Abstract
Like natural viruses, manmade protein cages for drug delivery are to be ideally formed by repetitive subunits with self-assembling properties, mimicking viral functions and molecular organization. Naturally formed nanostructures (such as viruses, flagella or simpler protein oligomers) can be engineered to acquire specific traits of interest in biomedicine, for instance through the addition of cell targeting agents for desired biodistribution and specific delivery of associated drugs. However, fully artificial constructs would be highly desirable regarding finest tuning and adaptation to precise therapeutic purposes. Although engineering of protein assembling is still in its infancy, arising principles and promising strategies of protein manipulation point out the rational construction of nanoscale protein cages as a feasible concept, reachable through conventional recombinant DNA technologies and microbial protein production.
Commentary
Targeted drug and nucleic acid delivery, in which biodistribution is achieved by ligand-receptor interactions, is a promising way to reduce drug toxicity and enhance effectiveness, what is of special relevance for the treatment of cancer and chronic viral diseases, among others. In gene therapy, the term 'artificial viruses' has been proposed to describe virus-like constructs exhibiting specific viral functions that are relevant to cell recognition, penetration and compartment-aimed release of cargo nucleic acids [1-3]. This nanoparticle-based concept, which can be also extended to chemical drug delivery, involves the use of refillable cages and the incorporation of functional agents for cell receptor binding, cellular uptake and eventually endosomal escape and nuclear delivery. Lipidic and polymeric nanoparticles have been under continuous development during the last four decades [4-6], while nanofibers, nanowires, carbon nanotubes and other types of nanosized cages are being progressively incorporated into the drug delivery scenario [7]. The desired molecular organization, size dispersion and geometry of these constructs is achieved by mechanical and chemical approaches, through fine micro- or nanofabrication procedures [8,9]. Most of these particles are blind; therefore they require to be functionalized with targeting agents for specific cell attachment, being proteins the most efficient, versatile and tunable tools for cell and tissue targeting.
Advances in proteomics and systems biology have permitted to expand the existing catalogues of molecular markers relevant in biomedicine, specially those of interest in cancer diagnosis and therapy [10-14]. In parallel, growing numbers of peptides, antibodies and other protein ligands suitable for cell targeted delivery are becoming available [15-25], supported by the efforts in medical-focused applications of peptide display technologies [20,26-29]. All these findings provide a growing spectrum of therapeutic opportunities in the context of innovative and personalized medicines.
Interestingly, protein-only entities are appealing candidates as building blocks of drug delivery cages [30,31]. Their biocompatibility, biological fabrication, functional diversity and versatility of design though protein engineering (assisted by in silico instruments) make them extremely plastic and powerful materials in comparison with liposomes and other types of artificial viruses. Importantly, modular approaches to protein engineering permit the accommodation of several virus-like functions in single (hybrid) polypeptide chains [16,25,32]. In addition, since many microbial and non microbial organisms are being used as cell factories for therapeutic proteins [33], a wide spectrum of biological platforms, molecular tools and comparative expertise in alternative production strategies is available [34]. In particular, bacterial cells are generically good producers of diverse nanomaterials of medical application, including polymers, metal particles and protein particles [35].
The microbial production of protein particles of biomedical interest has been mainly focused on bacteriophages for peptide display, virus-like particles as immunogens and flagella and filamentous phages as templates for micro- and nanofabrication [35,36]. Apart from peptide-displaying filamentous phages, which have been widely explored for targeted delivery of DNA and conjugated drugs in cell culture but also in vivo [37-39], these particles have been in general hardly adaptable to cell-targeted drug delivery.
