammalian family members, CRF and Ucn. Ucn II binds selectively to the type 2 CRF receptor (CRF-R2), with no appreciable activity on CRF-R1. Transcripts encoding Ucn II are expressed in discrete regions of the rodent central nervous system, including stress-related cell groups in the hypothalamus (paraventricular and arcuate nuclei) and brainstem (locus coeruleus). Central administration of 1–10 μg of peptide elicits activational responses (Fos induction) preferentially within a core circuitry subserving autonomic and neuroendocrine regulation, but whose overall pattern does not broadly mimic the CRF-R2 distribution. Behaviorally, central Ucn II attenuates nighttime feeding, with a time course distinct from that seen in response to CRF. In contrast to CRF, however, central Ucn II failed to increase gross motor activity. These findings identify Ucn II as a new member of the CRF family of neuropeptides, which is expressed centrally and binds selectively to CRF-R2. Initial functional studies are consistent with Ucn II involvement in central autonomic and appetitive control, but not in generalized behavioral activation.
CRF is a 41-aa peptide best known for its indispensable role in initiating pituitary-adrenal responses to stress, an effect mediated by type 1 CRF receptors (1). In addition, CRF is widely distributed in brain and has been shown repeatedly to participate in the mobilization of complementary autonomic and behavioral adjustments to a variety of threatening circumstances (2, 3). This has fostered the widely held hypothesis that CRF plays an important role in the integration of adaptive responses to stress. Rigorous testing of this idea has been impeded by the fact that a number of the cell groups identified as sites of peptide action in eliciting stress-like autonomic and behavioral responses have been found to be lacking or impoverished in the expression of requisite ligand(s), receptor(s), or both (4, 5). This has kindled the search for additional CRF-related signaling molecules, which currently number two ligands, G protein-coupled receptors derived from two distinct genes (CRF-R1 and CRF-R2), and a binding protein, whose function remains incompletely understood (6, 7).
A second mammalian CRF-related neuropeptide, urocortin (Ucn), was discovered recently by our group (8) and shown to be bound with high affinity by both known CRF receptor types, whereas CRF is bound in a highly preferential manner by CRF-R1. Centrally administered Ucn is more potent than CRF in suppressing appetite, but it is less so in generating acute anxiety-like effects and generalized behavioral activation (9). This has been taken to indicate that Ucn might mediate some stress-related effects attributed initially to CRF, at least in part by serving as an endogenous ligand for CRF-R2. This view has been challenged, however, by such observations as that the principal cellular seats of Ucn expression in brain are not recognized as integral components of central stress-related circuitry, and that most major sites of CRF-R2 expression are poorly innervated by Ucn-containing projections (10). These and other findings support the possible existence of one or more additional CRF receptor ligands in the mammalian brain.
The rapid advances in the deposition of sequence data for both the human and mouse genomes have provided an opportunity to identify new members of many protein families. Using sequence homology searching tools, we have identified a mouse gene encoding a 38-aa peptide that represents a new member of the CRF family of neuropeptides. This peptide, termed urocortin II (Ucn II), is distinct from the other known family members in that it binds with high selectivity to CRF-R2. In this report, we describe the cloning and characterization of this neuropeptide.
Materials and Methods
PCR Amplification.
Multiple sets of nested primers were designed on the basis of the sequence of a human genomic DNA fragment identified in database mining as exhibiting structural relatedness to genes encoding known CRF family members (see Results). RACE (rapid amplification of cDNA ends)-ready cDNA was prepared from mouse whole-brain poly(A)+ RNA using the SMART RACE cDNA amplification kit (CLONTECH). A touchdown PCR protocol was used (94°C 30 s, increment from 70°C to 55°C 30 s, 72°C 3 min) followed by a second round of amplification using the nested primers (94°C 20 s, 55°C 20 s, 72°C 3 min). Candidate PCR products were cloned into pCRII-TOPO (Invitrogen) for sequencing of both strands.
Peptide Synthesis.
Murine Ucn II and a human Ucn-related peptide were synthesized manually by using the solid phase approach, a methylbenzhydryl amine resin and the Boc-strategy (11). Trifluoroacetic acid, 60% in dichloromethane, was used to remove the Boc group. Main chain assembly was mediated by diisopropylcarbodiimide. The peptides were cleaved and deprotected in hydrofluoric acid and purified using RP-HPLC and three solvent systems (triethylammonium phosphate at pH 2.25 and 6.5 and/or 0.1% TFA) (12). Peptides were greater than 95% pure using independent HPLC and capillary zone electrophoresis criteria. Mass spectra confirmed the composition of the preparations.
Radioreceptor Assay.