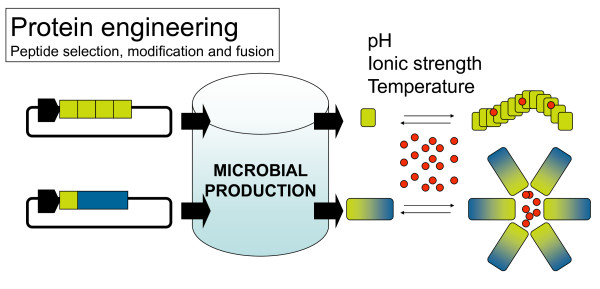
Ideally, polypeptides with specific functions relevant to drug delivery (mimicking those of viruses, [16]), should be engineered to self-assemble as nanoparticles with desired nanoscale properties, namely size and geometry, and produced in simple heterologous hosts. The construction of protein particles from their building blocks, as it occurs in natural viruses, can not be achieved by conventional nano-fabrication but through the selection of protein sequences promoting regular protein-protein interactions in absence of unspecific aggregation. In this regard, sets of both natural and non-natural amino acid sequences have been identified that promote peptide self-assembling in diverse patterns. Among them hydrogels, bilayers and nanofibers show important applications in tissue engineering but also in drug delivery [40-44]. Peptide self-assembling can be eventually controlled by pH, temperature and other environment parameters [45-47], and it often involves the formation of amyloid-like, beta-sheet cross-molecular interactions [41] similar to those governing the formation of bacterial inclusion bodies [48-51]. Amyloid-like architecture is probable driving also the assembling of protein fibers formed by misfolding prone recombinant proteins in the cytoplasm of recombinant bacteria [52,53]. Interestingly, some of these short peptides have been successfully produced in bacteria by means of conventional recombinant DNA approaches, fused to carrier proteins or as tandem repeats to enhance their stability [44,54-56] (Figure 1).
Figure 1.
Self-assembling peptide sequences (yellow boxes) are usually identified from nature, obtained by trial-and-error protein engineering approaches or isolated from combinatorial libraries. Peptides can be produced in microbial cells as tandem repeats or fused to carrier proteins for further recovery and isolation by proteolysis. Eventually, self-assembling peptides can be produced as fused to multifunctional proteins (blue), to generate self-organizing artificial viruses. Ideally, the assembling of both single peptides and more complex protein building blocks into supramolecular constructs should be controlled in vitro by simple physicochemical parameters, to permit the incorporation of the cargo drug (in red).
Very few of these peptides have been yet incorporated to larger, modular protein constructs for drug delivery to form multifunctional, virus-like protein cages. However, a couple of examples from our own experience prove the feasibility of the concept. In this regard, poly-lysine stretches, commonly used as cationic peptides for DNA condensation [57], confer architectonic properties to engineered beta-galactodidases, which already contained several functional motives for cell receptor binding and nuclear delivery. These chimerical enzymes formed amorphous nanoparticles [58,59], and have been proved to be excellent artificial viruses for in vivo gene delivery to the nervous system in ischemia models and for both histological and functional recovery of injured animals [60,61]. Also, poly-arginine peptides, again used by their DNA condensation properties promoted the self-assembling of scaffold green fluorescent proteins as monodisperse, highly regular 20 nm-nanoparticles useful for DNA and protein delivery [62]. These particles are formed irrespective of the solubility exhibited by the protein under different storage conditions [62,63], proving high regularity and consistence in their architectonic schemes. Interestingly, once exposure to cultured mammalian cells, the intracellular trafficking of these entities is extremely efficient, showing a fast and steady nuclear accumulation in fluorescent forms [62,64]. The sticky nature of cationic peptides, although not completely solved, does not seem to involve cross beta-sheet interactions.
These last results provide a valid proof of concept of the incorporation of self-assembling peptides to complex protein building blocks of artificial viruses (Figure 1), although this is still far from rational design. Moreover, steady conceptual advances in the biology of protein-protein interactions and the easy bioproduction of chimerical proteins in microbial hosts, permit to envisage further progresses in the design of protein cages for drug delivery based on both protein engineering and the exploitation of microbial cell factories.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
EV and AV have equally contributed to this work.
Contributor Information
Esther Vázquez, Email: esther.vazquez@uab.cat.
Antonio Villaverde, Email: antoni.villaverde@uab.cat.