Crude membrane fractions were prepared from Chinese hamster ovary cells stably expressing either cloned human CRF-R1 or murine CRF-R2β. Test peptides and radioligand, 125I-[Tyr0,Glu1,Nle17]-sauvagine, diluted in assay buffer (20 mM Hepes/2 mM EGTA/0.1% BSA/10% sucrose, pH 7.6) were combined with receptor in MAGV microtiter plates (Millipore) precoated with 0.1% polyethyleneimine. The reaction mixture was incubated for 90 min at room temperature followed by rapid washing twice with assay buffer and filtration. The radioligand complex was quantified by gamma-counting. Inhibitory binding constants were determined by using prism software.
cAMP Assay.
Stably transfected Chinese hamster ovary cells (cultured in DMEM/10% FBS) were plated into 48-well tissue culture dishes (Costar) and allowed to recover for 24 h. The medium was changed to DMEM/0.1% FBS at least 2 h before treatment. The cells were preincubated for 30 min with 0.1 mM 3-isobutyl-1-methylxanthine and then exposed to peptides for 20 min at 37°C. Intracellular cAMP was extracted and measured from triplicate wells using a radioimmunoassay kit (Biomedical Technologies).
Animals and Surgical Procedures.
Adult male Sprague-Dawley rats (250–300 g at start of experiments) and C57BL/6 mice (25–40 g) were housed in a colony room on a 12 h:12 h light/dark cycle, and with free access to food and water before experimentation. For intracerebroventricular (i.c.v.) injections, rats were anesthetized with ketamine/xylazine/acepromazine and stereotaxically implanted with a 26 ga guide cannula terminating in a lateral ventricle. For i.v. administration of peptides, animals were fitted with indwelling jugular venous catheters. Rats that received i.c.v. injections were also implanted intraabdominally with a transmitter to remotely monitor gross activity levels and body temperature (MiniMitter, Bend, OR). After surgery, animals were allowed to recover for 7 days before any experimentation, during which time they were handled daily. All procedures were approved by the Institutional Animal Care and Use Committee of the Salk Institute.
Experimental Procedures and Analysis.
To monitor induced patterns of Fos expression, rats were injected at 10 a.m., either i.c.v. or i.v. with synthetic Ucn II (1, 5, or 10 μg per animal in 2 μl of saline for i.c.v. injections or 200 μl for i.v. administration), or vehicle alone, and perfused 2 h later. To monitor the effect of peptide administration on food intake, animals were injected i.c.v. with synthetic mouse Ucn II, rat Ucn, or rat/human CRF 30 min before lights out. Consumption was then measured hourly for 6 h and at 12 h. Data were analyzed by using repeated measures ANOVA, with the Bonferoni correction for multiple comparisons applied as warranted.
Tissue Processing and Histology.
Animals were deeply anesthetized with chloral hydrate (350 mg/kg, i.p.) and perfused via the ascending aorta with saline followed by ice-cold 4% paraformaldehyde in 0.1% borate buffer (pH 9.5). Brains were postfixed for 16 h and cryoprotected overnight in 10% sucrose in 0.1 M phosphate buffer. Four (mice) or six (rats) series of 30 μm-thick frozen sections were cut with a sliding microtome, collected in cold ethylene glycol-based cryoprotectant, and stored at −20°C until histochemical processing.
Immuno- and Hybridization Histochemistry.
In situ hybridization was performed with 35S-labeled antisense and sense (control) cRNA probes (13), constructed by first linearizing the TOPO-II plasmid containing the mouse cDNA. Probes were labeled to specific activities of 1–3 × 109 dpm/μg, applied to slides at concentrations of about 107 cpm/ml, and hybridized overnight at 56°C under high stringency (50% formamide). Final washes were carried out in 15 mM NaCl/1.5 mM sodium citrate at 65–68°C. Slides were then dehydrated and exposed to x-ray film (β-Max; Kodak) for 16 h and then coated with Kodak NTB-2 liquid emulsion and exposed at 4°C for 21–28 days. For immunohistochemistry, tissue was pretreated sequentially with 0.3% hydrogen peroxide and 1% sodium borohydride. It was then permeabilized with PBS/0.2% Triton X-100, and incubated with primary antiserum for 48 h in PBS/2% blocking serum. Fos immunoreactivity was localized using a polyclonal antiserum raised in rabbit against an N-terminal synthetic fragment of human Fos protein (Santa Cruz Biotechnology, 1:5K). Localization was performed by using a conventional avidin-biotin immunoperoxidase method with nickel enhancement, as described (14).
Results
Cloning.
In an effort to identify novel CRF-R ligands, a hidden Markov model (HMM) was constructed from a Clustal W alignment of known CRF family proteins, including rat/human CRF, rat Ucn, human Ucn, frog sauvagine, and white-suckerfish urotensin I, using the hmmer software package (Sean Eddy, Department of Genetics, Washington University, St. Louis, MO; see ref. 15). This HMM was used to search the public human genome database and a BAC (GenBank accession no. AC005903) derived from chromosome 3p21.3–4 was identified that contained a 109 bp region that exhibited significant sequence homology but was not a part of a previously identified gene. This region was extended to 621 bp with the identification of a human expressed sequence tag (EST) clone that overlapped with this sequence (GenBank accession no. BE622276). The human sequence, however, lacks a consensus proteolytic cleavage site that would allow for C-terminal processing of the peptide, and we therefore refer to this as a urocortin-related peptide (URP) sequence.
Fragmentary cRNA probes based on the human gene sequence specifically cross-hybridized with rat tissue (brain), suggesting that a reasonable degree of homology existed between the two species. Based on this human sequence, primers were designed to identify the homologous mouse gene using RACE. PCR reactions were run under low stringency (low Tm) conditions in an effort to allow for the maximal heterologous priming. First round amplification was carried out by using a touchdown protocol, followed by a second low stringency amplification with multiple sets of nested primers. The mouse gene was identified from whole mouse brain poly(A)+ RNA. Candidate 5′ and 3′ reaction products were identified based on their predicted size (deduced from the human sequence), cloned, and sequenced. The predicted amino acid sequence for the mouse Ucn II is listed in Fig. 1A. The gene encodes a 112-aa precursor, and the C terminus includes the coding region for the putative 38-aa mature peptide, indicated in the boxed region (Fig. 1A). The C-terminal portion of the coding sequence is followed by a glycine and paired basic residues (R-R), presumed to be involved in amidation and cleavage from the precursor, respectively. Two other putative or known URPs exist: one in human, whose peptide sequence was deduced from the published human EST as well as a recently cloned (16) pufferfish URP (from Takifugu rubripes). Alignment with the human and fish URPs, rat Ucn, and rat/human CRF is shown in Fig. 1B. At the amino acid level, the coding region of mouse Ucn II displays 76% and 47% homology with the human and fish URPs, respectively. Mouse Ucn II is comparably related to known members of this peptide family, sharing 34% and 42% amino acid identity with rat CRF and rat Ucn, respectively. Allowing for conservative substitutions, relatedness increases to 58% (with CRF) and 55% (Ucn).
Figure 1.
(A) Predicted amino acid sequence of murine Ucn II. The start methionine, marked in bold, is located upstream of the peptide coding region, which is boxed. The complete nucleotide sequence has been deposited in GenBank (accession no. AF331517). (B) Alignment of mouse Ucn II with homologous human and fish peptides (URPs) and with rat Ucn and rat/human CRF; residues identical to the mouse Ucn II sequence are boxed. ■, Amidation site (putative for human URP).
Receptor Activation.
The affinity of Ucn II on the CRF-R1 and CRF-R2 was evaluated by using a radioreceptor assay (Table 1). Compared with urocortin, Ucn II was at least 1,000-fold less effective at competing for binding of labeled sauvagine to the CRF-R1, whereas it was nearly equipotent to Ucn in competing for binding to CRF-R2. This significant selectivity for the type 2 receptor was seen also in receptor activation as measured by accumulation of intracellular cAMP. In the cAMP assay, Ucn II displayed a comparable efficacy for CRF-R2 as did Ucn. The extremely low affinity of Ucn II for CRF-R1 precluded a determination of its efficacy on this receptor.
Table 1.
Binding properties and functional activities of select CRF receptor ligands
| Peptide | CRF-R1
|
CRF-R2
|
||
|---|---|---|---|---|
| Avg. Ki, nM (binding) | Avg. EC50, nM (cAMP) | Avg. Ki, nM (binding) | Avg. EC50, nM (cAMP) | |
| Ucn II (mouse) | >100 | >100 | 0.66 (0.13–3.3) | 0.14 (0.03–0.52) |
| URP (human) | >100 | >100 | 0.50 (0.22–1.16) | 0.42 (0.16–1.1) |
| Ucn (rat) | 0.32 (0.14–0.77) | 0.29 (0.12–0.70) | 0.62 (0.14–2.8) | 0.17 (0.043–0.68) |
The values were determined from three to six independent experiments using stably transfected Chinese hamster ovary cells or their membranes for each test peptide. EC50 and Ki values were determined by using prism software. Their log10 values were averaged (γ). The average EC50 or Ki was taken to be 10γ. The standard deviation of the log10 values was calculated (σ). The ranges given were taken to be [(10γ)10σ or 10γ/10σ].
Ucn II mRNA Expression.