Acknowledgements
We appreciate the financial support received for the design and microbial production of recombinant proteins for biomedical applications from FISS (PS09/00165), MEC (BFU2010-17450, ACI2009-0919, IT2009-0021, EUI2008-03610), AGAUR (2005SGR-00956), CIBER de Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), Spain, and from the European Science Foundation, which is also funded by the European Commission, Contract no. ERAS-CT-2003-980409 of the Sixth Framework Programme (ERANET-IB 08-007).
References
- Douglas KL. Toward Development of Artificial Viruses for Gene Therapy: A Comparative Evaluation of Viral and Non-viral Transfection. Biotechnol Prog. 2008;24:871–883. doi: 10.1021/bp070319o. [DOI] [PubMed] [Google Scholar]
- Mastrobattista E, van der Aa MA, Hennink WE, Crommelin DJ. Artificial viruses: a nanotechnological approach to gene delivery. Nat Rev Drug Discov. 2006;5:115–121. doi: 10.1038/nrd1960. [DOI] [PubMed] [Google Scholar]
- Wagner E. Strategies to improve DNA polyplexes for in vivo gene transfer: will "artificial viruses" be the answer? Pharm Res. 2004;21:8–14. doi: 10.1023/B:PHAM.0000012146.04068.56. [DOI] [PubMed] [Google Scholar]
- Parveen S, Sahoo SK. Polymeric nanoparticles for cancer therapy. J Drug Target. 2008;16:108–123. doi: 10.1080/10611860701794353. [DOI] [PubMed] [Google Scholar]
- Kaasgaard T, Andresen TL. Liposomal cancer therapy: exploiting tumor characteristics. Expert Opin Drug Deliv. 2010;7:225–243. doi: 10.1517/17425240903427940. [DOI] [PubMed] [Google Scholar]
- Malam Y, Loizidou M, Seifalian AM. Liposomes and nanoparticles: nanosized vehicles for drug delivery in cancer. Trends Pharmacol Sci. 2009;30:592–599. doi: 10.1016/j.tips.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Singh S. Nanomedicine-nanoscale drugs and delivery systems. J Nanosci Nanotechnol. 2010;10:7906–7918. doi: 10.1166/jnn.2010.3617. [DOI] [PubMed] [Google Scholar]
- Betancourt T, Brannon-Peppas L. Micro- and nanofabrication methods in nanotechnological medical and pharmaceutical devices. Int J Nanomedicine. 2006;1:483–495. doi: 10.2147/nano.2006.1.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldorera-Moore M, Guimard N, Shi L, Roy K. Designer nanoparticles: incorporating size, shape and triggered release into nanoscale drug carriers. Expert Opin Drug Deliv. 2010;7:479–495. doi: 10.1517/17425240903579971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H. Identification of biomarkers for colorectal cancer through proteomics-based approaches. Expert Rev Proteomics. 2010;7:879–895. doi: 10.1586/epr.10.81. [DOI] [PubMed] [Google Scholar]
- Deonarain MP, Kousparou CA, Epenetos AA. Antibodies targeting cancer stem cells: a new paradigm in immunotherapy? MAbs. 2009;1:12–25. doi: 10.4161/mabs.1.1.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoslahti E, Bhatia SN, Sailor MJ. Targeting of drugs and nanoparticles to tumors. J Cell Biol. 2010;188:759–768. doi: 10.1083/jcb.200910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Angulo AM, Hennessy BT, Mills GB. Future of personalized medicine in oncology: a systems biology approach. J Clin Oncol. 2010;28:2777–2783. doi: 10.1200/JCO.2009.27.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharali DJ, Mousa SA. Emerging nanomedicines for early cancer detection and improved treatment: current perspective and future promise. Pharmacol Ther. 2010;128:324–335. doi: 10.1016/j.pharmthera.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Dietz GP, Bahr M. Delivery of bioactive molecules into the cell: the Trojan horse approach. Mol Cell Neurosci. 2004;27:85–131. doi: 10.1016/j.mcn.