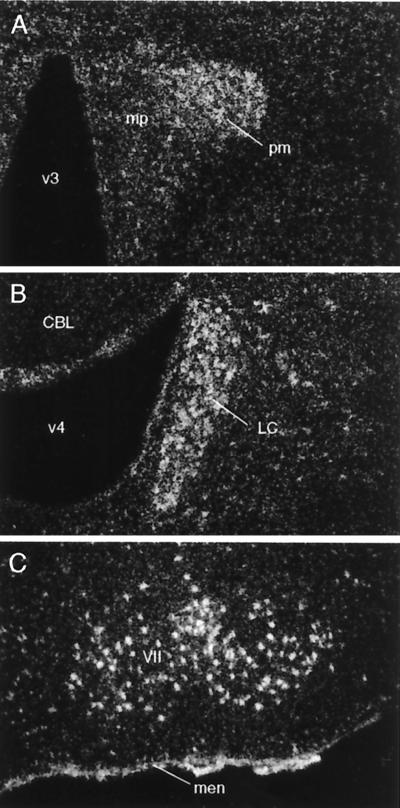
Hybridization histochemistry carried out with isotopically labeled cRNA probes revealed a consistent and restricted pattern of Ucn II mRNA expression in mouse and rat brain. Sense-strand runoffs labeled to similar specific activities as antisense probes failed to yield above-background hybridization signals. The observed distribution of Ucn II mRNA was seen to be predominantly subcortical, with major sites of expression including stress-related cell groups such as the paraventricular, supraoptic, and arcuate nuclei of the hypothalamus, and the locus coeruleus of the rostral pons (Fig. 2). Motor nuclei of the brainstem (trigeminal, facial, hypoglossal), as well of the spinal ventral horn, were also identified as sites of Ucn II mRNA expression. Among non-neuronal elements, positive hybridization signals were observed consistently over the meninges but not the choroid plexus or ependyma. No clear suggestion of Ucn II mRNA expression by glial elements was evident in our material.
Figure 2.
Ucn II mRNA expression in the rat brain. Darkfield photomicrographs showing labeling (white grains) observed over select regions using an isotopically labeled antisense cRNA probe generated from a mouse Ucn II cDNA. Positive hybridization signals are seen over the paraventricular nucleus of the hypothalamus (A), principally over its magnocellular division (pm), with more diffuse signal seen over the parvocellular aspect (mp), and broadly over the locus coeruleus (LC; B), facial motor nucleus (VII, C) and meninges (men) at the ventral surface of the brain. CBL, cerebellum; v3, third ventricle; v4, fourth ventricle. [Magnifications: ×75 (A and B); ×50 (C).]
Ucn II-Induced Fos Expression.
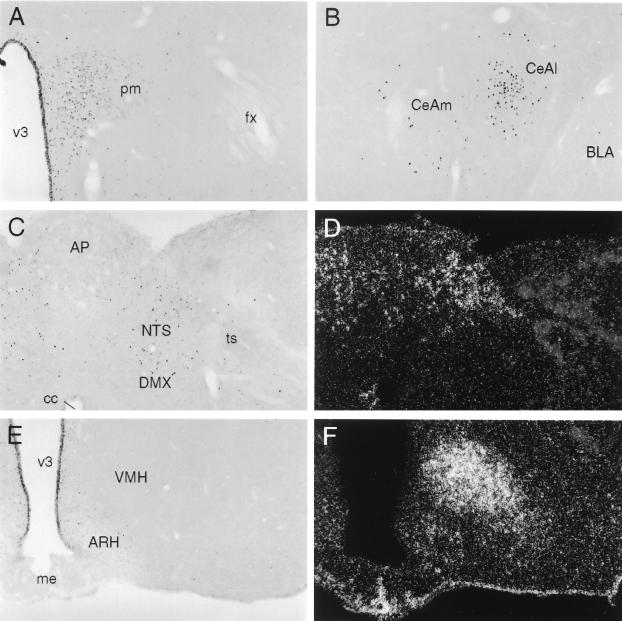
To identify cell groups responsive to central Ucn II administration, and to evaluate the extent to which these may conform to sites of CRF-R2 expression, we monitored the induced expression of the immediate-early gene product, Fos, in response to i.c.v. peptide administration. Injection of 1 μg of synthetic Ucn II gave rise to activational responses that were most salient in a group of interconnected structures involved in central autonomic control (17, 18). These included discrete aspects of the bed nucleus of the stria terminalis, the central nucleus of the amygdala, the paraventricular nucleus of the hypothalamus (PVH), parabrachial nucleus and nucleus of the solitary tract (NTS; Fig. 3). Of these, only the NTS has been described as a locus of CRF-R2 expression (19). Fos induction in other major sites of CRF-R2 expression, including the lateral septum, midbrain raphe nuclei, and the ventromedial nucleus of the hypothalamus (19, 20), was not distinguishable from that seen in saline-injected controls. Higher doses of peptide (5 or 10 μg) provoked more robust activational responses of similar distribution.
Figure 3.