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Aris A, Villaverde A. Modular protein engineering for non-viral gene therapy. Trends Biotechnol. 2004;22:371–377. doi: 10.1016/j.tibtech.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Ferrer-Miralles N, Vazquez E, Villaverde A. Membrane-active peptides for non-viral gene therapy: making the safest easier. Trends Biotechnol. 2008;26:267–275. doi: 10.1016/j.tibtech.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Vives E, Schmidt J, Pelegrin A. Cell-penetrating and cell-targeting peptides in drug delivery. Biochim Biophys Acta. 2008;1786:126–138. doi: 10.1016/j.bbcan.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Martin ME, Rice KG. Peptide-guided gene delivery. AAPS J. 2007;9:E18–E29. doi: 10.1208/aapsj0901003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aina OH, Liu R, Sutcliffe JL, Marik J, Pan CX, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Mol Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- Beck A, Klinguer-Hamour C, Bussat MC, Champion T, Haeuw JF, Goetsch L. et al. Peptides as tools and drugs for immunotherapies. Journal of Peptide Science. 2007;13:588–602. doi: 10.1002/psc.852. [DOI] [PubMed] [Google Scholar]
- Enback J, Laakkonen P. Tumour-homing peptides: tools for targeting, imaging and destruction. Biochem Soc Trans. 2007;35:780–783. doi: 10.1042/BST0350780. [DOI] [PubMed] [Google Scholar]
- Balestrieri ML, Napoli C. Novel challenges in exploring peptide ligands and corresponding tissue-specific endothelial receptors. Eur J Cancer. 2007;43:1242–1250. doi: 10.1016/j.ejca.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Fabre JW, Collins L. Synthetic peptides as non-viral DNA vectors. Curr Gene Ther. 2006;6:459–480. doi: 10.2174/156652306777934865. [DOI] [PubMed] [Google Scholar]
- Vazquez E, Ferrer-Miralles N, Mangues R, Corchero JL, Schwartz S Jr, Villaverde A. Modular protein engineering in emerging cancer therapies. Curr Pharm Des. 2009;15:893–916. doi: 10.2174/138161209787582084. [DOI] [PubMed] [Google Scholar]
- Brown KC. Peptidic tumor targeting agents: the road from phage display peptide selections to clinical applications. Curr Pharm Des. 2010;16:1040–1054. doi: 10.2174/138161210790963788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu SS. Phage display in pharmaceutical biotechnology. Curr Opin Biotechnol. 2000;11:610–616. doi: 10.1016/S0958-1669(00)00152-X. [DOI] [PubMed] [Google Scholar]
- Deutscher SL. Phage display in molecular imaging and diagnosis of cancer. Chem Rev. 2010;110:3196–3211. doi: 10.1021/cr900317f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Kokuryu Y, Matsunaga T. Novel method for selection of antimicrobial peptides from a phage display library by use of bacterial magnetic particles. Appl Environ Microbiol. 2008;74:7600–7606. doi: 10.1128/AEM.00162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida M, Klem MT, Allen M, Suci P, Flenniken M, Varpness Z. et al. Biological containers: Protein cages as multifunctional nanoplatforms. Adv Mater. 2007;19:1025–1042. doi: 10.1002/adma.200601168. [DOI] [Google Scholar]
- Maham A, Tang Z, Wu H, Wang J, Lin Y. Protein-Based Nanomedicine Platforms for Drug Delivery. Small. 2009;5:1706–1721. doi: 10.1002/smll.200801602. [DOI] [PubMed] [Google Scholar]
- Vazquez E, Ferrer-Miralles N, Villaverde A. Peptide-assisted traffic engineering for nonviral gene therapy. Drug Discov Today. 2008;13:1067–1074. doi: 10.1016/j.drudis.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Ferrer-Miralles N, Domingo-Espin J, Corchero JL, Vazquez E, Villaverde A. Microbial factories for recombinant pharmaceuticals. Microb Cell Fact. 2009;8:17. doi: 10.1186/1475-2859-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser B, Saloheimo M, Rinas U, Dragosits M, Rodriguez-Carmona E, Baumann K. et al. Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb Cell Fact. 2008;7:11. doi: 10.1186/1475-2859-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Carmona E, Villaverde A. Nanostructured bacterial materials for innovative medicines. Trends Microbiol. 2010;18:423–430. doi: 10.1016/j.tim.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Villaverde A. Nanotechnology, bionanotechnology and microbial cell factories. Microb Cell Fact. 2010;9:53. doi: 10.1186/1475-2859-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar H, Yacoby I, Benhar I. Killing cancer cells by targeted drug-carrying phage nanomedicines. BMC Biotechnol. 2008;8:37. doi: 10.1186/1472-6750-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca D, Burg MA, Jensen-Pergakes K, Ravey EP, Gonzalez AM, Baird A. Evolving phage vectors for cell targeted gene delivery. Curr Pharm Biotechnol. 2002;3:45–57. doi: 10.2174/1389201023378490. [DOI] [PubMed] [Google Scholar]
- Du B, Han H, Wang Z, Kuang L, Wang L, Yu L. et al. targeted drug delivery to hepatocarcinoma in vivo by phage-displayed specific binding peptide. Mol Cancer Res. 2010;8:135–144. doi: 10.1158/1541-7786.MCR-09-0339. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tanaka M, Kinoshita T, Higuchi M, Tan T. Self-assembling peptide nanofiber scaffolds for controlled release governed by gelator design and guest size. J Control Release. 2010;147:392–399. doi: 10.1016/j.jconrel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tanaka M, Kinoshita T, Higuchi M, Tan T. Nanofibrous scaffold from self-assembly of beta-sheet peptides containing phenylalanine for controlled release. J Control Release. 2010;142:354–360. doi: 10.1016/j.jconrel.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Gelain F, Unsworth LD, Zhang S. Slow and sustained release of active cytokines from self-assembling peptide scaffolds. J Control Release. 2010;145:231–239. doi: 10.1016/j.jconrel.2010.04.026. [DOI] [PubMed] [Google Scholar]
- Kyle S, Aggeli A, Ingham E, McPherson MJ. Production of self-assembling biomaterials for tissue engineering. Trends Biotechnol. 2009;27:423–433. doi: 10.1016/j.tibtech.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle S, Aggeli A, Ingham E, McPherson MJ. Recombinant self-assembling peptides as biomaterials for tissue engineering. Biomaterials. 2010;31:9395–9405. doi: 10.1016/j.biomaterials.2010.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Zhang Y, Kim KS, Yuan HK, Vo-Dinh T. Cellular uptake and photodynamic activity of protein nanocages containing methylene blue photosensitizing drug. Photochem Photobiol. 2010;86:662–666. doi: 10.1111/j.1751-1097.2009.00696.x. [DOI] [PubMed] [Google Scholar]
- Bessa PC, Machado R, Nurnberger S, Dopler D, Banerjee A, Cunha AM. et al. Thermoresponsive self-assembled elastin-based nanoparticles for delivery of BMPs. J Control Release. 2010;142:312–318. doi: 10.1016/j.jconrel.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Huang H, Sun XS. Rational design of responsive self-assembling peptides from native protein sequences (dagger) Biomacromolecules. 2010;11:3390–3394. doi: 10.1021/bm100894j. [DOI] [PubMed] [Google Scholar]
- Ventura S, Villaverde A. Protein quality in bacterial inclusion bodies. Trends Biotechnol. 2006;24:179–185. doi: 10.1016/j.tibtech.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Martinez-Alonso M, Gonzalez-Montalban N, Garcia-Fruitos E, Villaverde A. Learning about protein solubility from bacterial inclusion bodies. Microb Cell Fact. 2009;8:4. doi: 10.1186/1475-2859-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fruitos E, Villaverde A. Friendly production of bacterial inclusion bodies. Korean J Chem Eng. 2010;27:385–389. [Google Scholar]