Cellular activation patterns in response to central Ucn II microinjection. (A-C and E) Brightfield photomicrographs of immunoperoxidase preparations showing induced Fos expression in rats killed 2 h after i.c.v. injection of 1 μg of synthetic mouse Ucn II. Darkfield photomicrographs showing hybridization histochemical localization of CRF-R2 mRNA in regions corresponding to those illustrated in C and E are provided in D and F, respectively. Central Ucn injection provoked Fos induction primarily in a set of interconnected structures involved in central autonomic and neuroendocrine control, including the parvocellular division of the paraventricular nucleus (A), the central nucleus of the amygdala (B), and the nucleus of the solitary tract (NTS, C). Among these, only the NTS is a site of CRF-R2 expression (D). Other principal sites of CRF-R2 expression, including the ventromedial nucleus of the hypothalamus (F), failed to show Ucn II-induced Fos expression over the range of peptide doses examined (1–10 μg). (Magnification for all photomicrographs: ×75.)
To control for potential systemic effects of i.c.v. injections, a similar range of Ucn II doses was given intravenously to separate groups of rats. Only the highest (10 μg) dose gave rise to Fos induction that was clearly above control levels, and although the pattern was similar to that seen in response to central injections, neither the number of labeled cells nor their staining intensity approached that seen reliably following i.c.v. injections of 1 μg of Ucn II.
Behavioral Effects.
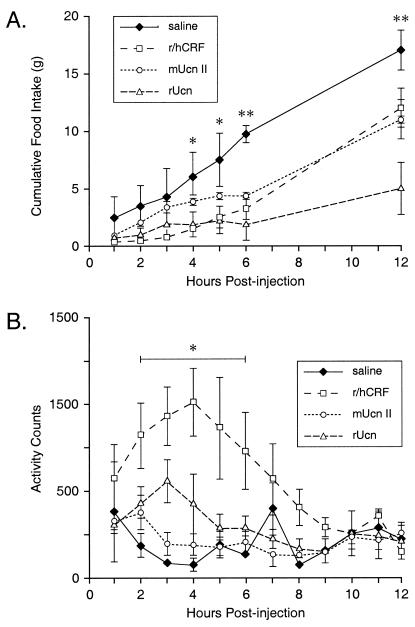
Like CRF and Ucn, Ucn II is also capable of acting centrally to inhibit food intake (Fig. 4A). Measures from separate groups of rats injected with these peptides (1 μg, i.c.v.) at the beginning of the nocturnal phase of their day-night cycle manifest a significant interaction between treatment and time point [F(18, 95) = 4.22, P < 0.0001], with both main effects also achieving reliability. All three peptides significantly reduced food intake over the 12-h interval, with the degree of suppression ranging from 30% (CRF) to 35% (Ucn II) to 70% (Ucn). These effects tended to be distributed differentially over time, with both Ucn- and CRF-treated animals eating significantly less than saline-injected controls earlier in the test period (4–5 h) than did Ucn II-treated rats (6 h).
Figure 4.
Effects of central Ucn II on food intake and gross motor activity. (A) Mean (± SEM; n = 3–6 per group) cumulative nighttime food intake (g) following i.c.v. administration of 1 μg of CRF, Ucn, or Ucn II. Both CRF and Ucn significantly reduced food intake compared with saline-injected controls, beginning at 4 h postinjection, whereas the effect of Ucn II was not manifest until 6 h after treatment. *, P < 0.002 (CRF and Ucn vs. saline); **, P < 0.002 (CRF, Ucn, and Ucn II vs. saline). (B) Telemetric measures of gross motor activity were significantly elevated in animals that received i.c.v. injections of CRF; neither Ucn nor Ucn II significantly affected motor activity. *, P < 0.001 (CRF vs. saline).
In these same subjects, gross motor activity and body temperature were monitored telemetrically (Fig. 4B). Analysis of activity data revealed a significant interaction between drug and time point [F(33, 110) = 1.94, P < 0.006], with both main effects also achieving significance. Posthoc comparisons revealed that animals that received CRF were significantly more active than vehicle-treated rats over the interval 2–6 h postinjection (P < 0.001). Neither Ucn nor Ucn II treatment provoked reliable alterations in this measure at any postinjection time point. Core body temperature was also recorded, with each peptide provoking comparably mild (0.5–1°C) and transient (2 h) hypothermic responses (data not shown).
Discussion
We have used genome-wide homology searching to identify a new member of the CRF family of neuropeptides. The new ligand, Ucn II, binds selectively to CRF-R2, is expressed in discrete areas of the rat central nervous system, and activates central neurons involved in the processing of visceral sensory information, and in modulating autonomic outflow. Further, Ucn II inhibits food intake, without any effect on gross motor activity.
In addition to a murine peptide that exhibits structural, binding, activity, and expression characteristics expected of a CRF family member, we have also identified a human URP (based on a publicly available EST sequence), which is 76% identical to the mouse sequence at the amino acid level. It is of interest to note that, in rats, central administration of synthetic human URP provokes a pattern of Fos induction in brain and a suppression of nighttime feeding that are comparable to those seen in response to similar doses of mouse Ucn II (unpublished observations). However, an important difference evident in the human peptide is the absence of any obvious proteolytic cleavage site that would provide for C-terminal processing of a human homolog. It remains to be determined whether and how any homologous human peptide may be generated from this protein. Nevertheless, whereas Ucn is bound with high affinity by, and signals potently through, both CRF-R1 and CRF-R2 (6–8), mouse Ucn II and human URP exhibit a high degree of CRF-R2 selectivity in these measures, and will doubtless be of value in dissociating functions mediated by the two receptor types.
Ucn II mRNA displays a limited subcortical distribution in rodent brain that is unique, although ostensibly overlapping in part with those of CRF (paraventricular nucleus; e.g., ref. 21) and Ucn (brainstem and spinal motor nuclei; e.g., ref. 10). Of particular interest is the fact that the transcript is expressed in cell groups involved in stress-related physiologic and behavioral functions (see ref. 5). This includes the locus coeruleus, which issues widespread projections to the cortical mantle and has been implicated in generating levels of arousal and anxiety (e.g., ref. 22), the paraventricular nucleus, which houses multiple relevant neurosecretory neuron populations and projects within the central nervous system to modulate sensory and motor traffic in central autonomic circuitry (e.g., ref. 23), and the arcuate nucleus, which has been identified as a pivotal component of an extended system subserving the regulation of food intake and energy balance (e.g., ref. 24). Although anatomical and functional data to define the new peptide's place in such contexts are as yet lacking, the central Ucn II system holds potential for participating in stress-related functions long implicated as the province of the broader central CRF network. This contrasts with Ucn, whose dominant seat of cellular expression in brain, the Edinger–Westphal nucleus, shows very limited capacities in this regard, largely by virtue of a paucity of documented projections to the forebrain (8, 10).
In view of its binding characteristics and activity, the failure of the pattern of cellular activation elicited by central Ucn II to closely mimic the CRF-R2 distribution was unexpected. A recent study comparing the distribution of Fos expression induced by i.c.v. CRF or Ucn documented activation patterns coarsely consistent with the binding affinities of these peptides for CRF-Rs encoded by the two known genes (25). That is, CRF at doses similar to those used here activated sites of CRF-R1 expression in a highly preferential manner, while Ucn provoked Fos induction mainly in subsets of cell groups that express each receptor. In addition, however, both peptides recruited the very same set of central autonomic structures that were seen here to be the dominant seats of Ucn II-induced activational responses in the rat brain. This is significant in that elements of the central autonomic system are among the best documented sites at which CRF-like peptides can act to elicit stress-related autonomic and behavioral responses. These findings would suggest that type 2, as well as type 1, receptor activation is capable of engaging this system, although the basis for this is unclear. Among the nodal points in the central autonomic network, only the parabrachial nucleus (R1) and the NTS (R2) have been identified as sites of CRF-R expression (19, 20, 25), and it remains to be determined whether receptor-mediated activation of either or both of these is sufficient to enlist the system as a whole. It is important to note that systemic injections of synthetic Ucn II failed to elicit comparably powerful activational responses within central autonomic cell groups over the same range of doses that were used for i.c.v. injection studies. This is an important control, as activation of peripheral CRF-R2 can yield a marked and persistent reduction in blood pressure (8, 9), and salient hypotensive challenges are capable of activating the very same central autonomic structures as are responsive to central Ucn II administration (26, 27).
The initial characterization of the effects of i.c.v. Ucn II on food intake and activity complements recent efforts to tease apart the roles of individual CRF-Rs in stress-related behaviors. For example, while mice bearing null mutations of either receptor display normal basal food intake, CRF-R1-deficient animals have been shown to be refractory to the anorexic effects of Ucn during the period immediately following injection, but not at later time points, while the converse is true of CRF-R2 mutant mice (28–30). This has been taken as suggesting that the early and later phase of Ucn-mediated feeding suppression may be CRF-R1- and CRF-R2-mediated events, respectively. Using a different paradigm (nighttime free-feeding rather than deprivation-induced refeeding), we have provided data supportive of such a parsing, as the R2-specific ligand did not reliably suppress food intake at the early time points, but did so beyond 6 h postinjection.