- Carrio M, Gonzalez-Montalban N, Vera A, Villaverde A, Ventura S. Amyloid-like properties of bacterial inclusion bodies. J Mol Biol. 2005;347:1025–1037. doi: 10.1016/j.jmb.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Schrodel A, de Marco A. Characterization of the aggregates formed during recombinant protein expression in bacteria. BMC Biochem. 2005;6:10. doi: 10.1186/1471-2091-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Alonso M, Gonzalez-Montalban N, Garcia-Fruitos E, Villaverde A. The functional quality of soluble recombinant polypeptides produced in Escherichia coli is defined by a wide conformational spectrum. Appl Environ Microbiol. 2008;101:1353–1358. doi: 10.1128/AEM.01446-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann BM, Kaar W, Falconer RJ, Zeng B, Middelberg AP. Expression and purification of a nanostructure-forming peptide. J Biotechnol. 2008;135:85–91. doi: 10.1016/j.jbiotec.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Hartmann BM, Kaar W, Yoo IK, Lua LH, Falconer RJ, Middelberg AP. The chromatography-free release, isolation and purification of recombinant peptide for fibril self-assembly. Biotechnol Bioeng. 2009;104:973–985. doi: 10.1002/bit.22447. [DOI] [PubMed] [Google Scholar]
- Riley JM, Aggeli A, Koopmans RJ, McPherson MJ. Bioproduction and characterization of a pH responsive self-assembling peptide. Biotechnol Bioeng. 2009;103:241–251. doi: 10.1002/bit.22274. [DOI] [PubMed] [Google Scholar]
- Saccardo P, Villaverde A, Gonzalez-Montalban N. Peptide-mediated DNA condensation for non-viral gene therapy. Biotechnol Adv. 2009;27:432–438. doi: 10.1016/j.biotechadv.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Aris A, Villaverde A. Molecular organization of protein-DNA complexes for cell-targeted DNA delivery. Biochem Biophys Res Commun. 2000;278:455–461. doi: 10.1006/bbrc.2000.3824. [DOI] [PubMed] [Google Scholar]
- Aris A, Villaverde A. Engineering nuclear localization signals in modular protein vehicles for gene therapy. Biochem Biophys Res Commun. 2003;304:625–631. doi: 10.1016/S0006-291X(03)00644-2. [DOI] [PubMed] [Google Scholar]
- Peluffo H, Acarin L, Aris A, Gonzalez P, Villaverde A, Castellano B. et al. Neuroprotection from NMDA excitotoxic lesion by Cu/Zn superoxide dismutase gene delivery to the postnatal rat brain by a modular protein vector. BMC Neurosci. 2006;7:35. doi: 10.1186/1471-2202-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluffo H, Aris A, Acarin L, Gonzalez B, Villaverde A, Castellano B. Nonviral gene delivery to the central nervous system based on a novel integrin-targeting multifunctional protein. Hum Gene Ther. 2003;14:1215–1223. doi: 10.1089/104303403767740759. [DOI] [PubMed] [Google Scholar]
- Vazquez E, Roldán M, Diez-Gil C, Unzueta U, Domingo-Espin J, Cedano J. et al. Protein nanodisk assembling and intracellular trafficking powered by an arginine-rich (R9) peptide. Nanomedicine. 2010;5:259–268. doi: 10.2217/nnm.09.98. [DOI] [PubMed] [Google Scholar]
- Toledo-Rubio V, Vazquez E, Platas G, Domingo-Espin J, Unzueta U, Steinkamp E. et al. Protein aggregation and soluble aggregate formation screened by a fast microdialysis assay. J Biomol Screen. 2010;15:453–457. doi: 10.1177/1087057110363911. [DOI] [PubMed] [Google Scholar]
- Vazquez E, Cubarsi R, Unzueta U, Roldan M, Domingo-Espin J, Ferrer-Miralles N. et al. Internalization and kinetics of nuclear migration of protein-only, arginine-rich nanoparticles. Biomaterials. 2010;31:9333–9339. doi: 10.1016/j.biomaterials.2010.08.065. [DOI] [PubMed] [Google Scholar]