Measures of motor activity also supported a dissociation of CRF-R involvement in this parameter. In line with recent evidence in knockout mice suggesting locomotor activation to be a CRF-R1-mediated event (31), we found that the R1-selective agonist, CRF, significantly increased gross motor activity, while Ucn II administration did not. Interestingly, treatment with Ucn, which is bound with high affinity by both receptors, resulted in a nonsignificant trend toward increased activity, with values being reliably lower than those seen in response to CRF. This is coarsely consistent with a growing body of evidence to support a functional antagonism between the two known receptor types. Whereas CRF-R1-deficient mice show reduced endocrine and anxiety-like responses to stress (32), CRF-R2 mutant lines display increases in these parameters (29, 30, 33), suggesting that basal activation of CRF-R2 may play a role in opposing CRF-R1-driven stress responses.
The identification of an endogenous CRF-R2-selective ligand will allow for more detailed analysis of the roles of individual CRF-related signaling molecules in stress-related physiologic and behavioral functions. We have demonstrated central expression of Ucn II mRNA, identified cell groups that respond to central administration of the peptide, and confirmed behavioral responses that are consistent with previously hypothesized consequences of CRF-R2 activation. Further insight into the place of this peptide in stress biology will require delineation of the central projections of Ucn II containing cells, and identification of the factors and circumstances that regulate gene expression and peptide release.
Acknowledgments
We gratefully acknowledge the scholarly contributions of A. P. Orth, L. M. Bilezikjian, J. C. Bittencourt, C. Donaldson, K. Kageyama, D. Kirby, R. Kaiser, A. Bloont, A. Craig, and C. Li. This work was supported by National Institutes of Health Grant DK-26741, The Adler Foundation, The Kleberg Foundation, and the Foundations for Research and Medical Research. W.W.V. and P.E.S. are investigators for the Foundation for Research and the Foundation for Medical Research, respectively. T.M.R. was supported by an National Research Service Award (DK-10135).
Abbreviations
- CRF
corticotropin-releasing factor
- CRF-R1 and CRF-R2
CRF receptor types 1 and 2
- i.c.v.
intracerebroventricular
- NTS
nucleus of the solitary tract
- Ucn
urocortin
- URP
urocortin-related peptide
Note Added in Proof.
We have identified (K.L. et al., unpublished work) a gene encoding the precursor of a second human URP. The putative 38 amino acid mature peptide has, at the amino acid level, 77, 41, 44, 23, and 33% identity to the pufferfish URP, human URP, mouse Ucn II, rat urocortin, and rat/human CRF sequences shown in Fig. 1.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AF331517 (mouse Ucn II)].
References
- 1.Vale W, Spiess J, Rivier C, Rivier J. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- 2.Menzaghi F, Heinrichs S C, Pich E M, Weiss R, Koob G F. Ann NY Acad Sci. 1993;697:142–154. doi: 10.1111/j.1749-6632.1993.tb49929.x. [DOI] [PubMed] [Google Scholar]
- 3.Sawchenko P E, Imaki T, Potter E, Kovacs K, Imaki J, Vale W. Ciba Found Symp. 1993;172:5–21. doi: 10.1002/9780470514368.ch2. [DOI] [PubMed] [Google Scholar]
- 4.Herkenham M. Neuroscience. 1987;23:1–38. doi: 10.1016/0306-4522(87)90268-5. [DOI] [PubMed] [Google Scholar]
- 5.Bittencourt J C, Sawchenko P E. J Neurosci. 2000;20:1142–1156. doi: 10.1523/JNEUROSCI.20-03-01142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behan D P, Grigoriadis D E, Lovenberg T, Chalmers D, Heinrichs S, Liaw C, De Souza E B. Mol Psychiatry. 1996;1:265–277. [PubMed] [Google Scholar]
- 7.Turnbull A V, Rivier C. Proc Soc Exp Biol Med. 1997;215:1–10. doi: 10.3181/00379727-215-44108. [DOI] [PubMed] [Google Scholar]
- 8.Vaughan J, Donaldson C, Bittencourt J, Perrin M H, Lewis K, Sutton S, Chan R, Turnbull A V, Lovejoy D, Rivier C, et al. Nature (London) 1995;378:287–292. doi: 10.1038/378287a0. [DOI] [PubMed] [Google Scholar]
- 9.Spina M, Merlo-Pich E, Chan R K, Basso A M, Rivier J, Vale W, Koob G F. Science. 1996;273:1561–1564. doi: 10.1126/science.273.5281.1561. [DOI] [PubMed] [Google Scholar]
- 10.Bittencourt J C, Vaughan J, Arias C, Rissman R A, Vale W W, Sawchenko P E. J Comp Neurol. 1999;415:285–312. [PubMed] [Google Scholar]
- 11.Miranda A, Koerber S C, Gulyas J, Lahrichi S L, Craig A G, Corrigan A, Hagler A, Rivier C, Vale W, Rivier J. J Med Chem. 1994;37:1450–1459. doi: 10.1021/jm00036a010. [DOI] [PubMed] [Google Scholar]
- 12.Miller C, Rivier J. Biopolymers. 1996;40:265–317. doi: 10.1002/(SICI)1097-0282(1996)40:3%3C265::AID-BIP2%3E3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 13.Simmons D M, Arriza J, Swanson L W. J Histotechnol. 1989;12:168–181. [Google Scholar]
- 14.Sawchenko P E, Cunningham E T, Jr, Mortrud M T, Gerfen C R. Methods Neurosci. 1990;3:247–260. [Google Scholar]
- 15.Eddy S R. Curr Opin Struct Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 16.Brunner B, Grutzner F, Yaspo M L, Ropers H H, Haaf T, Kalscheue V M. Chromosome Res. 2000;8:465–476. doi: 10.1023/a:1009263504671. [DOI] [PubMed] [Google Scholar]
- 17.Sawchenko P E. J Auton Nerv Syst. 1983;9:13–26. doi: 10.1016/0165-1838(83)90129-7. [DOI] [PubMed] [Google Scholar]
- 18.Saper C B. In: The Rat Nervous System. 2nd Ed. Paxinos G, editor. San Diego: Academic; 1995. pp. 107–128. [Google Scholar]
- 19.Van Pett K, Viau V, Chan R K W, Li H-Y, Arias C, Perrin M, Vale W, Sawchenko P E. J Comp Neurol. 2000;428:191–212. doi: 10.1002/1096-9861(20001211)428:2<191::aid-cne1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 20.Chalmers D T, Lovenberg T W, De Souza E B. J Neurosci. 1995;15:6340–6350. doi: 10.1523/JNEUROSCI.15-10-06340.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swanson L W, Sawchenko P E, Rivier J, Vale W W. Neuroendocrinology. 1983;36:165–186. doi: 10.1159/000123454. [DOI] [PubMed] [Google Scholar]
- 22.Bremner J D, Krystal J H, Southwick S M, Charney D S. Synapse. 1996;23:28–38. doi: 10.1002/(SICI)1098-2396(199605)23:1<28::AID-SYN4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 23.Swanson L W, Sawchenko P E. Annu Rev Neurosci. 1983;6:269–324. doi: 10.1146/annurev.ne.06.030183.001413. [DOI] [PubMed] [Google Scholar]
- 24.Elmquist J K, Maratos-Flier E, Saper C B, Flier J S. Nat Neurosci. 1998;1:445–450. doi: 10.1038/2164. [DOI] [PubMed] [Google Scholar]
- 25.Potter E, Sutton S, Donaldson C, Chen R, Perrin M, Lewis K, Sawchenko P E, Vale W. Proc Natl Acad Sci USA. 1994;91:8777–8781. doi: 10.1073/pnas.91.19.8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y W, Dampney R A L. Neuroscience. 1994;61:613–634. doi: 10.1016/0306-4522(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 27.Chan R K W, Sawchenko P E. J Comp Neurol. 1994;348:433–460. doi: 10.1002/cne.903480309. [DOI] [PubMed] [Google Scholar]
- 28.Bradbury M J, McBurnie M I, Denton D A, Lee K F, Vale W W. Endocrinology. 2000;141:2715–2724. doi: 10.1210/endo.141.8.7606. [DOI] [PubMed] [Google Scholar]
- 29.Bale T L, Contarino A, Smith G W, Chan R, Gold L H, Sawchenko P E, Koob G F, Vale W W, Lee K F. Nat Genet. 2000;24:410–414. doi: 10.1038/74263. [DOI] [PubMed] [Google Scholar]
- 30.Coste S C, Kesterson R A, Heldwein K A, Stevens S L, Heard A D, Hollis J H, Murray S E, Hill J K, Pantely G A, Hohimer A R, et al. Nat Genet. 2000;24:403–409. doi: 10.1038/74255. [DOI] [PubMed] [Google Scholar]
- 31.Contarino A, Dellu F, Koob G F, Smith G W, Lee K F, Vale W W, Gold L H. Endocrinology. 2000;141:2698–2702. doi: 10.1210/endo.141.7.7653. [DOI] [PubMed] [Google Scholar]
- 32.Smith G W, Aubry J M, Dellu F, Contarino A, Bilezikjian L M, Gold L H, Chen R, Marchuk Y, Hauser C, Bentley C A, et al. Neuron. 1998;20:1093–1102. doi: 10.1016/s0896-6273(00)80491-2. [DOI] [PubMed] [Google Scholar]
- 33.Kishimoto T, Radulovic J, Radulovic M, Lin C R, Schrick C, Hooshmand F, Hermanson O, Rosenfeld M G, Spiess J. Nat Genet. 2000;24:415–419. doi: 10.1038/74271. [DOI] [PubMed] [Google Scholar]